Abstract
Bacteriophages φ29 and Nf from Bacillus subtilis start replication of their linear genome at both DNA ends by a protein-primed mechanism, by which the DNA polymerase, in a template-instructed reaction, adds 5′-dAMP to a molecule of terminal protein (TP) to form the initiation product TP-dAMP. Mutational analysis of the 3 terminal thymines of the Nf DNA end indicated that initiation of Nf DNA replication is directed by the third thymine on the template, the recovery of the 2 terminal nucleotides mainly occurring by a stepwise sliding-back mechanism. By using chimerical TPs, constructed by swapping the priming domain of the related φ29 and Nf proteins, we show that this domain is the main structural determinant that dictates the internal 3′ nucleotide used as template during initiation.
Keywords: chimerical terminal protein, polymerase, linear genomes, protein-primed replication, sliding-back
Many organisms, such as bacteriophages; animal viruses, such as adenovirus, mitochondrial plasmids, linear chromosomes and plasmids of Streptomyces (1); and more recently virus infecting Archaea, such as halovirus (2, 3), possess replication origins constituted by inverted terminal repetitions (ITR) with a terminal protein (TP) linked to both 5′ ends of their linear chromosomes (4). In these cases, the location of the 2 replication origins allows both strands to be replicated continuously, without requiring asymmetric complexes of DNA polymerase with other accessory proteins to control the different mechanics of continuous and discontinuous DNA synthesis (5). Additionally, the TP provides the OH− group of a specific serine, threonine or tyrosine to prime initiation of DNA replication from the very ends of the linear chromosome, the TP remaining covalently linked to the 5′-DNA ends (parental TP) (1, 4, 6).
The development of an in vitro replication system with highly purified proteins and DNA from bacteriophage φ29 of B. subtilis has allowed to lay the foundations of the so-called protein-primed mechanism of DNA replication (4, 6). φ29 has a linear dsDNA, 19,285 bp long, containing a TP of 31 kDa covalently linked to each 5′ end [TP-DNA (7)] that, together with a 6-bp inverted terminal repeat (3′-TTTCAT-5′) (8, 9) form part of a minimal replication origin. Once the replication origins are specifically recognized by a heterodimer formed by the DNA polymerase and a free TP molecule (10, 11), the DNA polymerase catalyzes the formation of a covalent bond between dAMP and the hydroxyl group of Ser232 of the TP, a reaction directed by the second T at the 3′ end (12). Then, the TP-dAMP initiation product translocates backwards 1 position to recover the template information corresponding to the first T, the so-called sliding-back mechanism, which requires a terminal repetition of 2 bp (12) and provides a way to prevent mutations at the φ29 DNA ends during initiation, since the 3′-5′ exonuclease activity of φ29 DNA polymerase cannot proofread the TP-linked nucleotide (13).
The sliding-back mechanism, occurring also in the φ29-related phage GA-1 (14); Streptococcus pneumoniae phage Cp-1 (15); and Escherichia coli phage PRD1 (16) or variations of it, such as the jumping-back mechanism that takes place in adenovirus (17), seems to be a common theme of protein-priming replication systems to maintain full-length DNA (1, 6). In addition, RNA viruses, such as Poliovirus, characterized by the presence of a protein covalently linked to the 5′ end of the viral genome, have been also shown to initiate at internal positions with a further recovery of the terminal sequence by a sliding-back mechanism (18–20). Bacteriophage Nf belongs to the group of phages that infect Bacillus (21). It contains a linear, dsDNA, 18,754 bp long (GenBank accession number EU622808), with an 8-bp ITR (3′-TTTCATTC) (21, 22) and a TP of 31 kDa covalently linked to each 5′ DNA end (21). Previous in vitro analyses showed that Nf also replicates its genome (TP-DNA) by means of a protein-primed mechanism (23, 24).
To get further insights into this special way to initiate replication, we studied the initiation and the first elongation steps of Nf TP-DNA replication by using highly purified DNA polymerase and TP and template ssDNA oligonucleotides corresponding to either the natural replication origin sequence or variants of it. The analysis indicated that the TP-dAMP initiation product is template-instructed by the 3′ third T, a stepwise sliding-back mechanism being proposed as the most likely one to account for the maintenance of the DNA length. Moreover, by swapping specific TP regions between the φ29 and Nf TPs, we show that the priming domain of the TP is the main structural determinant that dictates the use of the internal 3′ nucleotide as template during protein-primed initiation of DNA replication.
