Abstract
Inwardly rectifying potassium (K+) channels gated by G proteins (Kir3.x family) are widely distributed in neuronal, atrial, and endocrine tissues and play key roles in generating late inhibitory postsynaptic potentials, slowing the heart rate and modulating hormone release. They are directly activated by Gβγ subunits released from G protein heterotrimers of the Gi/o family upon appropriate receptor stimulation. Here we examine the role of isoforms of pertussis toxin (PTx)-sensitive G protein α subunits (Giα1–3 and GoαA) in mediating coupling between various receptor systems (A1, α2A, D2S, M4, GABAB1a+2, and GABAB1b+2) and the cloned counterpart of the neuronal channel (Kir3.1+3.2A). The expression of mutant PTx-resistant Gi/oα subunits in PTx-treated HEK293 cells stably expressing Kir3.1+3.2A allows us to selectively investigate that coupling. We find that, for those receptors (A1, α2A) known to interact with all isoforms, Giα1–3 and GoαA can all support a significant degree of coupling to Kir3.1+3.2A. The M4 receptor appears to preferentially couple to Giα2 while another group of receptors (D2S, GABAB1a+2, GABAB1b+2) activates the channel predominantly through Gβγ liberated from GoA heterotrimers. Interestingly, we have also found a distinct difference in G protein coupling between the two splice variants of GABAB1. Our data reveal selective pathways of receptor activation through different Gi/oα isoforms for stimulation of the G protein-gated inwardly rectifying K+ channel.
Inwardly rectifying K+ channels gated by the direct action of G proteins are present in neurones, atrial myocytes, and endocrine cells and are responsible for mediating postsynaptic inhibitory effects, in slowing the heart rate in response to vagal nerve stimulation and in modulating hormone release. Their molecular counterparts have been identified and the channel has been shown to be a heteromultimeric structure comprised of members of the Kir3.x family of K+ channels (1–5). Co-expression of Kir3.1 with Kir3.2, Kir3.3, or Kir3.4 results in currents that show many of the basic characteristics of the native channels in neurones and atria (6–8). Channel activation is abolished by pertussis toxin (PTx) treatment, implicating the Gi/o family of G proteins (9–11). Although initially controversial, it is now well established that activation of these channels in native tissues and of the cloned counterparts in heterologous expression systems is via a membrane-delimited mechanism involving a direct interaction with the Gβγ dimer (12–14). Indeed the studies on this channel have become a paradigm of how Gβγ can be important in signaling to downstream effectors. Current studies have focused on domains on the channel important for binding Gβγ (15–20), trafficking of the channel complex (21–24), and the role of anionic phospholipids in regulating channel activity (25–29).
We have recently shown that the Gα subunit is the key determinant of specificity of channel activation for receptors coupling predominantly to Gi/o as against those that couple to Gs (30). In this study, we investigate the role of different Gi/oα variants in determining selective receptor coupling between receptors and the cloned G protein-gated inwardly rectifying K+ channel, Kir3.1+3.2A. The PTx-sensitive G protein family is made up of Gi, encoded by three separate gene products (Giα1, Giα2, and Giα3), and Go, made up of two splice variants (GoαA and GoαB) (31). It is apparent that some heptahelical receptors exhibit a preference for the type of Gα subunit they couple to within a G protein family—for example, evidence exists that suggests that the D2 dopamine receptor splice variants preferentially couple to some Gi/oα subunits rather than to others (32–35). In addition, it has been proposed that Gi/oα subunit variants, in particular Giα1, may have direct inhibitory actions on the G protein-gated K+ channel (36). However, the significance of this for receptor-mediated activation has not been addressed. In this study, we demonstrate that all Gi/oα variants are able to liberate Gβγ to mediate coupling between receptor and Kir3.1+3.2A channels, but that some receptors have a preference for the Gi/oα subunit variant with which they interact to activate the channel.
Methods
Molecular Biology, Cell Culture, and Transfection.
Standard molecular cloning and mutagenesis techniques were used throughout. Cell culture, generation of stable cell lines, construction of the bicistronic vector, and point mutations of Gα subunits were as described (30, 37). For this study we used a similar PCR-based strategy to introduce a C→G mutation at analogous positions in Giα2, Giα3, and GoαA. We transfected 400 ng of each receptor cDNA and 500 ng of each Gi/oα cDNA. We examined the effects of varying cDNA ratios for the α2A adrenergic receptor and the Giα2C352G and Giα3C351G mutants in the HKIR3.1/3.2 line treated with PTx. We found that reducing the cDNA concentration for the mutant G protein (from 500 ng to 125 ng) alters the magnitude but not the selectivity difference whereas decreasing the receptor concentration (from 400 ng to 100 ng) loses any response (data not shown). Increasing the amount of Gi/oα cDNA beyond 1 μg was toxic to cells. It should be noted that the IRES-containing vector we constructed does not ensure that translation starts in the optimal position from the end of the IRES element (38). It is thus likely that protein translation from the second cistron will be reduced. However, in our particular case, the expression of Kir3.1 and Kir3.2A from the IRES plasmid and from separate plasmids [pcDNA3 and pcDNA3.1(+)/Zeo (Invitrogen)] gave similar basal current levels (separate plasmids: 147 ± 22 pA/pF, n = 17; IRES vector: 112 ± 17 pA/pF, n = 41, P = 0.26).
Electrophysiology.
