Abstract
From bacterial viruses to humans, site-specific recombination and transposition are the major pathways for rearranging genomes on both long- and short-time scales. The site-specific pathways can be divided into 2 groups based on whether they are stochastic or regulated. Recombinases Cre and λ Int are well-studied examples of each group, respectively. Both have been widely exploited as powerful and flexible tools for genetic engineering: Cre primarily in vivo and λ Int primarily in vitro. Although Cre and Int use the same mechanism of DNA strand exchange, their respective reaction pathways are very different. Cre-mediated recombination is bidirectional, unregulated, does not require accessory proteins, and has a minimal symmetric DNA target. We show that when Cre is fused to the small N-terminal domain of Int, the resulting chimeric Cre recombines complex higher-order DNA targets comprising >200 bp encoding 16 protein-binding sites. This recombination requires the IHF protein, is unidirectional, and is regulated by the relative levels of the 3 accessory proteins, IHF, Xis, and Fis. In one direction, recombination depends on the Xis protein, and in the other direction it is inhibited by Xis. It is striking that regulated directionality and complexity can be conferred in a simple chimeric construction. We suggest that the relative ease of constructing a chimeric Cre with these properties may simulate the evolutionary interconversions responsible for the large variety of site-specific recombinases observed in Archaea, Eubacteria, and Eukarya.
Keywords: bacteriophage, λ recombinase, site-specific recombination, evolution
Cre and λ Int are virally encoded recombinases that belong to a large family of tyrosine recombinases. Family members in Archaea, Eubacteria, and Eukarya participate in a wide variety of biological functions such as chromosome segregation, chromosome copy number control, gene expression, conjugative transposition, gene dissemination, and viral integration and excision (for reviews, see ref. 1). Both recombinases have been widely exploited as powerful and flexible tools for genetic engineering: Cre primarily in vivo (2) and λ Int primarily in vitro (3). Preliminary steps have also been taken toward exploiting the λ system in eukaryotes (4, 5).
The Escherichia coli bacteriophage P1-encoded Cre reduces multimeric phage genomes to monomers and thereby enhances the probability of segregation to both daughter cells. It does so by promoting a site-specific intramolecular recombination between 34-bp lox sites that occur once on each P1 chromosome. Each lox site consists of a pair of inverted repeat Cre-binding sites separated by an 8-bp spacer sequence (6, 7).
The E. coli bacteriophage λ-encoded Int promotes site-specific recombination between viral and host chromosomes and thereby enables lysogenic or lytic phage growth in response to the physiological state of the host (8). Integrative recombination between the phage (attP) and bacterial (attB) targets generates an integrated prophage bounded by hybrid sites attL and attR. The latter are targets for the excisive recombination that regenerates attP and attB on their respective parental chromosomes. Although integrative and excisive recombination reactions superficially appear to be the reverse of one another, they are in fact 2 distinct unidirectional reactions that use different (partially overlapping) ensembles of proteins and protein-binding sites (9).
Cre and Int share with themselves, and other family members, the same chemistry of isoenergetic DNA cleavage and ligation reactions that are mediated by high-energy 3′-phosphotyrosine intermediates. A tetrameric complex of recombinases executes 2 ordered pairs of transesterification reactions: they first generate a 4-way DNA junction (Holliday recombination intermediate) and then resolve it to recombinant DNA products. The staggered DNA cleavage/ligation reactions carried out by Cre and Int are separated by an “overlap” region of 6 and 7 bp, respectively.
Cre consists of 2 domains that form a C-clamp structure around each of its 4 binding sites. The C-terminal domain harbors the active site on one side of the DNA helix, and the N-terminal domain interacts with the major groove on the other side of the helix (10, 11). Despite considerable dissimilarity in primary sequence, Int shares with Cre the same C-clamp arrangement of core domains (12). However, Int has an additional 65-residue N-terminal domain superimposed on this C-clamp core domain structure via a 10-residue protease-sensitive linker (13, 14). This N-terminal domain recognizes a family of DNA sequences that are unrelated to and quite distant from the sites where DNA strand cleavage and ligation are executed (15).
The λ att sites contain 5 binding loci for the N-terminal domain of λ Int (“N-domain”) in the DNA sequences (the P and P′ arms) flanking the regions of strand exchange. Interposed between the N-domain sites and the region of strand exchange are protein-binding sites for the accessory DNA-bending proteins IHF, Xis, and Fis. Overlapping subsets of these sites are used for integrative and excisive recombination (16). Both IHF and Fis are important transcription factors that play key roles in the global regulation of host physiology, and their recruitment as recombination accessory factors is likely to optimize the viral life cycle by linking it to the physiology of the host (17, 18). However, there appears to be considerable plasticity within the large group of heterobivalent recombinases possessing an N-terminal domain similar to that of λ Int and enjoying regulated directionality because the identity, number, and patterns of protein-binding sites in the distal arms of their respective att sites vary greatly (9).
