Abstract
The appearance of hair is one of the main evolutionary innovations in the amniote lineage leading to mammals. The main components of mammalian hair are cysteine-rich type I and type II keratins, also known as hard α-keratins or “hair keratins.” To determine the evolutionary history of these important structural proteins, we compared the genomic loci of the human hair keratin genes with the homologous loci of the chicken and of the green anole lizard Anolis carolinenis. The genome of the chicken contained one type II hair keratin-like gene, and the lizard genome contained two type I and four type II hair keratin-like genes. Orthology of the latter genes and mammalian hair keratins was supported by gene locus synteny, conserved exon–intron organization, and amino acid sequence similarity of the encoded proteins. The lizard hair keratin-like genes were expressed most strongly in the digits, indicating a role in claw formation. In addition, we identified a novel group of reptilian cysteine-rich type I keratins that lack homologues in mammals. Our data show that cysteine-rich α-keratins are not restricted to mammals and suggest that the evolution of mammalian hair involved the co-option of pre-existing structural proteins.
Keywords: cytokeratin, epidermis, evolution, reptiles, claw
One of the key steps in mammalian evolution was the emergence of hair, which served in protection from mechanical insults and also facilitated homeothermy (1, 2). Because hair is a defining feature of mammals and does not occur in other amniotes, it seems to have evolved after divergence of the therapsid lineage (leading to mammals) from the sauropsid lineage (reptiles, birds) approximately 310 to 330 million years ago (3). It has been proposed that hair originated by modification of scales (4–6); however, there is no paleontologic evidence for intermediate forms.
The main constituents of the hair shaft are the “hair keratins” and keratin-associated proteins. Hair keratins belong to the protein families of the type I (acidic) and type II (basic) α-keratins, with diverse members that are expressed in a wide variety of epithelia. Type I and type II keratins heterodimerize by coiled-coil formation of their α-helical central domains. The resulting dimers establish higher-order structures via hydrophobic and ionic interactions and ultimately form intermediate filaments (7). In contrast to soft keratins, hair keratins contain numerous cysteine residues, which are used for intermolecular disulfide bond formation during hardening of the hair shaft (8).
Hair keratin genes are nested within the type I and type II keratin gene clusters in the genomes of humans (type I keratin genes on chromosomes 17q21.2 and type II keratin genes as well as KRT18 on chromosome 12q13.13) and other mammals. The human genome contains 11 hair-type and 17 other type I keratins, as well as 6 hair-type and 20 other type II keratin genes (9). All hair keratins are expressed in the hair shaft, and some are also expressed in claws and nails, on the keratinized surface of the tongue, and within Hassall's bodies of the thymus (10, 11). The critical role in hair biology of hair keratins is revealed by the fact that mutations in single hair keratin genes suffice to cause the inherited hair fragility and alopecia syndrome monilethrix (12, 13).
Supporting a mammal-specific origin of hair, hair keratin genes have been reported to be absent from chicken and other nonmammalian species (14, 15). So-called β-keratins, a family of sauropsid-specific proteins structurally unrelated to α-keratins but similar to mammalian keratin-associated proteins (16, 17), are the main components of hard integumentary structures (scales, claws, beaks, feathers) of sauropsids. However, the role of α-keratins in sauropsid epidermis and epidermal appendages has not been explored in depth.
Here we performed a comparative genomics analysis to screen for new homologues of hair keratins in amniotes. We identified nonmammalian homologues of hair keratins in the newly available, unannotated draft genome sequence of the reptile Anolis carolinensis (green anole lizard) and a hard keratin gene orthologue of the chicken. In addition, we identified a group of sauropsid-specific cysteine-rich type I keratins. Our data suggest a nonmammalian origin of the main structural proteins of hair and, therefore, require a revision of the current concept of hair evolution.
Results
Identification of Nonmammalian Hair Keratin-like Genes.
