Abstract
Cytokinesis is the final step of cell division that completes the separation of two daughter cells. We found that the human discs large (hDlg) tumor suppressor homologue is functionally involved in cytokinesis. The guanylate kinase (GUK) domain of hDlg mediates the localization of hDlg to the midbody during cytokinesis, and over-expression of the GUK domain in U2OS and HeLa cells impaired cytokinesis. Mouse embryonic fibroblasts (MEFs) derived from dlg mutant mice contained an increased number of multinucleated cells and showed reduced proliferation in culture. A kinesin-like motor protein, GAKIN, which binds directly to the GUK domain of hDlg, exhibited a similar intracellular distribution pattern with hDlg throughout mitosis and localized to the midbody during cytokinesis. However, the targeting of hDlg and GAKIN to the midbody appeared to be independent of each other. The midbody localization of GAKIN required its functional kinesin-motor domain. Treatment of cells with the siRNA specific for hDlg and GAKIN caused formation of multinucleated cells and delayed cytokinesis. Together, these results suggest that hDlg and GAKIN play functional roles in the maintenance of midbody architecture during cytokinesis.
Keywords: hDlg, GAKIN, MAGUK, Cytokinesis, Midbody, HeLa cells, Fibroblasts
Introduction
Drosophila discs large (Dlg) tumor suppressor is an essential component for the proper cell polarity in epithelia [1, 2], and for the polarized asymmetric cell division of neuroblasts [3, 4]. Genetic studies revealed that Dlg works in the common pathway with two other tumor suppressors, Lethal giant larvae (Lgl) and Scribble (Scrib) [5–7]. These polarity regulators, Dlg, Lgl, and Scrib are conserved in mammals and have been shown to function in cell polarity pathways including astrocyte migration [8, 9] and immunological synapse formation of T-cells [10]. However, the precise mechanism of how the Dlg-Lgl-Scrib pathway regulates cell polarity is not clear. Dlg belongs to a family of scaffolding proteins called the membrane-associated guanylate kinase homologues (MAGUKs), which are characterized by the presence of the protein-protein interaction motifs including PDZ, SH3, and guanylate-kinase like (GUK) domains [11]. This scaffolding function mediated by the respective domains, capable of forming multiple protein-protein interactions, is considered to be the crucial aspect of Dlg as a polarity regulator.
Extensive efforts from various laboratories have identified a number of interacting partners for the human homologue of Dlg (hDlg), including a kinesin-like protein, GAKIN (guanylate kinase associated kinesin) [12]. GAKIN directly binds to the GUK domain of hDlg via a portion of its stalk region named MAGUK binding stalk (MBS) domain [13]. This interaction is conserved in the Drosophila Dlg and Khc-73, the Drosophila homologue of GAKIN [14]. The functional importance of the Dlg/Khc-73 complex has been demonstrated in the asymmetric cell division of Drosophila neuroblasts, where the Dlg/Khc-73 complex mediates the cortical polarization signal induced by the spindle microtubules [14]. GAKIN is also important for the neuronal axon-dendrite polarity formation by mediating PIP3 translocation [15]. Therefore, an intriguing question remains whether the hDlg/GAKIN complex regulates cell polarity in the mammalian cells.
It has been reported that hDlg redistributes dynamically in the dividing cells with highly concentrated localization at the midbody during cytokinesis [16]. Similar midbody localization of hDlg was also observed in the tissue samples obtained from human invasive cervical carcinoma [17]. Although this specific localization of hDlg at the midbody suggests its functional involvement in the process of cytokinesis, definitive experimental evidence is lacking. We are interested in the function of hDlg in cytokinesis because cytokinesis is considered as one form of cell polarity that requires localized deposition of signaling molecules and directed membrane transport [18, 19]. In this report, we present evidence showing the functional involvement of hDlg, GAKIN, and their respective domains in cytokinesis.
Materials and Methods
Cell culture and transfection
U2OS and HeLa cells were maintained in DMEM (GIBCO) containing 10% fetal bovine serum (FBS) (GIBCO). MEFs were cultured in DMEM containing 10% FBS, 2 mM L-Glutamine (Invitrogen), 0.1 mM MEM non-essential amino acids (GIBCO), 10 Units/ml Penicillin, and 10 µg/ml Streptomycin (GIBCO). DNA transfections were performed using Lipofectamine 2000 (Invitrogen).
Plasmids
The DsRed-hDlg-I2 and -I3, -GUK, -ΔGUK, -PDZ, -NT and –GUK (p55) constructs were generated by subcloning the respective cDNAs [12, 20] into the pDsRed2-C1 (Clontech). GFP-fused GAKIN constructs were made using the pEGFP-C1 vector (Clonetech). GFP-GAKIN and GFP-GAKIN (Δmotor) constructs were described previously [13]. GFP-GAKIN (ΔCAP-Gly) contains amino acids 1–1734 of GAKIN, lacking the C-terminal portion including the CAP-Gly domain. GFP-GAKIN (S110N) mutant was designed corresponding to the T93N mutation of KIF5, which has been characterized as an ATPase deficient rigor-motor [21]. The point mutation was introduced in GAKIN by the site-directed mutagenesis using QuickChange II site-directed mutagenesis kit (Stratagene).
Generation of MEFs
MEFs with the genotypes dlggt/dlggt, dlggt/+, and +/+ were generated from embryos produced by crossing dlggt/+ heterozygous males and females [22]. At E13.5, embryos were collected and MEFs were prepared from each embryo separately using the established methods (Hogan et al., 1994). The genotype of MEFs was established by Western blotting using an anti-hDlg rabbit polyclonal antibody (anti-SAP97, Affinity BioReagents) as previously described [22].
Cell proliferation assay
Cell proliferation was determined by staining the cells with crystal violet as well as direct counting. For crystal violet method, MEFs (passage 1) were seeded in 96 well plates at the density of 1×103 cells per well. After 3, 6, 9, and 12 days, cells were fixed with 4% paraformaldehyde, and stained with 0.5% crystal violet for 10 min at room temperature. After washing with PBS, stained cells were solubilized in 1% SDS (100 µl/well) for 30 min and absorbance was measured at 550 nm. For the direct counting by hemocytometer, MEFs (passage 2) were seeded in 6 well plates at 3 × 105 cells per well on the day 0. Cells were trypsinized and counted at every 3 days, and reseeded at 3 × 105 cells per well.
Immunofluorescence microscopy
U2OS, HeLa, and MEFs were grown on glass coverslips and fixed with 4% paraformaldehyde in PBS for 10 min. Following permeabilization with 0.1% Triton X-100 in PBS for 10 min, samples were blocked with 2% FBS for 1 h and treated for staining. Microtubules were stained with anti-β-tubulin mAb (TUB 2.1; Sigma) and with FITC-conjugated (Sigma) or Texas-Red-conjugated (Molecular Probe) anti-mouse IgG secondary Abs. Nuclear DNA was stained with 10 ng/ml of DAPI (4',6'-diamidino-2-phenylindole) (PIERCE). The specimens were observed using a TE2000E fluorescence microscope (Nikon) equipped with a 40X/1.30 objective at room temperature. Images were recorded by a CCD camera (CoolSnapHQ; Roper Scientific) and analyzed using MetaMorph software (Universal Imaging Corp.).
Knockdown of hDlg and GAKIN by RNAi
The siRNA for hDlg, (Sense; 5′-GCUCUACUGUGACUUCAGAdTdT-3′, Antisense; 5′-UCUGAAGUCACAGUAGAGCdTdT-3′) and for GAKIN, (Sense; 5′-GUGCCUUGGAGAGAAUAUCdTdT-3′, Antisense; 5′-GAUAUUCUCUCCAAGGCACdTdT-3′) were synthesized by QIAGEN. Negative control siRNA (Sense; 5′-UUCUCCGAACGUGUCACGUdTdT-3′, Antisense; 5′-ACGUGACACGUUCGGAGAAdTdT-3′) was also obtained from QIAGEN. The siRNAs were introduced into cells by a TransMessenger Transfection Reagent (QIAGEN) according to the vender’s instructions. The expression level of hDlg and GAKIN was determined by Western blot analysis of the transfected cell lysates using anti-hDlg mAb (2D11) [17] and anti-GAKIN mAb [15].