We have constructed a chimeric Cre recombinase (ch-Cre) by fusing it to the 74 N-terminal residues of λ Int. We generated hybrid recombination targets (lot sites) by replacing the inverted repeat λ Int-binding elements in the core region of attP with attenuated lox half-sites. ChCre promotes recombination between lotP and lotB only in the presence of IHF, and this recombination is strongly inhibited by Xis. However, recombination between lotL and lotR requires Xis in addition to IHF. In vivo recombination reactions, assayed by quantitative PCR (qPCR), exhibit the same regulated directionality and response to accessory proteins as predicted from the in vitro experiments. ChCre-mediated reactions proceed via the same pathway and generate recombinogenic complexes with the same structural configurations as those of λ Int. We suggest that the relative ease of constructing a chCre with regulated directionality may simulate the evolutionary interconversions responsible for the large variety of site-specific recombinases observed in nature.
Results
Construction of ChCre Recombinase and Hybrid Target Sites.
N-terminal fusions to Cre do not compromise its recombinase activity and have been used to great advantage (19). The 65 N-terminal residues of λ Int comprise a domain that recognizes and binds with high affinity to DNA sites distant from the region of strand exchange. It is joined to the larger core domain by a protease-sensitive linker region (residues 65–74) that is involved in intermolecular protein–protein interactions stabilizing the Int tetramer (14). Based on these results, we created a gene fusion encoding the first 74 residues of λ Int fused to Cre. The purified chCre is a stable protein that reacts with both Int-specific and Cre-specific antibodies, as predicted (data not shown).
To generate hybrid recombination targets, the inverted repeat-binding elements in the core regions of λ att sites were replaced with attenuated loxP sites (Fig. 1A). The λ attP arms (flanking the inverted repeat Int-binding sites) encode binding targets for the λ Int N-domain and the accessory DNA-bending proteins IHF, Xis, and Fis. To maintain the correct phasing of the accessory DNA sequences in the P and P′ arms with each other, and with the λ cleavage sites, the 6-bp overlap region of the lox site was replaced with the 7-bp overlap region from λ.
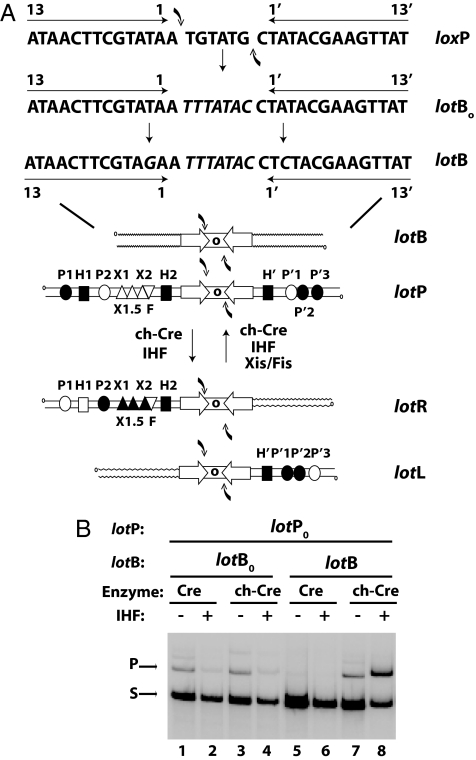
Fig. 1.
Construction and recombination of hybrid lot sites. (A) LotB was derived from loxP by changing the overlap region (lotBo) and mutating the Cre-binding sites (lotB). Replacement of the λ core regions with lotB yielded the 4 lot att sites. The arm DNA sequences encode binding sites for the N-domain of λ Int (N-domain) (circles), IHF (squares), Xis (triangles), and Fis (inverted triangles). Filled symbols indicate the subset of sites used for lotP by lotB and lotL by lotR as shown. (B) Cre- and chimeric Cre-promoted recombination between supercoiled lotP0 (6 nM) and either lotB0 or lotB (1.5 nM, [α-32P]dCTP-labeled), in the presence or absence of IHF. Recombination was analyzed by SDS/PAGE and visualized by autoradiography. The very faint bands above the product are likely the result of a second round of recombination.
Regulated Directionality of ChCre.
To identify and characterize regulated directionality in chCre-sponsored recombination, it is necessary to use recombination targets that do not respond to a latent Cre activity acting at the embedded lox sites. This was accomplished by introducing a mutation shown to weaken the interaction between Cre and its DNA-binding site (20). The base pair at position 2 in each of the inverted repeat Cre-binding sites was changed from T:A to G:C to generate the lot recombination targets (Fig. 1A). The corresponding recombination targets with wild-type Cre-binding sites are called lot0. Weakening the interaction between Cre and its DNA-binding site, along with the change in the overlap sequence, makes the Cre more λ-like, in that Int has a low affinity for its core sites on its own.