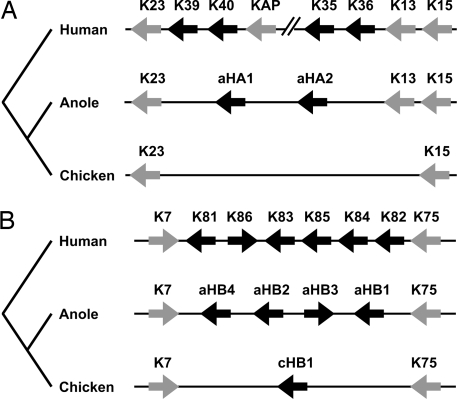
The keratin gene clusters of the anole and chicken were analyzed for the presence of orthologues of mammalian hair keratins. The keratin genes flanking the human hair keratins (KRT23 and KRT13/KRT15 at the type I keratin gene locus, as well as KRT7 and KRT75 at the type II keratin gene locus) were conserved and defined the borders of the hair keratin loci in all species investigated (Fig. 1). Six putative orthologues of mammalian hair keratins were identified in the anole genome and one orthologue in chicken. The novel A. carolinensis genes encoded two type I keratins, tentatively named aHA1 and aHA2 (for anole hard acidic keratins 1 and 2), which were most similar in sequence to keratin 36, and four type II keratins, tentatively named aHB1 through aHB4 (for anole hard basic keratins 1 through 4), which were most similar in sequence to keratins 84 among human keratins. The sequences of all new keratins, sequence alignments with human keratins, and sequence similarities among hard keratins are provided in supporting information (SI) Figs. S1–S3 and Tables S1–S4.
Fig. 1.
Comparison of the α-keratin gene family loci in amniotes. The chromosomal loci containing the type I (A) and type II (B) keratin genes of humans, anole, and chicken are schematically depicted. Genes are represented by arrows pointing in the 5′ to 3′ end direction. Black arrows represent homologues of hair keratins. Note that only 1 of a series of human keratin-associated protein (KAP) genes and 4 of 11 hair keratin genes at the human type I keratin gene locus are shown.
The chicken type I keratin locus lacked a hair keratin-like gene, whereas the chicken type II keratin locus contained one orthologue of mammalian hair keratins, which was most similar to keratin 84 among human keratins. This gene was also predicted by an automated algorithm of GenBank (Gene ID 426897, “similar to keratin, hair, basic, 4”). Here, we will refer to this gene as cHB1 (for chicken hard basic keratin 1) (Fig. 1B).
Because a high content in cysteine residues is an indicator of the ability of keratins to form hard, rigid structures (8), we used this criterion to screen the entire Anolis draft genome sequence for keratins with potential functional equivalence to mammalian hair keratins. Indeed, four more type I keratin genes encoding proteins with high numbers of cysteine residues were identified and tentatively named aHAS1 through aHAS4 (for anole hard acidic sauropsid keratins 1 through 4). These four genes were arranged in tandem at a locus upstream of the keratin 15 homologue of Anolis and were flanked by type I keratins most similar in sequence to human keratins 13 and 17, respectively (not shown). The homologous locus of the chicken genome contained two genes encoding cysteine-rich keratins (GenBank gene IDs 772080, “similar to type I hair keratin KA31” and 420039, “similar to Krt42 protein”), which we here refer to as cHAS1 and cHAS2.
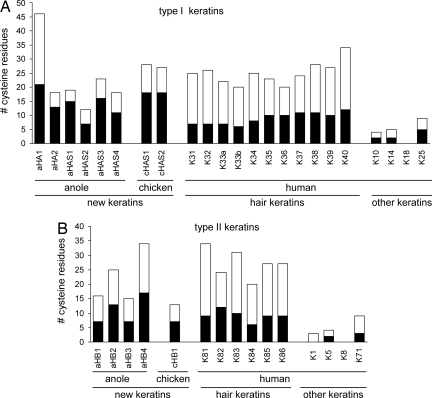
A comparison of the numbers of cysteine residues of the new keratins and of hard and soft human keratins is shown in Fig. 2. The proposed hard α-keratins of Anolis and chicken contained a similar total number of cysteine residues as human hair keratins. Sequence alignment showed that the positions of many cysteine residues were conserved between Anolis hair keratin-like proteins and human hair keratins (Figs. S2 and S3). This indicates that the intermolecular cross-linking in these reptilian keratins occurs in a manner similar to that of mammalian hair keratins.
Fig. 2.
(A and B) Numbers of cysteine residues in various keratins. Cysteine residues were counted in all new anole and chicken keratins and in all human hair keratins. For mammalian non-hair keratins, only representative examples are shown. The black portion of each column indicates the number of cysteines in the central domain of the keratin protein. Note that the cysteine counts of aHAS1, aHAS2, and aHAS4 do not contain the as-yet uncharacterized tail domains.