Mitotic synchronization
HeLa cells were synchronized at the M-phase using a double-thymidine block followed by nocodazole arrest [23]. Cells were plated on glass coverslips in 6 well plates at the density of 1 × 105 cells/well one day before the start of the thymidine treatment, and plasmid constructs were transfected during the second thymidine block. Following the treatment with 40 ng/ml nocodazole for 6 h, cells were washed to release them from the arrest. At certain time points after the release, cells were fixed and processed for tubulin and DAPI staining. Mitotic cells at metaphase, anaphase, cytokinesis stages, and the interphase cells still visibly connected by a cytokinesis bridge were counted by immunofluorescence microscopy. Two interphase cells connected by a cytokinesis bridge were counted as one cell for consistency throughout this study. To quantify the delay in cytokinesis, the percentage of cells not completing cytokinesis in three hours was calculated by dividing the percentage of cytokinesis attained at three hours by the percentage of the metaphase/anaphase cells at one hour. Cells treated with siRNAs were synchronized by essentially the same method, except that the cells were plated at the density of 5 × 104 cells/well one day before the addition of siRNAs, and the first thymidine block started 4 h after the siRNA treatment.
Results
GUK domain mediates the midbody localization of hDlg
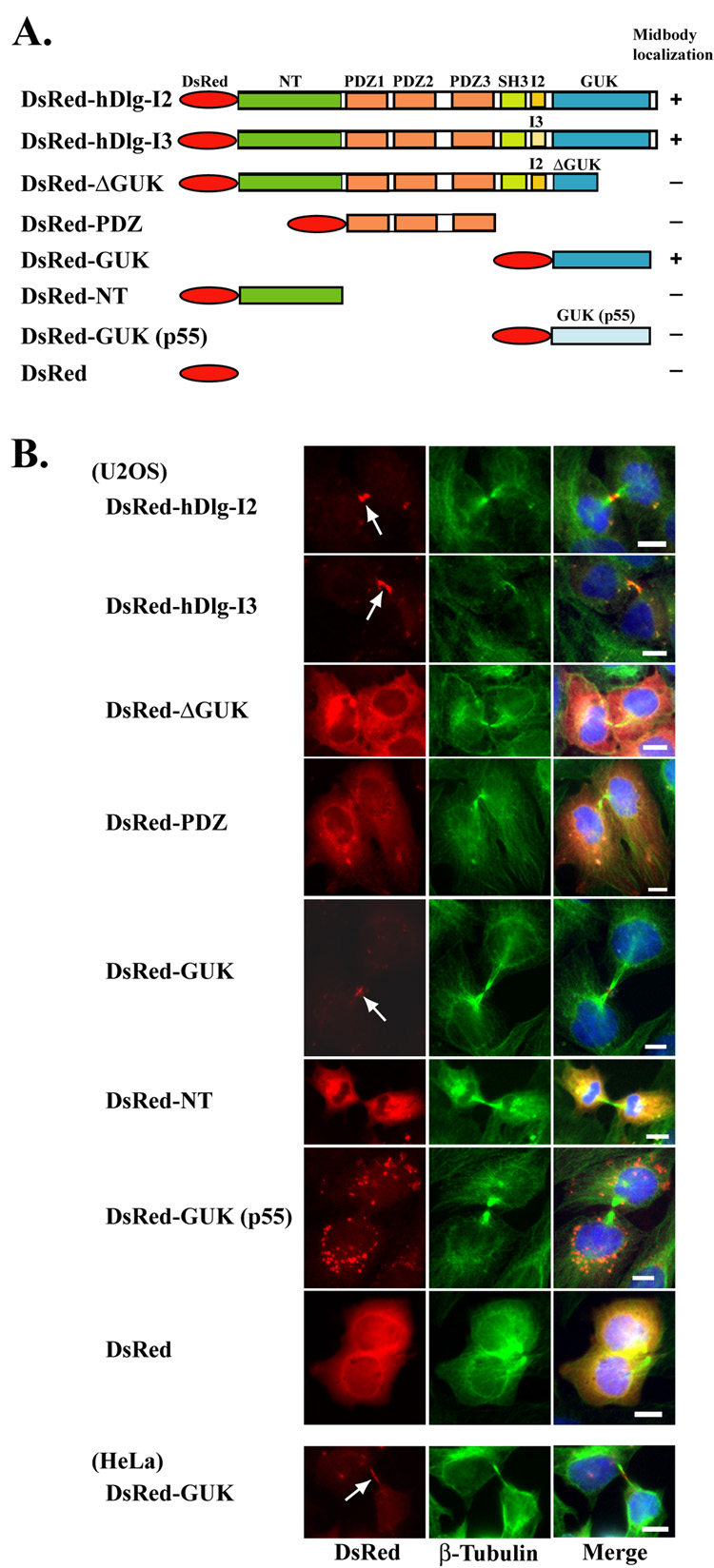
A previous study has shown that endogenous hDlg as well as the exogenously expressed full length hDlg are localized to the midbody in keratinocyte HaCaT cells and osteosarcoma U2OS cells during cytokinesis [16]. We used a series of truncated constructs of DsRed-hDlg (Fig. 1A) to identify the domain of hDlg that mediates its midbody localization. Expression of the protein products, corresponding to their expected size, was confirmed by Western blotting for all the constructs (data not shown). Consistent with the previous study [16], we observed that the DsRed-fused full length hDlg, in both I2 and I3 isoforms [20], appeared highly concentrated in the midbody structure when transiently expressed in U2OS cells (Fig. 1B). Among the truncated constructs, the DsRed-GUK domain localized efficiently to the midbody, whereas the DsRed-NT and DsRed- PDZ domains did not, suggesting the specific requirement of the GUK domain for such localization (Fig. 1B). Moreover, the DsRed-ΔGUK construct, which encodes a truncated GUK domain, failed to localize to the midbody. Interestingly, the GUK domain of p55, another MAGUK protein expressed in both erythroid and non-erythroid cells did not show any midbody localization. A similar midbody localization of the DsRed-GUK domain of hDlg was also observed in HeLa cells (Fig. 1B, bottom panels). These results demonstrate that the GUK domain is necessary and sufficient for the midbody localization of hDlg in these cells.
Fig. 1. Localization of various DsRed fusion constructs of hDlg during cytokinesis.
(A) Schematic representation of DsRed-hDlg constructs used in the study. The hDlg-I2 and -I3 are two naturally occurring isoforms, which were created by the inclusion of alternatively spliced exons, termed insertion 2 and insertion 3, respectively [20]. The GUK domain of another MAGUK, erythroid p55, was used as a negative control. Constructs that showed midbody localization were marked as (+). (B) Localization of DsRed-hDlg constructs during cytokinesis. U2OS and HeLa cells were transfected with DsRed-hDlg constructs, and the DsRed signal (red) was visualized along with β-tubulin (green) and DAPI (blue) staining. Arrows indicate the midbody localization of DsRed fusion constructs. Bar 10 µm.
