Abstract
Phosphorylation-based signaling pathways employ dephosphorylation mechanisms for signal termination. Histidine to aspartate phosphosignaling in the two-component system that controls bacterial chemotaxis has been studied extensively. Rhodobacter sphaeroides has a complex chemosensory pathway with multiple homologues of the Escherichia coli chemosensory proteins, although it lacks homologues of known signal-terminating CheY-P phosphatases, such as CheZ, CheC, FliY or CheX. Here, we demonstrate that an unusual CheA homologue, CheA3, is not only a phosphodonor for the principal CheY protein, CheY6, but is also is a specific phosphatase for CheY6-P. This phosphatase activity accelerates CheY6-P dephosphorylation to a rate that is comparable with the measured stimulus response time of approximately 1 s. CheA3 possesses only two of the five domains found in classical CheAs, the Hpt (P1) and regulatory (P5) domains, which are joined by a 794-amino acid sequence that is required for phosphatase activity. The P1 domain of CheA3 is phosphorylated by CheA4, and it subsequently acts as a phosphodonor for the response regulators. A CheA3 mutant protein without the 794-amino acid region lacked phosphatase activity, retained phosphotransfer function, but did not support chemotaxis, suggesting that the phosphatase activity may be required for chemotaxis. Using a nested deletion approach, we showed that a 200-amino acid segment of CheA3 is required for phosphatase activity. The phosphatase activity of previously identified nonhybrid histidine protein kinases depends on the dimerization and histidine phosphorylation (DHp) domains. However, CheA3 lacks a DHp domain, suggesting that its phosphatase mechanism is different from that of other histidine protein kinases.
Keywords: response regulator, signal termination, two-component, Rhodobacter sphaeroides, histidine protein kinase
Dephosphorylation is required for signal termination in phosphorylation-based signaling pathways. The most common phosphorylation-based signaling pathways in bacteria are two-component signal transduction systems, which can detect and mediate responses to a wide range of different environmental stimuli, with some bacteria having over 100 distinct systems (1, 2). These systems comprise sensor histidine protein kinases (HPKs) and response regulators (RRs). HPKs detect sensory stimuli; these regulate the rate at which the HPK autophosphorylates a conserved histidine residue. Subsequently, the phosphoryl group is transferred from the histidine residue of the HPK onto an aspartate residue in the receiver domain of the RR. Phosphorylation of the RR causes a conformational change, allowing it to mediate an output appropriate to the original stimulus, often a change in transcription (3). The phosphosignal is terminated by the hydrolysis of the aspartyl-phosphate residue of the RR.
Receiver domains have intrinsic autodephosphorylation activity, although in many systems a dedicated specific aspartyl-phosphate phosphatase is used to accelerate this process. Such phosphatases can be found in separate protein molecules, for example, RapA and CheZ dephosphorylate Spo0F-P and CheY-P, respectively (4, 5); alternatively, phosphatases can be integral parts of the HPKs. Hybrid HPKs, which are components of multistep phosphorelays, contain one or more receiver domains and all show phosphatase activity because of the autodephosphorylation activity of their receiver domains (6). Many nonhybrid HPKs also show phosphatase activity, for example, NtrB and EnvZ dephosphorylate their RRs, NtrC-P and OmpR-P, respectively (7, 8). The dimerization and histidine phosphorylation (DHp) domain of these HPKs has been implicated in the phosphatase reaction (8, 9). In this study, we report the discovery of an aspartyl-phosphatase activity within the chemotaxis protein, CheA3, of Rhodobacter sphaeroides. Interestingly, unlike all previously identified nonhybrid HPKs with phosphatase activity, CheA3 lacks a DHp domain and is not homologous to known phosphatases, suggesting that the CheA3 phosphatase activity is novel.
The two-component system controlling chemotaxis allows bacteria to move toward environments that are better for growth [reviewed in (10–12)]. Chemoreceptors modulate the autophosphorylation rate of CheA in response to chemotactic stimuli. In E. coli, unliganded receptors activate CheA, whereas attractant-occupied receptors inhibit CheA autophosphorylation. The phosphoryl group is transferred from CheA-P to specific aspartate residues on its cognate RRs, CheY and CheB. CheY-P binding to the FliM component of the flagellar motor brings about a change in flagellar rotation and therefore swimming direction, while CheB-P demethylates the chemoreceptors, mediating adaptation. Hydrolysis of the phosphoryl-aspartate residues in CheY-P and CheB-P allows signal termination. E. coli CheY-P autodephosphorylates with a half-time of approximately 14 s (13); however, the phosphatase CheZ can increase this rate by a factor of approximately 100 (14). Many bacteria lack CheZ homologues and some of these instead use CheY-P phosphatases belonging to the CheC/FliY/CheX family of proteins (15–17). Other bacteria, including Sinorhizobium meliloti and R. sphaeroides, do not have homologues of any of these CheY-P phosphatases but do have multiple CheYs. S. meliloti has two CheYs, one that can bind the motor, CheY2, and one that cannot bind the motor, CheY1. CheY1 mediates dephosphorylation of CheY2-P by functioning as a phosphate sink (18). Like S. meliloti CheY2 but unlike S. meliloti CheY1, all of the R. sphaeroides CheYs are capable of binding to FliM, suggesting that a phosphate sink may not be used for signal termination in the R. sphaeroides chemotaxis pathway (19).