Results and Discussion
In Vitro Formation of the TP-dAMP Initiation Complex with ssDNA Templates.
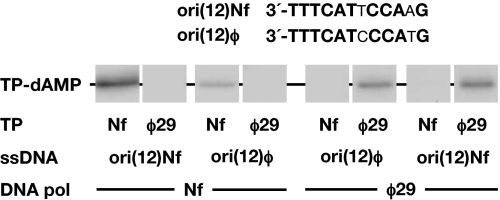
The inability of Nf DNA polymerase to catalyze DNA-independent formation of TP-dNMP products indicated that the specificity for dATP was provided by the Nf TP-DNA template whose replication origins, located at both ends, contain the 3′-TTT terminal sequence (24) (see in Fig. 1 the sequence of the right replication origin). To determine which 3′ terminal T of the template directed the Nf initiation reaction, we used as a template single-stranded oligonucleotides containing the Nf replication origin sequence or variants of it. To prevent exonucleolytic degradation of the template oligonucleotides, the catalytic Nf DNA polymerase Asp-66 residue, belonging to the Exo II motif and universally conserved in proofreading DNA polymerases (25) was mutated into alanine (see Materials and Methods). Initially, we chose as template 12-mer oligonucleotides based on previous analysis of deletion derivatives performed with the φ29 system, that indicated that the minimal replication origins were constituted by the terminal 12 bp at each φ29 DNA end (26). As shown in Fig. 1, in the presence of [α-32P]dATP, the single-stranded oligonucleotide corresponding to the 3′-sequence from the right end of Nf DNA (ori(12)Nf) could be used as template by the Nf DNA polymerase/TP heterodimer, leading to the formation of the TP-dAMP initiation complex. The labeled band is specific, because a control oligonucleotide lacking the origin sequence did not give signal using [α-32P]dATP as substrate (data not shown). As also shown in Fig. 1, only the DNA polymerase/TP homocomplexes from either φ29 or Nf gave detectable reaction. φ29 and Nf replication origins share the six 3′ terminal nucleotides, which could explain why ori (12)φ and ori(12)Nf oligonucleotides can direct the TP-dAMP formation by both, Nf and φ29, DNA polymerase/TP heterodimers (see Fig. 1).
Fig. 1.
ssDNA-dependent in vitro formation of the initiation complex TP-dAMP. The reaction mixtures contained, in addition to the indicated DNA polymerases (120 nM) and TPs (120 nM), 5.5 μM either ori(12)Nf or ori (12)φ oligonucleotides as templates. Reactions were started by adding 1 mM MnCl2 and, after incubation for 20 min at 30 °C, stopped, processed and analyzed by SDS-PAGE and autoradiography (see Materials and Methods for details). The various ssDNAs, DNA polymerases and TPs, and the mobility of the TP-dAMP complexes, are indicated. Only the area containing the relevant band is presented.
Nf DNA Initiation of Replication Is Directed by the Third Nucleotide at the 3′ End of the Template Strand.
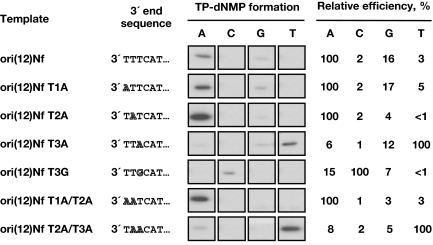
As shown in Fig. 2, when ori(12)Nf was used as template, the incorporation of dAMP to the TP was much more efficient than that of the other 3 dNMPs, supporting the idea that Nf TP-dAMP formation is directed by 1 of the 3 consecutive terminal Ts. To find out the T used as template of the initiation reaction, single changes at the first, second, or third position from the 3′-end of the template ori(12)Nf were introduced and assayed for the TP-primed initiation with each of the 4 [α-32P]dNTPs. As shown in Fig. 2, substitution of the first or second T into A [ori(12)Nf T1A, T2A and T1A/T2A] did not produce any significant change in the specificity for the nucleotide to be inserted, TP-dAMP being the initiation product preferentially formed. However, substitution of the third T either into A [ori(12)Nf T3A and ori(12)Nf T2A/T3A] or G [ori(12)Nf T3G], produced a drastic change in specificity, the nucleotide linked to the TP being mainly T and C, respectively. This result clearly indicated that Nf initiation mainly occurs opposite the third 3′ nucleotide of the template. Interestingly, the use of oligonucleotide ori(12)Nf T2A rendered the highest initiation activity. This could indicate that the sequence at the second 3′ position modulates the initiation activity, as it occurs in φ29 (27).