Dominant negative effect of GUK domain of hDlg on cytokinesis
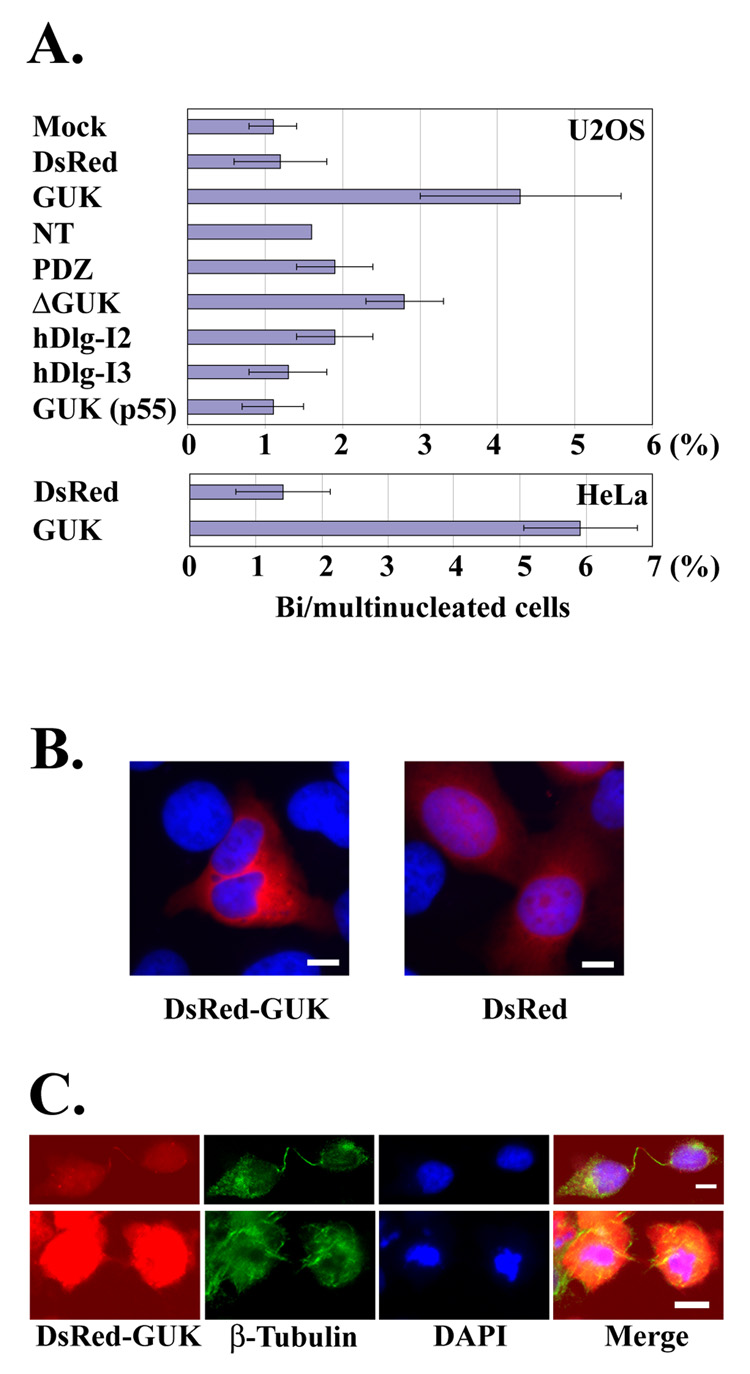
The presence of hDlg at the midbody suggests that hDlg may play a functional role in the process of cytokinesis. Failed cytokinesis generally results in the fusion of two daughter cells, thus leading to the formation of multinucleated cells that are used to assess the cytokinesis failure [24–26]. We speculated that the potent midbody localization of the DsRed-GUK domain might interfere with the function of endogenous hDlg at the midbody. Consistent with this idea, we observed an increase in the number of multinucleated cells after expression of DsRed-GUK domain in the U2OS and HeLa cells (Figs. 2A, B). Expression of the DsRed-ΔGUK domain also caused an increase in the number of multinucleated cells albeit to a significantly lesser extent. Full length hDlg constructs and other domains had negligible or no effect on multinuclearity. In addition to the multinucleated phenotype, the DsRed-GUK expressing cells often showed abnormal morphology, which is typically observed during cytokinesis failure. Two daughter cells connected with an unusually long and extended intracellular bridge (Figs. 2C, upper panels), and cells showing apoptotic nuclear morphology while still connected by the intercellular bridge (Fig. 2C, bottom panels), were frequently observed.
Fig. 2. Formation of multinucleated cells after over-expression of DsRed-GUK.
(A) Frequency of bi/multinucleated cells after the over-expression of DsRed-hDlg fusion proteins. U2OS and HeLa cells were transfected with various constructs, and the number of multinucleated cells among transfected cells was counted. The data are mean ± SD from three independent experiments. At least 116 transfected cells were analyzed for each sample. (B) Representative images of U2OS cells expressing DsRed-GUK and DsRed control. Note that the DsRed-GUK expressing cell contains two nuclei. Images are the merge of DsRed (red) and DAPI (blue) signals. (C) Unusual morphology of the HeLa cells expressing DsRed-GUK is noteworthy. Bar 10 µm.
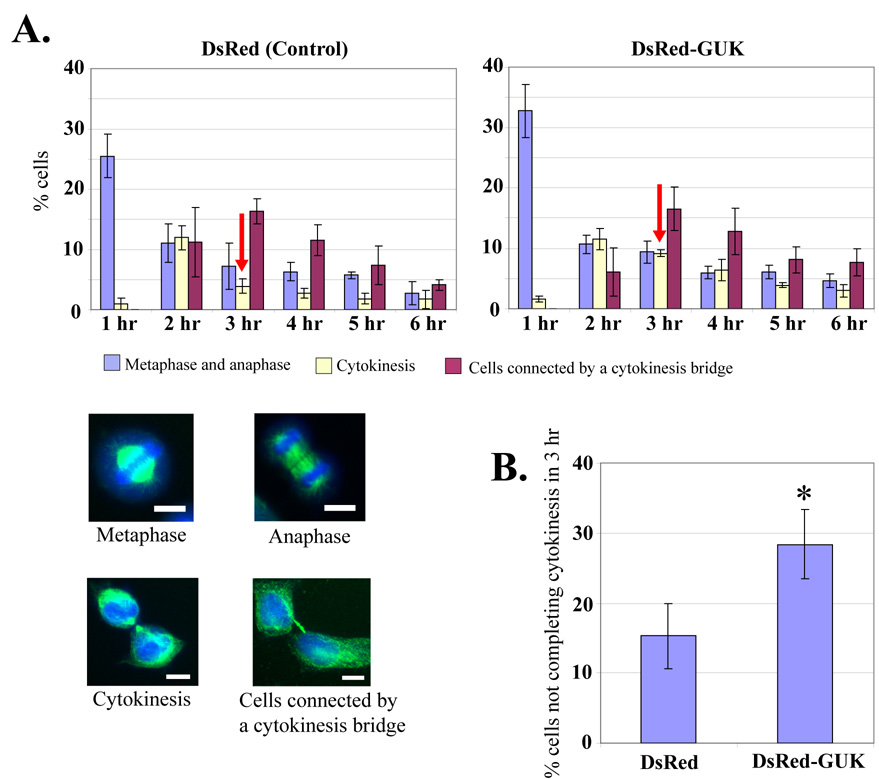
When the cytokinesis pathway is impaired, cells generally take longer time at the cytokinesis phase before they eventually either complete this step or fuse back [27]. We tested whether the overexpression of DsRed-GUK domain causes a delay in cytokinesis. To accomplish this, the DsRed-GUK and control DsRed were expressed in the HeLa cells and cells were arrested at the M-phase by double thymidine block and nocodazole treatment. At defined time points after the washout of the nocodazole, cells were fixed and stained for DNA and tubulin, and the number of cells at various phases of mitosis was quantified by fluorescence microscopy. Cells at the metaphase, anaphase, and cytokinesis stages were counted based on their morphology, as represented in Fig. 3A. Cytokinesis bridges often persist long after cells finished mitosis. Therefore we counted those interphase cells that appeared to have completed mitosis but were still visibly connected by midobody structure separately, as “cells connected by a cytokinesis bridge”. Under these conditions, about 25–30% of cells are synchronized at the M phase as shown by the percentage of mitotic cells seen one hour after the nocodazole washout (Fig. 3A). A relatively higher number of cells at the cytokinesis stage were observed two hours after the release in the control DsRed-expressing cells, and this number decreased three hours after the release, suggesting that the majority of control cells completed cytokinesis within three hours (Fig. 3A, left panel). In contrast, a significantly higher number of cells were observed at the cytokinesis stage three hours after the release in DsRed-GUK expressing cells (Fig. 3A, right panel). After three hours, the number of cells still at the cytokinesis stage was compared with the total number of mitotic cells observed at one hour and the values were represented as cells not completing cytokinesis within three hours (Fig. 3B). Significantly more DsRed-GUK domain expressing cells were arrested at the cytokinesis stage three hours after the release, as compared to the control (Fig. 3B). Together, these results demonstrate that the DsRed-GUK expressing cells exhibit a delay in cytokinesis under these conditions.
