Abstract
Cell wall peptidoglycan-anchored surface proteins are essential virulence factors in many gram-positive bacteria. The attachment of these proteins to the peptidoglycan is achieved through a transpeptidation reaction, whereby sortase cleaves a conserved C-terminal LPXTG motif and covalently attaches the protein to the peptidoglycan precursor lipid II. It is unclear how the sorting reaction is regulated spatially and what part sortase localization plays in determining the distribution of surface proteins. This is mainly the result of inadequate immunofluorescence techniques required to resolve these issues in certain bacterial pathogens. Here we describe the utilization of the phage lysin PlyC to permeabilize the cell wall of Streptococcus pyogenes to antibodies, thereby allowing the localization of sortase A using deconvolution immunofluorescence microscopy. We find that sortase localizes within distinct membranal foci, the majority of which are associated with the division septum and colocalize with areas of active M protein anchoring. Sortase distribution to the new septum begins at a very early stage, culminates during septation, and decays after division is completed. This implies that the sorting reaction is a dynamic, highly regulated process, intimately associated with cell division. The ability to study cytoplasmic and membrane antigens using deconvolution immunofluorescence microscopy will facilitate further study of cellular processes in S. pyogenes.
Keywords: immunofluorescence, microscopy, protein sorting, phage lysin, M protein
Cell wall anchored surface proteins play a vital role in the pathogenic process of many gram-positive bacteria (1). These proteins all have a conserved C-terminal anchor domain comprised of an LPXTG motif followed by a hydrophobic region and a few positively charged amino acids at the C terminus (2, 3). During protein export, the C-terminal anchor domain is stalled in the secretion channel, leaving the LPXTG motif exposed on the outer surface of the membrane. The transpeptidase sortase then cleaves this motif between the threonine and glycine residues (4) and attaches the freed threonine to the peptidoglycan precursor, lipid II (5). Lipid II then serves as substrate for peptidoglycan synthesis, leading to the covalent attachment of the protein to the cell wall.
The important human pathogen Streptococcus pyogenes (6) employs an impressive array of wall-anchored virulence factors that function in immune evasion, adherence, and invasion, among other roles (7). In S. pyogenes, sortase A is the housekeeping sortase and was shown experimentally to anchor M protein, protein F, C5a peptidase (ScpA), and protein G-related α2-macroglobulin-binding protein (GRAB) to the cell wall (8). In addition to sortase A, S. pyogenes harbors two specialized sortases, associated with the FCT (fibronectin-binding, collagen-binding T antigen) region. Of these, one is specific for T antigen, a major component of the streptococcal pili (8, 9), while the other recognizes an altered consensus sequence, QVPTGV and anchors a protein of unknown function (10).
While the biochemical aspects of the sorting reaction have been studied in detail in various organisms (1), much less is known about the spatial organization of this process. First clues that the sorting reaction is spatially controlled came from studies conducted in the 1960s that examined the regeneration of M protein on cells treated with trypsin (11, 12). These studies showed that M protein is actively anchored to the streptococcal cell wall solely at the division septum. On the other hand, protein F, a major fibronectin-binding protein, was subsequently found to be localized mainly at the old pole (13). A recent study showed that the signal sequence directs these two proteins to their respective positions on the cell surface and that switching the signal sequence between them results in altered localization (14).
Our current knowledge regarding the spatial organization of protein sorting in S. pyogenes is mainly derived from the study of anchored surface proteins, while the actual distribution of sortase remains unknown. Localization of this protein by immunofluorescence has so far been hindered by the fact that lysozyme, a muralytic enzyme commonly used to permeabilize bacterial cell walls to antibodies, has only a marginal effect on S. pyogenes (15, 16). In this report, we introduce the use of the phage lysin PlyC (17) as a tool to overcome this problem. We found that low-dose treatment of fixed cells with this highly efficient cell wall hydrolase, permeabilizes the bacterial cell wall to antibodies without causing adverse effects to the cellular morphology. Preservation of cellular morphology is largely dependent on the presence of M protein anchored to the cell wall. Using this method, we were able to determine the localization of sortase by deconvolution immunofluorescence microscopy.
Results
Production and Validation of Anti-Sortase A Antibodies.
Hexahistidine-tagged sortase A, lacking its N-terminal transmembrane domain, was purified (supporting information (SI) Fig. S1A) and determined to be catalytically active (data not shown). This protein was used for immunization of mice and affinity purification of resulting anti-sortase antibodies. We deleted sortase from D471 and found that the resulting strain, AR01, missorted M protein to the cytoplasm and supernatant in agreement with previous results (8). Complementation of AR01 with pAR107, a plasmid expressing sortase, restored the wild-type phenotype (Fig. S1B). We then analyzed the specificity of the anti-sortase antibodies by Western blot (Fig. S1C). These antibodies reacted with a band of the correct size in the wild-type strain D471, but not with the sortase mutant AR01. Recombinant sortase expressed from a plasmid restored reactivity to the antibodies. An additional faint band, present in both D471 and plasmid-complemented AR01, is likely to be a sortase cleavage product, as it is completely absent from AR01.