The results of recombination reactions between lotP0 (containing wild-type Cre-binding sites) and either lotB0 or lotB are shown in Fig. 1B. It is interesting that Cre is able to recombine targets with a 7-bp overlap region and that IHF suppresses recombination of lotB0 sponsored by either recombinase (lanes 1–4). As expected, IHF has no effect on Cre-promoted recombination between lox sites, which do not have binding sites for IHF (Fig. 2A). Evidently, the IHF-directed folding of lotB0 inhibits simple Cre recombination, and the tight binding of the chimeric recombinase at the Cre-binding sites interferes with, or distorts, proper binding of its N-domain.
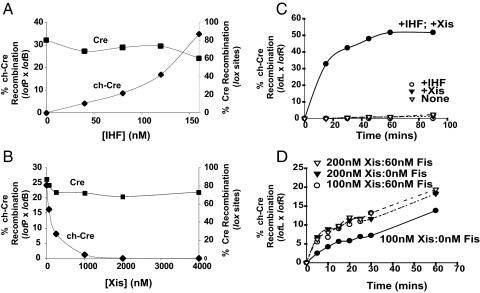
Fig. 2.
ChCre-promoted recombination depends on the relative concentrations of DNA-bending accessory proteins. (A) ChCre-promoted recombination between supercoiled lotP (48 nM) and [α-32P]dCTP-labeled lotB (12 nM) depends on IHF. (B) Xis inhibits chCre-promoted recombination between lotP (48 nM) and [α-32P]dCTP-labeled lotB (12 nM) in the presence of IHF (150 nM). (C) Effect of Xis and IHF on chCre-promoted recombination between supercoiled lotL (24 nM) and [α-32P]dCTP-labeled lotR (6 nM). Xis and IHF, when present, are at 200 and 25 nM, respectively. (D) Fis stimulates lotL and lotR recombination at limiting Xis concentrations. ChCre-promoted recombination between supercoiled lotL (6 nM) and [α-32P]dCTP-labeled lotR (6 nM) in the presence of 50 nM IHF and the indicated amounts of Xis and Fis. Cre-mediated recombination between 2 lox sites is not effected by IHF (A) or Xis (B). All recombination reactions were carried out in the presence of 100 nM recombinase, analyzed by SDS/PAGE, and quantified on a Typhoon PhosphorImager (GE Healthcare).
In contrast to the results with lotB0, chCre-mediated recombination of the lotB substrate (containing the position 2 mutation) was greatly stimulated by IHF (Fig. 1B, lanes 7 and 8). Cre-mediated recombination of this substrate was very poor and not stimulated by IHF (lanes 5 and 6). We also constructed another lot site, containing a Cre-binding defective mutation at position 6 of the inverted repeats (20), and it behaved the same as lotB (data not shown).
Based on these experiments we introduced the position 2 Cre-binding site mutation into the inverted repeats in each of the 4 hybrid att sites, to generate lotP, lotB, lotL, and lotR. ChCre-mediated recombination between lotP and lotB depends on IHF (Fig. 2A) and is inhibited by the Xis protein (Fig. 2B). Cre-mediated recombination of simple lox sites, which lack targets for the accessory proteins are neither stimulated by IHF nor inhibited by Xis to any significant extent, as expected (Fig. 2 A and B). The chCre-mediated recombination between lotL and lotR depends on both IHF and Xis (Fig. 2C). Furthermore, at limiting Xis concentrations, a small but reproducible stimulation by the host-encoded accessory protein, Fis, is observed (Fig. 2D).
Differential Requirements for N-Domain Binding Sites.
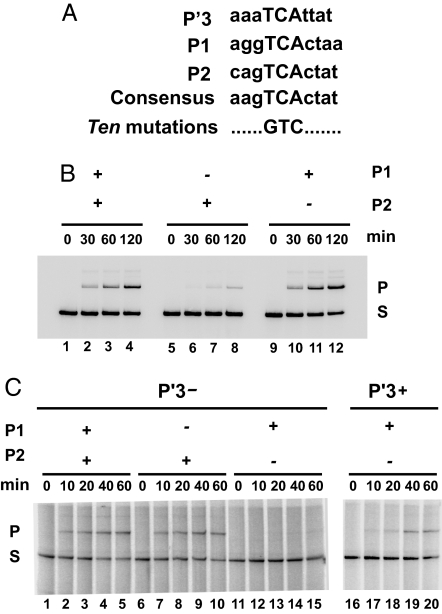
λ Int binding at N-domain sites is abolished by changing the conserved central 3 bases of the consensus recognition sequence from TCA to GTC (Fig. 3A) (16). The phenotypes of these “ten” mutants were consistent with earlier deletion studies (21, 22) and showed that the P1 but not the P2 site is required for λ integrative recombination. Concomitantly, the P2 site but not the P1 site is required for λ excisive recombination. A similar differential usage of N-domain sites was observed with the chCre. The chCre-mediated recombination of lotP by lotB requires the P1 but not the P2 site (Fig. 3B) whereas recombination of lotL by lotR has the reverse requirements (Fig. 3C).
