Abstract
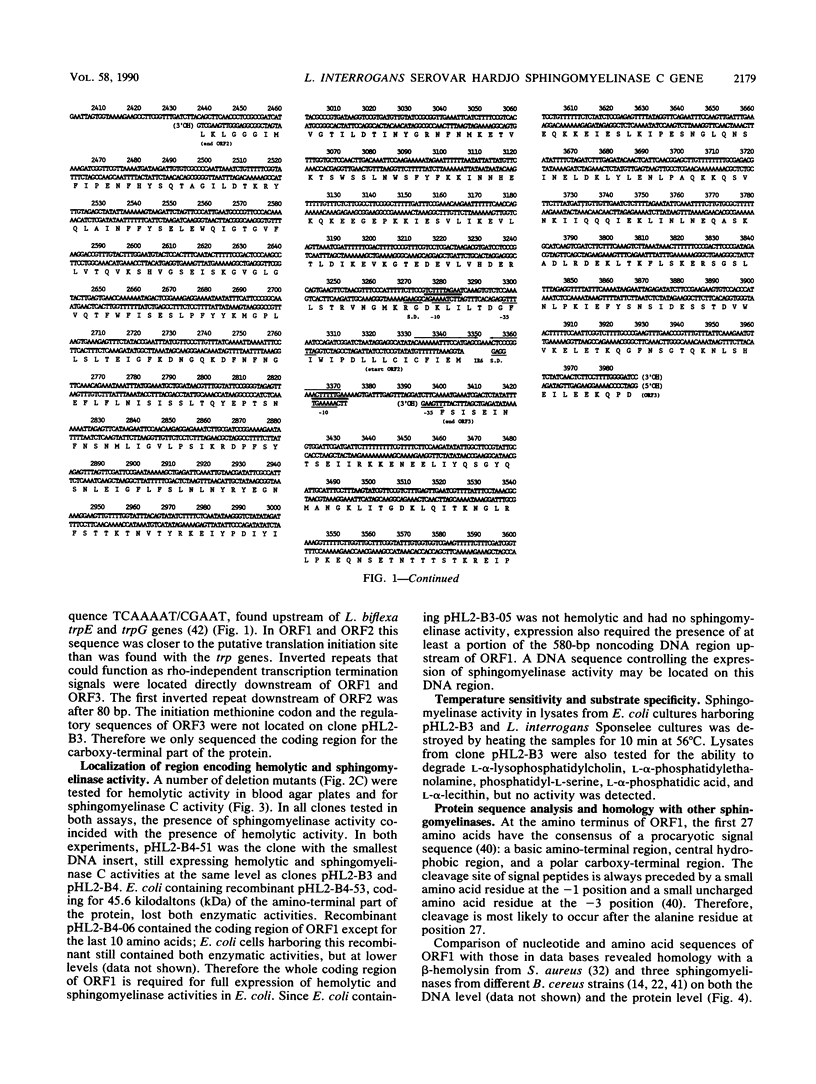
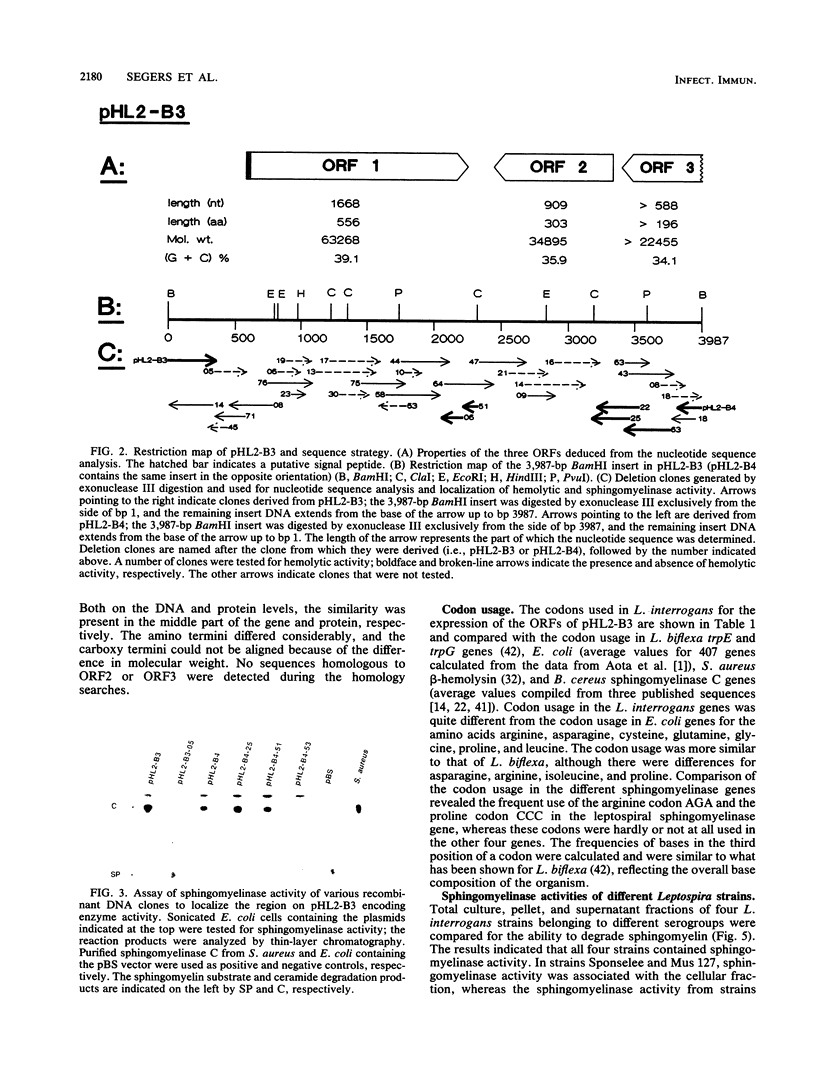
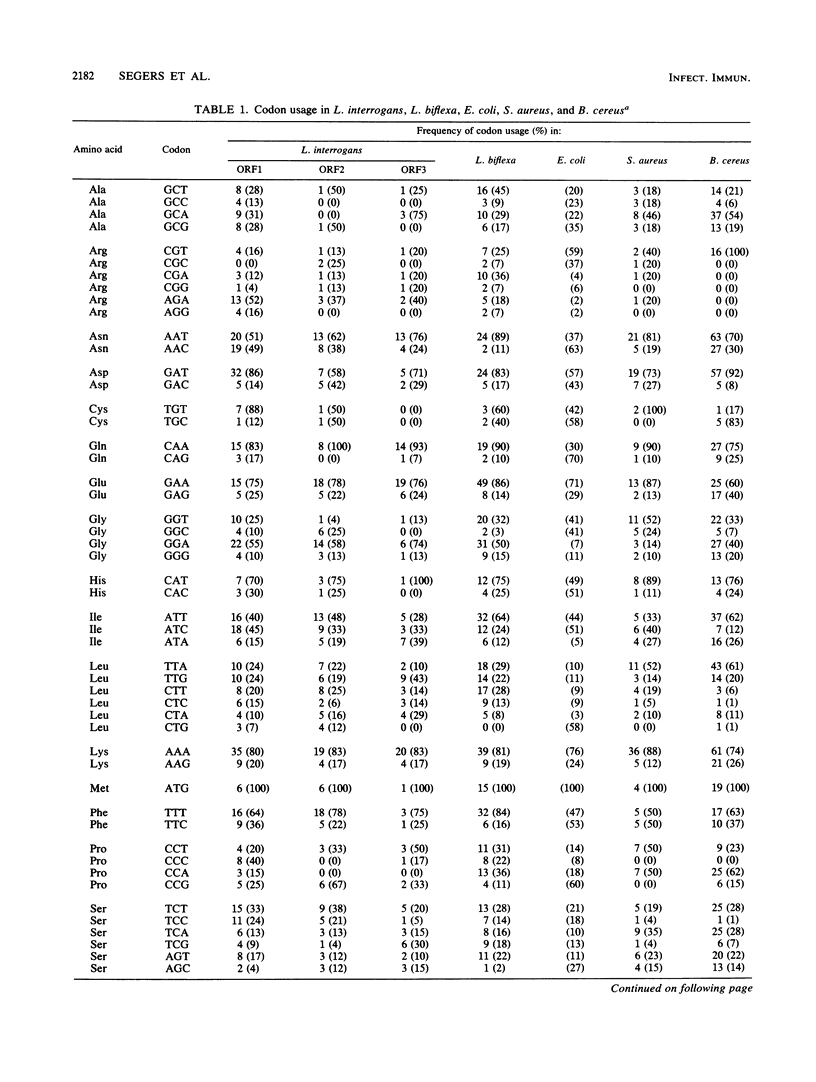
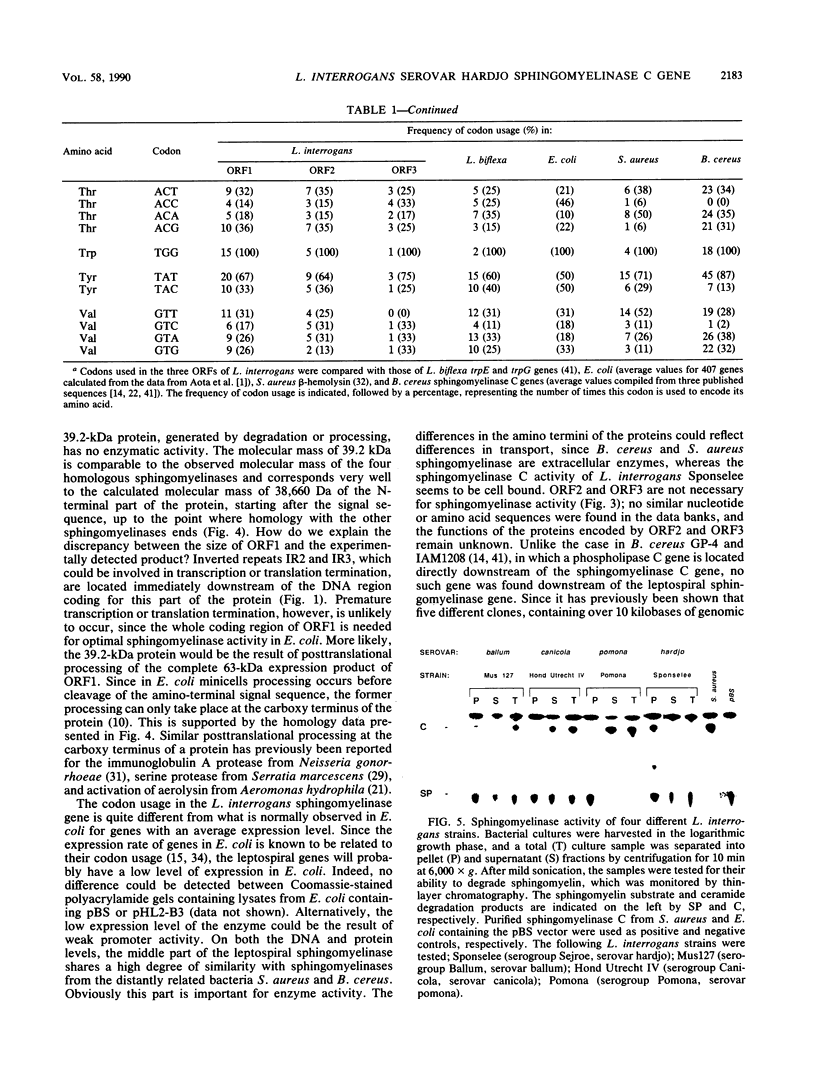
A thermolabile hemolysin from Leptospira interrogans serovar hardjo, strain Sponselee, was shown to specifically degrade sphingomyelin. Nucleotide sequence determination revealed that sphingomyelinase activity was encoded by an open reading frame of 1,668 nucleotides. Although a putative signal sequence could be identified, no evidence for protein export in either L. interrogans or Escherichia coli was obtained. The apparent molecular mass of the expression product in E. coli minicells was 41.2 kilodaltons, whereas open reading frame 1 encoded a protein of 63,268 daltons. The observed difference may be explained by processing at the carboxy-terminal part of the hemolysin in E. coli. A high degree of similarity on the DNA and protein levels with Staphylococcus aureus beta-hemolysin and sphingomyelinase C from three Bacillus cereus strains was observed. The presence of various sphingomyelinase genes within the L. interrogans species is demonstrated.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAUER D. C., EAMES L. N., SLEIGHT S. D., FERGUSON L. C. The significance of leptospiral hemolysin in the pathogenesis of Leptospira pomona infections. J Infect Dis. 1961 Mar-Apr;108:229–236. doi: 10.1093/infdis/108.2.229. [DOI] [PubMed] [Google Scholar]
- Bernheimer A. W., Bey R. F. Copurification of Leptospira interrogans serovar pomona hemolysin and sphingomyelinase C. Infect Immun. 1986 Oct;54(1):262–264. doi: 10.1128/iai.54.1.262-264.