The Effect of Mild PlyC Treatment on the Morphology of Fixed S. pyogenes Cells.
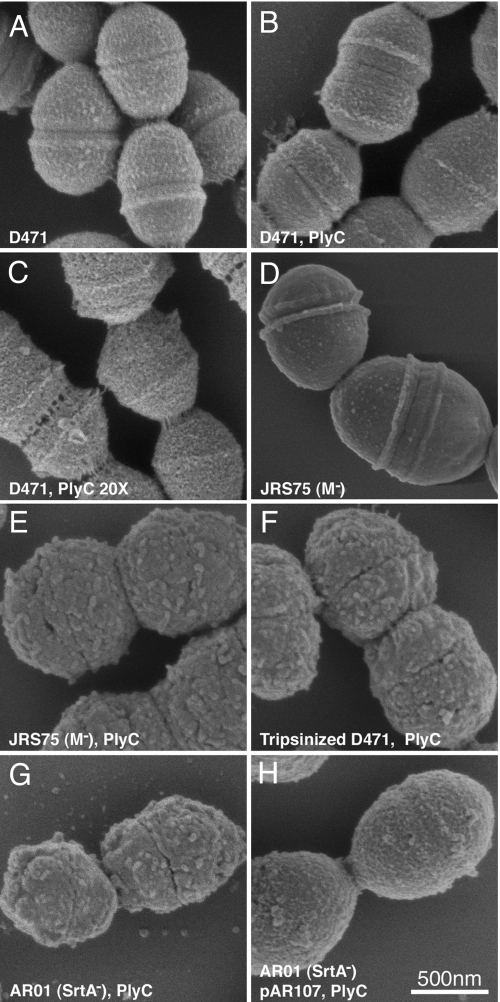
Permeabilization of the cell wall to antibodies is a prerequisite for the use of immunofluorescence microscopy for the study of subsurface antigens in bacteria. S. pyogenes cells were fixed using paraformaldehyde/glutaraldehyde, treated with methanol, and then treated with a low dose of the phage lysin PlyC. Membrane permeabilization with methanol equalizes the cellular and environmental osmotic pressures and is required to prevent membrane bulging through the PlyC-generated holes in the cell wall. Wild-type D471 cells treated in this manner display only minor morphological alterations compared to untreated cells (Fig. 1, compare A, untreated and B, treated). Furthermore, the application of 20× concentrated PlyC, which leads to visible perforation of the cell wall (not used for immunofluorescence), does not result in cell rupture (Fig. 1C). Close examination of the perforation pattern displayed by these cells reveals that PlyC is active throughout the cell wall, as was previously demonstrated by the binding pattern of fluorescent PlyC (17).
Fig. 1.
Morphology of S. pyogenes following PlyC treatment. Fixed cells were attached to glass slides, dipped in methanol, and washed with PBS. Unless otherwise noted, the cells were treated with 3 U/ml PlyC in PBS for 10 min at room temperature, before processing for scanning electron microscopy examination. (A) D471 untreated, (B) D471, PlyC treated. (C) D471, 20× PlyC treated (60 U/ml). (D) JRS75 untreated. (E) JRS75, PlyC treated. (F) Trypsinized D471, PlyC treated. (G) AR01, PlyC treated. (H) AR01 + pAR107, PlyC treated.
We observed that M protein, a fibrous coiled-coil molecule extending 60 nm from the cell surface, and readily seen as irregular structures on the wall of WT D471 (Fig. 1A), is pivotal in preserving cell wall integrity following PlyC treatment. Even mild PlyC treatment of the isogenic M protein knockout strain JRS75, causes the removal of the cell wall and the creation of spheroplasts (Fig. 1, compare D, untreated and E, treated). Furthermore, removal of M protein from D471 by trypsin digestion similarly predisposes the cell wall to removal by PlyC and formation of spheroplasts (Fig. 1F).
In addition, PlyC treatment of the sortase mutant, AR01, which does not anchor M protein to the cell wall (Fig. S1B), leads to a similar phenotype (Fig. 1G), while complementation of AR01 with pAR107 restores near-WT phenotype (Fig. 1H). We therefore propose that M protein may be forming a cross-linked mesh around the cell following fixation, and that this mesh preserves the gross overall integrity of the cell wall following gentle PlyC treatment, despite the partial degradation of the peptidoglycan.
