Abstract
In higher plants, the plane of cell division is faithfully predicted by the preprophase band (PPB). The PPB, a cortical ring of microtubules and F-actin, disassembles upon nuclear-envelope breakdown. During cytokinesis, the expanding cell plate fuses with the plasma membrane at the cortical division site, the site of the former PPB. The nature of the “molecular memory” that is left behind by the PPB and is proposed to guide the cell plate to the cortical division site is unknown. RanGAP is the GTPase activating protein of the small GTPase Ran, which provides spatial information for nucleocytoplasmic transport and various mitotic processes in animals. Here, we show that, in dividing root cells, Arabidopsis RanGAP1 concentrates at the PPB and remains associated with the cortical division site during mitosis and cytokinesis, requiring its N-terminal targeting domain. In a fass/ton2 mutant, which affects PPB formation, RanGAP1 recruitment to the PPB site is lost, while its PPB retention is microtubule-independent. RanGAP1 persistence at the cortical division site, but not its initial accumulation at the PPB requires the 2 cytokinesis-regulating kinesins POK1 and POK2. Depletion of RanGAP by inducible RNAi leads to oblique cell walls and cell-wall stubs in root cell files, consistent with cytokinesis defects. We propose that Arabidopsis RanGAP, a continuous positive protein marker of the plant division plane, has a role in spatial signaling during plant cell division.
Keywords: cytokinesis, Ran cycle, preprophase band, nuclear pore
Cell division in higher plants involves the construction of a new cell wall in the proper orientation of the division plane. Because plant cells are embedded in a rigid matrix of cell wall material, the spatial orientation of the new cell wall defines the geometry of the respective plant tissue (1). The plane of cell division is defined by the assembly of the preprophase band (PPB), a ring of microtubules and F-actin that appears during G2 phase, and the migration of the premitotic nucleus into the plane defined by the PPB. During mitosis, the site of the former PPB becomes the cortical division site (CDS), which remains “marked” in an unknown way and which is thought to guide the phragmoplast and the outwardly growing new plasma membrane.
The molecular nature of the CDS has long been enigmatic, and only recently have the first molecular markers been identified. A “negative marker” is the local depletion of cortical F-actin and of the kinesin KCA1 (2, 3). Several proteins appear and disappear with the PPB and reappear later at the CDS. These include the microtubule-associated protein AIR9, RSH, a hydroxyproline-rich glycoprotein, and T-PLATE, a protein resembling transport vesicle coat proteins (4–6). Only 1 protein is known that is recruited to the PPB and remains at the site of the future CDS throughout the cell division cycle. TANGLED was originally identified in maize, where in tan mutants cells divide in aberrant orientations, suggesting a requirement of TANGLED for proper division-plane establishment (7).
Arabidopsis TANGLED is recruited to the PPB in a microtubule- and kinesin-dependent manner, and persists at the CDS after PPB disassembly (8). Two related kinesins, PHRAGMOPLAST-ORIENTING KINESINS 1 and 2 (POK1 and POK2) were found to interact with TANGLED and a pok1 pok2 double mutant resembles the maize tan mutant in terms of misoriented division planes (9). Although the data suggest a role for kinesins and the pioneer protein TANGLED in division-plane definition, the molecular mechanism of the process remains unknown.
Ran is a small GTPase that in vertebrates controls multiple cellular processes including nucleocytoplasmic transport, spindle assembly, nuclear envelope reassembly, centrosome duplication, and cell-cycle control (ref. 10 and references therein). Crucial for its roles is the asymmetric distribution of RanGTP and RanGDP, enabled by specific locations of the RanGTPase activating protein RanGAP and the Ran nucleotide exchange factor RCC1. Although vertebrate RCC1 remains chromatin bound throughout cell cycle, RanGAP migrates from its interphase location at the outer surface of the nuclear pore to mitotic locations such as the kinetochores (11, 12). Unlike vertebrate RanGAP, Arabidopsis RanGAP1 was shown to associate with the phragmoplast and growing rim of the cell plate during cytokinesis (13, 14). The phragmoplast is a plant-specific array of microtubules, actin filaments and associated molecules that act as a framework for the future cell wall and might be analogous to the spindle midbody of animal cells (15). All subcellular targeting events of Arabidopsis RanGAP1 require an N-terminal domain (WPP domain, named after a highly conserved tryptophan-proline-proline motif), which is unique to plants.