Next we compared the exon–intron organization of the novel genes with that of human keratin genes. As shown in Fig. S4 (Upper), aHA1 and aHA2 consisted of seven exons and thereby resembled the human type I keratin genes KRT31 through KRT40 (11), as well as KRT18, which has been proposed to be the primordial type I keratin (18, 19). By contrast, anole aHAS3 and chicken cHAS1 and cHAS2 consisted of eight exons and thereby resembled the genes encoding mammalian type I soft keratins or inner root sheath keratins (11). All basic keratins analyzed, including aHB1, aHB2, and cHB1, contained nine exons (Fig. S4, Lower). As noted in a previous report that compared a smaller set of genes (20), most exon–intron borders were conserved between type I and II keratin genes (Fig. S4).
Phylogenetic Analysis Supports Hair Keratin Orthology of Some Sauropsid Keratins and Suggests the Existence of a Novel Clade of Sauropsid-Specific Type I Keratins.
Phylogenetic analyses were performed using the amino acid sequences of the central rod domain (pfam00038, intermediate filament domain). The N-terminal and C-terminal regions were excluded because low sequence complexity in these regions does not allow for unambiguous sequence alignment. The putative hard keratins of A. carolinensis and Gallus gallus were analyzed along with representative human hard and soft keratins. This analysis showed that, among type I keratins (Fig. 3A) and type II keratins (Fig. 3B), the soft keratin clusters were separated from the hard keratin clusters by the branches leading to the primordial keratins (18), keratin 18 and keratin 8, respectively. The anole keratins aHA1 and aHA2 clustered together with human type I hair keratins, and all anole and chicken HB keratins clustered together with human type II hair keratins, supporting a respective orthologous relationship (Fig. 3).
Fig. 3.
Phylogenetic analysis of sauropsid and mammalian keratins. Phylogenetic trees of selected type I (A) and type II (B) keratins were built using the neighbor-joining method. The branch lengths in the tree are proportional to the number of substitutions per site (scale: 0.1 substitutions per site). a, anole; c, chicken; h, human; m, mouse.
aHAS1 through aHAS4 and cHAS1 and cHAS2 formed a cluster that was separated from the branch leading to mammalian type I hair keratins and aHA1 and aHA2 (Fig. 3A), indicating that the ancestral HAS gene was different from the ancestral hair keratin gene. Orthologues of these sauropsid genes could not be detected in any mammalian genomes, including the recently published genome of the platypus (21).
Anolis Hard Keratin Gene Expression Patterns.
The expression of representative novel lizard hard keratin genes was analyzed by RT-PCR on RNAs extracted from various A. carolinensis organs. As shown in Fig. 4, mRNAs of aHA1 and aHA2, as well as aHB1, aHB2, and aHB3, were detected in digits, which contain, among nonepidermal components, scales, toe pad lamellae (22), and claw-forming epidermis. When the proximal part of the digits containing the pad lamellae was separated from the distal part containing the claw, strong expression of hard keratins was detected only in the distal region (not shown). Other than the digits, expression of aHA2, aHB2, and aHB3 was detected in abdominal skin and the tongue (Fig. 4 and data not shown). In contrast to the aforementioned keratins, aHAS3 was not preferentially expressed in the digits but showed a broad expression pattern, including abdominal skin, digits, tongue, esophagus, and stomach (Fig. 4).
Fig. 4.
RT-PCR analysis of anole hard keratin expression. RNA was prepared from various organs of A. carolinensis and analyzed by RT-PCR with keratin-specific primers. The ubiquitously expressed protease caspase-3 was amplified as a control (Con.) for the quality of the RNA preparations. PCRs without cDNA template were performed to control for purity of reagents (lane “control”).
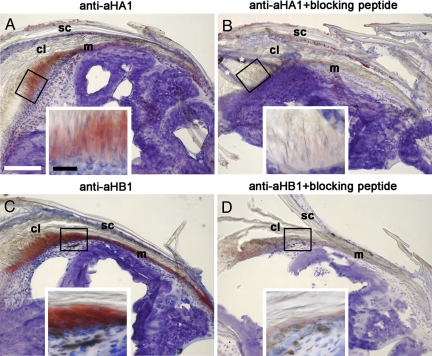
Next we raised antibodies against unique sequences of the C-terminal domains of aHA1 and aHB1 and used these antibodies for the immunohistochemical localization of aHA1 and aHB1. Both proteins were expressed in the claw-forming epidermis of the digit (Fig. 5), whereas expression in scales was very weak or negative (Fig. 5 and data not shown).
Fig. 5.