Fig. 2.
Internal initiation of Nf DNA replication. The formation of the 4 TP-dNMP complexes was assayed and analyzed. Standard initiation reactions were carried out with 0.1 μM each of the indicated labeled dNTP (A, C, G or T) under the conditions specified in Materials and Methods, in the presence of 60 nM Nf DNA polymerase, 120 nM Nf TP and 5.5 μM the indicated oligonucleotide as template. Reactions were started by adding 1 mM MnCl2 and, after incubation for 10 min at 10 °C, the reactions were stopped, processed, and analyzed by SDS/PAGE and autoradiography. The six 3′-terminal nucleotides of each ssDNA are also shown. The nucleotide changed in each template is indicated. For each template, the relative efficiency of the initiation reaction with each of the 4 possible substrates is indicated, considering as 100% the incorporation level of the preferred dNTP.
Recovery of the First and Second Nucleotides.
The TP-dAMP initiation complex formed using as template the third 3′ terminal nucleotide should not be directly elongated from the initiation site, because this would imply the lost of genetic information of the 2 first 3′ nucleotides. Thus, in virtue of the terminal repetitions, various terminal sequence recovery mechanisms have evolved: “sliding-back,” by which the initiation product translocates back 1 position, enabling the nucleotide used as template for the initiation reaction to direct also the insertion of the second nucleotide, as it has been described to occur in φ29 (12) and in the φ29-related bacteriophage GA-1 (14); “stepwise sliding-back,” that takes place in the S. pneumoniae phage Cp-1, which initiates at the 3′ third nucleotide of its terminal repetition (3′-TTT) (15), and by the E. coli phage PRD1 that initiates at the 4th nucleotide (3′-CCCC) (16), requiring 2 and 3 consecutive sliding-back steps, respectively, to recover the DNA end information; “jumping-back,” described to occur in adenovirus, in which the initiation product TP-CAT, initially synthesized using as template the GTA sequence at positions 4–6 in the template, jumps back to the sequence GTA at positions 1 to 3 (17). These mechanisms have been envisaged to increase the fidelity during the initiation reaction, because several base pairing checking steps have to occur before definitive elongation of the initiation product takes place (12, 13).
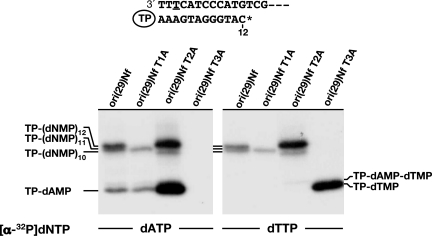
Once established that initiation of Nf DNA replication occurs opposite the third 3′ nucleotide of the template, it was necessary to ascertain whether the first 2 nucleotides are recovered during the first steps of replication. To address this question, elongation of the TP-dAMP complex was studied in the presence of ddCTP, using a 29-mer oligonucleotide containing the Nf replication origin sequence. Thus, if the 2 terminal nucleotides were recovered before elongation, the maximum length of the replication product in the presence of ddCTP would be TP-(dNMP)12 (see scheme in Fig. 3). On the contrary, if neither of the nucleotides were recovered, the elongation product would be TP-(dNMP)10. The use of the natural Nf TP-DNA as template rendered bands corresponding to all of the intermediate products (data not shown) and allowed us the proper assignment of each elongation band. As seen in Fig. 3, with the wild-type Nf ori sequence (ori(29)Nf), replication was truncated mainly at TP-(dNMP)12, although a faint band corresponding to TP-(dNMP)11 was also detected (Fig. 3 Left). This result clearly demonstrates that, despite initiating opposite the third nucleotide, all of the positions of the template, including the first and second nucleotides, are recovered during the first steps of Nf DNA replication. To establish the recovery mechanism that takes place in Nf, oligonucleotides containing a single mutation of each of the 3 terminal T were used. As seen in Fig. 3, when the mutated molecule ori(29)Nf T1A was used as template, the replicated product was TP-(dNMP)11, indicating that the mutation introduced precluded the recovery of the 3′ terminal position. Interestingly, with ori(29)Nf T2A, the elongated product corresponded mainly to TP-(dNMP)12, although a small amount of TP-(dNMP)10 was also produced. This result suggests that the presence of a single mutation in the second internal position does not preclude the recovery of the two 3′ terminal positions of the template. As expected, the use of ori(29)Nf T3A did not give any noticeable initiation product with [α-32P]dATP as substrate. Although such a labeled nucleotide could have been inserted in further elongation steps (see sequence in Fig. 3), the absence of elongation products would indicate that a mutation in the third internal position hinders not only the recovery of the ends, but also the elongation of the initiation complex. This hypothesis was supported by using [α-32P]dTTP as substrate (see Fig. 3 Right). As shown in Fig. 3, ori(29)Nf T3A gave rise to the initiation reaction, and no elongation products were detected. As expected, the elongation pattern obtained in the presence of the other 3 templates was similar to that described with [α-32P]dATP as substrate. Interestingly, a faint band likely corresponding to the TP–dAMP–dTMP product was detected with ori (29)Nf T2A. Additionally, although from the results presented here it could be inferred that only the first and third T appear to be absolutely required for recovering the terminal 3′ end sequence, because substitutions in either of them led to incomplete replication products whereas substitution at the second T gave rise to full replication, the terminal reiteration of 3 T residues seems optimal to favor the more efficient elongation of the initiation products.