R. sphaeroides has two sets of flagellar genes and three chemotaxis operons (20). The fla1 set encodes a single unidirectional flagellum that is controlled by the chemotaxis proteins encoded by cheOp2 and cheOp3 (21), whereas the fla2 set encodes polar flagella that are controlled by cheOp1 (22, 23). R. sphaeroides has four CheA homologues. Phosphosignaling from CheA2, CheA3, and CheA4 is essential for fla1 driven chemotaxis (24). CheA2 localizes with the transmembrane chemoreceptors to the cell poles, whereas CheA3 and CheA4 localize to a cytoplasmic chemotaxis cluster along with the cytoplasmic chemoreceptors (25). CheA1, CheA2, and E. coli CheA function as homodimers. Each protomer contains five domains (P1–P5) with the P3 domain mediating dimerization. In E. coli, the P5 domain has been shown to bind CheW and the chemoreceptors and to couple the rate of CheA autophosphorylation to receptor control. During the autophosphorylation reaction, the kinase domain (P4) phosphorylates the histidine residue within the Hpt (P1) domain using ATP as the phosphodonor. The phosphorylated P1 domain donates phosphoryl groups to aspartate residues on CheY and CheB. The P2 domain binds CheY and CheB and therefore increases their local concentrations, accelerating the phosphotransfer reaction mediated by the P1 domain. CheA3 and CheA4 from R. sphaeroides are atypical CheAs in that they each lack some of the domains found in E. coli CheA. CheA4 is a homodimeric protein containing only the P3, P4, and P5 domains whereas CheA3 has only the P1 and P5 domains, separated by a 794-amino acid sequence containing no identifiable domains. CheA3 and CheA4 both localize to the cytoplasmic chemotaxis cluster (25). Neither CheA3 nor CheA4 are capable of autophosphorylation; however, CheA4 can phosphorylate the P1 domain of CheA3 on residue H51. Subsequently CheA3-P acts as a phosphodonor for a specific subset of the chemotaxis RRs (24).
R. sphaeroides has eight chemotaxis RRs, six CheYs and two CheBs. While both CheBs are required for normal chemotaxis, only CheY6 plus either CheY3 or CheY4 are needed for control of the fla1 flagellum (21, 26). The CheAs all show different patterns of phosphotransfer to the RRs: CheA1-P phosphorylates CheY1, CheY2, CheY3, and CheY5; CheA2-P phosphorylates all eight chemotaxis RRs; and CheA3-P phosphorylates CheY1, CheY6, and CheB2 (24, 27). Here, we demonstrate an additional activity for CheA3, the ability to specifically catalyze the hydrolysis of the aspartyl-phosphate residue in CheY6-P.
Results
P1 Domain of CheA3 Is a Specific Phosphodonor for CheY1, CheY6, and CheB2.
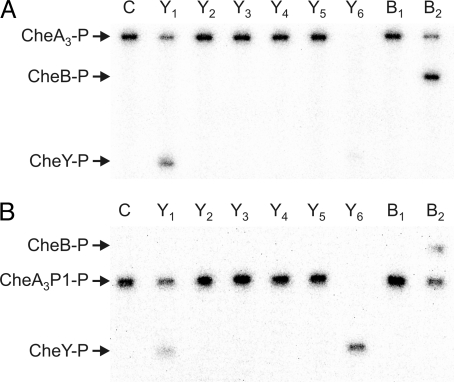
CheA3-P is a phosphodonor for only CheY1, CheY6, and CheB2 (24). To determine whether this specificity depends solely on the interactions between the RRs and the P1 domain of CheA3, phosphotransfer experiments were performed comparing the specificity of the isolated P1 domain of CheA3 (CheA3P1) with that of full-length CheA3. Transfer of phosphoryl groups to the eight chemotaxis RRs was measured under multiple turnover conditions in the presence of CheA4 and ATP, allowing either the CheA3 or CheA3P1 phosphorylation to continue throughout the course of the reactions. The progress of these phosphotransfer reactions after 30 s is shown in Fig. 1. Phosphotransfer occurred in reactions where a decrease in CheA3-P or CheA3P1-P levels was accompanied by an increase in CheY/B-P levels. CheA3-P and CheA3P1-P phosphorylated the same RRs (CheY1, CheY6, and CheB2) indicating that it is the interaction between the RRs and the P1 domain of CheA3-P that determines the phosphotransfer specificity of CheA3-P. Interestingly, despite using equimolar concentrations of phosphodonor, CheY6-P levels were greater when CheA3P1-P was used as the phosphodonor rather than CheA3-P. One possible explanation for this is that full-length CheA3 is both a phosphodonor and a phosphatase for CheY6-P. The isolated CheA3P1 domain would lack this phosphatase activity, retaining only phosphodonor function and allowing more CheY6-P to accumulate.
Fig. 1.
Phosphorimages of SDS/PAGE gels showing phosphotransfer from (A) CheA3 and (B) CheA3P1 (the isolated P1 domain of CheA3) to the R. sphaeroides chemotaxis response regulators. CheA3 (4 μM) and CheA4 (10 μM) were preincubated together with 0.5 mM [γ-32P] ATP for 30 min. RRs (5 μM) were then added. Ten-microliter reaction samples were removed after 30 s and quenched in 10 μl of 3× SDS/EDTA loading dye. The samples were analyzed by SDS/PAGE and detected by phosphorimaging. Lane C shows a control reaction in which an equal volume of buffer was added instead of the RRs. The remaining lanes are labeled according to which RR was used; for example, CheY1 was used in the lane labeled Y1. Phosphotransfer is indicated by the appearance of phosphorylated RR and a reduction in the amount of (A) CheA3-P or (B) CheA3P1-P.
CheA3 Is a Specific Phosphatase for CheY6-P.