Here, we show that Arabidopsis RanGAP1 positively labels the PPB and, like TANGLED, remains associated with the future site of division throughout cell cycle. RanGAP1 recruitment to the PPB depends on FASS/TONNEAU 2, a putative regulatory subunit of protein phosphatase 2A, which is necessary for PPB assembly (16). Its persistence at the CDS depends on POK1 and POK2. Inducible depletion of Arabidopsis RanGAP in seedling roots leads to misplaced cell walls similar to the Arabidopsis tan mutant alleles. Together, our data present RanGAP as a novel continuous positive protein marker of the plant division plane, dependent on known regulators of plant cytokinesis and poised to signal spatial information during plant cell division.
Results
RanGAP1 Positively Marks the Arabidopsis Division Plane Throughout Mitosis and Cytokinesis.
The mitotic localization pattern of Arabidopsis RanGAP1 was revealed using indirect immunofluorescence in root tip cells. During preprophase, RanGAP1 was concentrated at the PPB [Fig. 1 A and B and supporting information (SI) Movie S1]. Once the cell entered metaphase, the PPB disassembled, whereas RanGAP1 stayed at the position of the former PPB (cortical division site, CDS) until the end of cytokinesis (Fig. 1A). During metaphase, RanGAP1 also accumulated in several bright dots on chromosomes, resembling the kinetochore regions (Fig. 1 A and Fig. S1). As the cells progressed into anaphase, RanGAP1 was found enriched around the spindle midzone in addition to remaining at the CDS (Fig. 1 A and C). When the spindle midzone microtubules assembled into the phragmoplast, RanGAP1 was concentrated at the midline of the phragmoplast or the nascent cell plate and the CDS (Fig. 1A). None of the above-mentioned staining patterns was seen in a RanGAP1 null mutant (data not shown), confirming the specificity of the immunofluorescence signal. To monitor RanGAP1 localization throughout cell division, we performed real-time imaging of a 35S-promoter driven RanGAP1-GFP fusion protein in transgenic Arabidopsis roots. Except that RanGAP1-GFP accumulation on the phragmoplast midline was less evident, the fusion protein showed an essentially identical localization pattern compared with endogenous RanGAP1. In 43% of dividing cells, significant enrichment of RanGAP1 at the PPB and CDS was observed continuously throughout mitosis and cytokinesis (Table 1, Fig. S2, and Movie S2). Cells dividing without observable RanGAP1 concentration might either be below the detection limit of the assay, or suggest that the accumulation does not occur equally in all cells. The signal narrowed as the cells progressed from preprophase to metaphase and anaphase, similar to what has been observed for TANGLED (8). In summary, a concentration of RanGAP1 was seen at the division plane from preprophase to cytokinesis, making RanGAP1, after TANGLED, the second continuous positive protein marker of the plant division plane.
Fig. 1.
Arabidopsis RanGAP1 demarcates the PPB and CDS during mitosis and cytokinesis. (A) Concentration of RanGAP1 at the PPB (arrowheads in the topmost image), kinetochore (arrow), spindle midzone (bracket), phragmoplast midline (asterisk) and CDS (arrowheads in second, third and bottommost images) in dividing root cells, revealed by immunofluorescence. α-Tubulin (magenta) indicates the different mitotic stages listed on the left. (B) Four consecutive focal planes (increment of 0.3 μm) taken from a likely preprophase cell at the root tip, demonstrating near-continuous RanGAP1 enrichment at the PPB (arrowheads). (C) Four consecutive focal planes (increment of 0.3 μm) from a dividing root tip cell in anaphase, showing RanGAP1 association with both the spindle midzone (bracket) and CDS (arrowheads). DNA (magenta) was stained with SYTOX Orange in B and C. (Scale bars: 10 μm.)
Table 1.
Number of cells observed with RanGAP1 (or its derivatives) concentrated at the PPB and CDS in different backgrounds
| RanGAP1 |
RanGAP1WPP/AAP-GFP | RanGAP1ΔC-GFP | |||||
|---|---|---|---|---|---|---|---|
| WT | ton2–14 | pok1–1;2–1 | pok1–2;2–2 | tan-csh | |||
| PPB | 23 (55) | 0 (17) | 13 (24) | 11 (27) | 10 (25) | 0 (24) | 15 (28) |
| CDS | 28 (69) | 0 (35) | 0 (33) | 0 (25) | 17 (40) | 0 (26) | 14 (39) |
In each case, data collected from two independent experiments were summarized. Endogenous RanGAP1 was visualized using immunofluorescence with the anti-RanGAP1 antibody, whereas the anti-GFP antibody was used to stain both RanGAP1WPP/AAP-GFP and RanGAP1ΔC-GFP. The total number of dividing cells examined in the corresponding stages is shown in parentheses.