Whole-cell membrane currents were recorded by using an Axopatch 200B amplifier (Axon Instruments, Foster City, CA). Patch pipettes were pulled from filamented borosilicate glass (Clark Electromedical Instruments, Pangbourne, U.K.) and had a resistance of 1.5–2.5 MΩ when filled with pipette solution (see below). Before filling, tips of patch pipettes were coated with a Parafilm/mineral oil suspension. Records were filtered at 1 kHz and were digitized at 5 kHz, and data were acquired and analyzed by using a Digidata 1200B interface (Axon Instruments) and pclamp 6.0 software (Axon Instruments). Cell capacitance was approximately 15 pF, and series resistance (<10 MΩ) was at least 75% compensated. Recordings of membrane current were commenced after an equilibration period of approximately 5 min. Currents were measured at the end of each voltage step. Current densities were measured at −100 mV (unless otherwise stated), and all data are presented as mean ± SEM. Student's t tests were performed to examine statistical significance, and an asterisk in Figs. 2–4 indicates that P ≤ 0.05.
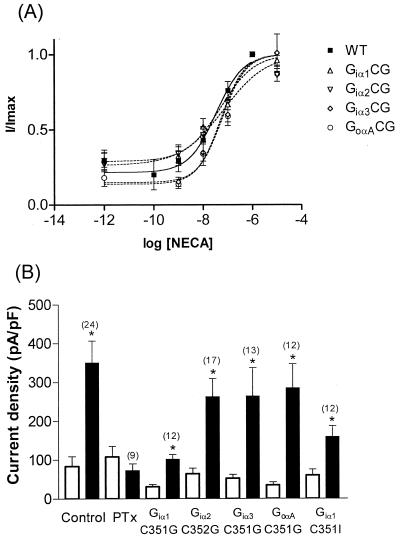
Figure 2.
The Gi/oα subunits have similar affinities in mediating signaling between A1 receptors and Kir3.1+3.2A. (A) Superimposed dose-response curves for NECA-induced activation of Kir3.1+3.2A channels in control, non-PTx treated cells (solid line) and in PTx-treated cells in which the mutant Gi/oα subunits (dashed lines), Giα1C351G, Giα2C352G, Giα3C351G, and GoαAC351G, have been co-expressed. (B) Bar chart summarizing the data obtained with the HKIR3.1/3.2/A1 cell line and expression of each of the Gi/o variants. Open bars represent basal currents, and solid bars represent current in response to receptor stimulation. Numbers in parentheses refer to the number of cells recorded from for each experiment. Current density was measured at −100 mV.
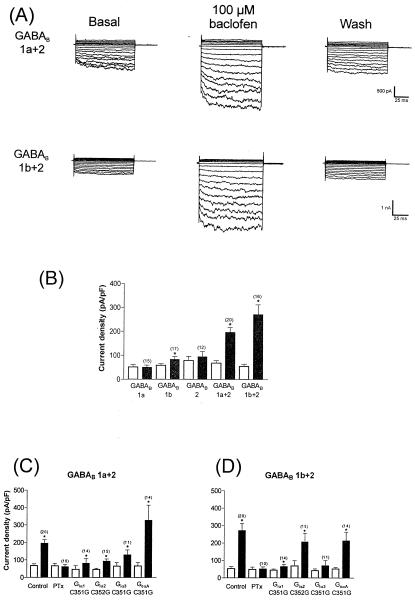
Figure 4.
The two forms of the GABAB receptor activate Kir3.1+3.2A channels through different Gi/oα subunits. (A) Stimulation of both the GABAB1a+2 and the GABAB1b+2 receptor (100 μM baclofen) led to robust activation of the Kir3.1+3.2A channels. Currents were elicited as described in Fig. 1B. (B) Summary of data obtained from expression of the GABAB1a, -1b, and -2 subunits alone, and when expressed as the dimeric receptors GABAB1a+2 and GABAB1b+2. (B and D) Bar charts summarizing the data obtained with the GABAB1a+2 receptor (C) and the GABAB1b+2 receptor (D) when the mutant Gi/oα mutants were co-expressed in PTx-treated cells. Open bars represent basal currents, and solid bars represent current in response to receptor stimulation. Numbers in parentheses refer to the number of cells recorded from for each experiment. Current density was measured at −100 mV.
Dose-response curves were constructed in the HKIR3.1/3.2/A1 cell line. It was not feasible to apply more than four concentrations of 5′-N-ethylcarboxyamidoadenasine (NECA) to individual cells because of receptor “desensitization” and a subsequent decline in response. Therefore, for each experimental condition (i.e., receptor coupling to either endogenous Gα or exogenously expressed Gα mutants), data were pooled from at least 12 cells, and the responses obtained by using different concentrations of NECA were normalized (I/Imax) to those obtained by using a maximal concentration (1 μM) that was applied to every cell recorded from. Concentrations of NECA were applied randomly, but 1 μM NECA was always applied twice to each cell at the start and end of the experiment. Curves were fitted by using nonlinear regression with prism 3.0 software (GraphPad, San Diego). Data were obtained from at least two independent transfections of each Gα/receptor combination.
Materials and Drugs.
Solutions were as follows (concentrations in mM): pipette solution, 107 KCl, 1.2 MgCl2, 1 CaCl2, 10 EGTA, 5 Hepes, 2 MgATP, 0.3 Na2GTP (KOH to pH 7.2, ≈140 mM total K+); bath solution, 140 KCl, 2.6 CaCl2, 1.2 MgCl2, 5 Hepes (pH 7.4). Cell culture materials were from GIBCO/BRL and Invitrogen. Molecular biology reagents were obtained from New England Biolabs or Roche Molecular Biochemicals, and oligonucleotides were from Genosys (Cambridge, U.K.). All chemicals were from Sigma or Calbiochem. Drugs were made up as concentrated stock solutions and were kept at −20°C or −80°C.