Fig. 3.
Individual N-domain binding sites have different roles in the 2 chimeric Cre-promoted recombination reactions. (A) The conserved triad core (TCA) in DNA sequences of the 5 N-domain binding sites (only 3 are shown) is altered to generate nonbinding ten mutations. (B) The P1 but not the P2 N-domain binding site is required for recombination between supercoiled lotP and [α-32P]dCTP-labeled lotB (6 nM). The P1 and P2 N-domain binding sites were either wild-type or contained the GTC ten mutation, as indicated. (C) The P2 but not the P1 N-domain binding site is required for recombination between lotL and lotR. [α-32P]dCTP-labeled lotL (6 nM) containing either a mutated P′3 N-domain site (lanes 1–15) or a wild-type P′3 N-domain binding site (lanes 16–20) was recombined with supercoiled lotR (6 nM) containing P1 and P2 N-domain binding sites that were either wild-type or contained the GTC ten mutation, as indicated. The very faint bands in lanes 1–4 and 9–12 (above the product) are likely the result of a second round of recombination.
In the recombination reactions shown in lanes 1–15 of Fig. 3C, the P′3 arm-type site, which is not required for excisive recombination, also carries a ten mutation. We used this configuration because in the λ excisive reaction a ten mutation in P′3 enhances the dependence on the P2 site (16, 23). Indeed, when the P′3 arm-type site is restored to wild type in the lotL by lotR reaction, the dependence on the P2 arm-type site is less pronounced (Fig. 3 C, lanes 16–20).
ChCre-Mediated Recombination in Vivo.
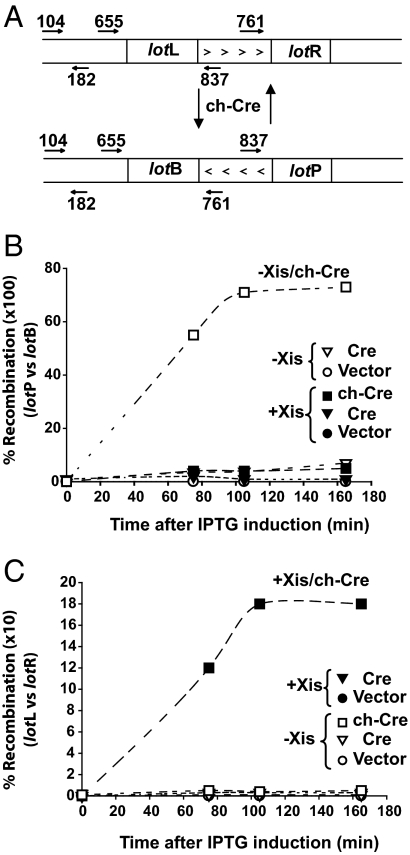
To assay recombination in vivo we constructed plasmids containing either a pair of lotP and lotB or lotL and lotR recombination partners separated by 4,000 bp. In each plasmid the recombination partners were oriented as inverted repeats so that intramolecular recombination produces an inversion of the intervening DNA flanked by the recombination products. As opposed to the more traditional genetic assays that look at the accumulation of recombination products after overnight growth, we wanted to examine the kinetics of recombination. We initiated recombination by transforming the appropriate cell lines with plasmids encoding recombinase and assayed the appearance of rearranged plasmid DNA as a function of time. Appropriate primer pairs were used to monitor the recombination reactions by qPCR (Fig. 4A). In addition to the recombination substrate plasmid, cells contained a compatible plasmid encoding IHF under the control of a regulated T7 polymerase promoter and another compatible plasmid, with or without the Xis gene, also under the control of a regulated T7 polymerase promoter. Cells containing these 3 plasmids were made competent and transformed with a compatible protein expression plasmid encoding the chCre, Cre, or no recombinase. Sixty minutes after transformation, the cells were pulsed with IPTG to induce protein expression, and aliquots of the cells were harvested at the indicated times for qPCR analysis of the recombination assay plasmid (see Experimental Procedures). A typical set of results is shown in Fig. 4. The results with the control plasmid show that in the absence of recombinase there is virtually no recombination, and there is very little stimulation of recombination when the plasmid encodes Cre (Fig. 4 B and C). More significantly, even the low levels of Cre-mediated recombination are not appreciably stimulated by Xis in either the lotP by lotB or the lotL by lotR reactions. The most salient aspects of these results are the strong Xis inhibition, ≈15-fold, of the chCre-mediated lotP by lotB recombination (Fig. 4B) and the strict dependence of chCre-mediated lotL by lotR recombination on Xis (an ≈30-fold effect) (Fig. 4C).
Fig. 4.