Molecular Requirement for RanGAP1 PPB and CDS Association.
Arabidopsis RanGAP1 has a plant-unique WPP domain that is necessary and sufficient for its targeting to the nuclear envelope during interphase and to the phragmoplast midline/cell plate during cytokinesis (13, 17). When a RanGAP1WPP/AAP-GFP (with key residues WPP mutated to AAP) fusion protein was tracked through mitosis in Arabidopsis roots, the protein was diffusely distributed throughout the cytoplasm without concentration at the PPB, kinetochore region, or CDS (Table 1 and Fig. S3A). This indicates that the WPP motif is necessary for the mitotic targeting of RanGAP1, likely through interaction with protein partners. To investigate whether the WPP domain is also sufficient for RanGAP1 targeting during mitosis, localization of a fusion protein between the WPP domain and GFP (RanGAP1ΔC-GFP) was monitored by immunofluorescence. Similar to full-length RanGAP1, RanGAP1ΔC-GFP was targeted to the PPB, kinetochore region, CDS, and cell plate (Table 1 and Fig. S3B), indicating that the WPP domain is the targeting domain for RanGAP1 throughout cell cycle.
RanGAP1 concentrates at the CDS after the PPB disassembles, indicating that its retention at the CDS does not require microtubules. Consistently, when PPB microtubules were disrupted with oryzalin, RanGAP1 accumulation at the PPB site persisted, pointing to a microtubule-independent mechanism for anchoring RanGAP1 at the PPB (Fig. 2A). Mutants of FASS/TONNEAU2, a putative regulatory subunit of protein phosphatase 2A, lack a preprophase band before mitosis and fail to guide the phragmoplast properly (16). In ton2-14 mutant cells, RanGAP1 still accumulated around the kinetochore, spindle midzone and phragmoplast midline/cell plate, while no RanGAP1 concentration could be detected corresponding to the PPB or CDS (Fig. 2B and Table 1). The lack of RanGAP1 concentration at the PPB/CDS in ton2-14 suggests that either FASS/TONNEAU2 plays a direct role in directing RanGAP1 targeting or that FASS/TONNEAU2 is required for PPB assembly and the PPB in turn both recruits RanGAP1 and sets up a mechanism for its subsequent retention.
Fig. 2.
RanGAP1 retention at the PPB is microtubule-independent, and its localization at the site of the PPB and CDS requires FASS/TONNEAU2. (A) A preprophase cell coexpressing RanGAP1-GFP and mCherry-TUB6 was imaged before and 10 min after treatment with 5 μM oryzalin. Concentration of RanGAP1-GFP at the PPB (arrowheads) remains after the disassembly of microtubules. (B) FASS/TONNEAU2 is required for RanGAP1 targeting to the site of the PPB and CDS, but not the kinetochore (white arrow), spindle midzone (bracket) or phragmoplast midline (asterisk). Note that the formation of perinuclear microtubules (yellow arrow) indicated a preprophase cell in the ton2-14 mutant without a detectable microtubular PPB. RanGAP1 (green) was immunostained in root tip cells, with α-tubulin (magenta) costaining to indicate the different mitotic stages listed on the left. (Scale bars: 10 μm.)
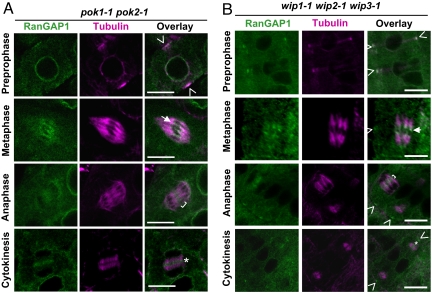
Two other players have been implicated in the spatial regulation of plant cytokinesis. POK1 and POK2 are 2 redundant kinesins whose mutations affect the guidance of the phragmoplast toward the CDS (9). In pok1-1 pok2-1 mutants (Fig. 3A and Table 1) and pok1-2 pok2-2 mutants (Table 1), RanGAP1 accumulated at the PPB, kinetochore region and spindle midzone. Significantly, however, no evident RanGAP1 concentration could be detected at the cell periphery corresponding to a CDS association (Table 1). These data show that RanGAP1 persistence at the CDS, but not its initial accumulation at the PPB requires POK1 and POK2.