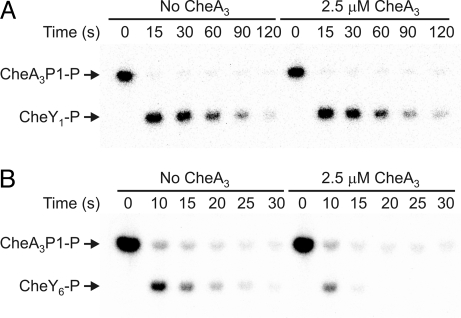
The ability of CheA3 to act as a chemotaxis RR phosphatase was measured in vitro by using a RR dephosphorylation assay that quantified the loss of 32P-labeled phosphoryl groups from RR-P as a function of time. Parallel time course experiments were performed where a phosphodonor (either CheA3P1-32P or CheA2-32P) was mixed with a vast excess of RR (400 μM) in the presence and absence of the putative phosphatase, CheA3 (Fig. 2 and Table 1). After phosphotransfer, no rephosphorylation of the phosphodonor could occur because ATP was not present. Under these conditions, phosphotransfer from phosphodonor to the RR occurred rapidly and was complete before the first time point. Therefore, the only reaction occurring after the first time point was the dephosphorylation of the phosphorylated RR that had been generated by the phosphotransfer reaction. By monitoring the subsequent decrease in RR-P levels over time it was possible to measure the dephosphorylation rate constant by fitting the time course data to a first order exponential decay model (Table 1). No significant change in CheY1–5-P or CheB1&2-P dephosphorylation rate constants was observed in the presence of CheA3 (Fig. 2A and Table 1), indicating that CheA3 is not a phosphatase for these RRs. In contrast, CheY6-P levels fell faster in the presence of CheA3 than in the absence of CheA3 indicating that CheA3 is a phosphatase for CheY6-P (Fig. 2B and Table 1). Under these reaction conditions, where the concentration of CheA3 used was 2.5 μM and the CheY6 concentration was 400 μM, giving a molar ratio of CheY6 to CheA3 of 160:1, CheA3 increased the dephosphorylation rate of CheY6-P by a factor of three. No CheA3-P was detected at any point in the assay (Fig. 2B), suggesting that the phosphatase mechanism does not involve reversed phosphotransfer to CheA3. The effect of varying [CheA3] on the rate of CheY6-P dephosphorylation is shown in the supporting information (SI) Text, and Fig. S1. In summary, CheA3 is an aspartyl-phosphate phosphatase that is specific for CheY6-P.
Fig. 2.
Phosphorimages of SDS/PAGE gels showing the response regulator dephosphorylation time courses. (A) 400 μM CheY1 was added to 2 μM CheA3P1-P in the absence (left half of gel) and presence of 2.5 μM CheA3 (right half of gel). (B) 400 μM CheY6 was added to 30 μM CheA3P1-P in the absence (left half of gel) and presence of 2.5 μM CheA3 (right half of gel). 10-μl reaction samples were taken at the time points indicated and quenched in 20 μl of 1.5× SDS/EDTA loading dye. The quenched samples were analyzed by SDS/PAGE and detected by phosphorimaging. ATP was not present in any of the reactions, so after the phosphotransfer reactions, which were completed before the first-time point, the only reaction occurring was RR-P dephosphorylation. As has been observed for E. coli CheA, a small fraction of CheA3P1-P (< 4%) failed to transfer phosphoryl groups to the RRs (42). Phosphatase activity is indicated by a reduction in CheY-P levels in the presence of 2.5 μM CheA3 when compared with those in the absence of CheA3 (seen in B but not in A).
Table 1.
The effect of CheA3 on the dephosphorylation rates of the R. sphaeroides CheY/Bs
| Protein | Dephosphorylation half-time, s* |
|
|---|---|---|
| 0 μM CheA3† | 2.5 μM CheA3‡ | |
| CheY1-P | 27 ± 1 | 27 ± 1 |
| CheY2-P | 63 ± 3 | 67 ± 4 |
| CheY3-P | 36 ± 3 | 40 ± 4 |
| CheY4-P | 38 ± 3 | 39 ± 2 |
| CheY5-P | 27 ± 1 | 30 ± 1 |
| CheY6-P | 4.1 ± 0.3 | 1.4 ± 0.1 |
| CheB1-P | 4000 ± 200 | 4100 ± 100 |
| CheB2-P | 52 ± 4 | 52 ± 7 |
*, Mean ± standard error (values rounded to two significant figures). Each experiment was performed six times.
†While most of these values are in good agreement with our previous estimates of dephosphorylation rate, some of these values differ considerably from our earlier estimates (27). The values in this table were derived from a direct assay of RR-P dephosphorylation and are more accurate. The previous estimates were based on an indirect assay that examined the steady state concentration of phosphorylated response regulator in a reaction mixture containing CheA2 and ATP, and consequently were very sensitive to small errors in measuring the CheA2 autophosphorylation rate and the steady state [CheY/B-P] and [CheA2-P].
‡The molar ratio of RR to CheA3 was 160:1.
Requirements for CheA3 Phosphatase Activity.
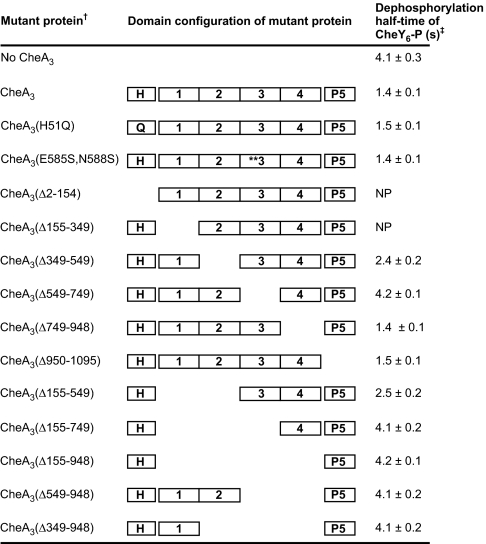
To determine whether the phosphorylation site (H51) of CheA3 has any involvement in phosphatase activity, as has been seen for other HPKs with intrinsic phosphatase activity, the phosphatase activity of CheA3(H51Q) was measured. The phosphatase activities of CheA3 and CheA3(H51Q) were indistinguishable, indicating that the phosphorylation site (H51) of CheA3 has no role in phosphatase activity (Fig. 3).