Time courses of chimeric Cre-initiated recombination in vivo highlight the requirement for IHF and the differentiating role of Xis. (A) The in vivo recombination assay plasmids each contain a pair of recombination partners oriented as inverted repeats so that recombination between them inverts the intervening 4-kb DNA segment. Inversion was measured by real-time qPCR as described in Experimental Procedures. Recombination was initiated by transforming cells with an expression plasmid encoding recombinase. Each experiment included a control in which cells were transformed with a plasmid not encoding any recombinase. (B) Xis inhibits chimeric Cre-mediated lotP by lotB recombination. Two cell lines each containing the lotP–lotB recombination substrate plasmid, the IHF expression plasmid, and another plasmid, with or without the Xis gene, were transformed at time 0 with protein expression vectors encoding Cre, chCre, or neither. (C) ChCre-mediated recombination between lotL and lotR requires Xis. The plasmids and protocol were the same as described in B except that the recombination substrate plasmid carried an invertible DNA segment flanked by lotL and lotR.
Discussion
ChCre exhibits regulated directionality analogous to that of λ Int. The lotP by lotB recombination depends on the IHF bending protein and it is inhibited by the Xis directionality factor (Fig. 2 A and B). In contrast, the lotL by lotR recombination depends on both IHF and Xis, and it is stimulated by Fis at limiting concentrations of Xis (Fig. 2 C and D). As predicted from the in vitro results, quantification of the in vivo reactions shows that the directionality factor Xis is required for chCre-mediated lotL by lotR recombination and inhibits the lotP by lotB reaction (Fig. 4 B and C).
The requirement for the P1, but not the P2, arm-type Int site for lotP by lotB recombination (Fig. 3B) and the requirement for the P2 but not the P1 site for lotL by lotR recombination (Fig. 3C) suggest that chCre-mediated recombination proceeds via the same pathway and generates structural intermediates of the same configuration as λ Int. ChCre even manifests one of the more subtle and poorly understood features of the λ Int reaction: the presence of either the P2 site in lotR or the P′3 site in lotL, but not both, is required for a modest level of recombination and loss of both sites abolishes lotL by lotR recombination (Fig. 3C). We propose that the regulated directionality of the λ Int pathway has been conferred on Cre by the appended 74 N-terminal residues.
In the chCre protein characterized here, the N-terminal domain of λ Int was fused directly to the N-terminal residue of Cre. Because the first 12 residues of Cre can be deleted without impairing function (24) and the first 20 residues of Cre are sufficiently unstructured that they do not appear in any of the Cre co-crystal structures (11), we also created a chCre in which the first 20 residues of Cre are deleted. We did not find any significant differences in the behavior of this chCre from the results reported here with the full length-Cre chimera. It seems likely that in the chCre characterized here, the first 20 residues of Cre are unstructured and looped out. It also seems likely that there are few if any specific interactions between the λ and Cre portions of the chimeric protein. These two observations suggest a reevaluation of the role of “allosteric control,” as proposed in models based on previous X-ray crystal structures (14).
A number of chimeric recombinases have been created to study site-specific recombination (25, 26). More generally, there has also been considerable success in generating chimeric enzymes with novel enzymatic activities such as new substrate specificities, altered catalytic properties, and enhanced expression levels (27). Here, regulated directionality and complexity are conferred in a simple chimeric construction.
The robust activity of Cre has made it an adaptable and malleable tool for genetic engineering (19, 28) and studies on topological selectivity (29, 30). The experiments described here additionally bear on the evolution of site-specific recombinases. According to a common view, the simple unregulated recombinases, such as Cre, preceded the more complex bivalent recombinases, such as λ Int. The data presented here suggest that 2 simple steps, in no specified order, would be required in this evolution: the acquisition of a small DNA-binding domain and a “detuning” of the Cre protein. The detuning (a reduction in the strength of DNA binding and/or synaptic protein–protein interactions) makes recombinase binding in the region of strand exchange dependent on both the additional DNA-binding domain and the facilitating accessory proteins. The detuning results from loss-of-function mutations and therefore could be a relatively frequent evolutionary step. This pathway is particularly easy to envisage for Cre because the first step, acquisition of a new N-terminal domain, did not significantly impair the parental Cre activity on canonical lox sites (data not shown). It seems reasonable to speculate, but difficult to implicate, the existence of such detuning steps in the evolution of many complex multielement regulatory pathways, especially those involving nucleic acid-binding proteins.
According to an alternative evolutionary trajectory, the simple and complex site-specific recombinases evolved in parallel from a common less efficient precursor. The Cre-like recombinases would have acquired a peptide in the N-terminal domain that, along with other mutations, conferred a greater propensity for tetramerization and/or a higher affinity for core-type DNA sites. The Int-like recombinases would have become more efficient by acquiring a small N-terminal domain comprising an additional DNA-binding pocket.