Fig. 3.
RanGAP1 localization patterns in pok and wip mutant backgrounds. (A) RanGAP1 association with the CDS but not the PPB is diminished in pok1-1 pok2-1. Similar results were also seen in pok1-2 pok2-2 (data not shown). RanGAP1 (green) was immunostained, and α-tubulin (magenta) was costained to indicate the different mitotic stages indicated on the left. Marked areas include PPB (arrowheads), kinetochore (arrow), spindle midzone (bracket) and phragmoplast midline (asterisk). (Scale bars: 10 μm.) (B) RanGAP1 targeting during mitosis and cytokinesis is independent of WIP1, WIP2, and WIP3. An essentially identical RanGAP1 mitotic localization pattern was seen in a wip1-1 wip2-1 wip3-1 triple mutant (18) compared with wild type. Labels were used to indicate the PPB (arrowheads in topmost image), kinetochore (arrow), spindle midzone (bracket), phragmoplast midline (asterisk) and CDS (arrow heads in second, third, and bottommost images). α-tubulin (magenta) was stained to indicate the different mitotic stages listed on the left. (Scale bars: 10 μm.)
To test whether this effect could be based on direct binding of RanGAP1 to the POK kinesins, we investigated their interaction after transient expression in N. benthamiana. Fig. S4A shows that both full-length RanGAP1 and the N-terminal WPP domain bind POK1C, the C-terminal fragment of POK1 also shown to bind to TANGLED (9). The RanGAP1WPP/AAP-GFP mutant, which does not accumulate at the CDS has significantly reduced affinity for POK1C. In addition, both POK1C and full-length POK1 bind the WPP-domain of RanGAP1 in a yeast 2-hybrid assay (Fig. S4 B and C).
Like RanGAP1, TANGLED was shown to positively label the PPB and CDS and to interact with the C-terminal domain of POK1 (9). When endogenous RanGAP1 was imaged in the TANGLED mutant allele tan-csh, mitotic targeting of RanGAP1 remained relatively normal (Fig. S5 and Table 1). It has to be noted, however, that all published tan mutant alleles have only very weak phenotypes. It is therefore possible that RanGAP1 localization might be affected in a stronger tan mutant background.
Arabidopsis RanGAP1 concentration at the nuclear envelope in nondifferentiated root tip cells is dependent on a group of nuclear pore-associated proteins (WIPs) (18). However, WIPs appear dispensable for RanGAP1 phragmoplast midline/cell plate targeting during cytokinesis (18). Similarly, the mitotic targeting of RanGAP1 to the PPB, kinetochore, or CDS does not depend on WIPs, supporting the notion that different mechanisms are involved in targeting plant RanGAP at different cell cycle stages (Fig. 3B).
Inducible RanGAP Depletion in Roots Leads to Incomplete and Misplaced Cell Walls.
The functional investigation of mitotic plant RanGAP is non-trivial for several reasons. First, depleting RanGAP likely affects interphase nucleocytoplasmic transport, which could conceivably lead to pleiotropic downstream effects. Second, Arabidopsis RanGAP is encoded by a family of 2 highly similar genes (63% identity and 80% similarity at the amino acid level) with similar expression patterns, expected to act redundantly. And third, RanGAP activity is likely essential, and a complete loss-of function mutant is therefore expected to be lethal.
We isolated T-DNA insertion mutants for RanGAP1 and RanGAP2 and applied 2 specific polyclonal antisera to investigate protein levels (see Materials and Methods for details). Both the RanGAP1 mutant allele rg1-1 and the RanGAP2 mutant allele rg2-3 are likely null alleles based on the absence of detectable protein (Fig. S6). Both rg1-1 and rg2-3 were phenotypically normal, suggesting that the 2 copies of RanGAP are indeed functionally redundant (data not shown). A cross between rg1-1 and rg2-3 showed that the rg1-1 rg2-3 double mutant is gametophyte lethal (T.R.-P., X.M.X., and I.M., unpublished data), suggesting that RanGAP activity is essential in plants.
