Abstract
Background
Persistent stimulation of cardiac β1-adrenergic receptors by endogenous norepinephrine promotes heart failure progression. Polymorphisms of this gene are known to alter receptor function or expression, as are polymorphisms of the α2C-adrenergic receptor, which regulates norepinephrine release from cardiac presynaptic nerves. The purpose of this study was to investigate possible synergistic effects of polymorphisms of these two intronless genes (ADRB1 and ADRA2C, respectively) on the risk of death/transplant in heart failure patients.
Methods
Sixteen sequence variations in ADRA2C and 17 sequence variations in ADRB1 were genotyped in a longitudinal study of 655 white heart failure patients. Eleven sequence variations in each gene were polymorphic in the heart failure cohort. Cox proportional hazards modeling was used to identify polymorphisms and potential intra- or intergenic interactions that influenced risk of death or cardiac transplant. A leave-one-out cross-validation method was utilized for internal validation.
Results
Three polymorphisms in ADRA2C and five polymorphisms in ADRB1 were involved in eight cross-validated epistatic interactions identifying several two-locus genotype classes with significant relative risks ranging from 3.02 to 9.23. There was no evidence of intragenic epistasis. Combining high risk genotype classes across epistatic pairs to take into account linkage disequilibrium, the relative risk of death or transplant was 3.35 (1.82, 6.18) relative to all other genotype classes.
Conclusion
Multiple polymorphisms act synergistically between the ADRA2C and ADRB1 genes to increase risk of death or cardiac transplant in heart failure patients.
Background
Congestive heart failure can be caused by a wide array of myocardial insults. Although this etiological heterogeneity has yet to be well characterized, the role of neurohormonal pathways in the progression of heart failure is well established [1-3]. Prejunctional α2-adrenergic receptors (α2A and α2C) regulate the release of norepinephrine from cardiac sympathetic nerves in a negative feedback manner. When these receptors are ablated in mouse models, uncontrolled norepinephrine release causes a lethal cardiomyopathy [4,5]. Moreover, the released norepinephrine activates β1-adrenergic receptors (β1AR) expressed on cardiac myocytes which are coupled to stimulatory G-proteins that propagate signals to downstream effectors such as adenylyl cyclase, ion channels, and phospholipases [6]. Prolonged activation of cardiomyocyte β1AR signaling by any number of pharmacologic or genetic means typically results in hypertrophy, ventricular dysfunction, ventricular remodeling, or frank failure [7]. Thus, sympathetic activation of the heart is coordinated in part by these two adrenergic receptors, in which genetic variation of expression or function could act to modify the progression of heart failure. And indeed, substantial variation in the progression, mortality, and treatment response of heart failure between otherwise similar individuals is well recognized [8-10].
Previous studies of a common single nucleotide polymorphism C->G in the β1AR gene (ADRB1) at codon 389, which results in an arginine for glycine substitution (Arg389Gly), have shown enhanced coupling of the Arg variant to Gs in recombinant cells [11]. Arg389 is associated with differential exercise capacity in heart failure patients [12], response to beta-blockers [13,14], hypertension [15], and risk of myocardial infarction [16]. In transgenic mice, the Arg variant has been associated with early enhanced cardiac function but in older mice a predisposition to heart failure [17].
A common insertion/deletion polymorphism in African-Americans resulting in a consecutive four amino acid loss (322–325) in the α2CAR gene (ADRA2C) has been found to reduce receptor function [18], leading to a loss of normal synaptic autoinhibitory feedback and concomitant enhanced release of norepinephrine [19]. Given the potential synergistic effects of these polymorphisms in the ADRB1 and ADRA2C genes, two previous studies have investigated and identified epistatic interactions affecting heart failure risk [20] and response to β-blockers in heart failure patients [21]. In both studies, only two putative functional polymorphisms (i.e., ADRB1 Arg389Gly and ADRA2C ins/del 322) were examined. Multiple additional polymorphisms in both of these two intronless genes have been recently identified and characterized in whole-gene transfection studies, some of which alter protein expression [22,23]. Here we examine whether there is evidence for more complex intragenic and intergenic epistatic effects on heart failure phenotypes and survival in these genes utilizing 16 DNA sequence variations in the ADRA2C and 17 DNA sequence variations in the ADRB1 genes.