Immunohistochemical localization of anole hard keratins aHA1 and aHB1. Cryosections of A. carolinensis digits were analyzed by immunohistochemistry using antisera specific for aHA1 (A and B) and aHB1 (C and D). Red staining marks expression of these keratins. Preincubation of the antisera with peptides corresponding to the respective immunization epitope (B and D) blocked the staining and confirmed the specificity of the antisera. Preincubation of the anti-aHA1 antiserum with the aHB1 peptide and vice versa did not block the reaction. In B, white asterisks indicate artefacts caused by folding of tissue fragments over the sectioned claw. (Insets) Larger magnifications of the boxed areas. cl, claw; m, matrix; sc, scale. [White scale bar, 100 μm; black bar, 20 μm.]
Evolutionary History of Hard Keratin Genes in Amniotes.
To determine the evolutionary implications of our data, the numbers of hard (cysteine-rich) keratins in various vertebrate species, as determined in this study and in previous reports (15, 23), were mapped onto the phylogenetic tree of vertebrates (Fig. 6). The identification of hair keratin-like genes in the anole and in chicken strongly suggests that the first hard keratins were innovations of a common ancestor of mammals and sauropsids (Fig. 6, black star). The presence of additional cysteine-rich type I keratins (aHAS1–4 and cHAS1–2) in anole and chicken (indicated by +4 and +2, respectively, in Fig. 6) suggest an additional hard keratin gene innovation after the divergence of sauropsids from the mammalian lineage (Fig. 6, white star).
Fig. 6.
Schematic overview of hard keratin gene evolution. The presence of hard keratins in various vertebrate species was mapped onto a phylogenetic tree of vertebrates. The number of functional type I and type II keratins is shown on the right. The number of sauropsid-specific cysteine-rich type I keratins (aHAS1–4 and cHAS1–2) is indicated separately (+4 and + 2, respectively). Stars indicate gene innovation events and a “strike” symbol indicates the loss of the type I hair keratin-like gene(s) in the lineage leading to chicken.
Discussion
Here we have identified cysteine-rich α-keratin genes in nonmammalian species. We provide three lines of evidence that some of the new Anolis keratins (aHA1, aHA2, aHB1–4) and one chicken keratin (cHB1) are orthologues of mammalian hair keratins, namely (i) syntenic gene location, (ii) conserved exon–intron organization, and (iii) maximum reciprocal sequence similarity. Another group of anole and chicken cysteine-rich keratins (aHAS1–4 and cHAS1–2) differed from hair keratins with regard to the criteria of gene locus synteny and exon–intron organization and, therefore, seem to be derived from a sauropsid-specific molecular innovation.
The high content of cysteine residues of the new sauropsid keratins argues for a functional similarity to mammalian hard keratins, that is, the ability to form hard rigid structures via intermolecular disulfide bonding. To date, the hard properties of sauropsid epidermis have been believed to be mainly mediated by β-keratins; however, an additional role of α-keratins in sauropsid cornification has not been excluded (6, 24). Our immunohistochemical study shows that the cysteine-rich Anolis keratins aHA1 and aHB1 are strongly expressed by keratinocytes that form the claw. Because mammalian claws also contain hair keratins, it is conceivable that the presence of hair keratins in claws is inherited from the last common ancestor of mammals and sauropsids. Other hair keratin-like genes of Anolis, namely aHA2, aHB2, and aHB3, were detected by RT-PCR not only in the digits but also in the skin of other body parts. This demonstrates that the expression of Anolis hair keratin-like genes is not strictly confined to claws and leaves open the possibility that the primordial hard keratins of ancestral sauropsids were also present in scales.
The identification of hard keratin genes in reptiles and in birds strongly argues against the concept that hair keratins were an evolutionary innovation that occurred in the mammalian lineage after the divergence from sauropsids (14, 15). Therefore, we propose the following revision of hair evolution at the molecular level: The last common ancestor of all extant amniotes contained cysteine-rich α-keratins, which served in the establishment of hard non-hair epidermal structures. These genes remained functional in sauropsids, such as A. carolinensis, and were co-opted for a role in hair formation in mammals.
Materials and Methods
Sequence Queries, Alignments, and Phylogenetic Analyses.
Genome sequences of A. carolinensis and G. gallus were retrieved from the GenBank (www.ncbi.nlm.nih.gov/) and the ENSEMBL (www.ensembl.org) databases. Basic local alignment search tool (BLAST) searches for homologues of human hard keratin were performed using the tBLASTn algorithm. Gene orthology was evaluated according to the criteria of reciprocal best hits in BLAST searches (25), conservation of the localization and phase of introns (26), and gene locus synteny (27, 28).