Site-specific recombinases are known to be engines of genomic rearrangements in an evolutionary pathway that draws on rare recombination events at loci scattered throughout the genome. These “secondary att sites” serve as inefficient recombination targets because they fortuitously possess some elements of similarity to a canonical att site. Their efficiencies, which are readily observable in E. coli over a 106-fold range, reflect the extent to which a particular secondary att site is compatible with the recombinase and the canonical recombination partner (31). Secondary att sites also occur fortuitously within recombinase genes. Recombination between one of these and a canonical att site (which most often is adjacent to the recombinase gene) leads to replacement of recombinase sequences by a “new att site.” Once such an att-containing recombinase gene has been created, it has the potential to be recombined (by a recombinase in trans) into other secondary att sites throughout the genome. This sets the stage for evolving “new” recombinases, not unlike the chCre protein characterized here. Such recombination events have been most intensively studied in bacteriophage λ and include one well-characterized secondary att site within the C-terminal region of the Int gene (32). A particularly dramatic example of this capacity for “self-promoted evolution” is found in the archaebacteria SSV family of recombinases in which the canonical viral att site already lies within the Int gene (33). As a consequence of this capacity for self-promoted evolution, it is not surprising that the site-specific recombinases are so numerous and so varied. We suggest it is plausible that conferring regulated directionality on Cre has simulated such a path and concurrently generated a potential platform for strategies in genetic engineering.
Experimental Procedures
Protein Expression and Purification.
All of the proteins were produced in E. coli BL21(DE3)RIL from expression plasmids under the control of the T7 promoter as described (34–39). The purity of all of the proteins was >90% as judged by Coomassie blue staining of overloaded SDS/polyacrylamide gels.
Chimeric Recombination Substrates and Proteins.
The chimeric recombination substrates were constructed as described in Fig. 1A. IHF binding at the H1 site in the P-arm inhibits excisive recombination and so was mutated in lotR substrates. The H1= (40), lox (20), and ten (16) mutations were introduced into their relevant sites by using the QuikChange site-directed mutagenesis kit (Stratagene). The chCre recombinase is a fusion of the first 74 aa of λ Int to the first methionine of the 343 aa of Cre.
In Vitro Recombination Assays.
A plasmid encoding the substrate to be labeled (lotB, lotL, or lotR) was linearized by digestion with HindIII, and the recessed ends were filled in with the Klenow fragment of DNA polymerase (NEB). The recovery of radiolabeled DNA was quantified by using agarose gel electrophoresis to determine the final concentration of substrate. LotP–lotB and lotR–lotL recombination reactions were carried out at 37 °C in 5 mM EDTA (pH 8), 6 mM spermidine, 25 mM Tris (pH 8), 0.5 mg/mL BSA, 2.5 mM DTT, 30 mM NaCl. The IHF and Xis titration experiments were carried out by serial 2-fold dilutions of the appropriate master mix. The reactions were stopped by the addition of SDS-containing loading solution and analyzed by electrophoresis through 1.2% agarose gels in Tris–acetate–EDTA buffer. The Cre recombination substrate plasmid contains a pair of loxP sites oriented as direct repeats so that recombination between them deletes the intervening DNA. Cre recombination reactions were carried out on 50 nM substrate DNA in 50 mM Tris·Cl (pH 7.4), 33 mM NaCl, and 10 mM MgCl2 at 25 °C.
In Vivo Recombination Assays.
The recombinases, Xis, and the lot recombination assay substrates were cloned into the Duet T7-controlled protein expression vectors pCDF, pRSF, and pACYC (Novagen), respectively, to generate the plasmids CDF-Cre, CDF-chCre, Rsf-Xis, pAW3236A (excisive) and pAW3236AH-PB (integrative). The parental vectors were designed with compatible replicons and different drug resistance genes for effective propagation and maintenance of up to 4 plasmids in a single cell (Novagen). The IHF expression plasmid (pPR204 from Phoebe Rice, University of Chicago) contains the ColE1 replicon and ampicillin resistance gene, making it compatible with the 3 Duet vectors. Competent ER2566 cells (NEB) were transformed with one of the lot recombination assay substrates (pAW3236A or pAW3236AH-PB for excisive and integrative recombination, respectively). A colony was picked for each, and competent cells were made by using the calcium chloride protocol. The procedure was repeated for the IHF plasmid. The double-transformed cell lines were then transformed with either pRSF or pRSF expressing Xis. These cells were then made competent with calcium chloride. The experiment was started by transforming the triple-transformed cells with CDF expression vectors expressing no protein, Cre, or chCre. Sixty minutes after transformation, the cells were pulsed with 100 mM IPTG for 30 min to induce protein expression; aliquots of the cells were harvested at the indicated times for qPCR analysis of the recombination assay plasmid.
Quantitative PCR Assays.