Fig. 3.
Truncated elongation on Nf wild-type and mutated single-stranded oligonucleotide templates. The TP-linked DNA molecule resultant of replicating ori(29)Nf in the presence of ddCTP is shown, assuming that the first and second nucleotides are recovered during elongation. Underlined T in the template molecule marks the initiation site. Reactions were carried out in the presence of 2.3 μM the indicated oligonucleotide, 60 nM Nf DNA polymerase, 120 nM Nf TP, 0.1 μM the corresponding labeled dNTP, 10 μM each dATP, dGTP and dTTP, and 100 μM ddCTP. Reactions were started by adding 1 mM MnCl2 and, after 30 min at 30 °C, were stopped and processed as indicated in Materials and Methods. In addition to the initiation complex (TP-dAMP or TP-dTMP), the position corresponding to different end-products (TP-(dNMP)n) of each truncated elongation assay is indicated.
Two terminal sequence recovery mechanisms could be contemplated for Nf DNA replication: (i) Stepwise sliding-back: In this model, the TP-dAMP formed opposite the third position is translocated 1 position back to pair with the second T, the third T directing again the formation of the TP-AA product. Similarly, this product slides-back 1 position again, recovering the first T of the template. Then, normal chain elongation can take place. This mechanism explains why elongation carried out with ori(29)Nf T1A oligonucleotide rendered an elongation product 1 base shorter, because the absence of full complementarity between the TP-AA product and the 3′-AT of the template precludes the second sliding-back event, forcing direct elongation after the first translocation step and, as a consequence, the loss of the terminal nucleotide. Additionally, the absence of elongation observed with ori(29)Nf T3A could be explained by dissociation of the TP-dTMP product after the first sliding-back step, caused by an incorrect base pairing with the penultimate T. However, the recovery of full length elongation products with ori(29)Nf T2A leads us to propose a second recovery mechanism, (ii) the jumping-back from the third to the first T of the template. Evidence that this mechanism is taking place with ori(29)Nf T2A is the presence of a faint band, that would correspond to TP-AT complex (see Fig. 3). Thus, once the TP-dAMP initiation product jumped-back opposite the first 3′ T, the second 3′ A of the mutated template directed the insertion of a [α-32P]dTMP molecule to render TP-AT. It is difficult to know the prevalent recovery mechanism during the first steps of Nf TP-DNA replication. Two arguments could be put forward to favor the stepwise sliding-back model. First, performing of the stepwise sliding-back mechanism exclusively relies on successive backwards motions of the TP-dAMP complex, the third 3′ templating nucleotide remaining at the same position at the catalytic site to direct consecutive incorporation of the second and third nucleotides. The same principle applies also for the more complex jumping-back mechanism described to occur in adenovirus DNA replication. In this case, the 4th to 6th positions of the template also should remain fixed respect to the catalytic site after the single jump of the TP-CAT initiation product, to direct the formation of the next CAT triplet. However, in the case of Nf DNA replication, the jumping-back model would require both, a backwards motion of the TP-dAMP product to be located opposite to the first 3′ T and the concomitant upwards movement of the template strand to allocate properly the second 3′ T at the DNA polymerase catalytic site to direct the insertion of the second dAMP. Second, from a fidelity point of view, the stepwise sliding-back would offer more fidelity checking points, aborting a misinsertion occurring opposite the second position, as hypothesized to occur in the case of Cp1 (15).
