Abstract
HIV-1 entry into CD4+ cells requires the sequential interactions of the viral envelope glycoproteins with CD4 and a coreceptor such as the chemokine receptors CCR5 and CXCR4. A plausible approach to blocking this process is to use small molecule antagonists of coreceptor function. One such inhibitor has been described for CCR5: the TAK-779 molecule. To facilitate the further development of entry inhibitors as antiviral drugs, we have explored how TAK-779 acts to prevent HIV-1 infection, and we have mapped its site of interaction with CCR5. We find that TAK-779 inhibits HIV-1 replication at the membrane fusion stage by blocking the interaction of the viral surface glycoprotein gp120 with CCR5. We could identify no amino acid substitutions within the extracellular domain of CCR5 that affected the antiviral action of TAK-779. However, alanine scanning mutagenesis of the transmembrane domains revealed that the binding site for TAK-779 on CCR5 is located near the extracellular surface of the receptor, within a cavity formed between transmembrane helices 1, 2, 3, and 7.
Protease and reverse transcriptase inhibitors of HIV-1 replication have had a major impact on the AIDS epidemic in the developed world (1). These drugs cannot, however, eradicate HIV-1 from infected people (2–4). Concerns about the long-term side effects of protease inhibitors and the increasing transmission of resistant variants emphasize the need to identify new classes of drugs able to suppress HIV-1 replication efficiently (5–7). The immune system then may be able to repair defects in CD4+ T cell production that are central to HIV-1 pathogenesis (8).
One way to inhibit HIV-1 replication is to prevent the virus entering its target cells (7). The potential of this approach is shown by T20, a peptide that prevents the conformational changes in the viral gp41 glycoprotein that drive membrane fusion (9). There are, however, other targets for entry inhibitors, notably the coreceptors CCR5 and CXCR4 (10, 11). The CC-chemokine receptor CCR5 is used by the most commonly transmitted HIV-1 strains, which persist in most individuals throughout the course of infection (10, 11). The lack of CCR5 expression in 1% of Caucasians is strongly protective against HIV-1 transmission, but is without any obvious adverse effect on health (12, 13). Furthermore, CCR5 knockout mice exhibit no overt pathology (14), although they have a reduced ability to resist Cryptococcal infections of the brain (15). The limited impact of a loss of CCR5 function renders this receptor an attractive target for new anti-HIV-1 drugs.
Among agents that prevent the coreceptor function of CCR5 in vitro are chemokine-based compounds (16, 17) and some mAbs (18–20). However, from the drug-development perspective, small molecules of less than 1,000 Da have significant advantages over protein-based inhibitors. Several CXCR4 inhibitors are known (21–23), but so far only one small molecule, TAK-779, has been reported to target CCR5 (24). Here, we show that TAK-779 inhibits HIV-1 replication by blocking the interaction of the viral surface glycoprotein gp120 with CCR5, thereby preventing virus–cell fusion. The binding site for TAK-779 is located near the CCR5 extracellular surface, within a cavity between transmembrane helices 1, 2, 3, and 7.
Materials and Methods
Compounds.
TAK-779 (N,N-dimethyl-N-(4-[[[2-(4-methylphenyl)-6,7-dihydro-5H-benzocyclohepten-8-yl]carbon-yl]amino]benzyl)- tetrahydro-2H-pyran-4-aminium chloride; Mr = 531.13) was synthesized by A. Bauer, M. Miller, S. Vice, and S. McCombie of Schering–Plough Research Institute, based on the structure in ref. 34. Anti-CCR5 mAbs gp120JR-FL and CD4-IgG2 were gifts from P. Maddon (Progenics, Tarrytown, NY) (19).
Inhibition of HIV-1 Infection.
Mitogen-stimulated peripheral blood mononuclear cells (2 × 105/100 μl) were incubated for 1 h with 50 μl of TAK-779 at 4× the final concentration before addition of HIV-1JR-FL (100 TCID50/50 μl). Production of p24 antigen was measured after 4–6 days (25). TAK-779 was present throughout the culture.
Inhibition of Env-Mediated Membrane Fusion.
Cell–cell fusion was measured by using the T7-luciferase system as described (26). Fusion between 2 × 104 Chinese hamster ovary (CHO)-K1-CD4-CCR5 cells and 4 × 104 HeLa cells expressing HIV-1JR-FL Env glycoproteins was measured after 2.5 h at 37°C by transactivation of a luciferase reporter gene (26). Luciferase activity in cell lysates was measured by using a standard kit (Promega) and is expressed in relative light units (r.l.u.). CHO-K1-CD4-CCR5 cells were made by transducing CHO-K1 cells with CD4- and CCR5-coding recombinant retrovirus particles, provided by D. Kabat (University of Oregon, Portland) (27, 28).
Binding of gp120 to CCR5.