In light of these findings, an inducible RNAi strategy was instead explored. Two constructs were made to specifically deplete RanGAP1 in the rg2-3 background (RanGAP1RNAi/rg2-3) or RanGAP2 in the rg1-1 background (RanGAP2RNAi/rg1-1). Identical phenotypes were achieved from both constructs (see below). The phenotype was dependent on the homozygosity of the corresponding T-DNA insertion background. A line that contained a truncated RanGAP1RNAi construct in rg2-3 that did not deplete RanGAP1 was used as a control against artifacts caused by the induction conditions (Fig. 4A). It behaved identical to the RNAi lines under uninduced conditions shown below (data not shown). Only data observed for RanGAP1RNAi/rg2-3 are described below.
Fig. 4.
Disorganized root cell files are observed after depletion of Arabidopsis RanGAP. (A) Immunoblot showing the effective depletion of RanGAP1 after 4-day Dex induction in RanGAP1RNAi/rg2-3 roots. Such depletion was not observed in the control line. Sections of Coomassie Brilliant Blue-stained replica gels below each blot serve as loading control. Un, uninduced; In, induced. (B) Root phenotype of 9-day-old RanGAP1RNAi/rg2-3 without (Left) or with Dex induction (Right). (C) In contrast to the uninduced control (Upper Left), oblique cell walls (arrowheads) and cell wall stubs (arrow) were observed frequently in induced RanGAP1RNAi/rg2-3 root tips (Upper Right and Lower). FM4–64-stained cortex cells are shown. (Scale bars: 20 μm.)
After 4–9 days of induction, there was a pronounced reduction of RanGAP1 level in the roots of RanGAP1RNAi/rg2-3 plants (≈70% reduction, Fig. 4A) compared with uninduced control plants, with a lesser reduction in shoots (≈40% reduction, data not shown). Immunofluorescence experiments with the anti-RanGAP1 antibody showed that the level of RanGAP1 was not depleted equally in all cells, with some cells, especially toward the root tip, still showing RanGAP1 signal at the nuclear envelope (data not shown). At this stage, induced plants had shorter roots with swollen root tips (Fig. 4B). Under higher magnification, this phenotype resembled the previously reported radially swollen mutants (19, 20). No obvious shoot phenotype was detected, possibly based on the less severe RanGAP1 depletion in shoots. The swollen root phenotype might conceivably be a currently not understood interphase effect on directional cell expansion, downstream of nucleocytoplasmic partitioning or other unknown functions of RanGAP.
When the root tips were examined closely, oblique cell walls and cell wall stubs were found frequently in induced RanGAP1RNAi/rg2-3 plants, suggesting aberrant cell division events (Fig. 4C). This was in contrast to the consistently orderly cell files in control plants. These effects were very similar to those observed in tan and pok1 pok2 mutants (8, 9). To further investigate whether mitotic figures were affected, we attempted to capture RanGAP-depleted cells during cell division and investigate the microtubule structures. However, this approach failed due to the near-absence of dividing cells in the growth-arrested roots in combination with the uneven RanGAP depletion levels. Although we can therefore—with the tools at hand—not exclude that the misplaced cell walls are downstream effects of interphase functions of RanGAP, we note the strong correlation between RanGAP1 localization at the PPB and CDS, its interaction with known cell plate positioning regulators, and the depletion phenotypes consistent with cell plate-positioning defects, all indicating a role for RanGAP during cytokinesis.
Discussion
An unsolved question in the process of plant cytokinesis is how the spatial memory of the PPB position is retained into cytokinesis, when the cell plate fuses with the plasma membrane at exactly the position of the former PPB. Until recently, no positive markers were known that labeled the division site after disassembly of the PPB. Arabidopsis TANGLED has recently been shown to be such a marker and tangled mutants show disruption of cell plate guidance (8). However, the molecular mechanism of the process is still unknown. A major player of delivering spatial information in animal cells is the small GTPase Ran. The specific localization of either RCC1 or RanGAP provides local information, through establishing a high or low concentration of RanGTP, enabling in turn local processes, such as microtubule plus end growth and microtubule attachment to kinetochores.