Results
The studies detailed here were performed on stably transfected HEK293 cells expressing Kir3.1 and Kir3.2A alone (HKIR3.1/3.2) or on another line (HKIR3.1/3.2/A1) that additionally stably expressed the A1 adenosine receptor together with Kir3.1 and Kir3.2A (30).
Characterization of PTx-Insensitive Mutants of Giα1, Giα2, Giα3, and Goα1.
PTx catalyzes the ADP ribosylation of the Gi/oα subunit at a cysteine residue four amino acids from the C-terminal end of the protein. The PTx-treated subunit is thus unable to participate in signaling. However, mutation of this residue to glycine or isoleucine renders the mutant subunit insensitive to the effects of PTx (34, 39). Such mutants have been shown to still functionally interact with receptors as determined by agonist-stimulated 35S[GTPγS] binding (37, 40, 41). We have previously shown that Giα1C351G is able to rescue coupling between Kir3.1+3.2A and the transiently transfected A1 and α2A receptors in the HKIR3.1/3.2 cell line after PTx treatment (30).
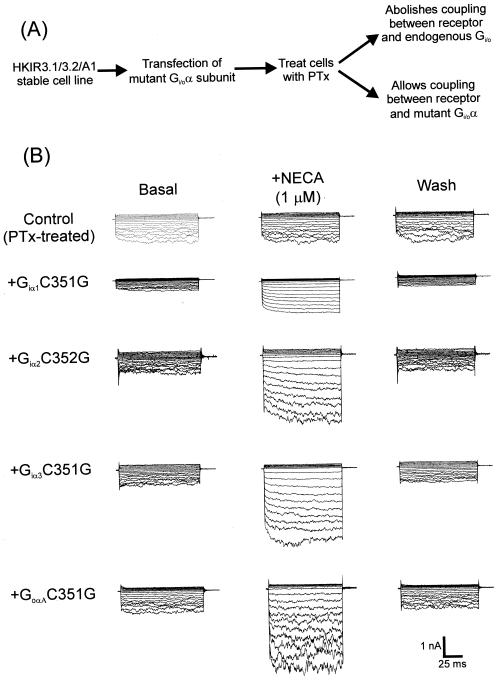
We now characterize the behavior of Giα1C351G, Giα1C351I, Giα2C352G, Giα3C351G, and GoαAC351G after transient expression and PTx treatment in the stable cell line HKIR3.1/3.2/A1 in which there are constant levels of expression of the A1 receptor and Kir3.1+3.2A channel current. The experimental protocol we used is shown in Fig. 1A. Expression of the mutants alone did not enhance membrane currents and in fact significantly reduced basal current density similarly to wild-type Gα (e.g., Giα1C351G: 34 ± 8 pA/pF, n = 8, P = 0.05; Giα2C352G: 18 ± 4 pA/pF, n = 10, P < 0.01). In PTx-treated cells, A1 receptor stimulation was unable to enhance Kir3.1+3.2A currents (Fig. 1B, top traces). However, when any of the mutant Gi/oα subunits were co-expressed, NECA stimulation of HKIR3.1/3.2/A1 cells led to a large enhancement of currents (Fig. 1B). Thus, the PTx-insensitive Gα subunits were able to rescue signaling between the A1 receptor and Kir3.1+3.2A in PTx-treated cells.
Figure 1.
The A1 adenosine receptor couples to Kir3.1+3.2A channels via Giα1–3 and GoαA. (A) This illustrates the experimental protocol used. The HKIR3.1/3.2/A1 monoclonal cell line stably expressing Kir3.1+3.2A channels together with the A1 receptor was transiently transfected with mutant PTx-insensitive Gi/oα subunits, and then cells were treated with PTx 1 day before electrophysiological recording. (B) These are examples of traces showing the effects of stimulating A1 receptors in the HKIR3.1/3.2/A1 cell line in PTx-treated cells (top traces) and when each of the mutated Gi/oα variants was co-expressed. Currents were elicited by holding cells at 0 mV and stepping to potentials between −100 and +50 mV in 10-mV increments for 100 ms. Traces indicate current responses before (Basal), during (+ NECA), and after (Wash) receptor stimulation.
We have quantitatively investigated the behavior of these Gα mutants by constructing dose-response curves for NECA in the HKIR3.1/3.2/A1 line. The dose-response curve obtained from control, non-PTx-treated cells expressing endogenous G protein is illustrated in Fig. 2A. From this curve, it can be seen that the logEC50 for NECA is −7.48 ± 0.20, equivalent to a concentration of 33.1 nM, and the Hill coefficient is 0.79 ± 0.27 (data pooled from 13 cells). We constructed dose-response curves to NECA for each of the Gα mutants. These are also illustrated in Fig. 2A, and the results are summarized in Table 1. To examine whether the EC50 values varied with the different Gα subunits, we compared the logEC50s measured with exogenous expression of each of the mutant Gα with endogenous Gα. No significant differences were observed when compared with endogenous Gi/o (P = 0.31–0.51). We also found no significant differences in the Hill coefficients when the Gα point mutants were compared with endogenous G protein (P = 0.3–0.95). These findings suggest that the mutant Gi/oα subunits (Giα1C351G, Giα1C351I, Giα2C352G, Giα3C351G, and GoαAC351G) are still able to couple the A1 receptor to the channel complex with approximately equal affinity and that this affinity is similar to that displayed by the endogenous Gi/o proteins present in the HEK293 cells.
Table 1.