A gp120-CD4-IgG2 complex (50 nM) formed from monomeric gp120JR-FL and biotinylated CD4-IgG2 was added to 1 × 106 L1.2-CCR5 cells in the presence of TAK-779 (19). The mean fluorescence intensity (m.f.i.) was measured by flow cytometry after addition of phycoerythrin-labeled streptavidin (19). Inhibition of gp120-CCR5 binding was calculated as follows: [(m.f.i. with TAK-779)/(m.f.i. without TAK-779)] × 100%. L1.2-CCR5 cells, made by transforming murine L1.2 cells with a retroviral vector encoding a C-terminal hemagglutinin-tagged human CCR5 (from D. Kabat) (27, 28) have similar properties to a line described previously (18).
Binding of mAbs to CCR5.
L1.2-CCR5 cells (1 × 106) were incubated with 50 nM of anti-CCR5 mAb with or without TAK-779 (100 nM) (19). mAb binding was detected by using a phycoerythrin-labeled goat anti-mouse antibody. The m.f.i. value was measured by flow cytometry. Inhibition of mAb binding was calculated as above.
HIV-1 Entry by CCR5 Mutants and Effect of TAK-779.
Entry of HIV-1JR-FL Env-pseudotyped viruses into U87-CD4 cells expressing wild-type or mutant CCR5 was determined by using a luciferase-based reporter assay (29–31). Briefly, 1 × 106 U87-CD4 cells were transiently lipofected with 10 μg of pcDNA3.1 expressing a CCR5 mutant. After 24 h, the cells were infected with NLenv-luc+ virus pseudotyped with HIV-1JR-FL Env, with or without 200 nM TAK-779, a concentration that causes ≈95% inhibition in this assay. The viral inoculum was equivalent to ≈100 ng of p24. After 48 h, the cells were lysed, and luciferase activity (r.l.u.) was measured (29–31).
Relative HIV-1 entry in the presence of TAK-779 was calculated as: [(r.l.u. with TAK-779)/(r.l.u. without TAK 779)] × 100%. All values are means ± SD of three independent experiments. A typical r.l.u. for wild-type CCR5 without TAK-779 was 50,000 ± 2,500. The uninhibited value, in each experiment, is defined as 100%. With TAK-779 present, the extent of HIV-1 entry was reduced to 3,500 ± 175 r.l.u. for wild-type CCR5. This residual entry was defined as 0%. For each CCR5 mutant, the extent of HIV-1 entry with and without TAK-779 was normalized in the range of 0 to 100%. CCR5 mutations that affect HIV-1 entry also affect the sensitivity of the entry assay in the presence of TAK-779. A few mutants, apparently more sensitive than the wild-type receptor to TAK-779, yielded entry levels <0%. These effects were small and may have no functional implication. For clarity, values <0% are plotted as = 0%.
Identifying CCR5 Mutants Significantly Insensitive to TAK-779.
We estimate that a variation of ±20% from the mean is not significant in luciferase-based assays of HIV-1 entry (29–31), so all mutants yielding entry levels <20% with TAK-779 were considered insignificantly different from wild-type CCR5. The normalized mean entry level (±SD) for this subset of mutant receptors was 4 ± 5%. Because a deviation from the mean of >2 SD is significant, a cut-off level of ≥14% entry in the presence of 200 nM TAK-779 was used.
Structural Model of the Transmembrane (TM) Domain of CCR5.
The model was based on homology with rhodopsin; NMR and mutational studies provided tight constraints for modeling the orientation of the TM helices and the location of the retinal prosthetic group (32). The amino acid sequence of CCR5 was aligned with that of rhodopsin. The CCR5 amino acid side chains were extended from the helical backbone of the rhodopsin model and energy-minimized with the program xplor, using Powell minimization for 10,000 cycles. Hydrogen-bonding restraints were applied between the backbone amide and carbonyl groups. This allowed the TM helices to maintain α-helical structure, but provided flexibility for kinks to be introduced at TM prolines unique to CCR5. Hydrogen-bonding restraints also were applied between the following pairs of residues: Asn48-Asp76, Trp153-Asn71, Tyr214-Arg235, Asp125-Arg126, and Asn293-Tyr297. These residues generally are conserved in G protein-coupled receptors and are thought to form key intramolecular interactions. The following residues are included in the CCR5 structural model: TM1 (Arg31-Asn57), TM2 (Lys62-His88), TM3 (Thr99-Ala133), TM4 (Thr143-Thr167), TM5 (Thr195-Cys224), TM6 (Lys228-Leu257), and TM7 (Met279-Glu302).
The TAK-779 structure, including chloride counterion, was energy-minimized by using the PM3 semiempirical method of the program hyperchem.
Results
Antiviral Activity of TAK-779.