Real-time qPCRs were performed with 25 μL of platinum SYBR Green qPCR Supermix-UDG with Rox master mix (Invitrogen) along with each primer (0.2 μM) in a final volume of 50 μL. Two microliters of DNA isolated from the transformed bacterial cells was mixed with PCR master mix in 96-well optical reaction plates (Applied Biosystems). The sequences of the primers were: primer 104, 5′-GTT TGA CAG CTT ATC ATC GAT ATA GTC AT-3′, and primer 182, 5′-AAA GCT GCT CGG GCA TCA-3′ (quantifies the total amount of recombination assay plasmid DNA); primer 655, 5′-GCT TTT GCC CGA CAT CAG ATG GCA GTA AT-3′, and primer 761, 5′-ATC ACC ATC ATC ACC ACA-3′ (quantifies the lotB product of lotL by lotR recombination); primer 837, 5′-AGT ATC GTG ATG ACA GAG GCA GG-3′, and primer 655 is used to quantify the lotL product of lotP by lotB recombination. The primer pair 655/761 fails to give a product from the lotL–lotR assay plasmid, and the primer pair 655/837 fails to give a product from the lotP–lotB assay plasmid confirming the specificity of the PCRs (see Fig. 4A). Thermal cycling and data analysis were carried out with an ABI Prism 7000 instrument with SDS software (Applied Biosystems). Water was used as a negative DNA control, and cloned plasmid products were used as positive DNA controls and to generate the standard curves. All real-time PCRs were performed in triplicate, and average cycle threshold (Ct) values for each sample were determined (the standard deviation on samples positive for recombination was typically <2%). The average Ct value was converted to moles of product by using the standard curves, and the amount of recombination product relative to the total amount of assay plasmid DNA was determined and reported as percentage recombination.
Acknowledgments.
We thank Tapan Biswas, Hideki Aihara, Tom Ellenberger, George Chaconas, Greg Van Duyne, and past and present members of the Landy laboratory for helpful discussions and suggestions. We also thank Christine Lank and Jo-Anne Nelson for technical assistance, Zishuo Hu for experimental contributions, and Joan Boyles for manuscript preparation. This work was supported by National Institutes of Health Grants GM33928 and GM62723 (to A.L.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Criag NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: Am Soc Microbiol; 2002. [Google Scholar]
- 2.Sauer B. Chromosome manipulation by Cre-lox recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: Am Soc Microbiol; 2002. pp. 38–58. [Google Scholar]
- 3.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolot M, Silberstein N, Yagil E. Site-specific recombination in mammalian cells expressing the Int recombinase of bacteriophage HK022. Mol Biol Rep. 1999;26:207–213. doi: 10.1023/a:1007096701720. [DOI] [PubMed] [Google Scholar]
- 5.Corona T, et al. Activation of site-specific DNA integration in human cells by a single chain integration host factor. Nucleic Acids Res. 2003;31:5140–5148. doi: 10.1093/nar/gkg711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abremski K, Hoess R, Sternberg N. Studies on the properties of P1 site-specific recombination: Evidence for topologically unlinked products following recombination. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 7.Van Duyne GD. A structural view of tyrosine recombinase site-specific recombination. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: Am Soc Microbiol; 2002. pp. 93–117. [Google Scholar]
- 8.Campbell AM. Episomes. In: Caspari EW, Thoday JM, editors. Advances in Genetics. New York: Academic; 1962. pp. 101–145. [Google Scholar]
- 9.Azaro MA, Landy A. λ Int and the λ Int family. In: Craig NL, Craigie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 118–148. [Google Scholar]
- 10.Guo F, Gopaul DN, Van Duyne GD. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 11.Van Duyne GD. A structural view of Cre-loxP site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 12.Aihara H, Kwon HJ, Nunes-Düby SE, Landy A, Ellenberger T. A conformational switch controls the DNA cleavage activity of λ integrase. Mol Cell. 2003;12:187–198. doi: 10.1016/s1097-2765(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 13.Moitoso de Vargas L, Pargellis CA, Hasan NM, Bushman EW, Landy A. Autonomous DNA binding domains of λ integrase recognize different sequence families. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 14.Biswas T, et al. A structural basis for allosteric control of DNA recombination by λ integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross W, Landy A. Bacteriophage λ Int protein recognizes two classes of sequence in the phage att site: Characterization of arm-type sites. Proc Natl Acad Sci USA. 1982;79:7724–7728. doi: 10.1073/pnas.79.24.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Numrych TE, Gumport RI, Gardner JF. A comparison of the effects of single-base and triple-base changes in the integrase arm-type binding sites on the site-specific recombination of bacteriophage λ. Nucleic Acids Res. 1990;18:3953–3959. doi: 10.1093/nar/18.13.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson JF, Moitoso de Vargas L, Koch C, Kahmann R, Landy A. Cellular factors couple recombination with growth phase: Characterization of a new component in the λ site-specific recombination pathway. Cell. 1987;50:901–908. doi: 10.1016/0092-8674(87)90516-2. [DOI] [PubMed] [Google Scholar]