Priming Domain of TP Dictates the Internal Nucleotide Used as Template of the Initiation Reaction.
In addition to the results presented above, φ29 and Nf DNA polymerase/TP heterodimers initiate replication opposite the second and third 3′-T, respectively, regardless whether the replication origin is Nf or φ29 (data not shown), indicating that the use of a specific internal nucleotide depends on a particular DNA polymerase/TP heterodimer. This prompted us to determine the role of the DNA polymerase and the TP in dictating such an initiation position.
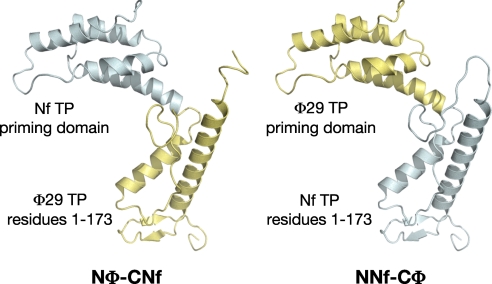
Based on the high degree of sequence identity between φ29 and Nf TPs (62.4%), we have made chimerical TPs by swapping the priming domains (residues 174–266) of both TPs (see Materials and Methods and Fig. 4) giving rise to the chimeras Nφ-CNf [φ29 TP N-terminal part, composed of intermediate and N-terminal domains (residues 1–173), linked to the Nf TP priming domain] and NNf-Cφ (Nf TP N-terminal part connected to the φ29 TP priming domain). Chimeras were overexpressed and purified as described in Materials and Methods, and further assayed in their ability to use a specific 3′ internal T as TP-primed initiation site.
Fig. 4.
Schematic representation of the chimerical TPs constructed for this study.
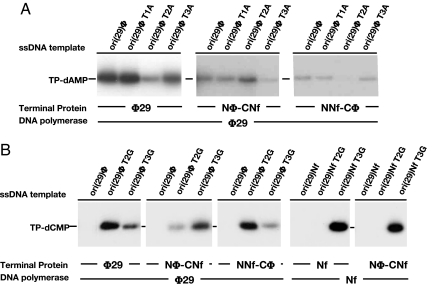
As shown in Fig. 5A, the initiation complex formation when the φ29 DNA polymerase/TP heterodimer used ori (29)φ T2A as template was much reduced compared with the case of the wild-type sequence or the T1A or T3A variants, in agreement with initiation occurring mainly opposite the penultimate 3′ T (12). Conversely, the heterodimer formed by φ29 DNA polymerase and chimera Nφ-CNf (bearing the Nf TP priming domain) clearly changed the preference in the use of the templating nucleotide: now the ori (29)φ T2A directed the initiation reaction more efficiently than ori (29)φ T3A, indicating that such a chimerical heterodimer initiated opposite the third 3′ T, as the Nf DNA polymerase/TP heterodimer did. The initiation reaction obtained with ori (29)φ T2A was higher than that obtained using the natural sequence (ori (29)φ), a hallmark of the Nf heterodimer, now mimicked simply by replacing the φ29 TP priming domain by the corresponding one of Nf TP. Interestingly, the presence of the φ29 TP priming domain in chimera NNf-Cφ produced the recovery of the pattern showed by the φ29 heterodimer, i.e., initiation at the second 3′ terminal position (see Fig. 5A). Altogether, the results presented in Fig. 5A strongly suggest that TP and, more specifically, its priming domain dictates the template position used to direct the initiation reaction. To further confirm this hypothesis, similar assays were performed using as templates oligonucleotides bearing a substitution of the second or third T into G (ori (29)φ T2G and ori (29)φ T3G, respectively) and [α-32P]dCTP as initiator nucleotide (Fig. 5B). As expected, the T2G, and in a lesser extent the T3G, variant directed the formation of the initiation product (TP-dCMP) by the φ29 heterodimer. Conversely, when the φ29 DNA polymerase/Nφ-CNf heterodimer was used, ori (29)φT3G was the main template that gave rise to initiation reaction (see Fig. 5B), in accordance with the initiation taking place preferentially at the third position, as it occurs with the Nf DNA polymerase/TP complex. Again, the presence of the φ29 TP priming domain in the heterodimer formed between φ29 DNA polymerase and chimera NNf-Cφ caused the recovery of the initiation pattern displayed by the φ29 heterodimer. In agreement with these findings, the heterodimer formed by Nf DNA polymerase and chimera Nφ-CNf (bearing the Nf TP priming domain) showed the same specificity pattern as the Nf heterodimer (see Fig. 5B). Nf DNA polymerase appeared to show a high stringency in the placement of the non-homologous φ29 TP priming domain as the complex formed with NNf-Cφ was not active (data not shown), as described to occur in the case of GA-1 DNA polymerase (28).