Fig. 3. Delayed cytokinesis in cells over-expressing the DsRed-GUK.
(A) HeLa cells expressing control DsRed or DsRed-GUK were arrested in the M phase by double thymidine block and nocodazole treatment. At the indicated hours after the washout of nocodazole, cells were fixed and stained for tubulin and DNA. Mitotic cells were counted at the metaphase, anaphase, cytokinesis, and interphase when they were still connected by a “cytokinesis bridge”. The data reflect mean ± SD from three independent experiments. The representative images are shown in the bottom panels. Bar 10 µm. (B) Percentage of cells not completing cytokinesis at three hours after nocodazole washout. The value is the percentage of cytokinesis at three hours divided by the percentage of metaphase and anaphase cells at one hour. Data are mean ± SD from three independent experiments. *P < 0.05.
Enhanced multinuclearity and reduced proliferation of MEFs from Dlg mutant mice
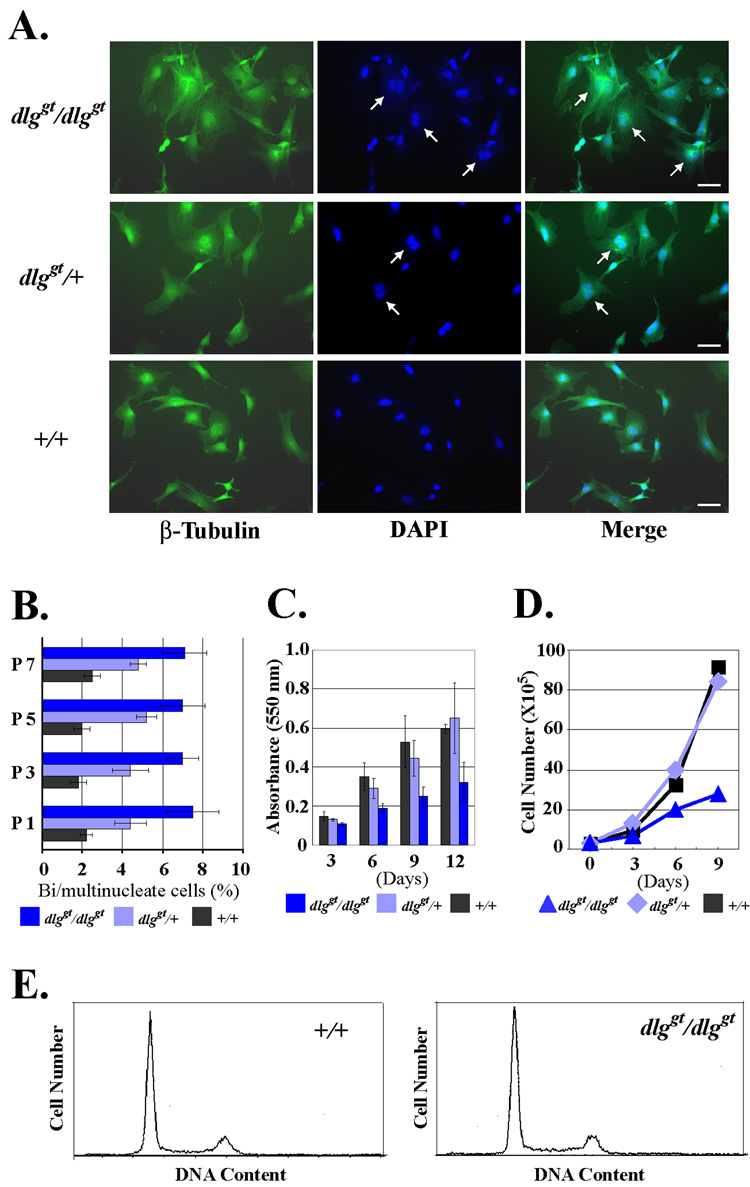
A genetically targeted mutant of the murine homologue of hDlg has been described previously [22]. The mutant mouse, dlggt/dlggt, has an insertion of β-geo at amino acid 549 of Dlg1, resulting in the expression of a fusion protein containing 1–549 residues of Dlg1 and β-geo. Since the resultant fusion protein of mouse Dlg is lacking the C-terminal GUK domain, it is predicted to be defective in the midbody localization. Homozygous dlg mutant mice are reported to exhibit growth retardation in utero, have hypoplasia of the premaxilla and mandible, show a cleft secondary palate, and die perinatally [22]. We isolated MEFs from dlggt/dlggt embryos to test whether they exhibit defects in cytokinesis. The MEFs from dlggt/dlggt mice contained a significantly increased number of multinucleated cells as compared to the MEFs from the wild type littermates, whereas the heterozygotes had an intermediate effect (Fig. 4A). The high frequency of multinucleated cells in dlggt/dlggt MEFs persisted through passages (Fig. 4B), suggesting that multinucleated cells were constantly produced in culture. We also noticed that the proliferation rate of dlggt/dlggt MEFs was significantly suppressed (Fig. 4C, D), although flow cytometry analysis did not reveal major difference in the cell cycle distribution such as a stop in G0 (Fig. 4E). Therefore, it is possible that the reduced proliferation rate is due to the frequent cytokinesis failure or the extended time required by dlggt/dlggt MEFs to complete cytokinesis. However, the increase of multinucleated cells was not detectable in the flow cytometry, possibly due to the relatively low frequency of multinucleated cells in the dlggt/dlggt MEFs. Together, these results demonstrate that mammalian Dlg is an important functional component for the completion of cytokinesis.
Fig. 4. Multinuclearity and reduced proliferation of dlg mutant MEFs.
(A) Multinucleated cells in dlggt/dlggt MEFs. Homozygous dlggt/dlggt, heterozygous dlggt/+, and wild type +/+ MEFs obtained from littermates were plated on glass coverslips, and stained for β-tubulin (green) and DNA with DAPI (blue). Multinucleated cells are indicated by arrows. Bar 50 µm. (B) Quantification of multinucleated cells. At the indicated number of passages, the MEFs were analyzed for the number of multinucleated cells. Three independent preparations of MEFs for homozygous, heterozygous, and wild type were used for the analysis. Data are mean ± SD. At least 225 cells for each sample were counted. (C) Proliferation assay of MEFs by crystal violet metod. Passage 1 MEFs were plated in the 96 well plates and the cell number was quantified by the crystal violet method at the indicated days after plating. Three independent preparations of MEFs for homozygous, three for heterozygous, and two for the wild type were derived from littermate embryos and the mean ± SD values are shown. (D) Proliferation of MEFs by direct cell counting. 3×105 MEFs were passaged every 3 days in a 6 well plate. The cell numbers were counted at each passage point. (E) Flow cytometry of the DNA content of the wild type and dlggt/dlggt MEFs.