Amino acid sequences were aligned using CLUSTALW, and phylogenetic trees were built with PHYLIP using the Jones-Taylor-Thornton matrix as distance algorithm and the neighbor-joining method for clustering. Trees were displayed using the TreeView program (29).
Nucleic Acid Preparation, PCR, and Sequence Analysis.
Genomic DNA was prepared from tissues according to a standard protocol (30). Tissue RNAs were extracted with the TRIzol reagent (Invitrogen) according to the manufacturer's instructions and reverse-transcribed and PCR-amplified according to standard protocols (31, 32). PCR products were either cloned into the pCR2.1 Topo vector (Invitrogen) according to the manufacturer's instructions and sequenced or purified and sequenced directly. Primer sequences are listed in Table S5.
Generation of Antibodies and Immunohistochemical Analyses.
Antisera against A. carolinensis proteins aHA1 and aHB1 were produced in mice by six injections of the synthetic oligopeptides CGPCPPGPRINTKICRM (corresponding to amino acid residues 509–525 of aHA1) and LSAASCIDLGSPGLP (corresponding to amino acid residues 511–525 of aHB1) (piChem) (100 μg per injection), respectively, coupled to keyhole limpet hemocyanin. The antisera were used at a dilution of 1:1,000 for immunohistochemical analyses of acetone-fixed A. carolinensis tissue cryosections. Endogenous peroxidase was quenched by preincubation with 0.3% H2O2/methanol. Biotinylated sheep anti-mouse Ig (1:200; GE) was used as secondary antibody. Ten percent sheep serum was added to the secondary antibody to prevent unspecific binding. The sections were then exposed to streptavidin–biotin–HRP complex, and 3-amino-9-ethylcarbazole (DakoCytomation) served as chromogen. Hematoxylin was used as a nuclear counterstain. The specificity of the immunoreactions was controlled using preimmune sera and antisera that were preabsorbed with 2 μg/ml of the peptide corresponding to the immunization epitope. Only those immunostainings for which controls yielded a negative or strongly reduced signal were considered specific.
Supplementary Material
Acknowledgments.
We thank Caterina Barresi, Heidemarie Rossiter, Minoo Schuch-Ghannadan, Michael Mildner, and Heinz Fischer for helpful discussions and technical support. The authors acknowledge all providers of public genome sequences, in particular the Broad Institute, Boston, MA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition footnote: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU755363–EU755367).
This article contains supporting information online at www.pnas.org/cgi/content/full/0805154105/DCSupplemental.
References
- 1.Maderson PFA. When? Why? And how? Some speculations on the evolution of the vertebrate integument. Am Zoologist. 1972;12:159–171. [Google Scholar]
- 2.Maderson PF. Mammalian skin evolution: A reevaluation. Exp Dermatol. 2003;12:233–236. doi: 10.1034/j.1600-0625.2003.00069.x. [DOI] [PubMed] [Google Scholar]
- 3.Donoghue PC, Benton MJ. Rocks and clocks: Calibrating the tree of life using fossils and molecules. Trends Ecol Evol. 2007;22:424–441. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Alibardi L. Adaptation to the land: The skin of reptiles in comparison to that of amphibians and endotherm amniotes. J Exp Zool B Mol Dev Evol. 2003;298:12–41. doi: 10.1002/jez.b.24. [DOI] [PubMed] [Google Scholar]
- 5.Alibardi L. Dermo-epidermal interactions in reptilian scales: Speculations on the evolution of scales, feathers, and hairs. J Exp Zool B Mol Dev Evol. 2004;302:365–383. doi: 10.1002/jez.b.20028. [DOI] [PubMed] [Google Scholar]
- 6.Wu P, et al. Evo-Devo of amniote integuments and appendages. Int J Dev Biol. 2004;48:249–270. doi: 10.1387/ijdb.041825pw. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernot KM, Lee CH, Coulombe PA. A small surface hydrophobic stripe in the coiled-coil domain of type I keratins mediates tetramer stability. J Cell Biol. 2005;168:965–974. doi: 10.1083/jcb.200408116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, et al. In vitro assembly and structure of trichocyte keratin intermediate filaments: A novel role for stabilization by disulfide bonding. J Cell Biol. 2000;151:1459–1468. doi: 10.1083/jcb.151.7.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schweizer J, et al. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langbein L, et al. Novel type I hair keratins K39 and K40 are the last to be expressed in differentiation of the hair: Completion of the human hair keratin catalog. J Invest Dermatol. 2007;127:1532–1535. doi: 10.1038/sj.jid.5700734. [DOI] [PubMed] [Google Scholar]