Summary of data from dose-response curves constructed to NECA in the HKIR3.1/3.2/A1 cell lines
| Endogenous | Giα1 C351G | Giα2 C352G | Giα3 C351G | GoαA C351G | |
|---|---|---|---|---|---|
| EC50 | 33.1 nM | 50.7 nM (P = 0.51) | 60.3 nM (P = 0.31) | 59.4 nM (P = 0.51) | 69.2 nM (P = 0.35) |
| Hill coefficient, nH | 0.79 ± 0.27 | 0.64 ± 0.15 (P = 0.64) | 0.5 ± 0.12 (P = 0.3) | 0.9 ± 0.51 (P = 0.85) | 0.82 ± 0.34 (P = 0.95) |
Numbers in brackets refer to level of significance in comparing each exogenously expressed Gi/oα with endogenous G protein.
The efficacy of the response was next investigated. We compared the responses obtained with a maximal concentration of NECA (1 μM) to see whether the efficacy of coupling had been altered by the mutant Gα subunits (Fig. 2B). In cells in which Giα1C351G had been expressed, the NECA-induced increase in current density was significantly smaller than in control non-PTx-treated cells (P = 0.01). Similarly, we found that, when Giα1C351I was expressed, the induced currents were also significantly smaller than in control cells (P = 0.01) and moreover were not significantly different from those obtained with Giα1C351G (P = 0.25). None of the other G protein mutants tested had any significant effects on NECA-induced currents (P = 0.28–0.83), suggesting that Giα2, Giα3, and GoαA all have similar efficacies.
It is likely that the mutant G proteins are all expressed to similar high levels as they are expressed in essentially the same plasmid under the control of the same cytomegalovirus promoter. This has been demonstrated for different mutations of Cys351in Giα1 (37) and also for C→G mutations in Giα1, Giα2, and Giα3 (40). The data presented above suggest that the levels of expression achieved are sufficient to functionally complement the response.
Delineating Different Patterns of Receptor Stimulation of Channel Activation.
Our studies with the A1 receptor establish that the mutant G protein subunits can substitute both qualitatively and quantitatively for the endogenous G proteins expressed in HEK293 cells. Given the broad ability of the A1 receptor to activate the isoforms of Gi/o, we next examined other Gi/o-coupled receptors, including the α2A, D2S, M4, and GABAB. We compared the ability of a concentration of standard, full agonist that would lead to maximal receptor occupancy (3 μM noradrenaline, 10 μM quinpirole, 10 μM carbachol, and 100 μM baclofen, respectively) to activate currents in the HKIR3.1/3.2 cell line transiently transfected with receptor and mutant G protein and treated with PTx.
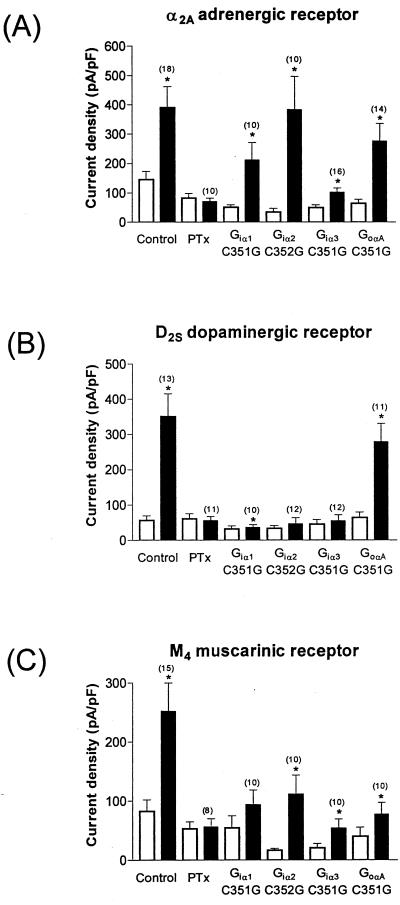
When the mutant Gα subunits were expressed in PTx-treated cells, the channel was still able to be activated via noradrenaline-mediated stimulation of the α2A receptor coupling to all of the Gα subunits tested. However, as observed with the A1 receptor, some Gα subunits appeared to be more efficient than others, although a different pattern was observed (Fig. 3A). Although there did not appear to be much difference between Giα1, Giα2, and GoαA, the responses obtained with expression of Giα3 were smaller. We also looked at the Giα1C351I mutant: the noradrenaline-induced currents obtained when this α subunit was expressed were not significantly different to those obtained with Giα1C351G (100.2 ± 33.6 pA/pF, n = 10, P = 0.33). We next investigated the coupling of D2S to Giα1–3 and GoαA. Interestingly, the expression of neither Giα1C351G, Giα2C352G, nor Giα3C351G was able to efficiently rescue signaling between the receptor and Kir3.1+3.2A. However, the D2S receptor was able to activate channels in PTx-treated cells to a similar level observed in control cells when GoαAC351G was co-expressed (Fig. 3B). Finally, we examined the coupling of the M4 muscarinic receptor to the channel whereupon we observed that co-expression of Giα2C352G was able to support channel activation. Giα3C351G could also support channel activation, but to a lesser extent, whereas Giα1C351G and GoαAC351G were not nearly as effective (Fig. 3C).
Figure 3.
The α2A adrenergic receptor, D2S dopaminergic receptor, and M4 muscarinic receptor exhibit different coupling profiles to Gi/oα subunits. Summary of data obtained from studying coupling between the α2A adrenergic receptor (A), the D2S dopaminergic receptor (B) and the M4 muscarinic receptor (C), and Kir3.1+3.2A channels via the Giα1, Giα2, Giα3, and GoαA C∏ G mutants. Open bars represent basal currents, and solid bars represent current in response to receptor stimulation. Numbers in parentheses refer to the number of cells recorded from for each experiment. Current density was measured at −100 mV.
Studies on Cloned GABAB Receptors.