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A. M., Yother J., Briles D. E., Hansman D., Paton J. C. Reduced virulence of a defined pneumolysin-negative mutant of Streptococcus pneumoniae. Infect Immun. 1989 Jul;57(7):2037–2042. doi: 10.1128/iai.57.7.2037-2042.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramley A. J., Patel A. H., O'Reilly M., Foster R., Foster T. J. Roles of alpha-toxin and beta-toxin in virulence of Staphylococcus aureus for the mouse mammary gland. Infect Immun. 1989 Aug;57(8):2489–2494. doi: 10.1128/iai.57.8.2489-2494.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P., Vicente M. F., Mengaud J., Baquero F., Perez-Diaz J. C., Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989 Nov;57(11):3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dain A. A., Rozinov M. N., Gol'tsmaier T. A., Gershanovich V. N., Chernukha Iu G. Klonirovanie i ékspressiia gena gemolizina Leptospira pomona pomona v Escherichia coli. Zh Mikrobiol Epidemiol Immunobiol. 1985 Jul;(7):7–10. [PubMed] [Google Scholar]
- Ellis W. A., O'Brien J. J., Bryson D. G., Mackie D. P. Bovine leptospirosis: some clinical features of serovar hardjo infection. Vet Rec. 1985 Aug 3;117(5):101–104. doi: 10.1136/vr.117.5.101. [DOI] [PubMed] [Google Scholar]
- Ellis W. A., Thiermann A. B., Montgomery J., Handsaker A., Winter P. J., Marshall R. B. Restriction endonuclease analysis of Leptospira interrogans serovar hardjo isolates from cattle. Res Vet Sci. 1988 May;44(3):375–379. [PubMed] [Google Scholar]
- FAINE S. Iron as a growth requirement for pathogenic Leptospira. J Gen Microbiol. 1959 Apr;20(2):246–251. doi: 10.1099/00221287-20-2-246. [DOI] [PubMed] [Google Scholar]