TAK-779 inhibits the replication of HIV-1JR-FL, a CCR5-using (R5) primary isolate, in peripheral blood mononuclear cells, with an IC50 of 10 nM (Fig. 1A). We find R5, but not X4, primary isolates to be sensitive to TAK-779 at 10–100 nM (data not shown), as reported previously for different test isolates (24). We also found that TAK-779 has no effect on entry of HIV-1, HIV-2, or SIVmac by CXCR4, CCR3, CCR8, V28, US28, APJ, BOB, Bonzo, or GPR1 in transfected GHOST-CD4 cells, but it efficiently inhibits entry of SIVrcm by means of CCR2 (Y.-J. Zhang and J.P.M., unpublished data). TAK-779 interacts with CCR2 (24), the major coreceptor for SIVrcm (33).
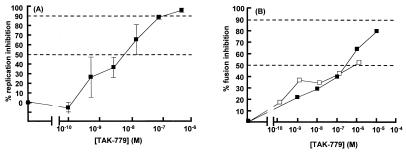
Figure 1.
Effect of TAK-779 on HIV-1 replication and Env-mediated membrane fusion. (A) Replication of HIV-1JR-FL in mitogen-activated peripheral blood mononuclear cells was measured in the presence of TAK-779 (■). (B) Fusion between CHO-K1-CD4-CCR5 cells and HeLa cells expressing the HIV-1JR-FL Env was measured in the presence of TAK-779 (■) or RANTES (□). The extent of inhibition of viral replication or cell–cell fusion at each inhibitor concentration is presented as a percentage of control (no inhibitor = 0%). p24 and r.l.u. values in the absence of inhibitor were typically 15 ± 3 ng/ml and 24,500 ± 9,000, respectively. Background r.l.u. values were 7 ± 2. Each data point represents the mean ± SD of three and seven replicates for the replication and fusion assays, respectively. The horizontal dashed lines in B indicate 50% and 90% inhibition.
The specificity of TAK-779 for CCR5 (and CCR2) suggests it targets the membrane-fusion stage of the HIV-1 life cycle. To confirm this, we performed a cell–cell fusion assay (Fig. 1B). Fusion between CHO-K1 cells expressing CD4 plus CCR5 and HeLa cells expressing HIV-1JR-FL Env was inhibited by TAK-779 (IC50, 200 nM). As a positive control, RANTES, a CC-chemokine ligand of CCR5, also inhibited fusion (Fig. 1B). Inhibition of cell–cell fusion generally requires higher antagonist concentrations than does virus–cell entry, because a greater number of Env-receptor interactions need to be blocked.
TAK-779 Inhibits gp120 Binding to CCR5.
To ascertain whether the fusion-inhibitory action of TAK-779 was by an effect on the gp120-CCR5 interaction, we measured the binding of gp120JR-FL (as a complex with CD4-IgG2) to the CD4-L1.2-CCR5 cell line (19). TAK-779 inhibited binding of gp120JR-FL to CCR5, with an IC50 of 15 nM (Fig. 2A). In contrast, TAK-779 (100 nM) had no effect on binding to L1.2-CCR5 cells of five mAbs to various epitopes in the CCR5 N-terminal tail (Nt) and/or the second extracellular loop (ECL-2) (Fig. 2B). Thus, TAK-779 does not cause CCR5 down-regulation, and, hence, the loss of cell surface gp120-binding sites.
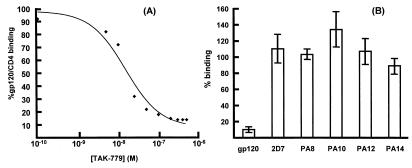
Figure 2.
Effect of TAK-779 on the binding of gp120 and mAbs to CCR5. (A) The extent of gp120JR-FL binding (as a CD4-IgG2 complex) to L1.2-CCR5 cells in the absence of TAK-779 was defined as 100% (m.f.i. 40 ± 5). Binding in the presence of TAK-779 is expressed as a percentage of control. When untransfected L1.2 cells were used, binding of the gp120-CD4-IgG2 complex was negligible (<10%; m.f.i. 2 ± 1). (B) Binding of the indicated mAbs (50 nM) or gp120JR-FL (50 nM plus 50 nM of CD4-IgG2) to L1.2-CCR5 cells was measured with and without 100 nM TAK-779. The extent of mAb binding in the absence of TAK-779 was defined as 100% (m.f.i. were 50–400, depending on the mAb). Binding in the presence of TAK-779 is expressed as a percentage of control. When untransfected L1.2 cells were used, mAb binding was negligible (m.f.i. ≈2). mAbs PA8 and PA12 bind to the CCR5 Nt; 2D7 to ECL-2; PA10 and PA14 to composite epitopes involving Nt and ECL-2 (19).
The binding of anti-CCR5 mAb 45531.111 (also described as mAb 31; ref. 20) to CCR5-expressing CHO cells is inhibited by TAK-779 (34). We have not been able to confirm this using L1.2-CCR5 cells, perhaps because of variation in CCR5 expression on the different cell lines.
Mutagenesis of the CCR5 Extracellular Domain.