Fig. 5.
In vitro protein-primed initiation with chimerical TPs. (A) Reaction mixtures contained, in addition to φ29 or Nf DNA polymerase (120 nM), and either the wild-type or chimerical TPs (240 nM), 3.6 μM the indicated oligonucleotide as template and 0.1 μM [α-32P]dATP as initiator nucleotide. Reactions were started by adding 1 mM MnCl2 and, after incubation for 1 h at 30 °C, were stopped, processed and analyzed by SDS-PAGE and autoradiography (see Materials and Methods for details). (B) The reactions were carried out essentially as described in A, using 0.1 μM [α-32P]dCTP as initiator nucleotide. Reactions were started by adding 1 mM MnCl2 and incubated at 30 °C for 1 h or 30 min in the case of φ29 or Nf DNA polymerase, respectively. The reactions were stopped, processed and analyzed by SDS-PAGE and autoradiography (see Materials and Methods for details). The various templates, DNA polymerases and TP variants, and the mobility of the TP-dAMP or TP-dCMP complexes are indicated.
Taking into account that in both, Nf and φ29, the priming Ser232 has to be placed at the polymerase catalytic site in an orientation competent for catalysis, it could be thought that the difference regarding the initiation site should rely on a distinct arrangement of the template strand at the catalytic site of the DNA polymerase. Binary structures of φ29 DNA polymerase with a 5 base long oligonucleotide showed that the 3′ end of the ssDNA molecule reached the DNA polymerase active site, while its 5′ terminus remained bound to the DNA polymerase in the tunnel that lies downstream of the active site in a non-sequence specific manner, suggesting that it alone cannot establish register of the template (29). Thus, our results together with the above mentioned structures involve the TP priming domain in the positioning of the templating T in the catalytic site, most probably by sterically excluding the template from the upstream duplex binding region of polymerase, implying that the TP priming domain acts as a structural barrier. Thus, once the 3′ end of the template contacts the TP priming domain located at the duplex binding region, the corresponding internal T would be allowed to be placed at the catalytic site to direct formation of the TP-dAMP product. Structural superposition of the φ29 binary complex with the DNA polymerase/TP heterodimer showed the TP priming loop [residues 227–233 (30)] as the region that most probably would contact the 3′ end of the template. There is a difference of only 1 amino acid residue between the priming loops of φ29 and Nf TPs (31). To analyze the role of this residue in the initiation reaction, we changed Glu-229 residue of φ29 TP into Tyr, the corresponding residue in Nf TP, and vice versa. The mutant TPs did not produce any change in the specificity of the internal T used as template (data not shown). These results lead us to propose that perhaps a difference in the interaction of each priming domain with each cognate DNA polymerase could provide the basis for the different initiation site. The work presented here represents a step further in the study of the initiation of protein-primed DNA replication. Before elongation of the initiation product formed by protein-primed DNA polymerases, the sliding-back mechanism has to take place. This implies both a backwards motion of the primer (TP) with respect to the fixed template molecule, the internal template T remaining at the catalytic site to direct the following insertion step, and a breakage of the pair TP-A:T (in the case of bacteriophages φ29, Cp1, Nf, and GA-1) or, most drastically, the triple base pairing TP-CAT:GTA, as in the case of adenovirus. This energetically unfavored step should be explained by a power stroke mechanism, by which the energy released after dissociation of the pyrophosphate could drive the backwards movement of the TP-dNMP initiation product with respect to the DNA polymerase and template strand with the consequent correct base pairing with the last or preceding 3′ nucleotide to reach a more energetically favored situation. The elucidation of the conformational changes that govern the sliding-back mechanism will give the clues to understand such a special way to initiate genome replication.
Materials and Methods
Nucleotides and DNAs.
Unlabeled nucleotides and dideoxynucleotides, and [α-32P]dATP (3,000 Ci/mmol), [α-32P]dTTP (3,000 Ci/mmol), [α-32P]dCTP (3,000 Ci/mmol), and [α-32P]dGTP (3,000 Ci/mmol), were supplied by Amersham Pharmacia. Single-stranded oligonucleotides containing the wild-type and mutated sequences of the Nf and φ29 DNA right replication origin, and used as template are shown in supporting information (SI) Table S1.