- 18.Ball CA, Osuna R, Ferguson KC, Johnson RC. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartung M, Kisters-Woike B. Cre mutants with altered DNA binding properties. J Biol Chem. 1998;273:22884–22891. doi: 10.1074/jbc.273.36.22884. [DOI] [PubMed] [Google Scholar]
- 21.Bushman W, Thompson JF, Vargas L, Landy A. Control of directionality in λ site-specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JF, Moitoso de Vargas L, Skinner SE, Landy A. Protein–protein interactions in a higher-order structure direct λ site-specific recombination. J Mol Biol. 1987;195:481–493. doi: 10.1016/0022-2836(87)90177-x. [DOI] [PubMed] [Google Scholar]
- 23.Hazelbaker D, Azaro MA, Landy A. A biotin interference assay highlights two different asymmetric interaction profiles for λ integrase arm-type binding sites in integrative versus excisive recombination. J Biol Chem. 2008;283:12402–12414. doi: 10.1074/jbc.M800544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rongrong L, Lixia W, Zhongping L. Effect of deletion mutation on the recombination activity of Cre recombinase. Acta Biochim Pol. 2005;52:541–544. [PubMed] [Google Scholar]
- 25.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc Natl Acad Sci USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaikh AC, Sadowski PD. Chimeras of the Flp and Cre recombinases: Tests of the mode of cleavage by Flp and Cre. J Mol Biol. 2000;302:27–48. doi: 10.1006/jmbi.2000.3967. [DOI] [PubMed] [Google Scholar]
- 27.Nixon AE, Ostermeier M, Benkovic SJ. Hybrid enzymes: Manipulating enzyme design. Trends Biotechnol. 1998;16:258–264. doi: 10.1016/s0167-7799(98)01204-9. [DOI] [PubMed] [Google Scholar]
- 28.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci USA. 2002;99:4185–4190. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akopian A, Gourlay S, James H, Colloms SD. Communication between accessory factors and the Cre recombinase at hybrid psi-loxP sites. J Mol Biol. 2006;357:1394–1408. doi: 10.1016/j.jmb.2006.01.050. [DOI] [PubMed] [Google Scholar]
- 30.Kilbride EA, Burke ME, Boocock MR, Stark WM. Determinants of product topology in a hybrid Cre-Tn3 resolvase site-specific recombination system. J Mol Biol. 2006;355:185–195. doi: 10.1016/j.jmb.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 31.Weisberg R, Landy A. Site-specific rcombination in phage λ. In: Stahl FW, Roberts J, Weisberg RA, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1983. pp. 211–250. [Google Scholar]
- 32.Ross W, Shulman M, Landy A. Biochemical analysis of att-defective mutants of the phage λ site-specific recombination system. J Mol Biol. 1982;156:505–529. doi: 10.1016/0022-2836(82)90263-7. [DOI] [PubMed] [Google Scholar]
- 33.She Q, Brügger K, Chen L. Archaeal integrative genetic elements and their impact on genome evolution. Res Microbiol. 2002;153:325–332. doi: 10.1016/s0923-2508(02)01331-1. [DOI] [PubMed] [Google Scholar]
- 34.Tirumalai RS, Kwon H, Cardente E, Ellenberger T, Landy A. The recognition of core-type DNA sites by λ integrase. J Mol Biol. 1998;279:513–527. doi: 10.1006/jmbi.1998.1786. [DOI] [PubMed] [Google Scholar]
- 35.Nash HA, Robertson CA, Flamm E, Weisberg RA, Miller HI. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J Bacteriol. 1987;169:4124–4127. doi: 10.1128/jb.169.9.4124-4127.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warren D, Lee SY, Landy A. Mutations in the amino-terminal domain of λ-integrase have differential effects on integrative and excisive recombination. Mol Microbiol. 2005;55:1104–1112. doi: 10.1111/j.1365-2958.2004.04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan CQ, et al. Structure of the Escherichia coli Fis–DNA complex probed by protein conjugated with 1,10-phenanthroline copper(I) complex. Proc Natl Acad Sci USA. 1994;91:1721–1725. doi: 10.1073/pnas.91.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoess R, Abremski K, Irwin S, Kendall M, Mack A. DNA specificity of the Cre recombinase resides in the 25-kDa carboxyl domain of the protein. J Mol Biol. 1990;216:873–882. doi: 10.1016/S0022-2836(99)80007-2. [DOI] [PubMed] [Google Scholar]
- 39.Shaikh AC, Sadowski PD. The Cre recombinase cleaves the lox site in trans. J Biol Chem. 1997;272:5695–5702. doi: 10.1074/jbc.272.9.5695. [DOI] [PubMed] [Google Scholar]
- 40.Thompson JF, et al. Mutations in an integration host factor-binding site: Effect on λ site-specific recombination and regulatory implications. J Bacteriol. 1986;168:1343–1351. doi: 10.1128/jb.168.3.1343-1351.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]