To define the TAK-779 binding site, we performed alanine-scanning mutagenesis of selected CCR5 residues. The aim was to identify CCR5 mutants that still supported HIV-1 entry, but that were less sensitive, or insensitive, to TAK-779 inhibition. We first evaluated well-characterized alanine-replacement mutants of the extracellular domain of CCR5 (30, 31), because the gp120-binding site maps to this region, in particular, the Nt (30, 31, 35–38). However, alanine substitutions of residues in the extracellular domain had little or no effect on the antiviral activity of TAK-779; the mutant receptors still supported HIV-1 entry and were still sensitive to TAK-779 (Fig. 3).
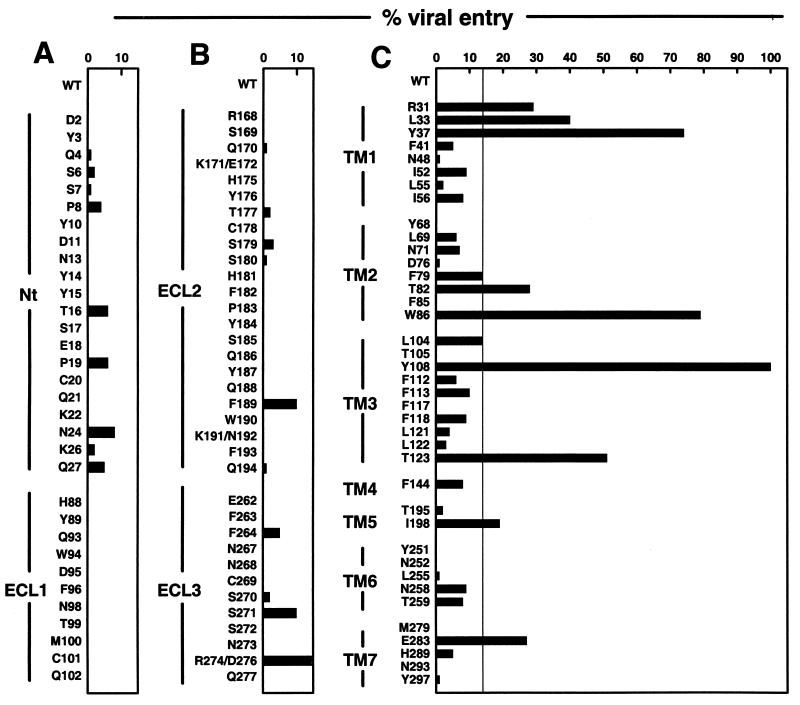
Figure 3.
Effects of alanine substitutions in CCR5 on inhibition of HIV-1JR-FL entry by TAK-779. (A) Alanine mutants of charged, polar and bulky, nonpolar amino acid residues in the Nt and ECL-1 of CCR5 were evaluated for their ability to mediate HIV-1 entry in the presence of 200 nM TAK-779. (B) The alanine mutants were located in ECL-2 and ECL-3. (C) The alanine mutants were located in the TM domain. The vertical line in each panel indicates the level of entry (14%) above which a CCR5 mutant was considered to have reduced sensitivity to TAK-779. Each data point represents the mean of three independent experiments. The different CCR5 mutants supported 10–120% of the level of entry for wild-type CCR5.
Mutagenesis of the CCR5 TM Domain Guided by Molecular Modeling.
The minimal effect of substitutions in the extracellular domains, combined with the failure of TAK-779 to inhibit binding of mAbs to the Nt and ECL-2 (Fig. 2B), focused our attention on the TM segments of CCR5 (Fig. 3). Our mutagenesis strategy was guided by a computer model of the CCR5 TM domain, based on the corresponding regions of rhodopsin (32). The CCR5 structure predicted by this model implied that a putative ligand binding pocket present within the TM helices might be a reasonable location for a small molecule antagonist of CCR5 function. We initially mutated residues around this pocket whose side chains were predicted to face inward, as opposed to those facing the membrane bilayer or predicted to be involved in helix–helix interactions. The positioning on this model of the first residues found to affect TAK-779 action guided further rounds of alanine substitutions at topologically proximal residues.
Nine TM mutants significantly resisted the action of TAK-779 (Fig. 3). Five were strongly resistant; L33A and Y37A from TM1, W86A from TM2, and Y108A and T123A from TM3. Four more mutants were moderately resistant; R31A from TM1, T82A from TM2, I198A from TM5, and E283A from TM7. Finally, two mutants had borderline resistance to TAK-779; F79A in TM2 and L104A in TM3. Mutants Y68A, F85A, Y251A, N252A, and N293A had <0.2% of wild-type CCR5 coreceptor activity because of their poor cell surface expression (data not shown), so TAK-779 inhibition could not be accurately evaluated.