Proteins.
Wild-type φ29 and Nf TPs were expressed in E. coli BL21(DE3) cells harboring the gene cloned into plasmid pT7–3 and further purified as described (24, 32). φ29 DNA polymerase mutant (D12A/D66A) and Nf DNA polymerase mutant D66A, both exonuclease deficient, were purified as described in ref. 33.
Construction, Expression, and Purification of Chimerical TPs Nφ-CNf and NNf-Cφ.
For details, see SI Text.
Construction, Expression, and Purification of Nf DNA Polymerase Exonuclease-Deficient Mutant D66A.
The D66A exo(−) Nf DNA polymerase mutant was obtained using the QuikChange site-directed mutagenesis kit provided by Stratagene. This mutation specifically inactivated the 3′ to 5′ exonuclease activity of the enzyme. The expression and purification of the protein was carried out as described for the wild-type Nf DNA polymerase (24).
Template-Dependent in Vitro Formation of the Initiation Complex TP-dNMP.
The assays were performed essentially as described in ref. 12, using the indicated amount of the specified single-stranded oligonucleotide in the presence of either 60 or 120 nM the corresponding DNA polymerase, the indicated amount of the specific TP, 40 mM ammonium sulfate, 4 μCi [α-32P]dNTP, 0.1 μM of the indicated dNTP, and 1 mM MnCl2. After incubation for the indicated times and temperatures, samples were stopped and processed as described in ref. 34. Quantification was done by densitometric analysis of the labeled band corresponding to the TP-dNMP complex.
Truncated Elongation Assays.
The reactions were carried out essentially in the same conditions described for the initiation assay, but in the presence of 10 μM each dATP, dGTP and dTTP, and 100 μM ddCTP. To allow separation of the different replication intermediates, the samples were analyzed in SDS/12% polyacrylamide gels (360 × 280 × 0.5 mm) as described in ref. 12.
Supplementary Material
Acknowledgments.
We thank Dr. Luis Blanco for helpful discussions and critical reading of the manuscript. This work was supported by Spanish Ministry of Education and Science Grant BFU 2005-00733 and Autonomous Community of Madrid (Grant P-MAT-0283-0505 (to M.S.) and by an Institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa.” E.L. was Predoctoral Fellow of the Spanish Ministry of Education and Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809882105/DCSupplemental.
References
- 1.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet Eng (N Y) 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 2.Bamford DH, et al. Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J Virol. 2005;79:9097–9107. doi: 10.1128/JVI.79.14.9097-9107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bath C, Cukalac T, Porter K, Dyall-Smith ML. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350:228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Salas M. Protein-priming of DNA replication. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 5.Blanco L, et al. Highly efficient DNA synthesis by the phage φ29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 6.Salas M, Miller J, Leis J, DePamphilis M. Mechanisms for Priming DNA Synthesis. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1996. [Google Scholar]
- 7.Salas M, Mellado RP, Viñuela E, Sogo JM. Characterization of a protein covalently linked to the 5′ termini of the DNA of Bacillus subtilis phage φ29. J Mol Biol. 1978;119:269–291. doi: 10.1016/0022-2836(78)90438-2. [DOI] [PubMed] [Google Scholar]
- 8.Escarmís C, Salas M. Nucleotide sequence of the early genes 3 and 4 of bacteriophage φ29. Nucleic Acids Res. 1982;10:5785–5798. doi: 10.1093/nar/10.19.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshikawa H, Ito J. Nucleotide sequence of the major early region of bacteriophage ϕ29. Gene. 1982;17:323–335. doi: 10.1016/0378-1119(82)90149-4. [DOI] [PubMed] [Google Scholar]
- 10.Blanco L, et al. Effect of NH4+ ions on φ29 DNA-protein p3 replication: Formation of a complex between the terminal protein and the DNA polymerase. J Virol. 1987;61:3983–3991. doi: 10.1128/jvi.61.12.3983-3991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutiérrez J, et al. Signals in the ϕ29 DNA-terminal protein template for the initiation of phage ϕ29 DNA replication. Virology. 1986;155:474–483. doi: 10.1016/0042-6822(86)90209-6. [DOI] [PubMed] [Google Scholar]