Localization of GAKIN to the midbody
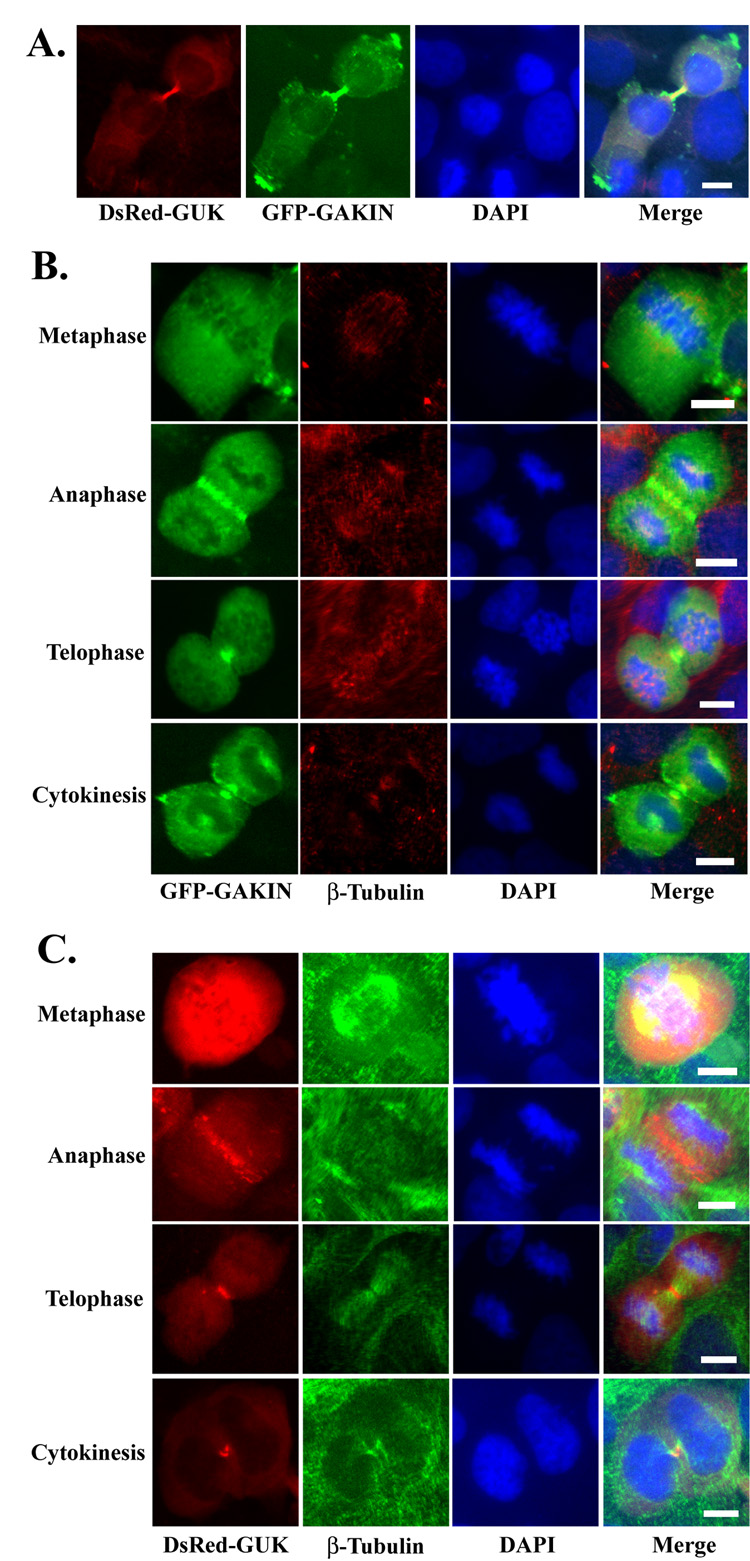
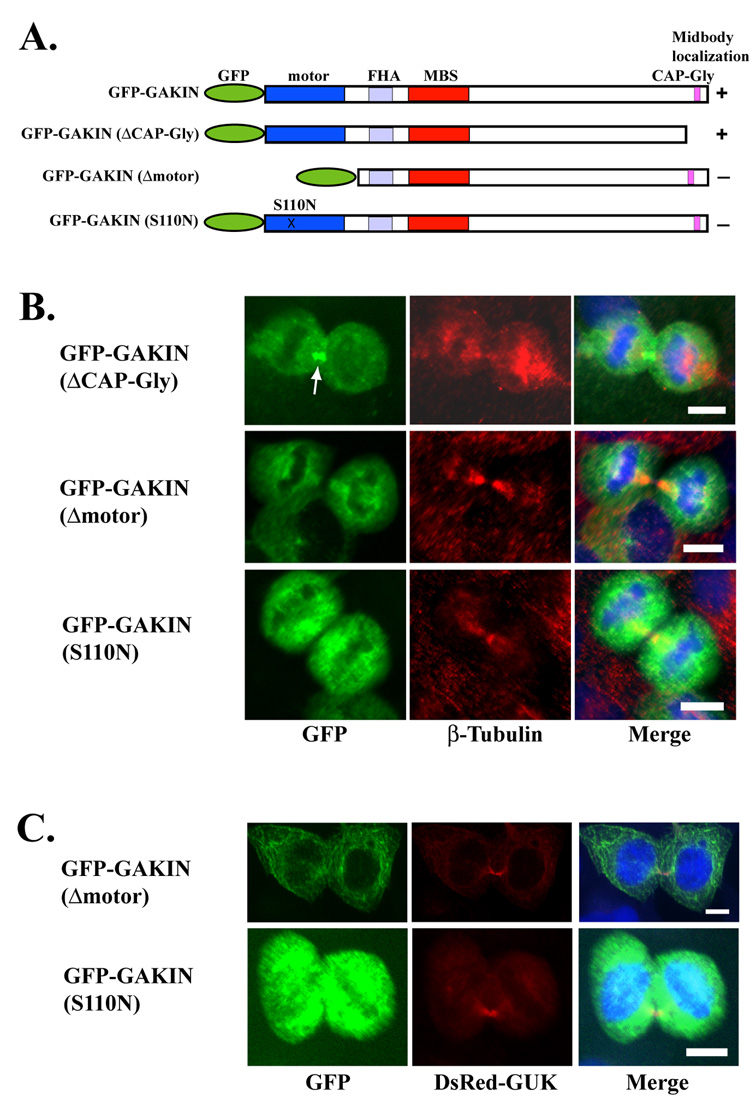
The importance of the GUK domain of hDlg for localization to the midbody prompted us to test the involvement of GAKIN, a kinesin-like motor protein. GAKIN binds directly to the GUK domain of hDlg, and co-accumulates with hDlg at the tip of cellular processes, possibly through a microtubule-based transport mechanism [13]. When the GFP-fused full length GAKIN was co-expressed with DsRed-GUK domain of hDlg, they co-localized intensely at the midbody during cytokinesis (Fig. 5A). When expressed singly, the GFP-GAKIN distributed diffusely throughout the cytoplasm at the metaphase, started to accumulate around the cleavage furrow at the anaphase, and became concentrated in the midbody at cytokinesis (Fig. 5B). This pattern of localization is strikingly similar to the DsRed-GUK domain of hDlg throughout cell division (Fig. 5C). Full length GAKIN has two domains that can interact with the microtubules. The motor domain, located in the N-terminus, and the CAP-Gly domain, located in the C-terminus (Fig. 6A). We tested if either of these domains can mediate the midbody localization of GAKIN. A truncated GAKIN (ΔCAP-Gly) was found to be concentrated at the midbody (Fig. 6B) suggesting that the C-terminus CAP-Gly domain is not required for the midbody localization of GAKIN under these conditions. The mutant GAKIN (S110N) has a point mutation in the motor domain, which is predicted to disrupt its ATPase activity [21]. Another GAKIN construct (Δmotor) is a truncation mutant lacking the motor domain. These two motor-defective mutants failed to concentrate in the midbody, although they appear to be localized along microtubules (Fig. 6B). These results suggest that a functional motor domain with the ATPase activity is essential for the localization of GAKIN to the midbody. If the targeting of hDlg to the midbody is mediated by the GAKIN-dependent transport process, then these motor-defective GAKIN mutants might function in the dominant negative fashion thus inhibiting the localization of hDlg to the midbody. However, the over-expression of either GAKIN (Δmotor) or GAKIN (S110N) did not inhibit the localization of DsRed-GUK domain to the midbody (Fig. 6C). These data suggest that the targeting of hDlg to the midbody appears to be independent from the motor activity of GAKIN under these conditions.