Fig. 3.
The effect of CheA3 mutant proteins on the dephosphorylation rate of CheY6-P. †Two and one-half μM of each CheA3 mutant protein was used in the phosphatase assays. The molar ratio of CheY6 to CheA3 mutant protein was 160:1. ‡Each experiment was performed six times and mean values ± standard error are shown (values are rounded to two significant figures). NP, could not be overexpressed and purified.
The CheA3(Δ155–948) mutant protein retains only the P1 and P5 domains and lacks the intervening 794-amino acid region. Like the isolated CheA3P1 domain, the CheA3(Δ155–948) mutant protein was phosphorylatable by CheA4 and functioned as a phosphodonor for the cognate RRs of CheA3 (data not shown). This mutant protein did not, however, show any CheY6-P phosphatase activity (Fig. 3), indicating that either the deleted 794-amino acid region is required for phosphatase activity or that this large deletion causes misfolding of the remainder of the protein such that phosphatase activity but not phosphodonor ability was abolished. The 794-amino acid region of CheA3 was then arbitrarily subdivided into four sub-regions (Fig. 3). Mutant CheA3 proteins with various nested deletions of these regions were purified and assayed for CheY6-P phosphatase activity. The P5 domain and sub-region 4 were not required for phosphatase activity. Sub-region 3 (residues 549–749) was essential for phosphatase activity, while deletion of sub-region 2 or sub-regions 1 and 2 together caused a partial reduction in phosphatase activity (Fig. 3). The sequence in sub-regions 1 and 2 may therefore either be required for correct folding of the phosphatase activity or may have a regulatory effect on the phosphatase activity. Sub-region 3 (residues 549–749) was the only segment of CheA3 that was been shown to be essential for phosphatase activity and is therefore presumed to contain the phosphatase activity. Within sub-region 3, there is a partial match to the consensus sequence of CheC-type phosphatases (SI Text). However, changing the predicted catalytic residues (E585 and N588) did not alter phosphatase activity (Fig. 3), indicating that CheA3 is not a homologue of CheC.
CheA3(Δ155–948) Localizes Correctly but Does Not Support Chemotaxis.
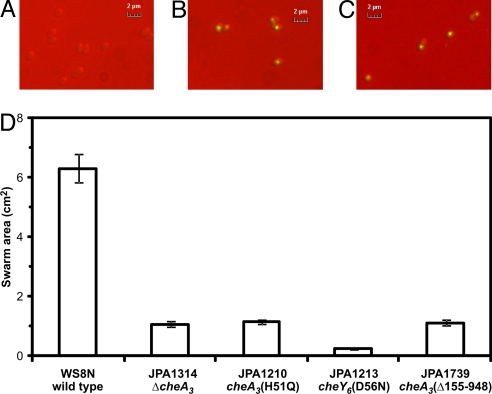
CheA3 has previously been shown to localize to the cytoplasmic chemotaxis cluster along with CheW4, CheA4 and the putative cytoplasmic chemoreceptors (25). Localization of CheA3 to this cluster may be required for it to function in the chemotaxis signal transduction system; therefore, the localization of CheA3(Δ155–948) was examined by replacing the wild-type cheA3 gene in the R. sphaeroides genome with yfp-cheA3(Δ155–948), generating strain JPA1741. YFP-CheA3(Δ155–948) localized to the cytoplasmic chemotaxis cluster in a pattern indistinguishable from that of YFP-CheA3 (Fig. 4 A–C), indicating that the localization determinants for CheA3 were still present and correctly folded in CheA3(Δ155–948). Western blotting using an antibody that recognizes YFP showed that expression levels of YFP-CheA3 and YFP-CheA3(Δ155–948) were similar (data not shown).
Fig. 4.
The 794-amino acid region between the P1 and P5 domains of CheA3 is not required for CheA3 localization but is required for chemotaxis. (A) YFP fluorescence image of wild-type cells (strain WS8N). (B) YFP fluorescence image of JPA1425 (yfp-cheA3). (C) YFP fluorescence image of JPA1741 [yfp-cheA3(Δ155–948)]. (D) Swarm plate chemotaxis assay comparing the chemotactic ability of JPA1739 [cheA3(Δ155–948)] with wild-type (WS8N), nonchemotactic (JPA1314 and JPA1210) and nonmotile (JPA1213) strains. The swarm plates contained 100 μM propionate and were incubated for 48 h under aerobic conditions. Error bars show the standard error of the mean obtained from nine independent experiments.