The stimulation of postsynaptic GABAB receptors in neurones and the subsequent activation of G protein-gated inwardly rectifying K+ channels is a major mechanism for generating late inhibitory postsynaptic potentials. It has recently been established that native GABAB receptors are a heterodimeric complex comprised of a combination of the two subunits GABAB1 and GABAB2 (42–45). There are now three splice variants of GABAB1, -1a, -1b, and -1c (46, 47). It has been reported that neither GABAB1a, GABAB1b, nor GABAB1c can express efficiently alone: the GABAB2 subunit is required to form a functional heterodimeric receptor.
We first established the behavior of the cloned receptors in our system. We transiently expressed the GABAB splice variants GABAB1a, -1b, or -2 alone and investigated whether these could activate the Kir3.1+3.2A channels. When either GABAB1a or GABAB2 were expressed, we observed no stimulation of Kir3.1+3.2A currents in response to 100 μM baclofen. However, expression and stimulation of the GABAB1b splice variant did lead to an enhancement of Kir3.1+3.2A currents in approximately one-third of cells recorded from, revealing a small but significant enhancement of currents (Fig. 4B). We then expressed receptors comprised of GABAB1a+2 and GABAB1b+2 (1:1 cDNA ratio) and investigated their coupling to Kir3.1+3.2A. Stimulation of GABAB1a+2 receptors significantly increased current density, and this was sensitive to PTx. Likewise, stimulation of receptors composed of GABAB1b+2 subunits also potentiated currents in a PTx-sensitive fashion (Fig. 4 A and B).
The GABAB1a+2 receptor was able to signal most prominently via GoαAC351G to Kir3.1+3.2A whereas signaling to the other Gα subunits was not so pronounced (Fig. 4C). Interestingly, GABAB1b+2 was able to signal to an equal extent through both Giα2C351G and GoαAC351G to a similar extent to that observed in control cells with coupling to endogenous Gα (Fig. 4D).
Discussion
The aims of the present study were to investigate the role of Gi/oα isoforms in coupling receptors to the G protein-gated inwardly rectifying K+ channel. Specificity of this phenomenon could lie at two levels: in the ability of receptors to couple to various Gα subunits or in the ability of liberated Gβγ from a particular heterotrimer to activate the channel. To address this, we have used a series of PTx-insensitive Gi/o point mutants in which a cysteine residue four amino acids from the C terminus of the α subunit is replaced by a glycine or isoleucine residue. In all cases, we could always rescue signaling between any of the receptors tested and the Kir3.1+3.2A channels in PTx-treated cells, and we observed different patterns of preferences between the different receptors and Gα subunits.
The first question is, are these mutants good reporters of the coupling between receptor and channel? We investigated this quantitatively by constructing dose-response curves in a stable line expressing both the A1 receptor and Kir3.1+3.2A channel complex. The data indicate that the EC50 for all Gα point mutants is similar and comparable to that displayed when the A1 receptor couples to endogenous G proteins. The lack of a statistically significant change in the EC50 and the Hill coefficient for channel stimulation via the A1 receptor is a strong result suggesting that the mutations, at these levels of expression, do not affect the ability of the receptor to interact with the G protein or the innate ability of these different G protein heterotrimers to liberate Gβγ for Kir3.1+3.2A channel activation.
Are all Gi/oα subunits, through the liberation of Gβγ, able to activate the channel to a similar extent? Schreibmayer et al. (36) showed inhibition of Gβ1γ2-induced currents by activated Giα1 (but not Giα2 or Giα3) added as purified proteins to inside-out patches containing cloned or native Kir3.x channels. The significance of this for receptor-mediated activation was not addressed, the implication being that stimulation would not occur through liberation of Gβγ from Giα1 heterotrimers due to simultaneous inhibition by the Gα subunit. Our studies suggest that it is not only possible to activate the channel via βγ released from Gi1 heterotrimers but that there are only moderate quantitative differences between the variants of Gi/o in the ability to mediate activation of the channel. The most profound response observed with Giα1C351G was in its coupling to the α2A receptor (mean current density: 211.51 ± 59.53 pA/pF, n = 10), a response not dissimilar to those observed with other Gi/oα/receptor combinations (α2A/Giα2C352G: 384.07 ± 113.31 pA/pF, n = 10, P = 0.19; A1/Giα3C351G: 263.85 ± 73.56 pA/pF, n = 13, P = 0.61; GABAB1a+2/GoαAC351G: 329.17 ± 86.69 pA/pF, n = 14, P = 0.32), suggesting that all Gi/oα variants can mediate channel activation to a similar extent.
To summarize, the Gi/oα point mutants are good reporters of receptor/channel coupling, and there are only moderate differences in the efficacy of their ability to activate currents from liberation of Gβγ. Thus, any major quantitative differences in the coupling pattern between Gi/oα variants and a particular Gi/o-coupled receptor is likely to be attributable to differences in the ability of the receptor to “talk” to a particular variant. Indeed, this experimental approach may be a useful model system to assay the specificity of such interactions.
It is apparent that the receptors we have studied exhibit different patterns of coupling to Gi/oα subunits to activate Kir3.1+3.2A channels. The receptors we investigated were the A1 adenosine receptor, the α2A adrenergic receptor, the D2S dopaminergic receptor, the M4 muscarinic receptor, and the two subtypes of the GABAB receptor, GABAB1a+2 and GABAB1b+2. The A1 receptor seemed to couple almost equally well to all Gi/oα subunits, although Giα1 appeared to be not quite as efficient. This is consistent with previous studies showing that recombinant A1 receptors have been shown to interact equally well with Giα1–3 (41). Differences do occur across species however, notably between human and bovine A1 receptors (48).