Fig. 5. Midbody localization of GAKIN during cytokinesis.
(A) Co-localization of DsRed-GUK and GFP-GAKIN in U2OS cells during cytokinesis. DsRed-GUK and GFP-GAKIN constructs were co-transfected in U2OS cells. DsRed-GUK and GFP-GAKIN co-localized intensely at the midbody during cytokinesis. (B) Distribution of GFP-GAKIN during mitosis. GFP-GAKIN was singly transfected in U2OS cells and its distribution in the dividing cells was observed along with β-tubulin (red) and DAPI staining (blue). (C) Distribution of DsRed-GUK. The U2OS cells were singly transfected with DsRed-GUK and its distribution was observed along with β-tubulin (green) and DAPI staining (blue). Bar 10 µm.
Fig. 6. Requirement of the functional motor domain of GAKIN for midbody localization.
(A) Schematic representation of GAKIN constructs used in this study. The motor designation in the N-terminus is the kinesin-like motor domain. The FHA (forkhead associated) domain is the binding site for PIP3BP [15]. The MBS domain is the binding site for hDlg [13]. The CAP-Gly domain is the microtubule binding domain. (B) Distribution of GAKIN mutants during cytokinesis. GFP-fused GAKIN mutant constructs were transfected in U2OS cells and their localization during cytokinesis was assessed by co-staining with β-tubulin (red) and DAPI (blue). GFP-GAKIN (ΔCAP-Gly) localized to the midbody (arrow). GFP-GAKIN (Δmotor) and (S110N) proteins did not localize to the midbody, although their co-localization with microtubules is noticeable. (C) Co-expression of GFP-GAKIN (Δmotor) and GFP-GAKIN (S110N) with DsRed-GUK in U2OS cells. Midbody localization of DsRed-GUK was not affected by the over-expression of GAKIN mutants. Bar 10 µm.
Impaired cytokinesis by siRNA knock down of hDlg and GAKIN
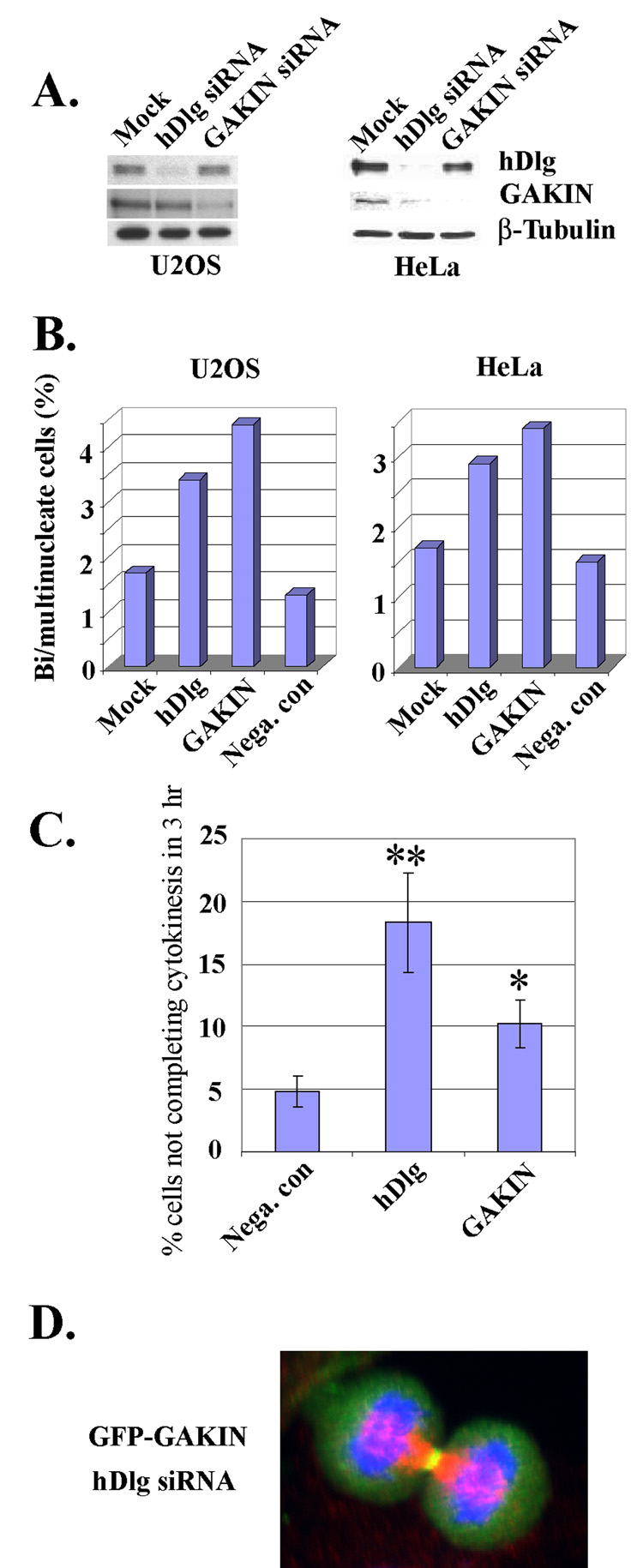
Finally, we tested the effect of siRNA knock down of hDlg and GAKIN on cytokinesis. Western blotting confirmed that the treatment of hDlg and GAKIN siRNAs efficiently suppressed the expression of hDlg and GAKIN, respectively, at the protein level in U2OS and HeLa cells (Fig. 7A). We observed an increased number of multinucleated cells after treatment with either hDlg or GAKIN siRNA in U2OS and HeLa cells (Fig. 7B). Mitotic synchronization experiments showed that both hDlg and GAKIN siRNA treated HeLa cells exhibit a delay in cytokinesis when observed three hours after the release (Fig. 7C). We also found that in the cells treated with hDlg siRNA, the GFP-GAKIN still localized dynamically in the mitotic cells, particularly around the cleavage furrow at the anaphase-telophase, and at the midbody during cytokinesis (a representative image shown in Fig. 7D), suggesting that the midbody localization of GAKIN is independent of hDlg. Therefore, the localization of hDlg and GAKIN to the midbody appears to be independent of each other under these conditions. Together, these results suggest that both GAKIN and hDlg functionally involve in the process of cytokinesis.
Fig. 7. Impaired cytokinesis by siRNA knock down of hDlg and GAKIN.
(A) Down regulation of hDlg and GAKIN by siRNA in U2OS and HeLa cells. 72 hrs (U2OS) or 48 hrs (HeLa) after transfection with siRNA, protein expression level of hDlg and GAKIN was determined by Western blotting using anti-hDlg mAb and anti-GAKIN mAb, respectively. The anti β-tubulin Western blot is shown to demonstrate the equal loading of the lysates. (B) Quantitative analysis of multinuclear phenotype formed after hDlg and GAKIN down-regulation by siRNA in the U2OS and HeLa cells. Cells were fixed at 72 hrs (U2OS) and 48 hrs (HeLa) after transfection with siRNA, and stained for β-tubulin and DAPI to visualize the cell shape and nucleus, respectively. The number of multinucleated cells was counted in a blind test where the researcher (K.U.) observed the stained slides under the fluorescence microscope without the knowledge of the identity of each sample. Data are the total of two independent experiments for U2OS, and three for the HeLa cells. At least 317 cells were analyzed for each sample. (C) Delayed cytokinesis of HeLa cells treated with hDlg and GAKIN siRNA. Percentage of the cells not completing cytokinesis after three hours is calculated as shown in Fig. 3B. The mean ± SD from three independent experiments is shown. *P < 0.05, **P < 0.005. (D) Localization of GFP-GAKIN in U2OS cells treated with hDlg siRNA. GFP-GAKIN (green), β-tubulin (red), and DAPI (blue). Localization of GFP-GAKIN around the cleavage furrow region is apparent in this U2OS cell at the telophase stage.