Localization of Sortase in S. pyogenes.
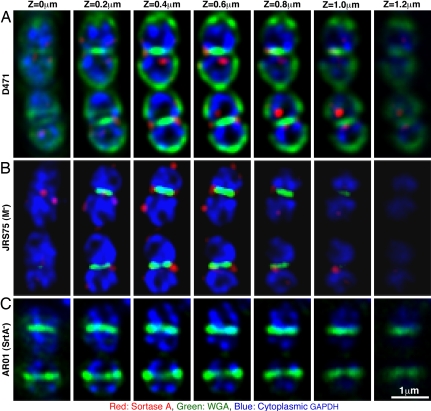
The application of mild PlyC treatment allowed us to study the localization of sortase A in S. pyogenes using deconvolution microscopy. The images in Fig. 2 are presented as serial Z-stack captures and represent a typical distribution of sortase in a short chain of streptococcal cells. Sortase was found to localize to a number of foci in D471 cells (Fig. 2A, Movie S1), while the sortase mutant AR01 did not react with the antibodies (Fig. 2C). Sortase foci were predominantly associated with the division septum, but were not always strictly confined to the division plane. Sortase foci could also be found at the equatorial rings (discussed below) and, to a lesser extent, at the poles. In these experiments, wheat germ agglutinin (WGA) was used to visualize the cell wall peptidoglycan and carbohydrate, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a cytoplasmic marker.
Fig. 2.
Sortase A localizes to distinct foci in S. pyogenes cells. D471 (A), JRS75 (B), and AR01 (C) were fixed and permeabilized with PlyC as described in Materials and Methods The cells were labeled for sortase (red) and cytoplasmic GAPDH (blue) using respective antibodies. Cell wall material was labeled with WGA (green). The data are presented as sequential Z-stacks captured with 0.2-μm intervals. To better visualize septal wall material, the WGA channel (green) of panels B andC is enhanced as compared to A.
Cytoplasmic GAPDH labeling was used to rule out the possibility that sortase localization in foci results from incomplete permeabilization of the cell wall. GAPDH is a glycolytic enzyme, which has also been found on the surface of several S. pyogenes strains, where it plays a role in binding various host factors (18). We found that log-phase D471 cells however, express very little surface GAPDH in comparison to the vast cytoplasmic pool (data not shown). This fact enabled us to use it as a cytoplasmic marker, applying labeling conditions under which surface GAPDH fluorescence is negligible. Effective labeling of cytoplasmic GAPDH demonstrates that antibodies have free access into the cell. Cytoplasmic areas from which GAPDH is excluded, coincide with the location of the nucleoid (data not shown), a phenomenon that was also observed when soluble GFP was expressed from a plasmid (data not shown) and is therefore not a labeling artifact. In addition, sortase distribution in the M protein negative strain JRS75, which loses its peripheral cell wall altogether during PlyC treatment (Fig. 1E), is similarly confined to foci (Fig. 2B, Movie S2). Taken together, these results confirm that uneven permeabilization of the cell wall could not account for the sortase localization pattern.
While peripheral cell wall is removed from the M protein negative JRS75 during PlyC treatment, cell wall labeling with WGA revealed that a small amount of wall material was trapped at the septal regions (Fig. 2B, green). This produced a labeling pattern that ranged from faint rings at early stages of the cell division to strong rings and disks at later stages. A similar WGA labeling pattern was observed in the sortase mutant AR01 (Fig. 2C), which does not anchor M protein to the cell wall (Fig. S1B). Our observation that the M protein negative JRS75 is labeled with the anti-sortase antibodies to the same extent as the WT strain D471, suggests that that removal of the cell wall does not account for AR01's lack of reactivity with these antibodies.
Sortase Localizes to Membrane Foci.
Sortase A has an N-terminal membrane anchor, which suggests that it is localized to the cell membrane. To examine sortase localization in relation to the membrane, we labeled D471 cells for sortase, the membrane [with nonyl acridine orange (NAO)] and DNA, and determined the distribution of sortase in the middle section of the cells, as the image resolution over the xy plane is better than that of the z-axis (Fig. S2 A−G). Sortase foci consistently localized to the membrane and no sortase labeling was observed in the cytoplasm. Control cells (without anti-sortase antibodies) showed no sortase labeling (Fig. S2H). It is important to note that while NAO labeling shows a membranal microdomain enriched for anionic lipids in untreated S. pyogenes (19), this fine lipid structure is disrupted by the methanol treatment required for membrane permeabilization.
Localization of Sortase as a Function of Cell-Cycle Stage.