CheA3(Δ155–948) retains all of the known activities of full-length CheA3 except for the phosphatase activity, that is, it can be phosphorylated by CheA4, is a specific phosphodonor for the cognate RRs of full-length CheA3, is expressed at wild-type levels in R. sphaeroides, and localizes to the cytoplasmic chemotaxis cluster. To assess the importance of the CheY6-P phosphatase activity in vivo, the cheA3 gene in the R. sphaeroides genome was replaced with cheA3(Δ155–948). As expected, and unlike the ΔcheA3 strain, the cheA3(Δ155–948) strain was capable of responding to step changes in chemoeffector concentration (1 mM to 0 mM sodium propionate) in tethered cell assays (data not shown), suggesting that CheA3(Δ155–948) is capable of transducing signals in vivo. However, the cheA3(Δ155–948) strain was nonchemotactic in swarm plate assays (Fig. 4D), suggesting that although cells lacking the phosphatase activity of CheA3 are capable of responding to changes in chemoeffector levels, they do so in a time frame that is too slow to allow migration up a chemoeffector gradient. The lack of chemotaxis exhibited by the cheA3(Δ155–948) strain suggests that the phosphatase activity of CheA3 may be required for chemotaxis.
Discussion
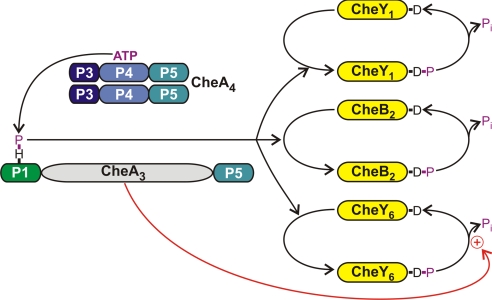
In this study, we have shown that CheA3 is a aspartyl-phosphate phosphatase that is specific for CheY6-P. CheA3 localizes with its partner protein CheA4 to the cytoplasmic chemotaxis cluster (25), where CheA4 phosphorylates CheA3 (24). CheA3-P is a phosphodonor for CheY1, CheY6, and CheB2 (Fig. 5). Chemotactic stimuli, possibly reflecting the metabolic state of the cell, are presumed to control the rate at which CheA4 phosphorylates CheA3 (12). Both CheA3 and CheA4 contain a P5 (regulatory) domain, and the detection of phosphatase activity within CheA3 raises the intriguing possibility that the phosphatase activity of CheA3 and the kinase activity of CheA4 could be reciprocally regulated by chemotactic stimuli. These results may also explain why the activities of CheA3 and CheA4 are encoded within separate proteins rather than within a single polypeptide chain. The chemotactic response of R. sphaeroides has been shown to depend on growth conditions (12). Variation of the expression levels of CheA3 relative to that of CheA4 would alter the phosphatase to kinase ratio and could allow tuning of the CheY6-P output of the signaling pathway according to growth conditions. Consistent with this hypothesis, an internal promoter has recently been discovered within cheOp3 that could allow CheA3 and CheA4 expression levels to be independently regulated (M. Gould, M.A.J.R., and J.P.A., unpublished work).
Fig. 5.
The phosphorylation reactions involving CheA3. The domain structures of CheA3 and CheA4 are shown. The P1 domain of CheA3 is phosphorylated by a CheA4 dimer. CheA3-P then acts as a phosphodonor for either CheY1, CheY6, or CheB2. These RRs all autodephosphorylate. However, CheA3 acts as a phosphatase on CheY6-P (red arrow) and can accelerate the rate of dephosphorylation by at least a factor of 3 over the rate of autodephosphorylation.
CheA3 Phosphatase Activity.
Outside of chemotaxis, many nonhybrid histidine protein kinases have built in aspartyl-phosphatase activity (28, 29); some of the most extensively studied examples are HPKs, NtrB, EnvZ, and PhoR. The phosphatase activity of these proteins has been shown to depend on their dimerization and histidine phosphorylation (DHp) domains (8, 9, 29). Interestingly, the DHp domain is not present in CheA3, suggesting that the phosphatase activity of CheA3 differs substantially from those found in previously characterized HPKs.
The ability of the CheA3 phosphorylation site mutant protein, CheA3(H51Q) to act as a phosphatase indicates that the CheY6-P dephosphorylation mechanism used by CheA3 does not require reversed phosphotransfer from CheY6-P to CheA3. Deletion of the 794-amino acid region between the P1 and P5 domains of CheA3 abolished the CheY6-P phosphatase activity (Fig. 3) but not the ability of the protein to be phosphorylated by CheA4 and subsequently act as a phosphodonor for its RRs. Nested deletion analysis showed that residues 549–749 (sub-region 3) are essential for phosphatase activity, and because no other region of CheA3 was shown to be essential for phosphatase activity, the phosphatase activity presumably resides within this sequence. The entire 794-amino acid region contains no known conserved domains although homologues have been identified in Roseovarius sp. TM1035 (RefSeq accession: ZP_01878577) and in a marine metagenomic sample obtained from a hypersaline lagoon in the Galapagos Islands (CAMERA accession: JCVI_PEP_1105096654245) (30), suggesting that a range of different bacteria may employ this phosphatase activity in their signal transduction pathways.
Relevance of the CheA3 Phosphatase to Chemotactic Signaling.
E. coli CheY-P autodephosphorylates with a half-time of approximately 14 s (13), and its phosphatase, CheZ, can accelerate this by a factor of 100 (14) to approximately 0.14 s. This dramatic stimulation of CheY-P dephosphorylation is required because the stimulus response time of E. coli is approximately 0.2 s (31) and signal termination needs to occur on a comparable time scale (32). When comparing the phosphatase activity of CheA3 toward CheY6-P with E. coli CheZ toward CheY-P, the phosphatase activity of CheA3 appears modest giving a 3-fold versus a 100-fold stimulation, respectively. However, R. sphaeroides may not need such a potent phosphatase because CheY6-P already has one of the fastest known dephosphorylation rates with an approximate half time of 4.1 s versus the 14-s half-time of E. coli CheY. In addition, the measured stimulus response time is slower for R. sphaeroides (∼1 s) than E. coli (∼0.2 s) (31, 33), possibly reflecting the requirement for transport and partial metabolism in R. sphaeroides chemotaxis (12). The enhancement of CheY6-P dephosphorylation by CheA3 reduces the half-time to 1.4 s, which is comparable with the stimulus response time of 1 s (33). The modest phosphatase activity of CheA3 may therefore be critical for chemotaxis because this would bring the rate of CheY6-P dephosphorylation within the physiological range required for efficient gradient sensing and chemotactic signaling. Consistent with this, R. sphaeroides strains without the 794-amino acid region of CheA3 and therefore lacking phosphatase activity were nonchemotactic even though the remainder of the CheA3 protein, CheA3(Δ155–948), was expressed at wild-type levels and localized correctly to the cytoplasmic chemotaxis cluster (Fig. 4). These observations suggest that the phosphatase activity of CheA3 is required to achieve a rapid rate of signal termination that is compatible with the chemotactic migration of cells in chemoeffector gradients.