Intergenic epistasis, or interaction between two genes, occurs when the phenotypic effects of a variation in one gene is affected by a variation in a second gene. This could result from conformational changes that prevent physical interaction between the two gene products, or from a change in the ability of one gene to regulate the expression of the other or, the pathologic pathway involves both genes in a manner that their downstream effects converge to alter critical events. Intragenic epistasis occurs when a variation in one location of a gene influences the phenotype differently depending on other variations within the same gene. This type of epistasis is most notable when two different amino acid substitutions within a gene result in differentially functioning the resulting protein. However, it is also seen to have an effect on gene expression and processing.
Methods
Study Population
The 655 Caucasian heart failure patients were identified and enrolled in the study at the University of Cincinnati, Cincinnati, OH, between 1999 and 2004. The study was restricted to Caucasians because of the significant differences in the allele frequencies between those of African- and European-descent in these two genes [22,23], and a smaller number of potential black enrollees. Other enrollment criteria were: age of 18 to 80 years, left ventricular ejection fraction (LVEF) of less than 40%, and New York Heart Association heart failure class II-IV. The primary study endpoint was the combined event of death or cardiac transplantation. Descriptive statistics of this cohort are given in Table 1. The human study protocols were approved by the institutional review board of the University of Cincinnati, and subjects provided written informed consent.
Table 1.
Descriptive statistics for the heart failure cohort
| Variable | N | Mean ± SD |
| Age at onset of heart failure (yrs) | 655 | 53.79 ± 12.73 |
| Follow-up time (yrs) | 655 | 3.16 ± 2.70 |
| Height (cm) | 554 | 172.2 ± 10.06 |
| Weight (kg) | 560 | 85.82 ± 20.73 |
| Left Ventricular Ejection Fraction | 415 | 27.72 ± 13.72 |
| Left Ventricular Mass indexed to Body Surface Area | 456 | 189.14 ± 69.04 |
| Fractional Shortening | 492 | 21.97 ± 11.16 |
| Variable | N | % |
| Males | 453 | 69.2 |
| Hypertension | 310 | 47.9 |
| Beta Blocker Use | 455 | 69.6 |
| ACE Inhibitor Use | 550 | 84.0 |
| Had Endpoint: | ||
| Death | 127 | 19.4 |
| Cardiac Transplant | 171 | 26.1 |
| Heart Failure Etiology: | ||
| Dilated Cardiomyopathy | 382 | 58.3 |
| Ischemic Cardiomyopathy | 259 | 39.5 |
| Other | 14 | 2.1 |
Genotyping
Sequence variants have been previously identified for ADRA2C [22] and ADRB1 [23] and the nomenclature used in these papers is retained in the current work to maintain consistency. Both genes are intronless, and the A of the initiation codon is denoted as +1, proceeding in the positive direction 5' -3' through the coding and 3' UTR. In the 5' -flanking region the most 3' non-coding nucleotide prior to the ATG is denoted as -1 and the numbering proceeds in the negative direction. The genotyping was performed on the variant sites shown in Table 2. For orientation purposes +1 of ADRA2C is nucleotide 2638 of AY605898, and +1 of ADRB1 is nucleotide 36085 of AL355543. The variants are deposited in the PharmGKB (ADRA2C) and SeattleSNPs (ADRB1) public databases (http://www.pharmgkb.org/ and http://pga.gs.washington.edu/, respectively). Genotyping was performed on genomic DNA derived from blood samples, by sequencing PCR products spanning one or two variant positions, using an ABI 3730 sequencer. Variants detected by alignment to a reference were verified by visual electropherogram examination.
Table 2.