- 11.Rogers MA, Winter H, Langbein L, Bleiler R, Schweizer J. The human type I keratin gene family: Characterization of new hair follicle specific members and evaluation of the chromosome 17q21.2 gene domain. Differentiation. 2004;72:527–540. doi: 10.1111/j.1432-0436.2004.07209006.x. [DOI] [PubMed] [Google Scholar]
- 12.Winter H, et al. A new mutation in the type II hair cortex keratin hHb1 involved in the inherited hair disorder monilethrix. Hum Genet. 1997;101:165–169. doi: 10.1007/s004390050607. [DOI] [PubMed] [Google Scholar]
- 13.Winter H, et al. Mutations in the hair cortex keratin hHb6 cause the inherited hair disease monilethrix. Nat Genet. 1997;16:372–374. doi: 10.1038/ng0897-372. [DOI] [PubMed] [Google Scholar]
- 14.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 15.Zimek A, Weber K. Terrestrial vertebrates have two keratin gene clusters; striking differences in teleost fish. Eur J Cell Biol. 2005;84:623–635. doi: 10.1016/j.ejcb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Alibardi L, Toni M, Dalla Valle L. Hard cornification in reptilian epidermis in comparison to cornification in mammalian epidermis. Exp Dermatol. 2007;16:961–976. doi: 10.1111/j.1600-0625.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- 17.Toni M, Dalla Valle L, Alibardi L. Hard (beta-)keratins in the epidermis of reptiles: Composition, sequence, and molecular organization. J Proteome Res. 2007;6:3377–3392. doi: 10.1021/pr0702619. [DOI] [PubMed] [Google Scholar]
- 18.Blumenberg M. Concerted gene duplications in the two keratin gene families. J Mol Evol. 1988;27:203–211. doi: 10.1007/BF02100075. [DOI] [PubMed] [Google Scholar]
- 19.Schaffeld M, Bremer M, Hunzinger C, Markl J. Evolution of tissue-specific keratins as deduced from novel cDNA sequences of the lungfish Protopterus aethiopicus. Eur J Cell Biol. 2005;84:363–377. doi: 10.1016/j.ejcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Tyner AL, Eichman MJ, Fuchs E. The sequence of a type II keratin gene expressed in human skin: Conservation of structure among all intermediate filament genes. Proc Natl Acad Sci USA. 1985;82:4683–4687. doi: 10.1073/pnas.82.14.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruibal R, Ernst V. The structure of the digital setae of lizards. J Morphol. 1965;117:271–294. doi: 10.1002/jmor.1051170302. [DOI] [PubMed] [Google Scholar]
- 23.Zimek A, Weber K. The organization of the keratin I and II gene clusters in placental mammals and marsupials show a striking similarity. Eur J Cell Biol. 2006;85:83–89. doi: 10.1016/j.ejcb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Alibardi L, Toni M. Cytochemical, biochemical and molecular aspects of the process of keratinization in the epidermis of reptilian scales. Prog Histochem Cytochem. 2006;40:73–134. doi: 10.1016/j.proghi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 25.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 26.Roy SW, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc Natl Acad Sci USA. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng XH, et al. Using shared genomic synteny and shared protein functions to enhance the identification of orthologous gene pairs. Bioinformatics. 2005;21:703–710. doi: 10.1093/bioinformatics/bti045. [DOI] [PubMed] [Google Scholar]
- 28.Eckhart L, et al. Identification of novel mammalian caspases reveals an important role of gene loss in shaping the human caspase repertoire. Mol Biol Evol. 2008;25:831–841. doi: 10.1093/molbev/msn012. [DOI] [PubMed] [Google Scholar]
- 29.Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 30.Strauss WM. In: Current Protocols in Molecular Biology. Ausubel FM, et al., editors. New York: John Wiley & Sons; 1998. p. 2.2. [Google Scholar]
- 31.Dalla Valle L, Toffolo V, Belvedere P, Alibardi L. Isolation of a mRNA encoding a glycine-proline-rich beta-keratin expressed in the regenerating epidermis of lizard. Dev Dyn. 2005;234:934–947. doi: 10.1002/dvdy.20581. [DOI] [PubMed] [Google Scholar]
- 32.Eckhart L, Ban J, Ballaun C, Weninger W, Tschachler E. Reverse transcription-polymerase chain reaction products of alternatively spliced mRNAs form DNA heteroduplexes and heteroduplex complexes. J Biol Chem. 1999;274:2613–2615. doi: 10.1074/jbc.274.5.2613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.