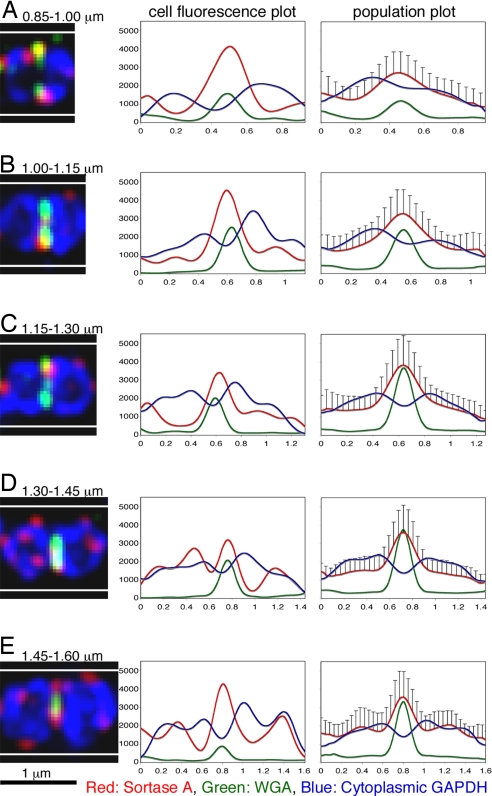
Streptococcal cells grow in chains and divide asynchronously within the chain, a fact that complicates determination of the cell-cycle stage of individual cells. We took advantage of the fact that WGA specifically labels the septa of PlyC treated, M protein negative JRS75, to detect the active division septa. JRS75 cells were stained for sortase, septal wall material, and cytoplasmic GAPDH. Cells containing a single defined septum were divided into five size groups (small to large), representing consecutive stages in the cell cycle. Two-dimensional projections were made from the Z-stack data, and the average fluorescence intensity for each antigen was plotted as a function of the distance from one of the cell poles. In the smaller size groups (Fig. 3 A–C) the cell orientation in the chain often made it possible to determine which pole is the result of the previous cell division. The fluorescence plots for these cells were aligned so that the younger pole is located to the left. For each size group at least 30 cells were analyzed, and the WGA signal, which regularly gave a sharp peak indicating the septum location, was used to align them to one another for the creation of population distribution plots (Fig. 3, right column).
Fig. 3.
Localization of sortase A in JRS75 as a function of the cell cycle stage. JRS75 cells were fixed and permeabilized with PlyC as described in Materials and Methods. The cells were labeled for sortase (red) and cytoplasmic GAPDH (blue) using respective antibodies. Septal wall material was labeled with WGA (green). Images of cells in different stages of the cell cycle (A–E) are presented as two-dimensional projections of the 3D data. Fluorescence intensity distribution of the antigens (from left to right, analyzed area is confined by the top and bottom white lines) is presented in a graph by the cells, the x-axis denotes distance from the left pole in micrometers, and the y-axis denotes arbitrary fluorescence units. The population plots to the right contain averaged data from at least 30 cells for each division stage; error bars represent one standard deviation.
The population distribution plots reveal a clear preference for sortase localization to the septum throughout the cell cycle. Localization to the septum increases gradually following septation (Fig. 3 A–B) and peaks at mid-division (Fig. 3C). In addition to the septum, sortase foci can also be found distributed to other locations in the cell, particularly to the equatorial rings and to a lesser extent, the poles. The equatorial rings are peptidoglycan features formed following division. They are located on both sides of the septum, at the border between new and old peptidoglycan, and are particularly visible in the M protein negative JRS75 cells (Fig. 1D). In time, the division sites of the daughter cells would be placed at those locations (20). Sortase localization to the equatorial rings increases during late division stages and is manifested in the population plot as secondary peaks on both sides of the septum (Fig. 3E). Assembly of sortase at these sites, before septation in the daughter cells can be detected by WGA staining, illustrates sortase recruitment as an early and gradual event in the division cycle.
Sortase localization to the poles is not uncommon, but is less pronounced. Following conclusion of a division cycle, sortase foci often linger at the closed division septum, seen at the population level as a small peak at the young (left) pole (Fig. 3A). Localization to the poles however, is not restricted to very young cells and can be detected on some cells throughout the cell cycle.
Sortase Foci Are Predominantly Associated with Sites of Active M-Protein Anchoring.