- Gilmore M. S., Cruz-Rodz A. L., Leimeister-Wächter M., Kreft J., Goebel W. A Bacillus cereus cytolytic determinant, cereolysin AB, which comprises the phospholipase C and sphingomyelinase genes: nucleotide sequence and genetic linkage. J Bacteriol. 1989 Feb;171(2):744–753. doi: 10.1128/jb.171.2.744-753.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J., Hof H., Emödy L., Goebel W. Influence of cloned Escherichia coli hemolysin genes, S-fimbriae and serum resistance on pathogenicity in different animal models. Microb Pathog. 1986 Dec;1(6):533–547. doi: 10.1016/0882-4010(86)90039-2. [DOI] [PubMed] [Google Scholar]
- Hannun Y. A., Bell R. M. Functions of sphingolipids and sphingolipid breakdown products in cellular regulation. Science. 1989 Jan 27;243(4890):500–507. doi: 10.1126/science.2643164. [DOI] [PubMed] [Google Scholar]
- Hathaway S. C., Marshall R. B. Haemolysis as a means of distinguishing between Leptospira interrogans serovars balcanica and hardjo. J Med Microbiol. 1980 Aug;13(3):477–481. doi: 10.1099/00222615-13-3-477. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Howard S. P., Buckley J. T. Activation of the hole-forming toxin aerolysin by extracellular processing. J Bacteriol. 1985 Jul;163(1):336–340. doi: 10.1128/jb.163.1.336-340.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Haugli F. B., Ikezawa H., Little C. Bacillus cereus strain SE-1: nucleotide sequence of the sphingomyelinase C gene. Nucleic Acids Res. 1988 Nov 11;16(21):10370–10370. doi: 10.1093/nar/16.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima T., Yanagihara Y., Mifuchi I. Characterization of inhibitor to leptospiral hemolysin present in bovine serum. Microbiol Immunol. 1984;28(3):291–302. doi: 10.1111/j.1348-0421.1984.tb00681.x. [DOI] [PubMed] [Google Scholar]