Summary of the SNP frequency distributions
| SNP | Genotype (N) | Minor Allele | Minor Allele Freq | H-W P-value |
| ADRA2C(-2579 T/C) | 568 | C | 0.013 | 0.000 |
| ADRA2C(-2416 C/G) | 613 | G | 0.001 | 1.000 |
| ADRA2C(-2357 C/T) | 615 | T | 0.001 | 1.000 |
| ADRA2C(-2280 G/T) | 612 | T | 0.002 | 1.000 |
| ADRA2C(-2069 C/T) | 602 | T | 0.111 | 0.151 |
| ADRA2C(-1926 G/A) | 608 | A | 0.031 | 1.000 |
| ADRA2C(-1692 T/G) | 624 | G | 0.069 | 0.066 |
| ADRA2C(-1513 T/G) | 626 | G | 0.018 | 1.000 |
| ADRA2C(-965 G/C) | 634 | C | 0.001 | 1.000 |
| ADRA2C(-940 G/A) | 634 | A | 0.136 | 0.615 |
| ADRA2C(-933 C/A) | 634 | A | 0.028 | 1.000 |
| ADRA2C(-696 C/G) | 426 | G | 0.058 | 0.640 |
| ADRA2C(-241 C/G) | 644 | G | 0.001 | 1.000 |
| ADRA2C(-230 T/C) | 644 | C | 0.019 | 1.000 |
| ADRA2C(+964 ins/del) | 587 | del | 0.066 | 0.010 |
| ADRA2C(+1736 G/C) | 651 | C | 0.235 | 0.387 |
| ADRB1(-4415 T/C) | 572 | C | 0.287 | 0.477 |
| ADRB1(-4267 ins/del) | 573 | ins | 0.001 | 1.000 |
| ADRB1(-3641 C/T) | 593 | T | 0.146 | 0.073 |
| ADRB1(-3598 C/T) | 593 | T | 0.066 | 1.000 |
| ADRB1(-3255 A/C) | 608 | C | 0.141 | 0.243 |
| ADRB1(-2915 G/A) | 643 | A | 0.001 | 1.000 |
| ADRB1(-2853 G/A) | 644 | A | 0.001 | 1.000 |
| ADRB1(-2827 C/A) | 644 | A | 0.030 | 0.447 |
| ADRB1(-2639 T/C) | 637 | C | 0.433 | 0.568 |
| ADRB1(-2297 T/G) | 607 | G | 0.151 | 0.082 |
| ADRB1(-2142 T/C) | 608 | C | 0.146 | 0.250 |
| ADRB1(-1294 G/A) | 579 | A | 0.003 | 1.000 |
| ADRB1(-1121 T/C) | 580 | C | 0.003 | 1.000 |
| ADRB1(-517 T/C) | 627 | C | 0.131 | 0.015 |
| ADRB1(+145 A/G) | 584 | G | 0.146 | 0.018 |
| ADRB1(+315 G/T) | 587 | T | 0.002 | 1.000 |
| ADRB1(+1165 G/C) | 557 | G | 0.268 | 0.665 |
Statistical Methods
Allele frequencies were estimated using standard gene counting methods. Hardy-Weinberg disequilibrium was tested using Weir's method [24]. Linkage disequilibrium was assessed using the r2 statistic [24]. Cox Proportional Hazards modeling [25]. was used to test for significant effects for each polymorphism (separately) and pairwise epistatic effects after adjustment for age at initial diagnosis, β-blocker usage, hypertension status, and sex. Genotypes for each polymorphism were coded using two dummy variables where the most frequent genotype was considered the reference group. Epistasis was assessed by including interaction terms into the Cox models.
To adjust for multiple testing we used Storey's modification of the false discovery rate (FDR) method of Benjamini and Hochberg [26] that takes into account the correlation among tests due to linkage disequilibrium. We used an FDR cutpoint of 0.30. To assess whether the information from the Cox proportional hazards modeling of genetic effects provides a useful prediction of a new patient's risk of death or cardiac transplant, we implemented a leave-one-out cross-validation approach [27]. Each individual was sequentially left out and a Cox proportional hazards model for time from study enrollment to death or cardiac transplant was estimated. Using the coefficients estimated with the n-1 individuals, a survival risk was calculated for the individual left out. These risks were then used as the predictor in a new Cox proportional hazards model. Because each individual was omitted from the model used to calculate their survival risk, the performance of a model using these risks as predictors approximates the predictive ability of the association in an independent sample drawn from the same population.
In order to assess the overall impact of high-risk two-locus genotype classes on risk of death or cardiac transplant, a pooling procedure was performed. If an individual possessed one or more two-locus genotype class that had a relative risk whose 95% confidence interval was greater than 1.0, they were assigned the label "has high-risk genotype." All other individuals were assigned the label "does not have high-risk genotype." Cox proportional hazards modeling was then performed with this label as the explanatory variable.