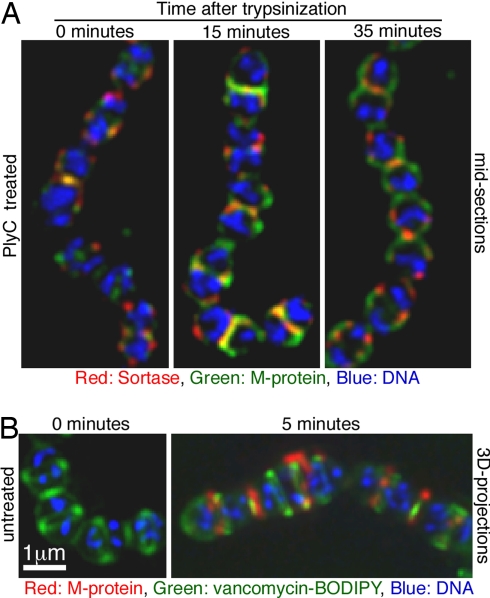
M protein, one of the most abundant surface proteins, is anchored exclusively at the division septum (12). To test the localization of sortase in relation to areas of active M protein anchoring, M protein was removed from D471 cells by trypsin digestion, and the cells were either fixed immediately or washed and incubated in media without trypsin before fixation and permeabilization. Using the 3B8 monoclonal (21), directed to the N-terminus of the M protein molecule, which is distal from the cell surface, only a small amount of M protein is detected in cells fixed immediately after trypsin digestion (Fig. 4A, time 0). This small amount, primarily trapped at the septum, was not accessible to trypsin, but could be reached by antibodies after permeabilization of the cell wall with PlyC. Following 15 min in media without trypsin, M protein was regenerated at the septal regions of cells, and sortase foci were regularly seen associated with these areas of newly anchored M protein. Following 35 min incubation, more extensive M protein anchoring is seen. By this time, septation often began in the daughter cells, and sortase associated with these newly forming septa can be seen at the flanks of areas to which M protein has been anchored. Note that the cell wall is removed during the PlyC treatment from those areas not containing M protein, because M protein is required to maintain the cell wall integrity.
Fig. 4.
Sortase foci preferentially localize to sites of active M protein anchoring. (A) D471 cells were grown to OD600 0.5 in media containing 0.05% trypsin and either fixed immediately or washed and incubated in media without trypsin for 15 or 35 min before fixation. The cells were permeabilized, stained for sortase (red), and subsequently labeled for M protein using 3B8-FITC conjugated (green) and for DNA using DAPI (blue). (B) D471 cells were trypsinized as in A and allowed to regenerate M protein for 5 min, but were not permeabilized. The cells were stained for M protein (red) using the 10B6 monoclonal and Alexa Fluor 647 conjugate, and for DNA (blue). Vancomycin-BODIPY (green) was used to detect lipid II export regions. Images are presented as two-dimensional projection of the 3D data.
M-Protein Anchoring Is Localized to Sites of Lipid II Export.
Following cleavage of the LPXTG motif in the C terminus of surface proteins, sortase attaches the protein to the peptidoglycan precursor lipid II (5), leading to its covalent linkage to the cell wall. To study the relative localization of active M protein anchoring and lipid II export, D471 cells were trypsinized as described above, allowed to regenerate M protein for 5 min, and then fixed but not permeabilized. These cells were labeled with the 10B6 monoclonal (22), which binds close to the base of the M protein molecule (at the cell wall surface) and better reflects the location of the actual anchoring sites, and vancomycin-BODIPY, which binds the D-ala−D-ala motif found in lipid II and newly deposited peptidoglycan (23). Vancomycin-BODIPY labeled a strong septal band and weaker bands at the equatorial rings at some stages of the cell cycle (Fig. 4B), similar to the pattern observed in S. pneumoniae (23, 24). Newly anchored M protein initially appeared in foci, closely associated with sites of vancomycin-BODIPY labeling. M protein was often detected simultaneously at the closing primary division septum and the mature equatorial rings/daughter cell division sites. This observation is in agreement with the early and gradual migration of sortase to the new septum (Fig. 3).
A more detailed view of M protein regeneration following trypsin treatment is presented in Fig. S3. While M protein is initially observed in septum-associated foci, these coalesce to form continuous patches at the 15- and 30-min time points (Fig. S3A) and cover the surface of the organism when no trypsin is applied (Fig. S3B). However, the distribution of M protein may not be completely even, as some variations in fluorescence intensity are visible.