- Kozaki S., Kato K., Asao T., Kamata Y., Sakaguchi G. Activities of Aeromonas hydrophila hemolysins and their interaction with erythrocyte membranes. Infect Immun. 1987 Jul;55(7):1594–1599. doi: 10.1128/iai.55.7.1594-1599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M., Kathariou S., Goebel W. Hemolysin supports survival but not entry of the intracellular bacterium Listeria monocytogenes. Infect Immun. 1988 Jan;56(1):79–82. doi: 10.1128/iai.56.1.79-82.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFebvre R. B., Thiermann A. B., Foley J. Genetic and antigenic differences of serologically indistinguishable leptospires of serovar hardjo. J Clin Microbiol. 1987 Nov;25(11):2094–2097. doi: 10.1128/jcm.25.11.2094-2097.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki H., Yanagida N., Horinouchi S., Beppu T. Characterization of the precursor of Serratia marcescens serine protease and COOH-terminal processing of the precursor during its excretion through the outer membrane of Escherichia coli. J Bacteriol. 1989 Dec;171(12):6566–6572. doi: 10.1128/jb.171.12.6566-6572.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner J., Halter R., Beyreuther K., Meyer T. F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. 1987 Jan 29-Feb 4Nature. 325(6103):458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. L., Eisner W., Novick R. P. Nucleotide sequence: the beta-hemolysin gene of Staphylococcus aureus. Nucleic Acids Res. 1989 Apr 25;17(8):3305–3305. doi: 10.1093/nar/17.8.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalheim O. H. Virulent and avirulent Leptospires: biochemical activities and survival in blood. Am J Vet Res. 1971 Jun;32(6):843–849. [PubMed] [Google Scholar]