We have now found that in Arabidopsis RanGAP1 is a positive marker of the PPB to CDS memory. RanGAP1 persists at the future division site after disassembly of the PPB and consistently persists in the presence of the microtubule-depolymerizing drug oryzalin. Its association with the PPB site requires FASS/TONNEAU2, suggesting that, although RanGAP can persist after PPB disassembly, it requires microtubules for its primary targeting to the PPB. Alternatively, FASS/TONNEAU2 might act upstream of both PPB assembly and a microtubule-independent process of RanGAP targeting to the PPB. Interestingly, RanGAP1 persistence at the division site, but not RanGAP1 association with the PPB also requires the kinesins POK1 and POK2, which have been shown to be required for efficient association of TANGLED with PPB and CDS (9). We found evidence that the WPP-domain of RanGAP1 physically interacts with the C-terminal coiled-coil domain of POK1 and with full-length POK1. POK1 interaction and CDS association require the same 3-amino acid motif within the WPP domain (Figs. S3 and S4A), indicating a close correlation between the ability to bind POK1 (and possibly other coiled-coil proteins) and retention at the CDS.
The finding that RanGAP1 still associates with the PPB in pok1 pok2 mutants suggests that POK1 and POK2 are not responsible for the delivery of RanGAP1 to the PPB per se. However, the absence of a RanGAP1 signal at the CDS in pok1 pok2 mutants suggests a defect in maintaining an aspect of CDS identity necessary for RanGAP1 association in pok1 pok2 mutants. The ability of the WPP-domain to bind POK1 might suggest that RanGAP1 interacts with either POK1 or POK2 directly, or with another member of the large family of Arabidopsis kinesins. The Arabidopsis genome encodes for 23 kinesins that are in some way implicated in mitosis-related processes. POK1 and POK2 belong to a group of 8 mitotic kinesins that are specific to plants. This group contains AtPAKRP1 and AtPAKRP2, which associate with the phragmoplast, and several as yet uncharacterized kinesins (3). The possibility that RanGAP1 interacts in vivo with a different kinesin might explain why the original recruitment of RanGAP1 to the PPB is not disrupted in the absence of POK1 and POK2. POK1 and POK2 might then be required to deliver a factor to the CDS that is essential for RanGAP retention after PPB disappearance. It will be important to determine the currently unknown subcellular location of POK1 and POK2, specifically during cell division, and the affinity of RanGAP for other kinesins to begin to address these scenarios.
Interestingly, we found no effect of depleting the WIP coiled-coil protein family involved in anchoring RanGAP1 at the NE on the association of RanGAP1 with the PPB, CDS, or other mitotic locations. Nevertheless, introducing the WPP to AAP mutation that disrupts binding to the WIP family disrupted all cellular targeting events. This suggests that different, currently unknown interaction partners of the WPP domain are involved in anchoring RanGAP1 to the cell cortex, the kinetochores, and the cell plate. Of these events, only targeting to the cell cortex requires POK1 and POK2, suggesting that the different mitotic targeting events are again accomplished by different mechanisms.
Induced depletion of RanGAP leads to incomplete and irregularly positioned cell walls. Although we cannot formally exclude downstream effects of impaired interphase RanGAP functions, we favor the hypothesis that RanGAP plays a direct role in Arabidopsis cytokinesis, consistent with its continuous association with the division site. What could such a role be? Several lines of evidence point at important functions of the Ran cycle in microtubule biology during animal mitosis and cytokinesis. The general theme is that a high local concentration of RanGTP promotes microtubule growth. This is based on the recently recognized function of karyopherins of the importin beta type to act as inhibitors of microtubule growth by sequestering NLS-containing spindle assembly factors (SAFs), such as TPX2 and HURP. Like in nuclear import, RanGTP dissociates the karyopherin-cargo complex, thereby releasing SAFs from inhibition. Downstream events involve TPX2-dependent phosphorylation of Aurora-A kinase, which in turn is proposed to activate microtubule nucleation factors (ref. 10 and references therein). One proposition in line with these activities is that RanGAP could assist in the disassembly of the PPB by keeping local RanGTP levels low and thereby favoring depolymerization over polymerization of microtubules at this site.
In addition to microtubule stability, RanGTP has been shown to affect the polarity of microtubule motor activities. Wilde et al. (21) showed that RanGTP increases plus end-directed motor activity and decreases minus-end directed activity. During plant cytokinesis, the microtubule arrays of the phragmoplast are oriented with the plus ends toward the growing cell plate and deliver vesicles presumably by plus end-directed motor activity (22). The association of RanGAP1 with the CDS and growing rim of the cell plate/phragmoplast midline in plants might indicate reduced RanGTP at these sites. In analogy to the animal systems, this might cause reduced plus end growth of the phragmoplast microtubules and/or reduced plus-end motor activity. Such a regulatory function could conceivably fine-tune phragmoplast and vesicle delivery dynamics and thereby contribute to the overall precision of the processes underlying cell plate synthesis and positioning.