A completely different pattern was observed with the α2A adrenergic receptor. In other studies examining the coupling of α2A to Gi/o proteins, it was found to equally activate Giα1–3 (40). However, we found that Giα3 was much less effective than the other Gα subunits. Our observation that the D2S dopamine receptor couples exclusively to GoαA but not any of the Gα subunits is an interesting one and one that is in contrast to some studies by other investigators. A point to note is the disagreement between different studies concerning D2S and Gα coupling. Some investigators report that D2S couples to both Giα2 and Giα3 (33), others report selective coupling to Giα2 (34), and others report preferred coupling to Giα1 rather than Giα2 (35). We found that the M4 muscarinic receptor, in agreement with previous findings (49), preferentially coupled to the Giα2 subunit. Some coupling to Giα3 was also observed, but this was less effective. A number of issues should be considered when comparing these differences and similarities. Studies done by other groups have been concerned with effector mechanisms linked to the Gα subunit: e.g., the inhibition of forskolin- or receptor-stimulated cAMP accumulation or agonist-induced [35S]GTPγS binding. In contrast, the reporter system we are using (i.e., Kir3.1+3.2A channel activation) is one that is mediated directly by the Gβγ subunit. In addition, there is less amplification in this system than, for example, looking at cAMP accumulation. Furthermore, we have made a comparative study of a much larger number of Gi/oα family members (Giα1–3 and GoαA) whereas, in some of the studies above, only a select number were examined.
A further finding is the different G protein coupling profiles observed with two different heterodimers of GABAB receptors. The GABAB receptors belong to the class 3 family of seven-pass receptors and as such share most homology with the metabotropic glutamate receptors. Members of this family have extended N termini that bear some sequence similarity to periplasmic amino acid binding proteins found in bacteria (50) that are thought to be involved in ligand binding (51, 52) but not G protein coupling (53). The splice variants GABAB1a and GABAB1b differ by a 117-aa stretch that is missing at the N terminus of GABAB1b: this region contains amino acid motifs, termed “Sushi repeats,” thought to be involved in protein-protein interactions (54). The regions involved in coupling the metabotropic glutamate receptors to G proteins are thought to be the second and third intracellular loops (55–57), so it might be likely that the same applies to GABAB receptors (58). However, we have clearly shown that GABAB1a+2 receptors have a different Gα coupling profile to the GABAB1b+2 receptors. This is unlikely to be attributable to differences in binding affinity of the splice variants for baclofen because the two variants have identical pharmacological agonist/antagonist profiles (58). The most likely mechanism we envisage is that the agonist-occupied active state is different between the two splice variants and that this is reflected by different conformation and G protein preferences in the receptor/Gα coupling domain, despite the only differences between the two receptor splice variants being at the proximal N terminus. However, we cannot exclude differences in trafficking or the possibility that one, but not the other, splice variant may interact with a protein that influences the above functional property.
In conclusion, we have examined the role of different Gi/o isoforms in coupling receptors to the activation of G protein-gated inwardly rectifying K+ channels. Our data indicate that PTx-insensitive point mutants of Gi/o are able to report these interactions meaningfully and that there are only minor differences in the ability of these variants to activate the channel. Different receptors appear to prefer different Gα subunits to couple to the channels: indeed, the N-terminal splice variants of the GABAB heterodimeric receptor show different patterns of selectivity. Thus, we have revealed a mechanism for selective receptor activation of the channel that lies at the interface between the receptor and Gi/o variant. Differential or localized expression of Gi/o variants (59) could lead to selective pathways of channel activation.
Acknowledgments
We thank the following people for providing us with cDNAs: B. Conklin (Giα2, Giα3, GoαA), L. Y. Jan (α2A receptor and Kir3.1), M. Lazdunski (Kir3.2A), F. Marshall (GABAB clones), G. Milligan, (G iα1C351G and Giα1C351I), T. Palmer (A1 receptor), E. G. Peralta (M4), and W. Xu (D2S receptor) and the members of the Human Frontiers Science Program collaboration for fruitful discussions (L. Y. Jan, Y. Kurachi, and E. Reuveny). We also thank G. Milligan for extensive discussions concerning the PTX-insensitive variants, Z. Hafeez and other members of the lab for technical help and discussion, and L. Clapp for helpful comments. This work was supported by the Human Frontiers Science Program and the Wellcome Trust.