Discussion
In this study, we demonstrate that hDlg is functionally involved in the process of cytokinesis based on the use of dominant-negative, over-expression, knock down by siRNA, and the use of mouse embryo fibroblasts from genetically targeted mutant mice. Genetic and biochemical approaches have previously identified a number of proteins that localize to the midbody and are essential for cytokinesis [19]. Among them, components of central spindle, acto-myosin, RhoA signaling pathway, and membrane fusion machinery are conserved throughout evolution and are considered to be the core components of cytokinesis [19]. However, there are several other proteins that are also involved in this process but are not considered as essential for cytokinesis. A relatively low frequency of the multinucleated cells formed (4–7%) upon inhibition of hDlg suggests that hDlg may not be an essential component of cytokinesis. Indeed, the development of dlggt/dlggt mouse embryos up to day 13.5, the stage where the MEFs were obtained, indicates that a sufficient number of cells must have completed proper cytokinesis before the embryo reached this stage. Given the functional involvement of hDlg in the regulation of efficient cytokinesis, it is tempting to speculate that the accumulating effect of failed cytokinesis might contribute to the growth retardation and perinatal lethality phenotype observed in the dlggt/dlggt mice at a later stage of development [22].
An increasing number of proteins involved in cytokinesis have been identified recently, and many of them seem to play a functional role that is distinct from the involvement of the classic “core components of cytokinesis”. For example, the BRCA2 tumor suppressor localizes to the midbody and the BRCA2 mutant MEFs show a cytokinesis defect, which might explain the aneuploidy frequently observed in the BRCA2-deficient tumor cells [27]. Similarly, CD2AP, a protein implicated in the endocytic degradation pathway and remodeling of the actin cytoskeleton, also localizes to the midbody and is functionally involved in cytokinesis [28]. These proteins, although clearly involved in the process of cytokinesis, are not considered as essential components since the majority of cells lacking these proteins can still complete cytokinesis as revealed by the phenotype of the gene targeted mice. Thus, further knowledge of the exact molecular mechanisms of these proteins will be essential in delineating the complex process of cytokinesis in vivo.
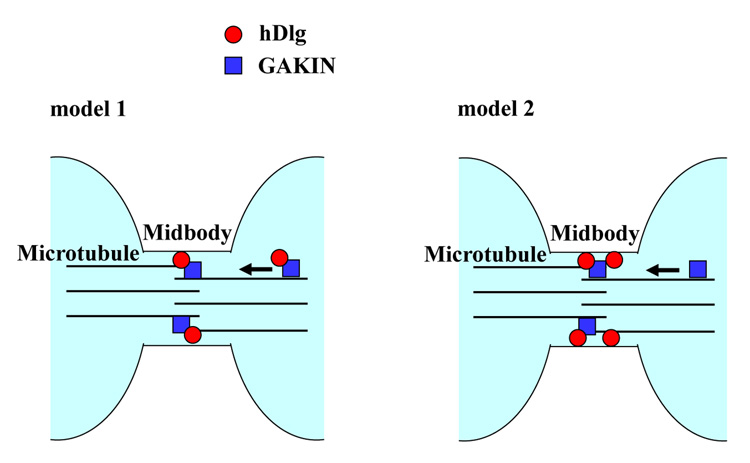
The midbody localization of the GUK domain-binding kinesin-like protein, GAKIN, and the functional requirement of the GUK domain of hDlg for midbody targeting suggested a simple mechanism whereby the GAKIN transports hDlg along microtubules to the midbody (Fig. 8, model 1). However, our data suggest otherwise. The over-expression of motor-deficient GAKIN did not affect the midbody targeting of DsRed-GUK (Fig. 6); and in the GAKIN siRNA treated cells, the hDlg was still found in the midbody (our unpublished data). Although a possibility remains that the endogenous GAKIN has not been completely inhibited either by the motor-deficient mutant or siRNA treatment, we consider at this stage that the localization of hDlg to the midbody is likely to be independent of the GAKIN-mediated transport mechanism. Similarly, experiments using hDlg siRNA suggested that the localization of GAKIN is also independent of the presence of hDlg (Fig. 7D). Therefore, an alternative mechanism may account for the GUK-domain dependent targeting of hDlg to the midbody, which likely involves the protein-protein interactions mediated by the GUK domain. Such mechanism may operate redundantly or cooperatively with the GAKIN mediated pathway. In fact, this view is consistent with the recent model proposed for the function of the Drosophila Dlg/Khc-73 complex in asymmetric cell division of neuroblasts [14]. Khc-73, a Drosophila ortholog of GAKIN, regulates the proper positioning of the polarized cortical localization of Dlg, but this phenotype is apparent only in the Inscuteable null background because the Inscuteable pathway functions partially redundantly with the Khc-73 pathway [14]. The authors of this study proposed that the Khc-73 mediated polarized localization of Dlg might be a transport-independent phenomenon. In their model, the Dlg first localizes to the cortex in mitosis that is independent of the activity of Khc-73. The Khc-73 localizes to the distal ends of spindle microtubules possibly via the CAP-Gly domain, a mechanism that is similar to the plus-tip tracking protein, CLIP-170 with two CAP-Gly domains [29]. Khc-73 meets and forms a complex with Dlg when the microtubule ends reach the cortical sites, and induces “clustering” of Dlg, thus facilitating its polarized cortical localization [14]. In the case of hDlg during cytokinesis, the hDlg seems to reach the midbody independent of GAKIN. The GAKIN arrives at this site through a motor-based mechanism along microtubules, that does not involve the participation of the CAP-Gly domain, and both GAKIN and hDlg accumulate at the midbody where they have a chance to interact (Fig. 8, model 2). In future studies, it would be important to determine how much of the Dlg/Khc-73 functional mechanisms that regulate the asymmetric cell division of Drosophila are conserved in the hDlg/GAKIN regulating the normal cell division of mammalian cells.
Fig. 8. Two models for the midbody targeting of hDlg and GAKIN.
Model 1, GAKIN and hDlg form a complex in the cytoplasm. GAKIN then transports hDlg along microtubules to mediate the accumulation of hDlg at the midbody. Model 2, hDlg localizes at the midbody by the GUK domain mediated mechanism independent of GAKIN. GAKIN then localizes to the midbody via its motor dependent translocation along the microtubules. Finally, the GAKIN and hDlg form a complex at the midbody.
Several RNAi based screens have identified kinesin-like motor proteins, including KIF4, MKlp1, and MKlp2, as important factors for cytokinesis, [30]. In addition, KIF14, which belongs to the kinesin-3 family, was also identified as a factor required for cytokinesis [26]. Neither GAKIN, which is a kinesin-3 family protein, nor its Drosophila homologue, Khc-73, have been identified as cytokinesis components in the published RNAi based screens [30, 31]. One explanation of why such high-throughput screening approaches failed to identify the involvement of GAKIN in cytokinesis could be due to a relatively small number of multinucleated cells formed after the treatment of GAKIN siRNA (Fig. 7). Nonetheless, our RNAi and localization data suggest a functional involvement of GAKIN in the process of cytokinesis. The question that remains to be resolved is whether GAKIN functionally interacts and cooperates with hDlg at the midbody to mediate the process of cytokinesis. If GAKIN is not responsible for intracellular targeting of hDlg to the midbody, then how does GAKIN cooperate with hDlg? One possibility is that it might be through the ability of hDlg/GAKIN complex to connect the multiprotein scaffolding complex to the microtubule structure, and provide adequate mechanical force to sustain the cytoskeletal integrity within the restricted space of the midbody. Alternatively, as suggested by the Drosophila neuroblast system, a conformational change of hDlg is induced through direct binding with GAKIN, thus leading to the formation of specialized scaffolding structure [14]. It is also possible that the conformation of GAKIN is altered through direct binding with hDlg, as reported recently [32]. Still, despite their strikingly similar dynamic localization patterns and their ability to form tight binding interaction, we can not at this stage exclude the possibility that GAKIN and hDlg may act totally independently in cytokinesis regulation pathway.