Further work will be required to establish the spatial distribution of RanGTP and RanGDP during plant mitosis and its relationship to microtubule biology, and to develop more precise tools to disturb the Ran cycle specifically in mitotic cells. The data presented here connect a spatial organization problem of the plant cell (cell plate positioning) with a well-established spatial regulator of animal mitosis, the Ran cycle.
Materials and Methods
Plant Materials.
Arabidopsis seedlings (Columbia and Wassilewskija ecotype) were grown in soil under standard long-day condition (16 h light and 8 h dark) or on MS (Caisson Laboratories) plates under constant light. Mutant rg1-1 (SALK_058630, with a T-DNA inserted ≈720 bp downstream of the RanGAP1 start codon) was acquired from the Arabidopsis Biological Resource Center (ABRC, The Ohio State University, Columbus, OH). Mutant rg2-3 (FLAG_184A06, with the T-DNA insertion ≈190 bp upstream of the RanGAP2 start codon) was obtained from the Versailles T-DNA lines collection (23). Homozygous insertion plants were identified by PCR genotyping (http://signal.salk.edu/tdnaprimers.2.html). The mutant line wip1-1 wip2-1 wip3-1 and transgenic plants expressing RanGAP1-GFP, RanGAP1WPP/AAP-GFP, and RanGAP1ΔC-GFP were described in ref. 18. Mutant or marker lines ton2-14 (16), pok1 pok2 (9), and HTR12-GFP (24) were previously described.
Inducible RNAi.
For Dex-inducible RNAi, the first 334 nt of the RanGAP1 coding region and the last 478 nt of the RanGAP2 coding region were cloned into pENTR3C and pENTR/D-TOPO, respectively. The target fragments were subsequently recombined into destination vector pOpOff2(hyg) (25). Transgenic Arabidopsis plants were selected on MS plates containing Hygromycin (35 μg/ml) and CEFOTAXIM (125 μg/ml) after Agrobacterium transformation. A line recovered from this transformation that carried a truncated transgene and did not lead to RanGAP depletion was used as a control (Fig. 4A). For induction, seedlings were either germinated directly on or transferred to MS plates containing Dex (10 μM; Sigma), whereas ethanol (solvent) was used for control treatments.
Immunolabeling and Confocal Microscopy.
Immunolabeling and confocal microscopy were performed as described in ref. 26 and as detailed in SI Materials and Methods.
Antibody Development.
Development of the anti-RanGAP1 antibody has been described in ref. 27. For the anti-RanGAP2 antibody, full length RanGAP2 protein was expressed as a His-tag fusion protein using the pDEST17 vector (Invitrogen). After purification of the recombinant protein with a Ni-NTA resin column and excision from a preparative SDS/PAGE gel, a guinea pig antiserum was produced by Cocalico Biologicals (Reamstown, PA).
Supplementary Material
Acknowledgments.
We thank SIGnAL and ABRC for sequence-indexed T-DNA insertion mutants; Annkatrin Rose and members of the Meier lab for fruitful discussions; Biao Ding for use of his confocal microscope; Paula Monsma for technical help with microscopy; Jessica Lucas for ton2-14; Laurie Smith for pok1 pok2, tan-csh, and pAD-POK1C; and Masayoshi Nakamura and Takashi Hashimoto for the mCherry-TUB6 line. This work was supported by the National Science Foundation (I.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806157105/DCSupplemental.