Chemotaxis Phosphatase Localization.
In this study, we have shown that CheA3 is not only a phosphodonor for CheY6, but is also a specific phosphatase for CheY6-P. However, this colocalization of phosphotransfer and phosphatase functions is not restricted to CheA3 and its close homologues. Methanospirillum hungatei JF-1 has a CheA-CheC fusion protein (RefSeq accession: YP_501607) that sequence analysis suggests would show CheY-P phosphatase activity (34). The CheY-P phosphatases, CheC from B. subtilis and CheX from Treponema denticola, both interact with CheA in two-hybrid assays, suggesting that the principle of colocalizing phosphotransfer and phosphatase activities is also used by these bacteria (35, 36). In E. coli, the chemotaxis phosphatase, CheZ, localizes to the polar chemoreceptor cluster via its interaction with CheAshort. This colocalization of the E. coli chemotaxis kinase and phosphatase at the cell poles prevents the formation of steep spatial gradients of CheY-P concentration that would otherwise form if the phosphatase was not localized at the cell poles and was instead distributed throughout the cytoplasm (37). This is important because steep gradients of CheY-P would expose each flagellar motor to different concentrations of CheY-P depending on their proximity to the chemoreceptor cluster and therefore motor switching would become a function of the distance between the motor and the chemoreceptor cluster (38, 39). In CheA3, R. sphaeroides appears to have extended this network design principle by colocalizing phosphotransfer and phosphatase activities within the same protein molecule.
Methods
Plasmids and Strains.
The plasmids and strains used are shown in Table S1. E. coli strains were grown in LB medium at 37°C. R. sphaeroides strains were grown in succinate medium at 30°C under aerobic conditions with shaking. Where required antibiotics were used at concentrations of 100 μg ml−1 for ampicillin and 25 μg ml−1 for kanamycin and nalidixic acid.
Molecular Genetic Techniques.
All standard genetic techniques were performed as described (40). Pfu polymerase (Promega) was used for all PCRs. All primers were synthesized by Sigma-Genosys. DNA sequencing was performed by Geneservice (Department of Biochemistry, Oxford University).
Mutagenesis of cheA3 in the R. sphaeroides Genome.
Overlap extension PCR was used to generate constructs for (i) overexpressing mutant CheA3 proteins and (ii) for replacing either cheA3 in strain WS8N or yfp-cheA3 in strain JPA1425 in the R. sphaeroides genome with mutant versions of cheA3 (41).
Behavioral Analysis.
The swarm plate and tethered cell responses to propionate of the R. sphaeroides strains were characterized as described previously (21). Swarm plates were used to assess motility by reference to known nonmotile, nonchemotactic, and chemotactic strains; nonmotile cells form smaller colonies on swarm plates than do motile but nonchemotactic cells that, in turn, form smaller colonies than chemotactic cells. Nine datasets were obtained.
Fluorescence Microscopy.
DIC and fluorescence images of YFP fusion expressing R. sphaeroides strains were acquired as described previously (25). At least seven fields of view each containing at least 30 cells from independent cultures were analyzed for each strain.
Protein Purification.
His-tagged and GST-tagged R. sphaeroides CheA, CheY, and CheB proteins were purified as described previously (24, 27). All CheA3 derivatives were purified in the same way as wild-type CheA3 (24). Protein purity and protein concentrations were measured as described (27). Purified proteins were stored at −20°C.
Phosphotransfer from CheA3-P and CheA3P1-P to the Response Regulators.
Phosphotransfer assays were performed at 20°C in TGMNKD buffer (50 mM Tris·HCl, 10% (vol/vol) glycerol, 5 mM MgCl2, 150 mM NaCl, 50 mM KCl, and 1 mM DTT, pH 8.0). Reaction mixtures contained 10 μM CheA4 and either 4 μM CheA3 or CheA3P1. The reactions mixtures were incubated at 20°C for 1 h before addition of 0.5 mM [γ-32P] ATP (specific activity 14.8 GBq mmol−1; PerkinElmer). The ATP dependent phosphorylation of CheA3/CheA3P1 was allowed to proceed for 30 min and then the phosphotransfer reactions were initiated by the addition of 5 μM RR. Reaction aliquots of 10 μl of were taken after 30 s and quenched immediately in 5 μl of 3× SDS/PAGE loading dye (7.5% (wt/vol) SDS, 90 mM EDTA, 37.5 mM Tris·HCl, 37.5% glycerol, and 3% (vol/vol) β-mercaptoethanol, pH 6.8). Quenched samples were analyzed by using SDS/PAGE and phosphorimaging as described previously (27).
Preparation of CheA3P1-32P and CheA2-32P.