Abbreviations
- PTx
pertussis toxin
- NECA
5′-N-ethylcarboxyamidoadenasine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080572297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080572297
References
- 1.Dascal N, Schreibmayer W, Lim N F, Wang W, Chavkin C, DiMagno L, Labarca C, Kieffer B L, Gaveriaux-Ruff C, Trollinger D, et al. Proc Natl Acad Sci USA. 1993;90:10235–10239. doi: 10.1073/pnas.90.21.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubo Y, Reuveny E, Slesinger P A, Jan Y N, Jan L Y. Nature (London) 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 3.Lesage F, Duprat F, Fink M, Guillemare E, Coppola T, Lazdunski M, Hugnot J-P. FEBS Lett. 1994;353:37–42. doi: 10.1016/0014-5793(94)01007-2. [DOI] [PubMed] [Google Scholar]
- 4.Krapivinsky G, Gordon E A, Wickman K, Velimirovic B, Krapivinsky L, Clapham D E. Nature (London) 1995;374:135–141. doi: 10.1038/374135a0. [DOI] [PubMed] [Google Scholar]
- 5.Hedin K E, Lim N F, Clapham D E. Neuron. 1996;16:423–429. doi: 10.1016/s0896-6273(00)80060-4. [DOI] [PubMed] [Google Scholar]
- 6.Lesage F, Guillemare E, Fink M, Duprat F, Heurteaux C, Fosset M, Romey G, Barhanin J, Lazdunski M. J Biol Chem. 1995;270:28660–28667. doi: 10.1074/jbc.270.48.28660. [DOI] [PubMed] [Google Scholar]
- 7.Velimirovic B, Gordon E A, Lim N F, Navarro B, Clapham D E. FEBS Lett. 1996;379:31–37. doi: 10.1016/0014-5793(95)01465-9. [DOI] [PubMed] [Google Scholar]
- 8.Corey S, Krapivinsky G, Krapivinsky L, Clapham D E. J Biol Chem. 1998;273:5271–5278. doi: 10.1074/jbc.273.9.5271. [DOI] [PubMed] [Google Scholar]
- 9.Breitwieser G E, Szabo G. Nature (London) 1985;317:538–540. doi: 10.1038/317538a0. [DOI] [PubMed] [Google Scholar]
- 10.Pfaffinger P J, Martin J M, Hunter D D, Nathanson N M, Hille B. Nature (London) 1985;317:536–538. doi: 10.1038/317536a0. [DOI] [PubMed] [Google Scholar]
- 11.Kurachi Y, Nakajima T, Sugimoto T. Am J Physiol. 1986;251:H681–H684. doi: 10.1152/ajpheart.1986.251.3.H681. [DOI] [PubMed] [Google Scholar]
- 12.Logothetis D E, Kurachi Y, Galper J, Neer E J, Clapham D E. Nature (London) 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 13.Yamada M, Inanobe A, Kurachi Y. Pharmacol Rev. 1998;50:723–757. [PubMed] [Google Scholar]
- 14.Reuveny E, Slesinger P A, Inglese J, Morales J M, Iniguez-Lluhi J A, Lefkowitz R J, Bourne H R, Jan Y N, Jan L Y. Nature (London) 1994;370:143–146. doi: 10.1038/370143a0. [DOI] [PubMed] [Google Scholar]
- 15.Huang C L, Slesinger P A, Casey P J, Jan Y N, Jan L Y. Neuron. 1995;15:1133–1143. doi: 10.1016/0896-6273(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 16.Huang C L, Jan Y N, Jan L Y. FEBS Lett. 1997;405:291–298. doi: 10.1016/s0014-5793(97)00197-x. [DOI] [PubMed] [Google Scholar]
- 17.Inanobe A, Morishige K-I, Takahashi N, Ito H, Yamada M, Takumi T, Nishina H, Takahashi K, Kanaho Y, Katada T, Kurachi Y. Biochem Biophys Res Commun. 1995;212:1022–1028. doi: 10.1006/bbrc.1995.2072. [DOI] [PubMed] [Google Scholar]
- 18.Krapivinsky G, Krapivinsky L, Wickman K, Clapham D E. J Biol Chem. 1995;270:29059–29062. doi: 10.1074/jbc.270.49.29059. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel M T, Peralta E G. Cell. 1995;83:443–449. doi: 10.1016/0092-8674(95)90122-1. [DOI] [PubMed] [Google Scholar]
- 20.Krapivinsky G, Kennedy M E, Nemec J, Medina I, Krapivinsky L, Clapham D E. J Biol Chem. 1998;273:16946–16952. doi: 10.1074/jbc.273.27.16946. [DOI] [PubMed] [Google Scholar]
- 21.Morishige K-I, Inanobe A, Yoshimoto Y, Kurachi H, Murata Y, Tokunaga Y, Maeda T, Maruyama Y, Kurachi Y. J Biol Chem. 1999;274:7969–7974. doi: 10.1074/jbc.274.12.7969. [DOI] [PubMed] [Google Scholar]
- 22.Stevens E B, Woodward R, Ho I H M, Murrel-Lagnardo R. J Physiol. 1997;503:547–562. doi: 10.1111/j.1469-7793.1997.547bg.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vivaudou M, Chan K W, Sui J L, Jan L Y, Reuveny E, Logothetis D E. J Biol Chem. 1997;272:31553–31560. doi: 10.1074/jbc.272.50.31553. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy M E, Nemec J, Corey S, Wickman K, Clapham D E. J Biol Chem. 1999;274:2571–2582. doi: 10.1074/jbc.274.4.2571. [DOI] [PubMed] [Google Scholar]
- 25.Huang C L, Feng S Y, Hilgemann D W. Nature (London) 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 26.Kim D, Bang H. J Physiol. 1999;517:59–74. doi: 10.1111/j.1469-7793.1999.0059z.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, He C, Yan X, Mirshahi T, Logothetis D E. Nat Cell Biol. 1999;1:183–188. doi: 10.1038/11103. [DOI] [PubMed] [Google Scholar]
- 28.Sui J L, PetitJacques J, Logothetis D E. Proc Natl Acad Sci USA. 1998;95:1307–1312. doi: 10.1073/pnas.95.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logothetis D E, Zhang H. J Physiol. 1999;520:630. doi: 10.1111/j.1469-7793.1999.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leaney J L, Milligan G, Tinker A. J Biol Chem. 2000;275:921–929. doi: 10.1074/jbc.275.2.921. [DOI] [PubMed] [Google Scholar]