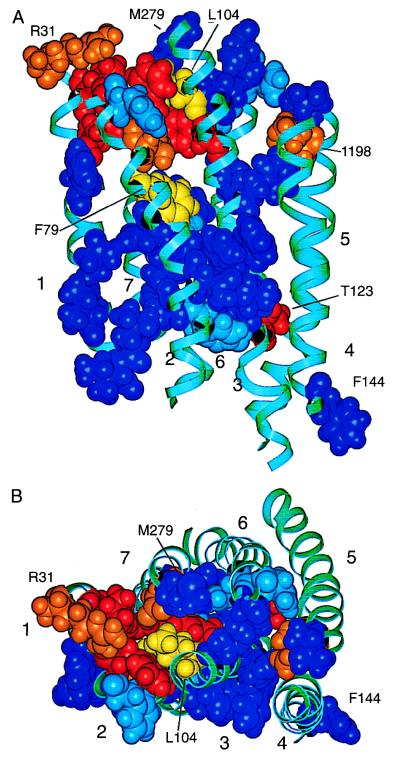
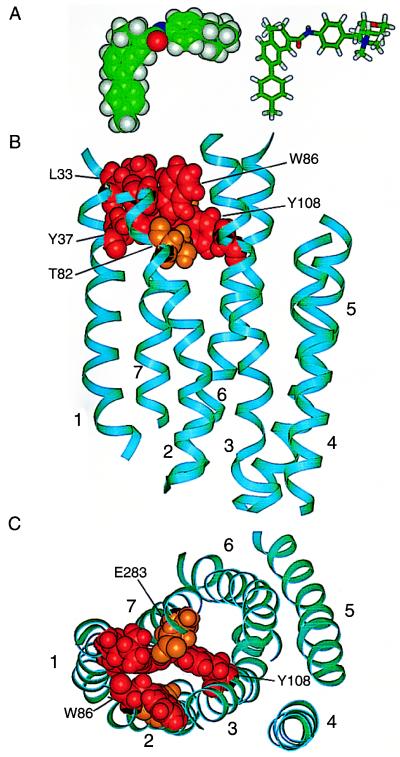
The positions of the TM domain residues implicated in TAK-779 action are displayed on the CCR5 model (Figs. 4 and 5). The side chains of Leu33, Tyr37, Thr82, Trp86, Tyr108, and Glu283 form a cluster bordered by TM helices 1, 2, 3, and 7 (Fig. 5). These residues are located within about two helical turns of the extracellular surface of CCR5. Residue Arg31 is situated at the interface between TM helix 1 and the Nt. Of the sites that had significant effects on TAK-779 binding, only Thr123 and Ile198 are not clustered with the other residues. The model suggests that Thr123 and Ile198 participate in interhelical interactions that stabilize the TAK-779 binding pocket at a distance. This remains to be confirmed experimentally.
Figure 4.
Structural model of the TM domain of the CCR5 receptor. TM helical segments, labeled 1–7, are shown as cyan-colored ribbons. The amino acid residues substituted by alanine are shown with space-filling atoms and are color-coded as follows: alanine substitutions of red-colored residues had a strong inhibitory effect on the antiviral activity of TAK-779 (Leu33, Tyr37, Trp86, Tyr108, Thr123); alanine substitutions of orange-colored residues had an intermediate effect (Arg31, Thr82, Ile198, Glu283); alanine substitutions of yellow-colored residues had a borderline effect (Phe79, Leu104); alanine substitutions of dark blue-colored residues had no effect (Phe41, Asn48, Ile52, Leu55, Ile56, Leu69, Asn71, Asp76, Thr105, Phe112, Phe113, Phe117, Phe118, Leu121, Leu122, Phe144, Thr195, Leu255, Asn258, Thr259, Met279, His289, Tyr297). Light blue-colored residues indicate mutations that caused poor expression of CCR5 (Tyr68, Phe85, Tyr251, Asn252, Asn293). These receptors could not be evaluated for HIV-1 entry. (A) View of CCR5 from within the plane of the membrane. The extracellular surface is toward the top of the figure, the cytoplasmic surface toward the bottom. For orientation, Arg31 is at the upper left in orange, and Phe144 is at the lower right in blue. (B) View of CCR5 from its extracellular surface. The model is rotated by approximately 90° from the orientation in A.
Figure 5.
Structural models of the TAK-779 inhibitor and the CCR5 receptor. (A) Space-filling and stick representations of minimized TAK-779 structure. Atoms are color coded: carbon, green; oxygen, red; nitrogen, blue; hydrogen, gray. TAK-779 has two roughly planar segments connected by an amide bond. Its hydrophobic 4-methylphenyl ring is thought to interact with critical residues on CCR5, whereas the positively charged aminium tetrahydro-2H-pyran end of TAK-779 is oriented along the extracellular surface of CCR5, where it may block the binding of chemokine ligands and the gp120-CD4 complex. (B) The CCR5 structural model viewed from within the plane of the membrane. The CCR5 color-coding scheme is the same as in Fig. 4. Amino acid residues nearest the extracellular surface include Leu33, Trp86, and Glu283. Residues Tyr37, Thr82, and Tyr108 are deeper within the receptor. The indicated cluster of amino acids in the TAK-779 binding site includes several aromatic residues (Tyr37, Trp86, and Tyr108) that may form favorable interactions with the aromatic rings of TAK-779. (C) CCR5 viewed from the extracellular side of the receptor to show the orientation of TAK-779 binding. The colors are the same as in B, but the model is rotated by 90°. The proline residues at positions 34, 35, and 84 on TM helices 1 and 2 may facilitate the opening of the binding pocket for TAK-779. The scale is the same in A–C.