Proteins were phosphorylated in reactions performed at 20°C in TGMNKD buffer. The final reaction volumes were 4.5 ml. For production of CheA3P1-32P, reaction mixtures contained 300 μM CheA3P1 (His-tagged) and 20 μM CheA4 (GST-tagged), while for production of CheA2-32P, reaction mixtures contained 60 μM CheA2 (His-tagged). Reactions were initiated by addition of 0.5 mM [γ-32P] ATP (specific activity 14.8 GBq mmol−1). After a 1-h incubation, samples were purified by using Ni-NTA columns (Qiagen) as described previously for unphosphorylated His-tagged CheA2 and CheA3 (24, 27). This purification step removed the unincorporated ATP from the CheA2-32P and CheA3P1-32P preparations and also removed the GST-tagged CheA4 protein from the CheA3P1-P preparation as judged by Coomassie-stained SDS/PAGE gels of the eluted proteins. Purified proteins were stored at −20°C.
Dephosphorylation Assays.
CheY/B-P dephosphorylation rates were measured by using a modification of the method previously described (42, 43). All assays were performed at 20°C in TGMNKD buffer. The final reaction volume was 150 μl. An excess of CheY/B (400 μM final concentration) was added to either CheA3P1-32P (for CheY1, CheY6, and CheB2) or CheA2-32P (for CheY2, CheY3, CheY4, CheY5, and CheB1); the phosphodonor used for each RR was chosen on the basis of fastest phosphotransfer rate. For all RRs except CheY6, the concentration of phosphodonor used in these assays was 2 μM, although because of the rapid autodephosphorylation of CheY6-P it was necessary to increase the concentration of CheA3P1-32P to 30 μM to obtain detectable levels of CheY6-P throughout the 30-s time course. For assessing CheA3 phosphatase activity, parallel reaction mixtures were set up with and without 2.5 μM CheA3, allowing RR dephosphorylation rates to be compared in the presence and absence of CheA3. After addition of the RR to the reaction mixture, 10-μl aliquots were removed at regular time intervals and quenched immediately in 20 μl of 1.5× SDS/PAGE loading dye. Six samples were taken for each time course. The duration of the time course was optimized according to the rate of dephosphorylation of each RR-P; for CheY6 the time course covered 30 s, for CheB1 the time course covered 3,600 s, and for all other RRs the time course covered 120 s. Quenched samples were analyzed by using SDS/PAGE and phosphorimaging as described previously (41). Owing to the vast molar excess of CheY/B used in these assays, the phosphotransfer reactions were completed within 10 s of mixing. Because ATP was not present in the reaction mixtures, no rephosphorylation of the phosphodonor occurred, so once the phosphotransfer reaction had completed (before the first time point), the only reaction occurring was CheY/B-P dephosphorylation. Consequently, by measuring the decrease in CheY/B-P levels over time, it was possible to directly calculate the dephosphorylation rate. All dephosphorylation reactions displayed kinetics that gave a good fit to single exponential decay (R2 > 0.998) allowing the rate constants to be determined by using Microcal Origin software.
SI.
Supplementary Material
Acknowledgments.
This work was funded by the Biotechnology and Biological Sciences Research Council and Lincoln College, Oxford.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808010105/DCSupplemental.
References
- 1.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich LE, Zhulin IB. MiST: A microbial signal transduction database. Nucl Acids Res. 2007;35:D386–D390. doi: 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galperin MY. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perego M, et al. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 5.Hess JF, Oosawa K, Kaplan N, Simon MI. Phosphorylation of three proteins in the signalling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 6.West AH, Stock AM. Histidine kinases and response regulator proteins in two- component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 7.Russo FD, Silhavy TJ. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol. 1991;222:567–580. doi: 10.1016/0022-2836(91)90497-t. [DOI] [PubMed] [Google Scholar]
- 8.Jiang P, Atkinson MR, Srisawat C, Sun Q, Ninfa AJ. Functional dissection of the dimerization and enzymatic activities of Escherichia coli Nitrogen Regulator II and their regulation by the PII protein. Biochemistry. 2000;39:13433–13449. doi: 10.1021/bi000794u. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Qin L, Yoshida T, Inouye M. Phosphatase activity of histidine kinase EnvZ without kinase catalytic domain. Proc Natl Acad Sci USA. 2000;97:7808–7813. doi: 10.1073/pnas.97.14.7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Bio. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 11.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the Bacteria and Archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porter SL, Wadhams GH, Armitage JP. Rhodobacter sphaeroides: Complexity in chemotactic signalling. Trends Microbiol. 2008;16:251–260. doi: 10.1016/j.tim.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Appleby JL, Bourret RB. Proposed signal transduction role for conserved CheY residue Thr87, a member of the response regulator active-site quintet. J Bacteriol. 1998;180:3563–3569. doi: 10.1128/jb.180.14.3563-3569.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silversmith RE, Levin MD, Schilling E, Bourret RB. Kinetic characterization of catalysis by the chemotaxis phosphatase CheZ: Modulation of activity by the phosphorylated CheY substrate. J Biol Chem. 2008;283:756–765. doi: 10.1074/jbc.M704400200. [DOI] [PubMed] [Google Scholar]