Results
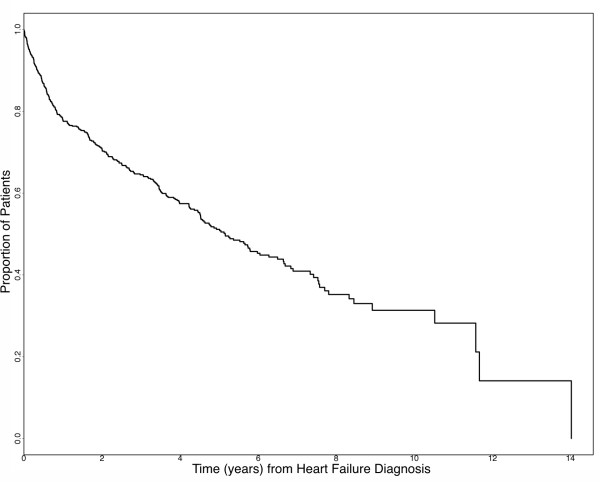
The 655 subject heart failure cohort was 69.2% male, with 47.9% having a history of hypertension, 84.0% receiving ACE inhibitor therapy and 69.6% receiving β-blocker therapy (Table 1). Ischemic cardiomyopathies made up 39.5% of the cohort, idiopathic dilated cariomyopaties comprised 58.3% of the cohort, and the remaining 2.1% of the cohort had other causes of heart failure, such as primary valvular defects. They had an average age of heart failure onset of 53.8 years and an average follow-up time of 3.16 years. Figure 1 shows a Kaplan-Meier curve of time from heart failure diagnosis to death or cardiac transplant for this cohort. The minor alleles and the frequencies of the 16 DNA sequence variations in the ADRA2C and 17 DNA sequence variations in the ADRB1 genes are displayed in Table 2. Eleven of the 16 variants genotyped in the ADRA2C gene and 11 of the 17 in the ADRB1 gene had frequencies greater than 1% and are considered polymorphic. Four polymorphisms, ADRA2C(-2579 T/C), ADRA2C(+964/965 ins/del), ADRB1(-517 T/C), and ADRB1(+145 A/G), were significantly out of Hardy Weinberg equilibrium (HWE) (P < 0.05). The linkage disequilibrium between polymorphisms is illustrated in Figure 2 and indicates that several polymorphisms within each gene have significant frequency correlations.
Figure 1.
Kaplan-Meier curve of time from heart failure diagnosis to death or cardiac transplant.
Figure 2.
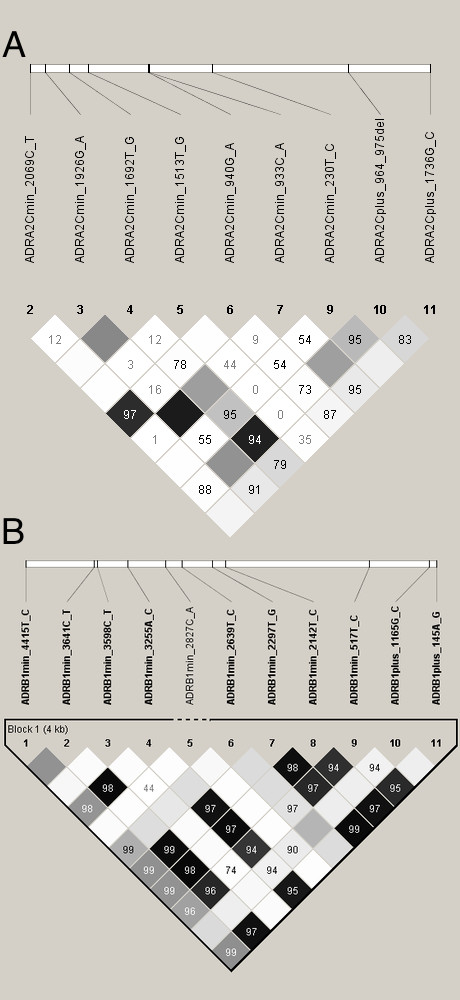
A. Linkage disequilibrium among ADRA2C polymorphisms B. Linkage disequilibrium among ADRB1 polymorphisms.
Given the large number of tests for main effects and epistatic effects using these 22 polymorphic loci we used both the false discovery rate and cross-validation methods to reduce the probability of false positive results. Table 3 provides a summary of the tests of association between the polymorphism and age of onset, LVEF, LV mass, and survival. Overall, only two polymorphisms had significant main effects on the survival distribution and 8 pairs of polymorphisms had epistatic effects on survival that were significant (after adjustment by FDR) and cross-validated.
Table 3.