Then, how does hDlg possibly contribute functionally at the midbody? The potent dominant negative effect of the DsRed-GUK domain (Fig. 2) suggests that protein-protein interactions mediated by domains other than GUK domain are essential for its function at the midbody. One possible interaction partner may be the exocyst complex, an evolutionarily conserved multi-protein complex involved in the vesicle transport. The exocyst complex localizes to the midbody, and is required for cytokinesis [33]. SAP102, a close homologue of hDlg, binds to a component of exocyst complex, Sec8, through the PDZ domain mediated interaction, and regulates the targeting of NMDA receptors in neuronal cells [34]. It is possible that similar interactions are conserved between hDlg and Sec8 at the midbody during normal cell division. Other possible partners are the homologues of the components of Drosophila polarity pathways, which might perform an evolutionarily conserved function. For example, Lgl2, a mammalian homologue of the Drosophila Lgl, is involved in the process of normal cell division by forming a complex with LGN, which is a homologue of the Drosophila partner-of inscuteable (Pins) [35]. Interestingly, the LGN is also localized in the midbody suggesting that it might function in the common pathway with hDlg [36]. Lgl and Pins are both essential components for the asymmetric cell division of Drosophila neuroblasts. Our findings that both hDlg and GAKIN are involved in cytokinesis add a new example of the functional involvement of Drosophila polarity factors in the regulation of mammalian cell division.
Acknowledgements
This research was supported by the National Institutes Grants CA 94414 and HL60755. Toshihiko Hanada is the recipient of a Campus Research Board Award from the University of Illinois at Chicago (2004–2005). We thank Dena Inempolidis for editorial assistance and final assembly of the manuscript, and Dr. Anwar Khan for help with the preparation of figures. We are grateful to Dr. Georgina Caruana for sharing the Dlg mutant mice.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Woods DF, Bryant PJ. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 2.Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- 4.Ohshiro T, Yagami T, Zhang C, Matsuzaki F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:593–596. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- 5.Bilder D, Schober M, Perrimon N. Integrated activity of PDZ protein complexes regulates epithelial polarity. Nat Cell Biol. 2003;5:53–58. doi: 10.1038/ncb897. [DOI] [PubMed] [Google Scholar]
- 6.Albertson R, Doe CQ. Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 7.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 8.Osmani N, Vitale N, Borg JP, Etienne-Manneville S. Scrib controls Cdc42 localization and activity to promote cell polarization during astrocyte migration. Curr Biol. 2006;16:2395–2405. doi: 10.1016/j.cub.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 9.Etienne-Manneville S, Manneville JB, Nicholls S, Ferenczi MA, Hall A. Cdc42 and Par6-PKCzeta regulate the spatially localized association of Dlg1 and APC to control cell polarization. J Cell Biol. 2005;170:895–901. doi: 10.1083/jcb.200412172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludford-Menting MJ, Oliaro J, Sacirbegovic F, Cheah ET, Pedersen N, Thomas SJ, Pasam A, Iazzolino R, Dow LE, Waterhouse NJ, Murphy A, Ellis S, Smyth MJ, Kershaw MH, Darcy PK, Humbert PO, Russell SM. A network of PDZ-containing proteins regulates T cell polarity and morphology during migration and immunological synapse formation. Immunity. 2005;22:737–748. doi: 10.1016/j.immuni.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Dimitratos SD, Woods DF, Stathakis DG, Bryant PJ. Signaling pathways are focused at specialized regions of the plasma membrane by scaffolding proteins of the MAGUK family. Bioessays. 1999;21:912–921. doi: 10.1002/(SICI)1521-1878(199911)21:11<912::AID-BIES3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 12.Hanada T, Lin L, Tibaldi EV, Reinherz EL, Chishti AH. GAKIN, a novel kinesin-like protein associates with the human homologue of the Drosophila discs large tumor suppressor in T lymphocytes. J Biol Chem. 2000;275:28774–28784. doi: 10.1074/jbc.M000715200. [DOI] [PubMed] [Google Scholar]
- 13.Asaba N, Hanada T, Takeuchi A, Chishti AH. Direct interaction with a kinesin-related motor mediates transport of mammalian discs large tumor suppressor homologue in epithelial cells. J Biol Chem. 2003;278:8395–8400. doi: 10.1074/jbc.M210362200. [DOI] [PubMed] [Google Scholar]
- 14.Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 15.Horiguchi K, Hanada T, Fukui Y, Chishti AH. Transport of PIP3 by GAKIN, a kinesin-3 family protein, regulates neuronal cell polarity. J Cell Biol. 2006;174:425–436. doi: 10.1083/jcb.200604031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Massimi P, Gardiol D, Roberts S, Banks L. Redistribution of the discs large tumor suppressor protein during mitosis. Exp Cell Res. 2003;290:265–274. doi: 10.1016/s0014-4827(03)00317-3. [DOI] [PubMed] [Google Scholar]
- 17.Lin HT, Steller MA, Aish L, Hanada T, Chishti AH. Differential expression of human Dlg in cervical intraepithelial neoplasias. Gynecol Oncol. 2004;93:422–428. doi: 10.1016/j.ygyno.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 18.Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 20.Hanada T, Takeuchi A, Sondarva G, Chishti AH. Protein 4.1-mediated membrane targeting of human discs large in epithelial cells. J Biol Chem. 2003;278:34445–34450. doi: 10.1074/jbc.M305209200. [DOI] [PubMed] [Google Scholar]
- 21.Nakata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caruana G, Bernstein A. Craniofacial dysmorphogenesis including cleft palate in mice with an insertional mutation in the discs large gene. Mol Cell Biol. 2001;21:1475–1483. doi: 10.1128/MCB.21.5.1475-1483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura K, Tsuji T, Takada Y, Miki T, Narumiya S. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J Biol Chem. 2000;275:17233–17236. doi: 10.1074/jbc.C000212200. [DOI] [PubMed] [Google Scholar]
- 24.Matuliene J, Kuriyama R. Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol Biol Cell. 2002;13:1832–1845. doi: 10.1091/mbc.01-10-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill E, Clarke M, Barr FA. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. Embo J. 2000;19:5711–5719. doi: 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruneberg U, Neef R, Li X, Chan EH, Chalamalasetty RB, Nigg EA, Barr FA. KIF14 and citron kinase act together to promote efficient cytokinesis. J Cell Biol. 2006;172:363–372. doi: 10.1083/jcb.200511061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–879. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 28.Monzo P, Gauthier NC, Keslair F, Loubat A, Field CM, Le Marchand-Brustel Y, Cormont M. Clues to CD2-associated protein involvement in cytokinesis. Mol Biol Cell. 2005;16:2891–2902. doi: 10.1091/mbc.E04-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mimori-Kiyosue Y, Tsukita S. "Search-and-capture" of microtubules through plus-end-binding proteins (+TIPs) J Biochem (Tokyo) 2003;134:321–326. doi: 10.1093/jb/mvg148. [DOI] [PubMed] [Google Scholar]
- 30.Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell. 2005;16:3187–3199. doi: 10.1091/mbc.E05-02-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Sans N, Prybylowski K, Petralia RS, Chang K, Wang YX, Racca C, Vicini S, Wenthold RJ. NMDA receptor trafficking through an interaction between PDZ proteins and the exocyst complex. Nat Cell Biol. 2003;5:520–530. doi: 10.1038/ncb990. [DOI] [PubMed] [Google Scholar]
- 35.Yasumi M, Sakisaka T, Hoshino T, Kimura T, Sakamoto Y, Yamanaka T, Ohno S, Takai Y. Direct binding of Lgl2 to LGN during mitosis and its requirement for normal cell division. J Biol Chem. 2005;280:6761–6765. doi: 10.1074/jbc.C400440200. [DOI] [PubMed] [Google Scholar]
- 36.Blumer JB, Chandler LJ, Lanier SM. Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. J Biol Chem. 2002;277:15897–15903. doi: 10.1074/jbc.M112185200. [DOI] [PubMed] [Google Scholar]