Discussion
Fluorescence microscopy is one of the most widely used techniques in cell biology. Immunofluorescence uses specific antibodies to localize native antigens in the cell, and therefore requires permeabilization of the cell wall, which in model organisms such as Escherichia coli and Bacillus subtilis is routinely performed through the use of the muralytic enzyme lysozyme (25, 26). However, because of the high resistance of S. pyogenes to lysozyme (15, 16), we used the phage lysin PlyC to permeabilize the cell wall. Treatment of fixed S. pyogenes cells with a low concentration of PlyC does not result in complete degradation of the cell wall, but rather in weakening and fragmentation of the peptidoglycan. This treatment renders the peptidoglycan permeable to large molecules, such as antibodies, allowing the use of immunofluorescence to study intracellular antigens in the context of surface molecules. Our observations suggest that M protein is required to preserve the cell wall integrity following PlyC treatment. We propose that following fixation, the fibrillar coiled-coil M protein forms a cross-linked mesh at the cell surface, which tethers the fragmented cell wall, and prevents its dissociation. In the absence of M protein, the cell wall fragments are removed in subsequent washing steps, leaving behind spheroplasts that retain only a small amount of wall material trapped at the septum, which can be used for precise localization of the septum using fluorescent WGA (concentrated PlyC removes this material as well). Compared to the available immuno-EM method, immunofluorescence offers higher processivity and the ability to look at cells as a whole, rather than as thin sections. The use of immunofluorescence may be expanded to other bacteria refractory to standard procedures, by the use of phage lysins specific to these organisms, a number of which have already been cloned and characterized (27).
Proteins are anchored to the cell wall of S. pyogenes in two known patterns. M protein is anchored concomitantly with peptidoglycan synthesis at the septum and is therefore distributed throughout the cell periphery (11, 12). Protein F on the other hand, is primarily anchored at the poles (13, 14). We observed that the majority of sortase foci localize to the septal region, where they colocalize with areas of active M protein anchoring and where lipid II is exported. This suggests that the bulk of anchoring activity in the cell is coupled to peptidoglycan synthesis at the septum. Sortase recruitment to the septa is a gradual process. Sortase foci first appear at the equatorial rings of the mother cell, which then mature into division sites displaying deposition of peptidoglycan. At those initial stages it is not uncommon to find sortase distributed both to the closing mother cell septum and the forming daughter cells septa. Simultaneous protein anchoring at both locations seems possible, as both M protein regeneration experiments, and vancomycin BODIPY labeling, commonly show simultaneous labeling at these locations. Sortase foci observed at mature poles tended to be smaller and less frequent than the septal ones, but may explain anchoring of protein F at this location. The relative localization of sortase and protein F however, remains to be determined. It is also not clear whether sortase is actively recruited to the poles or is only passively retained there following division. The factors that control the recruitment and distribution of sortase are completely unknown at present. However, given the intimate connection between sortase and the division septum, the growing knowledge base regarding the factors orchestrating the division process may provide useful hints (28).
The question of whether the translocation of LPXTG proteins across the plasma membrane and protein sorting are coordinated, is still open. Localization of SecA, an ATPase associated with the Sec channel, through which LPXTG proteins are translocated, has been addressed by two immuno-EM studies. In one report SecA was found to localize to a single membranal microdomain (29), while in another, SecA was randomly distributed in the membrane (14). The reason for this difference is not clear. In a different immuno-EM study, Streptococcus mutans SecA and sortase A were found to colocalize in a single microdomain (30). While our observations suggest that S. pyogenes sortase generally localizes to more than one focus per cell, the relative locations of sortase and the secretion apparatus in this organism will have to be addressed by future studies.
A recent study found that the LPXTG-anchored protein A is attached to the cell wall of Staphylococcus aureus in 2–4 discrete foci per cell (31). These foci are distributed in a ring-like structure associated with, but not necessarily parallel to, the division septum. This distinct pattern prompts the question as to whether S. aureus sortase also localizes in foci.
In addition to sortase, S. pyogenes harbors a membrane-localized, LPXTG-specific peptidase termed LPXTGase, a unique glycosylated enzyme that contains amino acids in both D and L conformations and noncanonical amino acids (32, 33). Studying the relative localization of LPXTGase as compared to sortase may serve as a first step in understanding the relationship between these two LPXTG-specific enzymes. Further studies on possible factors that cooperate with sortase to support efficient protein sorting, and the mechanisms controlling sortase localization, could be greatly aided by the use of immunofluorescence microscopy using PlyC. Better understanding of these mechanisms may, in turn, lead to the development of new anti-infective agents given the crucial role surface proteins play in the survival of gram-positive pathogens in vivo.
Materials and Methods
Bacterial Strains.
The E. coli strains DH5α and BL21 were used for molecular cloning and recombinant protein expression, respectively. The S. pyogenes M6-type strain D471 was from the Rockefeller University collection. JRS75 is an isogenic M protein knockout mutant (34).
Culture Conditions.