The mutagenesis and modeling data strongly suggest that the TAK-779 binding site is a pocket surrounded by TM helices 1, 2, 3, and 7. The depth of the pocket is approximately equal to one-third of the membrane span and is about the same length as the methylphenyl-benzocycloheptenyl group of TAK-779. This conjugated hydrophobic group is planar and connected to the positively charged benzyl-pyran-aminium moiety by an amide bond. The covalent amide linkage is “meta” to the cyclobenyzl group on the heptenyl ring, producing a notable bend of approximately 90° in the middle of the minimized TAK-779 structure (Fig. 5A). Because this structure has distinctive hydrophobic and polar ends, we envision that the methylphenyl-benzocycloheptenyl group inserts into the hydrophobic TM pocket, allowing the charged moiety to protrude and presumably interact with the polar extracellular domain.
Discussion
We performed this study to gain information on how a small molecule, TAK-779, interferes with the replication of HIV-1 isolates that use CCR5 to enter their target cells. We have determined the mechanism by which TAK-779 inhibits HIV-1 entry and identified its binding site on CCR5. Whether this particular compound will be developed into a clinically useful drug will depend on its pharmacological properties, but its binding site within the CCR5 TM domain represents a viable target for a range of small molecule inhibitors of CCR5 function. Such compounds may antagonize not only CCR5's subverted role as a HIV-1 coreceptor, but also its natural functions as a chemokine receptor, because TAK-779 inhibits ligand-induced signaling (24). CCR5-targeted small molecules may be useful in other clinical conditions, such as inflammatory or autoimmune diseases or asthma (39, 40).
How TAK-779 prevents HIV-1 entry probably is revealed by the observation that it inhibits the binding of gp120 to CCR5. The failure of the viral envelope glycoproteins to properly engage CCR5 would prevent them undergoing the conformational changes that drive the membrane fusion reaction (41). It was unexpected that TAK-779 inhibits gp120 binding without also interfering with the interactions of mAbs with the extracellular domain, because all of the test mAbs are themselves able to block gp120-CCR5 binding (19). In contrast, small molecule inhibitors of HIV-1 entry by means of CXCR4 block the binding of mAb 12G5 to ECL-2 of CXCR4 (21–23). A further complexity is that mutagenesis studies have implicated the CCR5 Nt as being critical for gp120 binding (30, 31, 35–38), yet alanine substitutions in the Nt have, paradoxically, no effect on the antiviral action of TAK-779. One interpretation of the ligand binding experiments is that there is more overlap between the binding sites for TAK-779 and gp120 than there is between the TAK-779 binding site and the mAb epitopes. Another is that the binding of TAK-779 to the CCR5 TM domain might induce conformational changes within the receptor that directly or indirectly perturb the gp120- and RANTES-binding sites, without affecting most of the mAb epitopes.
A clue as to how TAK-779 blocks gp120 binding is that it also inhibits the binding of the natural CCR5 ligand, RANTES (24). It has been proposed that chemokines bind to their receptors at two sites, one in the Nt and the other within the TM domain, and that the latter interaction is critical for signal transduction (39, 42, 43). Perhaps gp120 has adopted a similar twin-site strategy for its binding to CCR5; for a stable, high-affinity interaction, an initial contact may be made between gp120 and the CCR5 Nt, followed by a secondary insertion of some gp120 residues into the TM pocket. The coreceptor binding site covers a significant area of the gp120 surface involving both variable and conserved regions of the molecule (41, 44). This may permit a multistage binding mechanism.
Because chemokine receptors are structurally conserved, a similar pocket in the TM domain could be a target for antagonists of other members of this pharmacologically important receptor family. Small molecule inhibitors of CCR1 and CXCR2 are known, but their binding sites are not (45, 46). There may be both generality and specificity to how small molecule inhibitors interact with CC-chemokine receptors, exemplified by TAK-779. This is an antagonist of CCR2 and CCR5 but has no effect on several other chemokine receptors. However, there is significant conservation of the TM domain residues involved in the TAK-779 binding pocket among the chemokine receptor family. Of the residues at which an alanine substitution conferred strong TAK-779 resistance, Tyr37, Trp86, and Tyr108 are conserved in all CC-chemokine receptors (except in CCR8 where residue 86 is Gln); a conservative substitution of Val for Leu33 or Ser for Thr123 also occurs in some receptors. Positions in CCR5 where a mutation resulted in moderate resistance to TAK-779 are more variable. However, the residue at position 82 is exclusively Thr or Ser, and residue 283 is always Glu (except in CCR7 where it is Tyr). The aligned sequence of human CXCR4 shows the same conservation pattern.