References
- 1.Van Damme D, Vanstraelen M, Geelen D. Cortical division zone establishment in plant cells. Trends Plants Sci. 2007;12:458–464. doi: 10.1016/j.tplants.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 2.Hoshino H, Yoneda A, Kumagai F, Hasezawa S. Roles of actin-depleted zone and preprophase band in determining the division site of higher-plant cells. Propoplasma. 2003;222:157–165. doi: 10.1007/s00709-003-0012-8. [DOI] [PubMed] [Google Scholar]
- 3.Vanstraelen M, et al. Cell cycle-dependent targeting of a kinesin at the plasma membrane demarcates the division site in plant cells. Curr Biol. 2006;16:308–314. doi: 10.1016/j.cub.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 4.Buschmann H, et al. Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr Biol. 2006;16:1938–1943. doi: 10.1016/j.cub.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 5.Hall Q, Cannon MC. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Damme D, et al. Somatic cytokinesis and pollen maturation in Arabidopsis depend on TPLATE, which has domains similar to coat proteins. Plant Cell. 2006;18:3502–3518. doi: 10.1105/tpc.106.040923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith LG, Hake S, Sylvester AW. The tangled-1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development. 1996;122:481–489. doi: 10.1242/dev.122.2.481. [DOI] [PubMed] [Google Scholar]
- 8.Walker KL, Muller S, Moss D, Ehrhardt DW, Smith LG. Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr Biol. 2007;17:1827–1836. doi: 10.1016/j.cub.2007.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller S, Han S, Smith LG. Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr Biol. 2006;16:888–894. doi: 10.1016/j.cub.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 10.Ciciarello M, Mangiacasale R, Lavia P. Spatial control of mitosis by the GTPase Ran. Cell Mol Life Sci. 2007;64:1891–1914. doi: 10.1007/s00018-007-6568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol. 2004;14:611–617. doi: 10.1016/j.cub.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 12.Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong SY, Rose A, Joseph J, Dasso M, Meier I. Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 2005;42:270–282. doi: 10.1111/j.1365-313X.2005.02368.x. [DOI] [PubMed] [Google Scholar]
- 14.Pay A, et al. Plant RanGAPs are localized at the nuclear envelope in interphase and associated with microtubules in mitotic cells. Plant J. 2002;30:699–709. doi: 10.1046/j.1365-313x.2002.01324.x. [DOI] [PubMed] [Google Scholar]
- 15.Otegui MS, Verbrugghe KJ, Skop AR. Midbodies and phragmoplasts: Analogous structures involved in cytokinesis. Trends Cell Biol. 2005;15:404–413. doi: 10.1016/j.tcb.2005.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camilleri C, et al. The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell. 2002;14:833–845. doi: 10.1105/tpc.010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose A, Meier I. A domain unique to plant RanGAP is responsible for its targeting to the plant nuclear rim. Proc Natl Acad Sci USA. 2001;98:15377–15382. doi: 10.1073/pnas.261459698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu XM, Meulia T, Meier I. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol. 2007;17:1157–1163. doi: 10.1016/j.cub.2007.05.076. [DOI] [PubMed] [Google Scholar]
- 19.Bannigan A, Wiedemeier AM, Williamson RE, Overall RL, Baskin TI. Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol. 2006;47:949–958. doi: 10.1093/pcp/pcj067. [DOI] [PubMed] [Google Scholar]
- 20.Wiedemeier AM, et al. Mutant alleles of Arabidopsis RADIALLY SWOLLEN 4 and 7 reduce growth anisotropy without altering the transverse orientation of cortical microtubules or cellulose microfibrils. Development. 2002;129:4821–4830. doi: 10.1242/dev.129.20.4821. [DOI] [PubMed] [Google Scholar]
- 21.Wilde A, et al. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- 22.Jurgens G. Cytokinesis in higher plants. Annu Rev Plant Biol. 2005;56:281–299. doi: 10.1146/annurev.arplant.55.031903.141636. [DOI] [PubMed] [Google Scholar]
- 23.Bouchez D, Camilleri C, Caboche M. A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C Roy Acad Sci Paris Sci de la vie/Life Sci. 1993;316:1188–1193. [Google Scholar]
- 24.Fang Y, Spector DL. Centromere positioning and dynamics in living Arabidopsis plants. Mol Biol Cell. 2005;16:5710–5718. doi: 10.1091/mbc.E05-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wielopolska A, Townley H, Moore I, Waterhouse P, Helliwell C. A high-throughput inducible RNAi vector for plants. Plant Biotechnol J. 2005;3:583–590. doi: 10.1111/j.1467-7652.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- 26.Friml J, Benkova E, Mayer U, Palme K, Muster G. Automated whole mount localisation techniques for plant seedlings. Plant J. 2003;34:115–124. doi: 10.1046/j.1365-313x.2003.01705.x. [DOI] [PubMed] [Google Scholar]
- 27.Jeong S, Rose A, Joseph J, Dasso M, Meier I. Plant-specific mitotic targeting of RanGAP requires a functional WPP domain. Plant J. 2005;42:270–282. doi: 10.1111/j.1365-313X.2005.02368.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.