- 31.Downes G B, Gautam N. Genomics. 1999;62:544–552. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- 32.Guiramand J, Montmayeur J-P, Ceraline J, Bhatia M, Borrelli E. J Biol Chem. 1995;270:7354–7358. doi: 10.1074/jbc.270.13.7354. [DOI] [PubMed] [Google Scholar]
- 33.Ghahremani M H, Cheng P, Lembo P M C, Albert P R. J Biol Chem. 1999;274:9238–9245. doi: 10.1074/jbc.274.14.9238. [DOI] [PubMed] [Google Scholar]
- 34.Senogles S E. J Biol Chem. 1994;269:23120–23127. [PubMed] [Google Scholar]
- 35.Grunewald S, Reilander H, Michel H. Biochemistry. 1996;35:15162–15173. doi: 10.1021/bi960757w. [DOI] [PubMed] [Google Scholar]
- 36.Schreibmayer W, Dessauer C W, Vorobiov D, Gilman A G, Lester H A, Davidson N, Dascal N. Nature (London) 1996;380:624–627. doi: 10.1038/380624a0. [DOI] [PubMed] [Google Scholar]
- 37.Bahia D S, Wise A, Fanelli F, Lee M, Rees S, Milligan G. Biochemistry. 1998;37:11555–11562. doi: 10.1021/bi980284o. [DOI] [PubMed] [Google Scholar]
- 38.Rees S, Coote J, Stables J, Goodson S, Harris S, Lee M G. Biotechniques. 1996;20:102–110. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- 39.Hunt T W, Carroll R C, Peralta E G. J Biol Chem. 1994;269:29565–29570. [PubMed] [Google Scholar]
- 40.Wise A, Watson-Koken M-A, Rees S, Lee M, Milligan G. Biochem J. 1997;321:721–728. doi: 10.1042/bj3210721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wise A, Sheehan M, Rees S, Lee M, Milligan G. Biochemistry. 1999;38:2272–2278. doi: 10.1021/bi982054f. [DOI] [PubMed] [Google Scholar]
- 42.Kuner R, Kohr G, Grunewald S, Eisenhardt G, Bach A, Kornau H-C. Science. 1999;283:74–77. doi: 10.1126/science.283.5398.74. [DOI] [PubMed] [Google Scholar]
- 43.Jones K A, Borowsky B, Tamm J A, Craig D A, Durkin M M, Dai M, Yao W-J, Johnson M, Gunwaldsen C, Huang L-Y, et al. Nature (London) 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 44.White J A, Wise A, Main M J, Green A, Fraser N J, Disney G H, Barnes A A, Emson P, Foord S M, Marshall F H. Nature (London) 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 45.Kaupmann K, Malitschek B, Schuler V, Heid J, Froesti W, Beck P, Mosbacher J, Bischoff S, Kulik A, Shigemoto R, et al. Nature (London) 1998;396:683–687. doi: 10.1038/25360. [DOI] [PubMed] [Google Scholar]
- 46.Kaupmann K, Schuler V, Mosbacher J, Bischoff S, Bittiger H, Heid J, Froestl W, Leonhard S, Pfaff T, Karschin A. Proc Natl Acad Sci USA. 1998;95:14991–14996. doi: 10.1073/pnas.95.25.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfaff T, Malitschek B, Kaupmann K, Prezeau L, Pin J P, Bettler B, Karschin A. Eur J Neurosci. 1999;11:2874–2882. doi: 10.1046/j.1460-9568.1999.00704.x. [DOI] [PubMed] [Google Scholar]
- 48.Jockers R, Linder M E, Hohenegger M, Nanoff C, Bertin B, Strosberg A D, Marullo S, Freissmuth M. J Biol Chem. 1994;269:32077–32084. [PubMed] [Google Scholar]
- 49.Migeon J C, Nathanson N M. J Biol Chem. 1994;269:9767–9773. [PubMed] [Google Scholar]
- 50.O'Hara P J, Sheppard P O, Thögersen H, Venezia D, Haldeman B A, McGrane V, Houamed K M, Thomsen C, Gilbert T L, Mulvihill E R. Neuron. 1993;11:41–52. doi: 10.1016/0896-6273(93)90269-w. [DOI] [PubMed] [Google Scholar]
- 51.Galvez T, Parmentier M-L, Joly C, Malitschek B, Kaupmann K, Kuhn R, Bittiger H, Froestl W, Bettler B, Pin J P. J Biol Chem. 1999;274:13362–13369. doi: 10.1074/jbc.274.19.13362. [DOI] [PubMed] [Google Scholar]
- 52.Malitschek B, Schweizer C, Keir M, Heid J, Froestl W, Mosbacher J, Kuhn R, Henley J, Joly C, Pin J P, et al. Mol Pharmacol. 1999;56:448–454. doi: 10.1124/mol.56.2.448. [DOI] [PubMed] [Google Scholar]
- 53.Parmentier M-L, Joly C, Restituito S, Bockaert J, Grau Y, Pin J P. Mol Pharmacol. 1998;53:778–786. [PubMed] [Google Scholar]
- 54.Hawrot E, Xiao Y, Shi Q L, Norman D, Kirkitadze M, Barlow P N. FEBS Lett. 1998;432:103–108. doi: 10.1016/s0014-5793(98)00794-7. [DOI] [PubMed] [Google Scholar]
- 55.Gomeza J, Joly C, Kuhn R, Knöpfel T, Bockaert J, Pin J P. J Biol Chem. 1996;271:2199–2205. doi: 10.1074/jbc.271.4.2199. [DOI] [PubMed] [Google Scholar]
- 56.Francesconi A, Duvoisin R M. J Biol Chem. 1998;273:5615–5624. doi: 10.1074/jbc.273.10.5615. [DOI] [PubMed] [Google Scholar]
- 57.Pin J P, Joly C, Heinemann S H, Bockaert J. EMBO J. 1994;13:342–348. doi: 10.1002/j.1460-2075.1994.tb06267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaupmann K, Huggel K, Heid J, Flor P J, Bischoff S, Mickel S J, McMaster G, Angst C, Bittiger H, Froestl W, Bettler B. Nature (London) 1997;386:239–246. doi: 10.1038/386239a0. [DOI] [PubMed] [Google Scholar]
- 59.Milligan G. Br J Pharmacol. 1999;128:501–510. doi: 10.1038/sj.bjp.0702824. [DOI] [PMC free article] [PubMed] [Google Scholar]