Summary of the significant, false discovery rate adjusted, and cross-validated ADRA2C and ADRB1 polymorphism effects and interactions on age of onset, LVmass, LVEF, and heart failure survival
| ED (N = 655) | ||||||||
| Age of Onset | LVMass Adj* | LVEF Adj** | Survival | |||||
| SNP Main Effects | SNP-SNP Interaction | SNP Main Effects | SNP_SNP Interaction | SNP Main Effects | SNP_SNP Interaction | SNP Main Effects | SNP_SNP Interaction | |
| Number of tests | 22 | 209 | 22 | 200 | 22 | 204 | 22 | 231 |
| P < 0.10 | 2 | 24 | 1 | 8 | 2 | 16 | 4 | 50 |
| FDR (<0.30) | 0 | 0 | 0 | 0 | 2 | 0 | 4 | 44 |
| Cross Validation*** | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 10 |
| FDR and Cross-validation | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 6 |
* LVMass was adjusted for age of onset, sex, height, weight, hypertension
** LVEF was adjusted for age of onset, sex
*** For survival analysis, Risk Index was used to cross-validate, P-value for Cox model with Risk Index <0.1 is considered as crossvalidated. For continuous outcomes, 4-fold CV was used. Press-Rsquared >0.005 (SNP univ model) or Press-Rsquared for full model >0 and Press-Rsquared Difference between full model and submodel >0.005 (Epistasis) is considered to be crossvalidated for interaction.
The relative risks (RR) associated with these polymorphisms are listed in Table 4. The two polymorphisms with single locus effects were ADRA2C(-1692 T/G) and ADRA2C(+964/965 ins/del). However, both of these polymorphisms were involved in significant epistatic interactions making the interpretation of the relative risks of their main effects impossible. Even though we equally tested for interactions within and among the two genes, somewhat surprisingly, we only found evidence of intergenic epistasis. The interaction between ADRA2C(+964/965 ins/del) and ADRB1(+145 A/G) arises from the 10:AG genotype class having elevated risk of death or transplant with RR = 5.12 (2.66,9.88). With respect to the interaction between ADRA2C(-230 T/C) and ADRB1(-517 T/C), the TC:CT genotype class has much higher risk of death or transplant with RR = 8.31 (3.32, 20.75) compared to the reference genotype class of TT:TT. The interaction between ADRA2C(-230 T/C) and ADRB1(-2297 T/G) appears to be due to the TC:TG genotype class (RR = 9.23 (3.67,23.16)), and the interaction between ADRA2C(-1692 T/G) and ADRB1(+145 A/G) is associated with an increased risk of death or transplant in the TG:AG genotype class with RR = 4.59 (2.31, 9.12). Likewise, the interaction between ADRA2C(-1692 T/G) and ADRB1(-3641 C/T) identifies the TG:CT genotype as the high risk group with RR = 3.56 (1.80, 7.05). Finally, the ADRA2C(-1692 T/G) and ADRB1(-3255 A/C) polymorphism appear to be interacting and again the TG genotype of the ADRA2C(-1692 T/G) gene has a significant increase risk in a particular ADRB1 genotype background – namely, AC genotype – such that the TG:AC genotype class has a relative risk of 3.02(1.53,5.97).
Table 4.