E. coli strains were grown in Luria–Bertani (LB) medium, and S. pyogenes were grown in Todd–Hewitt medium (Oxoid) supplemented with 1% yeast extract (Fisher) at 37 °C. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml for E. coli; erythromycin, 200 μg/ml for E. coli, and 15 μg/ml for S. pyogenes; spectinomycin, 20 μg/ml for E. coli, and 120 μg/ml for S. pyogenes. When applicable, 0.05% trypsin was added to the media, and the cells were washed four times before incubation in fresh media without trypsin.
Reagents and Antibodies.
The 10B6 monoclonal antibody (22) was used at a 1:30,000 dilution for Western blot, and 1:2,000 for immunofluorescence. The 3B8 monoclonal (21), was FITC labeled according to manufacturer instructions (PIERCE) and used at 1:10. Affinity-purified rabbit anti-GAPDH serum (18) was used at 1:3,000 for Western blots and 1:2,000 for immunofluorescence. Affinity-purified mouse anti-sortase A antibodies were used at 1:50 for Western blotting and 1:10 for immunofluorescence. Goat anti-mouse IgG, Rhodamine Red (Jackson ImmunoResearch), and Alexa Fluor 647 (Invitrogen) conjugates, were used at 1:500. Goat anti-rabbit IgG, FITC conjugate (Sigma) was used at 1:2,000 and pseudocolored blue for figure clarity. WGA Marina Blue conjugate (Invitrogen) was used at 5 μg/ml and pseudocolored green. Vancomycin BODIPY FL, and NAO (Invitrogen) were used at 1 μg/ml, and 10 μM, respectively. All other reagents were purchased from Sigma unless otherwise noted.
Light Microscopy.
Light microscopy procedures were adjusted for S. pyogenes from Levin et al. (35). Paraformaldehyde and glutaraldehyde were added to the culture medium to final concentrations of 2.6% and 0.012%, respectively, and phosphate buffer pH 7.4 was added to 30 mM. The cells were incubated for 15 min at room temperature, and 30 min on ice, washed with PBS, and attached to polylysine-coated cover slips. The slides were washed with PBS, dipped in ice-cold methanol for 10–60 seconds, and dipped in PBS. The cells were then treated with 3 U/ml PlyC in PBS for 10 min at room temperature, washed, and blocked for 15 min with normal goat serum (Zymed) supplemented with 1% gelatin from cold-water fish skin (Sigma). Antibodies and dyes were diluted in PBS containing 2% BSA and 1% gelatin, and incubated with the cells in a moist chamber for 1 hour at room temperature. Between incubation steps the cells were washed thoroughly with PBS. To reduce bleaching of the fluorochromes, the slides were mounted in 50% glycerol and 0.1% p-phenylenediamine in PBS pH 8. Images were obtained using a DeltaVision image restoration microscope (Applied Precision/Olympus) equipped with CoolSnap QE cooled CCD camera (Photometrics). An Olympus 100×/1.40 NA, UPLS Apo oil immersion objective was used in conjunction with a 1.5× optovar. Z-stacks were taken at 0.1-μm intervals. Images were deconvolved using the SoftWoRx software (Applied Precision/DeltaVision), and corrected for chromatic aberrations. ImageJ (http://rsb.info.nih.gov/ij/) was used to analyze the fluorescence distribution profiles, and raw data were transferred to Microsoft Excel for the creation of average distribution plots. Adobe Photoshop version 7 was used for the preparation of the figures.
Scanning Electron Microscopy.
For morphology studies, slides were prepared as described for light microscopy. Following PlyC treatment, the cells were fixed in 2.5% glutaraldehyde at 4°C overnight. The cells were then treated with 1% osmium tetroxide in 0.1 M cacodylate buffer pH 7.4 for 1 hour, dehydrated using graded ethanol solutions, and critical-point dried. The slides were coated with a thin gold-palladium layer using a Desk IV coater (Denton Vacuum). Images were obtained using a LEO 1550 scanning electron microscope, with field-emission electron gun.
Description of cloning procedure, sortase purification, the production of specific antibodies, and Western blot analysis is provided as SI Methods.
Supplementary Material
Acknowledgments.
We are grateful to Sung Lee for useful discussion throughout the preparation of this manuscript, and Patricia Ryan and Anu Daniel for helpful comments. We thank Eleana Sphicas and Alison North from the Rockefeller University Bio-Imaging resource center for the capturing of EM images and advice concerning deconvolution immunofluorescence microscopy, respectively. We thank Erec Stebbins for the gift of a modified pET21a vector. This work was supported by U.S. Public Health Service Grant AI11822 (to V.A.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808301105/DCSupplemental.