Specificity in the interactions of small molecule inhibitors may be dictated by the interactions of a relatively hydrophilic moiety of the inhibitor with the extracellular domain of the chemokine receptor. It may be no coincidence that TAK-779 also antagonizes the functions of CCR2 because the sequence conservation in the extracellular domain is greater between CCR2 and CCR5 than among other members of the CC-chemokine receptor family (39). In this respect, the two apparent structural halves of TAK-779 may have functional significance; it is conceivable that the hydrophobic moiety inserts directly into the TM pocket, whereas the rest of the molecule contacts the extracellular surface.
The binding sites on CXCR4 for small molecule or peptide-based inhibitors have been localized to the extracellular domain, particularly anionic residues within ECL-2 (21, 23, 47). However, these CXCR4 inhibitors are all strongly cationic, and their interactions with CXCR4 are probably substantially electrostatic in nature. CXCR4 is unusual among chemokine receptors in having a strongly negative surface charge (48), so the properties of this class of CXCR4-targeted inhibitors may be atypical.
An emphasis on the TM domains of G protein-coupled receptors as targets for inhibitors is supported by a report that peptide mimics of the TM helices can disrupt receptor function and the ability of CXCR4 to mediate HIV-1 entry (49). Further mutagenesis studies on CCR5, including the use of nonalanine substitutions, should refine the confines of the TAK-779 binding site, as will the performance of computational docking studies between CCR5 and TAK-779. In particular, changing the benzyl-pyran-aminium moiety of TAK-779 by rational combinatorial synthetic approaches might result in a library of related compounds with different receptor subtype specificities. The accrued information may facilitate the design of superior inhibitors of HIV-1 entry by means of CCR5 and other coreceptors.
Acknowledgments
We thank Annette Bauer, Michael Miller, Susan Vice, Bahige Baroudy, and Stuart McCombie for TAK-779; Paul Maddon for gp120JR-FL and CD4-IgG2; David Kabat for Ψ2 ecotropic- and PA12 amphotropic-packaging cells; and Robert Doms and Bernard Moss for the T7-luciferase system and recombinant vaccinia viruses. This work was supported by National Institutes of Health Grants RO1 AI 41420 (to J.P.M.), RO1 AI 43847 (to T.D.), RO1 GM 41412 (to S.O.S.), and R01 DK 54718 (to T.P.S.); by the Allene Reuss Memorial Trust (to T.P.S. and S.W.L.); and by the Pediatric AIDS Foundation, of which J.P.M. is an Elizabeth Glaser Scientist.
Abbreviations
- r.l.u.
relative light units
- CHO
Chinese hamster ovary
- m.f.i.
mean fluorescence intensity
- Nt
N-terminal tail
- ECL
extracellular loop
- TM
transmembrane
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090576697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090576697
References
- 1.Fauci A S. N Engl J Med. 1999;341:1046–1050. doi: 10.1056/NEJM199909303411406. [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 3.Furtado M R, Callaway D S, Phair J P, Kunstman K J, Stanton J L, Macken C A, Perelson A S, Wolinsky S M. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 4.Chun T W, Fauci A S. Proc Natl Acad Sci USA. 1999;96:10958–10961. doi: 10.1073/pnas.96.20.10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lucas G M, Chaisson R E, Moore R D. Ann Intern Med. 1999;131:81–87. doi: 10.7326/0003-4819-131-2-199907200-00002. [DOI] [PubMed] [Google Scholar]
- 6.Yerly S, Kaiser L, Race E, Bru J P, Clavel F, Perrin L. Lancet. 1999;354:729–733. doi: 10.1016/S0140-6736(98)12262-6. [DOI] [PubMed] [Google Scholar]
- 7.Michael N L, Moore J P. Nat Med. 1999;5:740–742. doi: 10.1038/10462. [DOI] [PubMed] [Google Scholar]
- 8.Rowland-Jones S. Lancet. 1999;354:5–7. doi: 10.1016/S0140-6736(99)90017-X. [DOI] [PubMed] [Google Scholar]
- 9.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, et al. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 10.Berger E A, Murphy P M, Farber J M. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 11.Moore J P, Trkola A, Dragic T. Curr Opin Immunol. 1997;9:551–562. doi: 10.1016/s0952-7915(97)80110-0. [DOI] [PubMed] [Google Scholar]
- 12.Liu R, Paxton W, Choe S, Ceradini D, Martin S, Horuk R, MacDonald M, Stuhlmann H, Koup R, Landau N. Cell. 1996;86:367–378. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 13.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, Kurihara T, Ryseck R P, Yang Y, Ryan C, Loy J, Warr G, Bravo R. J Immunol. 1998;160:4018–4025. [PubMed] [Google Scholar]