FDR Significant and Cross-Validated Epistatic Effects of ADRA2C and ADRB1 polymorphisms on survival
| SNP1 | SNP2 | Genotype1 | Genotype2 | Relative Risk | N | Model P-value | Cross-Validation P-value |
| Main Effects | |||||||
| ADRA2C(-1692 T/G) | TT | 1 | 544 | 0.029 | 0.031 | ||
| TG | 1.34(0.91, 1.95) | 74 | |||||
| GG | n/a | 6 | |||||
| ADRA2C(+964 ins/del) | 11 | 1 | 516 | 0.065 | 0.018 | ||
| 10 | 0.51(0.07, 3.64) | 65 | |||||
| 00 | 1.58(1.07, 2.32)* | 6 | |||||
| Epistatic Effects | |||||||
| ADRA2C(+964 ins/del) | ADRB1(+145 A/G) | 00 | AA | n/a | 5 | <0.001 | 0.069 |
| 10 | AA | 1.20 (0.74,1.95) | 39 | ||||
| 10 | AG | 5.12 (2.66, 9.88)* | 17 | ||||
| 11 | AA | 1 | 349 | ||||
| 11 | AG | 0.86 (0.59, 1.25) | 108 | ||||
| 11 | GG | 0.70 (0.31, 1.61) | 15 | ||||
| ADRA2C(-230 T/C) | ADRB1(-517 T/C) | TC | CT | 8.31(3.32, 20.75)* | 7 | <0.001 | <0.001 |
| TC | TT | 0.46(0.15, 1.42) | 16 | ||||
| TT | CC | 0.51(0.21,1.24) | 17 | ||||
| TT | CT | 0.88 (0.62,1.26) | 121 | ||||
| TT | TT | 1 | 455 | ||||
| ADRA2C(-230 T/C) | ADRB1(-2297 T/G) | TC | TG | 9.23(3.67,23.16)* | 7 | 0.001 | 0.014 |
| TC | TT | 0.61(0.22, 1.64) | 16 | ||||
| TT | GG | 0.61(0.27,1.38) | 18 | ||||
| TT | TG | 0.99 (0.71,1.36) | 137 | ||||
| TT | TT | 1 | 422 | ||||
| ADRA2C(-1692 T/G) | ADRB1(+145 A/G) | GG | AA | n/a | 5 | 0.001 | 0.097 |
| TG | AA | 0.99 (0.62,1.61) | 44 | ||||
| TG | AG | 4.59 (2.31,9.12)* | 17 | ||||
| TG | GG | n/a | 2 | ||||
| TT | AA | 1 | 371 | ||||
| TT | AG | 0.85(0.59, 1.22) | 111 | ||||
| TT | GG | 0.61(0.27, 1.38) | 17 | ||||
| ADRA2C(-1692 T/G) | ADRB1(-3641 C/T) | GG | CC | n/a | 6 | 0.005 | <0.001 |
| TG | CC | 0.95 (0.60, 1.53) | 52 | ||||
| TG | CT | 3.56 (1.80,7.05)* | 18 | ||||
| TT | CC | 1 | 369 | ||||
| TT | CT | 0.84 (0.58,1.21) | 116 | ||||
| TT | TT | 0.62 (0.27, 1.42) | 17 | ||||
| ADRA2C(-1692 T/G) | ADRB1(-3255 A/C) | GG | AA | n/a | 6 | 0.010 | <0.001 |
| TG | AA | 1.08 (0.68,1.71) | 52 | ||||
| TG | AC | 3.02(1.53,5.97)* | 19 | ||||
| TT | AA | 1 | 379 | ||||
| TT | AC | 0.83(0.58,1.21) | 116 | ||||
| TT | CC | 0.73 (0.32,1.66) | 16 |
* RR is significant at α < 0.05
Given some degree of non-independence among these epistatic effects, we performed an a posteriori pooling of all high risk two locus genotype classes with RR's whose 95% confidence interval exceed 1.0 to attempt to identify the extent of overlap between the effects from the pairs of loci. These were compared to all other genotype classes combined, which provides a way to interpret these very specific sets of polymorphisms as if being used clinically. Here we found that the combined relative risk was 3.35 (1.82, 6.18).
Discussion
In this study, we identified multiple polymorphisms within the ADRA2C and ADRB1 genes that interact to increase risk of death or transplant in heart failure patients. We investigated these genes because numerous studies in human patient populations [20,12,14], animal models [17], and in vitro cell systems [18,11,22,23] have demonstrated their relevance to heart failure phenotypes or receptor expression/function.
As noted in the Results, four of the polymorphism significantly deviated from Hardy-Weinberg equilibrium. However, a previous study that genotyped the ADRA2C polymorphisms in a cohort of unaffected individuals did not show evidence of deviations from HWE [22]. Similarly, genotyping of the ADRB1 polymorphisms in unaffected individuals for the Seattle SNPs database http://pga.gs.washington.edu/ do not show significant deviations from HWE. The HW deviations we observed could be due to an underlying association with heart failure [28] since our sample is exclusively heart failure patients and these genes have been previously associated with this disease, but random chance and genotyping error cannot be excluded.