- Thiermann A. B. Leptospirosis: current developments and trends. J Am Vet Med Assoc. 1984 Mar 15;184(6):722–725. [PubMed] [Google Scholar]
- Thompson J. C., Marshall R. B. In vitro studies of haemolysis by Leptospira interrogans serovars pomona and ballum. Vet Microbiol. 1986 Mar;11(3):285–292. doi: 10.1016/0378-1135(86)90030-1. [DOI] [PubMed] [Google Scholar]
- Trowbridge A. A., Green J. B., 3rd, Bonnett J. D., Shohet S. B., Ponnappa B. D., McCombs W. B., 3rd Hemolytic anemia associated with leptospirosis. Morphologic and lipid studies. Am J Clin Pathol. 1981 Oct;76(4):493–498. doi: 10.1093/ajcp/76.4.493. [DOI] [PubMed] [Google Scholar]
- Volina E. G., Levina L. F., Soboleva G. L. Phospholipase activity and virulence of pathogenic leptospirae. J Hyg Epidemiol Microbiol Immunol. 1986;30(2):163–169. [PubMed] [Google Scholar]
- Yamada A., Tsukagoshi N., Udaka S., Sasaki T., Makino S., Nakamura S., Little C., Tomita M., Ikezawa H. Nucleotide sequence and expression in Escherichia coli of the gene coding for sphingomyelinase of Bacillus cereus. Eur J Biochem. 1988 Aug 1;175(2):213–220. doi: 10.1111/j.1432-1033.1988.tb14186.x. [DOI] [PubMed] [Google Scholar]
- Yelton D. B., Peng S. L. Identification and nucleotide sequence of the Leptospira biflexa serovar patoc trpE and trpG genes. J Bacteriol. 1989 Apr;171(4):2083–2089. doi: 10.1128/jb.171.4.2083-2089.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Real G., Segers R. P., van der Zeijst B. A., Gaastra W. Cloning of a hemolysin gene from Leptospira interrogans serovar hardjo. Infect Immun. 1989 Aug;57(8):2588–2590. doi: 10.1128/iai.57.8.2588-2590.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]