- 15.Huffnagle G B, McNeil L K, McDonald R A, Murphy J W, Toews G B, Maeda N, Kuziel W A. J Immunol. 1999;163:4642–4646. [PubMed] [Google Scholar]
- 16.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 17.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N C, Proudfoot A E. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, et al. J Exp Med. 1997;186:1373–1881. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson W C, Rabut G E, Nagashima K A, Tran D N, Anselma D J, Monard S P, Segal J P, Thompson D A, Kajumo F, Guo Y, et al. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill C M, Kwon D, Jones M, Davis C B, Marmon S, Daugherty B L, DeMartino J A, Springer M S, Unutmaz D, Littman D R. Virology. 1998;248:357–371. doi: 10.1006/viro.1998.9283. [DOI] [PubMed] [Google Scholar]
- 21.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, et al. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doranz B J, Grovit-Ferbas K, Sharron M P, Mao S H, Goetz M B, Daar E S, Doms R W, O'Brien W A. J Exp Med. 1997;186:1395–1400. doi: 10.1084/jem.186.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 24.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, et al. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trkola A, Gordon C, Matthews J, Maxwell E, Ketas T, Czaplewski L, Proudfoot A E, Moore J P. J Virol. 1999;73:6370–6379. doi: 10.1128/jvi.73.8.6370-6379.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1154. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 27.Kozak S L, Platt E J, Madani N, Ferro F E, Jr, Peden K, Kabat D. J Virol. 1997;71:873–882. doi: 10.1128/jvi.71.2.873-882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trkola A, Matthews J, Gordon C, Ketas T, Moore J P. J Virol. 1999;73:8966–8974. doi: 10.1128/jvi.73.11.8966-8974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 30.Dragic T, Trkola A, Lin S W, Nagashima K A, Kajumo F, Zhao L, Olson W C, Wu L, Mackay C R, Allaway G P, et al. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabut G E, Konner J A, Kajumo F, Moore J P, Dragic T. J Virol. 1998;72:3464–3468. doi: 10.1128/jvi.72.4.3464-3468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh T, Han M, Sakmar T P, Smith S O. J Mol Biol. 1997;269:373–384. doi: 10.1006/jmbi.1997.1035. [DOI] [PubMed] [Google Scholar]
- 33.Chen Z, Kwon D, Jin Z, Monard S, Telfer P, Jones M S, Lu C Y, Aguilar R F, Ho D D, Marx P A. J Exp Med. 1998;188:2057–2065. doi: 10.1084/jem.188.11.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiraishi M, Aramaki Y, Seto M, Imoto H, Nishikawa Y, Kanzaki N, Okamoto M, Sawada H, Nishimura O, Baba M, Fujino M. 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA. 1999. pp. A911–A914. [Google Scholar]
- 35.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, et al. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 36.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, et al. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 39.Premack B A, Schall T J. Nat Med. 1996;2:1174–1178. doi: 10.1038/nm1196-1174. [DOI] [PubMed] [Google Scholar]
- 40.Hall I P, Wheatley A, Christie G, McDougall C, Hubbard R, Helms P J. Lancet. 1999;354:1264–1265. doi: 10.1016/s0140-6736(99)03425-x. [DOI] [PubMed] [Google Scholar]
- 41.Wyatt R, Kwong P D, Desjardins E, Sweet R, Robinson J, Hendrickson W, Sodroski J. Nature (London) 1998;393:705–710. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 42.Siciliano S J, Rollins T E, DeMartino J, Konteatis Z, Malkowitz L, Van Riper G, Bondy S, Rosen H, Springer M S. Proc Natl Acad Sci USA. 1994;91:1214–1218. doi: 10.1073/pnas.91.4.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monteclaro F S, Charo I F. J Biol Chem. 1996;271:19084–19092. doi: 10.1074/jbc.271.32.19084. [DOI] [PubMed] [Google Scholar]
- 44.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 45.Hesselgesser J, Ng H P, Liang M, Zheng W, May K, Bauman J G, Monahan S, Islam I, Wei G P, Ghannam A, et al. J Biol Chem. 1998;273:15687–15692. doi: 10.1074/jbc.273.25.15687. [DOI] [PubMed] [Google Scholar]
- 46.White J R, Lee J M, Young P R, Hertzberg R P, Jurewicz A J, Chaikin M A, Widdowson K, Foley J J, Martin L D, Griswold D E, Sarau H M. J Biol Chem. 1998;273:10095–10098. doi: 10.1074/jbc.273.17.10095. [DOI] [PubMed] [Google Scholar]
- 47.Labrosse B, Brelot A, Heveker N, Sol N, Schols D, De Clercq E, Alizon M. J Virol. 1998;72:6381–6388. doi: 10.1128/jvi.72.8.6381-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loetscher M, Geiser T, O'Reilly T, Zwahlen R, Baggiolini M, Moser B. J Biol Chem. 1994;269:232–237. [PubMed] [Google Scholar]
- 49.Tarasova N I, Rice W G, Michejda C J. J Biol Chem. 1999;274:34911–34915. doi: 10.1074/jbc.274.49.34911. [DOI] [PubMed] [Google Scholar]