The α2ARs expressed on the presynaptic cardiac sympathetic nerves inhibit the release of norepinephrine when they are bound by the neurotransmitter [5], and thus provide a mechanism to regulate release of the neurotransmitter as sympathetic nervous system activity markedly increases in progressive heart failure. And indeed, in studies of gene-targeted mice where α2ARs have been ablated, severe cardiomyopathy results due to norepinephrine cardiotoxicity exerted through myocyte β1ARs [5]. The human ADRA2C ins/del polymorphism previously denoted Del322-325, and here denoted ADRA2C(+964/965 ins/del) significantly reduces the function of α2CAR receptors in transfected cells [18]. While Caucasians have a very low prevalence of this allele [20] it appears that other common polymorphisms within this racial group, in the promoter and 3' UTR region of the gene, affect receptor expression [22]. Of note, evidence suggests that there are few "spare receptors" in the complement of α2CARs expressed in cardiac presynaptic nerves since heterozygous α2CAR knock-out mice (~50% less receptor) also develop cardiomyopathy under pressure overload [29]. Thus relatively small changes in expression due to polymorphisms may have physiologically relevant effects on the heart during heart failure progression. For the β1AR, the most frequently studied polymorphic variation is at nucleotide 1165, where Arg or Gly can be commonly found at amino acid 389. This lies within a G-protein coupling domain, and in transfected cells the Arg variant exhibits enhanced coupling to adenylyl cyclase [11]. In transgenic mice, with matched cardiac expression of the human β1AR Arg or Gly389 receptors, Arg hearts have enhanced contractility, but progress to failure by 9-months of age, while Gly hearts show less contractile enhancement and no pathologic effects [17]. The Arg389 phenotype also revealed a gene dose-response, and given that promoter SNPs of the ADRB1 gene alter expression in cell-based systems [23], the potential for cardiac relevance of non-coding β1AR SNPs is apparent.
The previous studies of interactions between one ADRA2C and one ADRB1 polymorphism illustrate that the physiological effects of variations in these two genes act synergistically to increase risk of heart failure [20] and influences patient responsiveness to beta-blocker therapy as assessed by short-term improvements in LVEF [21]. Our study indicates that there may be several other polymorphisms within these genes that have important physiological and clinical consequences for heart failure patients. Specifically, the ADRA2C(-1692 T/G) was involved in three epistatic interactions with ADRB1 polymorphisms (+145 A/G, -3641 C/T, and -3255 A/C) and the ADRA2C(-230 T/C) polymorphism was involved in two epistatic interactions with ADRB1 polymorphisms (-2297 T/G and -517 T/C). Only one ADRB1 polymorphism (+145 A/G) was involved in more than one epistatic interaction with the ADRA2C polymorphisms (-1692 T/G and +964/965 ins/del). In total, three polymorphisms in the ADRA2C gene and five polymorphisms in the ADRB1 gene appear to be implicated in epistatic effects on heart failure survival.
Because of the relatively large number of polymorphisms investigated in each gene one natural analytical approach might have been to investigate haplotypes within each gene and to test for potential interaction between haplotypes in their influence on risk. However, one of the main drawbacks of such an approach is reducing the haplotype space to the relevant set with phenotypic effects that could potentially interact within or across genes. Since we did not find any evidence of significant intragenic epistasis, it becomes even more difficult to analyze the diverse set of haplotypes – many with low frequency – to identify potentially relevant interactions across genes.
One of the short-comings of genetic association studies is that they have often failed to replicate and Manly [30] suggests that internal validation, common to good experimental practices, is one way to avoid the publication of spurious findings. In our study, we used cross-validation methods to significantly reduce the chance of false positives. Cross-validation methods were developed as a way to incorporate a measure of predictive accuracy (and correspondingly, a measure of prediction error) for an estimated model based on its performance predicting the outcome for independent test cases [31]. During the last decade, cross-validation methods have been used widely for everything from robust variable selection in gene expression array studies [32] to reducing false positives in gene-gene interaction studies [33,34] to evaluating the predictive accuracy of molecular or genetic classifiers of disease before clinical implementation [35]. It has become a standard in the field of metabolomic [36], proteomic [37,38], and transcriptomic [39] studies because of its ease of execution and its emphasis on prediction in independent test cases as a method of discriminating between true associations and false associations.