- 12.Méndez J, Blanco L, Esteban JA, Bernad A, Salas M. Initiation of φ29 DNA replication occurs at the second 3′ nucleotide of the linear template: A sliding-back mechanism for protein-primed DNA replication. Proc Natl Acad Sci USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esteban JA, Salas M, Blanco L. Fidelity of φ29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J Biol Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 14.Illana B, Blanco L, Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J Mol Biol. 1996;264:453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- 15.Martín AC, Blanco L, García P, Salas M, Méndez J. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: A stepwise sliding-back mechanism. J Mol Biol. 1996;260:369–377. doi: 10.1006/jmbi.1996.0407. [DOI] [PubMed] [Google Scholar]
- 16.Caldentey J, Blanco L, Bamford DH, Salas M. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993;21:3725–3730. doi: 10.1093/nar/21.16.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King AJ, van der Vliet PC. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: Regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994;13:5786–5792. doi: 10.1002/j.1460-2075.1994.tb06917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paul AV, Peters J, Mugavero J, Yin J, van Boom JH, Wimmer E. Biochemical and genetic studies of the VPg uridylylation reaction catalyzed by the RNA polymerase of poliovirus. J Virol. 2003;77:891–904. doi: 10.1128/JVI.77.2.891-904.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul AV, et al. A “slide-back” mechanism for the initiation of protein-primed RNA synthesis by the RNA polymerase of poliovirus. J Biol Chem. 2003;278:43951–43960. doi: 10.1074/jbc.M307441200. [DOI] [PubMed] [Google Scholar]
- 20.Yin J, Paul AV, Wimmer E, Rieder E. Functional dissection of a poliovirus cis-acting replication element [PV-cre(2C)]: Analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J Virol. 2003;77:5152–5166. doi: 10.1128/JVI.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshikawa H, Ito J. Terminal proteins and short inverted terminal repeats of the small Bacillus bacteriophage genomes. Proc Natl Acad Sci USA. 1981;78:2596–2600. doi: 10.1073/pnas.78.4.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikawa H, Garvey KJ, Ito J. Nucleotide sequence analysis of DNA replication origins of the small Bacillus bacteriophages: Evolutionary relationships. Gene. 1985;37:125–130. doi: 10.1016/0378-1119(85)90264-1. [DOI] [PubMed] [Google Scholar]
- 23.González-Huici V, Lázaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J Biol Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 24.Longás E, de Vega M, Lázaro JM, Salas M. Functional characterization of highly processive protein-primed DNA polymerases from phages Nf and GA-1, endowed with a potent strand displacement capacity. Nucleic Acids Res. 2006;34:6051–6063. doi: 10.1093/nar/gkl769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernad A, Blanco L, Lázaro JM, Martín G, Salas M. A conserved 3′-5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 26.Gutiérrez J, Garmendia C, Salas M. Characterization of the origins of replication of bacteriophage φ29 DNA. Nucleic Acids Res. 1988;16:5895–5914. doi: 10.1093/nar/16.13.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González-Huici V, Salas M, Hermoso JM. Sequence requirements for protein-primed initiation and elongation of phage φ29 DNA replication. J Biol Chem. 2000;275:40547–40553. doi: 10.1074/jbc.M007170200. [DOI] [PubMed] [Google Scholar]
- 28.Pérez-Arnaiz P, et al. Involvement of phage ϕ29 DNA polymerase and terminal protein subdomains in conferring specificity during initiation of protein-primed DNA replication. Nucleic Acids Res. 2007;35:7061–7073. doi: 10.1093/nar/gkm749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman AJ, et al. Structures of phi29 DNA polymerase complexed with substrate: The mechanism of translocation in B-family polymerases. EMBO J. 2007;26:3494–3505. doi: 10.1038/sj.emboj.7601780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamtekar S, et al. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leavitt MC, Ito J. Nucleotide sequence of Bacillus phage Nf terminal protein gene. Nucleic Acids Res. 1987;15:5251–5259. doi: 10.1093/nar/15.13.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaballos A, Salas M. Functional domains in the bacteriophage φ29 terminal protein for interaction with the φ29 DNA polymerase and with DNA. Nucleic Acids Res. 1989;17:10353–10366. doi: 10.1093/nar/17.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage φ29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 34.Peñalva MA, Salas M. Initiation of phage φ29 DNA replication in vitro: Formation of a covalent complex between the terminal protein, p3, and 5′-dAMP. Proc Natl Acad Sci USA. 1982;79:5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.