- 15.Szurmant H, Bunn MW, Cannistraro VJ, Ordal GW. Bacillus subtilis hydrolyzes CheY-P at the location of its action: The flagellar switch. J Biol Chem. 2003;278:48611–48616. doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- 16.Muff TJ, Foster RM, Liu PJY, Ordal GW. CheX in the three-phosphatase system of bacterial chemotaxis. J Bacteriol. 2007;189:7007–7013. doi: 10.1128/JB.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Motaleb MA, et al. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J Bacteriol. 2005;187:7963–7969. doi: 10.1128/JB.187.23.7963-7969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sourjik V, Schmitt R. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry. 1998;37:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- 19.Ferre A, de la Mora J, Ballado T, Camarena L, Dreyfus G. Biochemical study of multiple CheY response regulators of the chemotactic pathway of Rhodobacter sphaeroides. J Bacteriol. 2004;186:5172–5177. doi: 10.1128/JB.186.15.5172-5177.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackenzie C, et al. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth Res. 2001;70:19–41. doi: 10.1023/A:1013831823701. [DOI] [PubMed] [Google Scholar]
- 21.Porter SL, Warren AV, Martin AC, Armitage JP. The third chemotaxis locus of Rhodobacter sphaeroides is essential for chemotaxis. Mol Microbiol. 2002;46:1081–1094. doi: 10.1046/j.1365-2958.2002.03218.x. [DOI] [PubMed] [Google Scholar]
- 22.Poggio S, et al. A complete set of flagellar genes acquired by horizontal transfer coexists with the endogenous flagellar system in Rhodobacter sphaeroides. J Bacteriol. 2007;189:3208–3216. doi: 10.1128/JB.01681-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.del Campo AM, et al. Chemotactic control of the two flagellar systems of Rhodobacter sphaeroides is mediated by different sets of CheY and FliM proteins. J Bacteriol. 2007;189:8397–8401. doi: 10.1128/JB.00730-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter SL, Armitage JP. Chemotaxis in Rhodobacter sphaeroides requires an atypical histidine protein kinase. J Biol Chem. 2004;279:54573–54580. doi: 10.1074/jbc.M408855200. [DOI] [PubMed] [Google Scholar]
- 25.Wadhams GH, Warren AV, Martin AC, Armitage JP. Targeting of two signal transduction pathways to different regions of the bacterial cell. Mol Microbiol. 2003;50:763–770. doi: 10.1046/j.1365-2958.2003.03716.x. [DOI] [PubMed] [Google Scholar]
- 26.Porter SL, et al. The CheYs of Rhodobacter sphaeroides. J Biol Chem. 2006;281:32694–32704. doi: 10.1074/jbc.M606016200. [DOI] [PubMed] [Google Scholar]
- 27.Porter SL, Armitage JP. Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J Mol Biol. 2002;324:35–45. doi: 10.1016/s0022-2836(02)01031-8. [DOI] [PubMed] [Google Scholar]
- 28.Raivio TL, Silhavy TJ. Transduction of envelope stress in Escherichia coli by the Cpx two- component system. J Bacteriol. 1997;179:7724–7733. doi: 10.1128/jb.179.24.7724-7733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carmany DO, Hollingsworth K, McCleary WR. Genetic and biochemical studies of phosphatase activity of PhoR. J Bacteriol. 2003;185:1112–1115. doi: 10.1128/JB.185.3.1112-1115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yooseph S, et al. The Sorcerer II global ocean sampling expedition: Expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segall JE, Manson MD, Berg HC. Signal processing times in bacterial chemotaxis. Nature. 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 32.Berg HC, Purcell EM. Physics of chemoreception. Biophys J. 1977;20:193–219. doi: 10.1016/S0006-3495(77)85544-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berry RM, Armitage JP. Response kinetics of tethered Rhodobacter sphaeroides to changes in light intensity. Biophys J. 2000;78:1207–1215. doi: 10.1016/S0006-3495(00)76678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wuichet K, Alexander RP, Zhulin IB. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Method Enzymol. 2007;422:3–31. doi: 10.1016/S0076-6879(06)22001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirby JR, et al. CheC is related to the family of flagellar switch proteins and acts independently from CheD to control chemotaxis in Bacillus subtilis. Mol Microbiol. 2001;42:573–585. doi: 10.1046/j.1365-2958.2001.02581.x. [DOI] [PubMed] [Google Scholar]
- 36.Sim JH, Shi W, Lux R. Protein–protein interactions in the chemotaxis signalling pathway of Treponema denticola. Microbiology. 2005;151:1801–1807. doi: 10.1099/mic.0.27622-0. [DOI] [PubMed] [Google Scholar]
- 37.Vaknin A, Berg HC. Single-cell FRET imaging of phosphatase activity in the Escherichia coli chemotaxis system. Proc Natl Acad Sci USA. 2004;101:17072–17077. doi: 10.1073/pnas.0407812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipkow K, Andrews SS, Bray D. Simulated diffusion of phosphorylated CheY through the cytoplasm of Escherichia coli. J Bacteriol. 2005;187:45–53. doi: 10.1128/JB.187.1.45-53.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao CV, Kirby JR, Arkin AP. Phosphatase localization in bacterial chemotaxis: Divergent mechanisms, convergent principles. Physical Biology. 2005;2:148–158. doi: 10.1088/1478-3975/2/3/002. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Russell DW. Molecular Cloning: A laboratory manual. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 41.Porter SL, Wadhams GH, Armitage JP. In vivo and in vitro analysis of the Rhodobacter sphaeroides chemotaxis signaling complexes. Method Enzymol. 2007;423:392–413. doi: 10.1016/S0076-6879(07)23018-6. [DOI] [PubMed] [Google Scholar]
- 42.Thomas SA, Brewster JA, Bourret RB. Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol. 2008;69:453–465. doi: 10.1111/j.1365-2958.2008.06296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silversmith RE, Appleby JL, Bourret RB. Catalytic mechanism of phosphorylation and dephosphorylation of CheY: Kinetic characterization of imidazole phosphates as phosphodonors and the role of acid catalysis. Biochemistry. 1997;36:14965–14974. doi: 10.1021/bi9715573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.