References
- 1.Marraffini LA, DeDent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischetti VA, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins of Gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 3.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the Staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 4.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 5.Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham MW. Pathogenesis of group A Streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- 8.Barnett TC, Scott JR. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J Bacteriol. 2002;184:2181–2191. doi: 10.1128/JB.184.8.2181-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mora M, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnett TC, Patel AR, Scott JR. A novel sortase, SrtC2, from Streptococcus pyogenes anchors a surface protein containing a QVPTGV motif to the cell wall. J Bacteriol. 2004;186:5865–5875. doi: 10.1128/JB.186.17.5865-5875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole RM, Hahn JJ. Cell wall replication in Streptococcus pyogenes. Science. 1962;135:722–724. doi: 10.1126/science.135.3505.722. [DOI] [PubMed] [Google Scholar]
- 12.Swanson J, Hsu KC, Gotschlich EC. Electron microscopic studies on streptococci: I. M antigen. J Exp Med. 1969;130:1063–1091. doi: 10.1084/jem.130.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ozeri V, et al. De novo formation of focal complex-like structures in host cells by invading Streptococci. Mol Microbiol. 2001;41:561–573. doi: 10.1046/j.1365-2958.2001.02535.x. [DOI] [PubMed] [Google Scholar]
- 14.Carlsson F, et al. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- 15.Krause RM, McCarty M. Studies on the chemical structure of the streptococcal cell wall: I. The identification of a mucopeptide in the cell walls of groups A and A-variant streptococci. J Exp Med. 1961;114:127–140. doi: 10.1084/jem.114.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallis HA, Miller SE, Wheat RW. Degradation of 14C-labeled streptococcal cell walls by egg white lysozyme and lysosomal enzymes. Infect Immun. 1976;13:1459–1466. doi: 10.1128/iai.13.5.1459-1466.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: A multimeric bacteriophage lysin. Proc Natl Acad Sci USA. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pancholi V, Fischetti VA. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosch JW, Hsu FF, Caparon MG. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol. 2007;189:801–806. doi: 10.1128/JB.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasz A. Streptococcus pneumoniae: Functional anatomy. In: Tomasz A, editor. Streptococcus pneumoniae: Molecular Biology and Mechanism of Disease. New York: Mary Ann Liebert; 2000. pp. 9–21. [Google Scholar]
- 21.Jones KF, Fischetti VA. The importance of the location of antibody binding on the M6 protein for opsonization and phagocytosis of group A M6 streptococci. J Exp Med. 1988;167:1114–1123. doi: 10.1084/jem.167.3.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones KF, et al. Location of variable and conserved epitopes among the multiple serotypes of streptococcal M protein. J Exp Med. 1985;161:623–628. doi: 10.1084/jem.161.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 24.Ng W-L, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol. 2004;53:1161–1175. doi: 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- 25.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 26.Harry EJ, Pogliano K, Losick R. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischetti VA. Bacteriophage lytic enzymes: Novel anti-infectives. Trends Microbiol. 2005;13:491–496. doi: 10.1016/j.tim.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 28.Vicente M, Rico AI, Martinez-Arteaga R, Mingorance J. Septum enlightenment: Assembly of bacterial division proteins. J Bacteriol. 2006;188:19–27. doi: 10.1128/JB.188.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosch J, Caparon M. A microdomain for protein secretion in gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 30.Ping H, Zhuan B, Mingwen F, Meijing H, Ping Z. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol. 2008;53:150–154. doi: 10.1016/j.archoralbio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 31.DeDent AC, McAdow M, Schneewind O. Distribution of protein A on the surface of Staphylococcus aureus. J Bacteriol. 2007;189:4473–4484. doi: 10.1128/JB.00227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SG, Pancholi V, Fischetti VA. Characterization of a unique glycosylated anchor endopeptidase that cleaves the LPXTG sequence motif of cell surface proteins of gram-positive bacteria. J Biol Chem. 2002;277:46912–46922. doi: 10.1074/jbc.M208660200. [DOI] [PubMed] [Google Scholar]
- 33.Lee SG, Fischetti VA. Presence of D-alanine in an endopeptidase from Streptococcus pyogenes. J Biol Chem. 2003;278:46649–46653. doi: 10.1074/jbc.M307378200. [DOI] [PubMed] [Google Scholar]
- 34.Norgren M, Caparon MG, Scott JR. A method for allelic replacement that used the conjugative transposon Tn916: Deletion of the emm6.1 allele in Streptococcus pyogenes JRS4. J Infect Immun. 1989;57:3846–3850. doi: 10.1128/iai.57.12.3846-3850.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levin PA. Light microscopy techniques for bacterial cell biology. In: Sansonetti P, Zychlinksy A, editors. Methods in Microbiology. Vol 31. London: Academic; 2002. pp. 115–132. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.