The epistatic interactions identified here occur between -1692, -230, and the +964/965 polymorphisms of ADRA2C and -3641, -3255, -2297, -517 and +145 polymorphisms in ADRAB1. We have previously generated constructs that mimic these polymorphisms and by transient transfection of cells ascertained expression phenotypes (5' and 3' -flanking regions) or signaling phenotypes (nonsynonymous coding polymorphisms) [11,22,23,18]. All but one of the ADRA2C 5' promoter and 5' UTR polymorphisms found here that interact with ADRAB1 have been found to influence α2CAR expression in these model systems. (The ADRA2C(-1692) polymorphism has not been studied in this manner.) And, the +964/965 deletion polymorphism results in depressed agonist-promoted function [18]. For the ADRAB1, the aforementioned polymorphisms of the promoter region (except for -1294 which has not been studied) have also been shown to alter β1AR expression [23]. The nonsynonymous polymorphism at nucleotide position +145 (representing Gly49) results in a β1AR that undergoes enhanced agonist-promoted downregulation compared to the Ser49 receptor [40], an important phenotype since downregulation is a protective mechanism in heart failure. Taken together, then, there is biologic plausibility in the epistatic interactions that were observed. The fact that there is a relatively large fraction of the patients that do not have these specific combinations, and yet they do display variability in heart failure progression, indicates additional genetic (and likely non-genetic) causes for heterogeneity. Within the axis which we are currently exploring, there are a number of other genes to be considered. As additional information from fine mapping and cell-based studies is obtained, there is the possibility for enhanced predictive power and an assignment of risk alleles for a greater percentage of heart failure patients. Nevertheless, the current work shows that epistatic interactions between α2CAR and β1AR polymorphisms affect heart failure survival, and further confirm the notion that this complex syndrome is modified by multiple polymorphisms in multiple genes.
Conclusion
Although we did not observe intragenic epistatic interactions between the ADRA2C and ADRB1 genes, we did observe multiple polymorphisms acting synergistically between the ADRA2C and ADRB1 genes to increase risk of death or cardiac transplant in heart failure patients. This underscores the complexity of the genetic factors that affect the progression of this syndrome.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SLRK performed statistical analysis and drafted the manuscript. RK performed statistical analysis and assisted in drafting the manuscript. MK, BA, and GG developed genotyping methods for the ADRA2C and ADRB1 polymorphisms and performed the genotyping of the subjects. HH and LW recruited subjects and interpreted echocardiogram results. KC managed the subject clinical and endpoint data. GWD designed the study and assisted in drafting the manuscript. SL identified the ADRA2C and ADRB1 polymorphisms and their haplotypes, developed genotyping methods, participated in the design of the study and interpretation of results, and assisted in drafting the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
The present study was funded by NHLBI P50 HL77101, Genetic and Molecular Signaling in Heart Failure.
Contributor Information
Sharon LR Kardia, Email: skardia@umich.edu.
Reagan J Kelly, Email: reagank@umich.edu.
Mehdi A Keddache, Email: mehdi.keddache@cchmc.org.
Bruce J Aronow, Email: bruce.aronow@cchmc.org.
Gregory A Grabowski, Email: greg.grabowski@cchmc.org.
Harvey S Hahn, Email: hahnhs@ucmail.uc.edu.
Karen L Case, Email: casekl@ucmail.uc.edu.
Lynne E Wagoner, Email: LWagoner@cinci.rr.com.
Gerald W Dorn, II, Email: gdorn@im.wustl.edu.
Stephen B Liggett, Email: sligg001@umaryland.edu.
References
- Bristow MR. Why does the myocardium fail? Insights from basic science. Lancet. 1998;352:SI8–14. doi: 10.1016/S0140-6736(98)90311-7. [DOI] [PubMed] [Google Scholar]
- Bristow MR. Mechanistic and clinical rationales for using beta-blockers in heart failure. J Card Fail. 2000;6:8–14. doi: 10.1054/jcaf.2000.9501. [DOI] [PubMed] [Google Scholar]
- Liggett SB. Beta-adrenergic receptors in the failing heart: the good, the bad, and the unknown. J Clin Invest. 2001;107:947–948. doi: 10.1172/JCI12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A, Esler MD, Socratous F, Kaye DM. Evidence for functional presynaptic alpha-2 adrenoceptors and their down-regulation in human heart failure. J Am Coll Cardiol. 2001;37:1246–1251. doi: 10.1016/S0735-1097(01)01121-4. [DOI] [PubMed] [Google Scholar]
- Hein L, Altman JD, Kobilka BK. Two functionally distinct alpha2-adrenergic receptors regulate sympathetic neurotransmission. Nature. 1999;402:181–184. doi: 10.1038/46040. [DOI] [PubMed] [Google Scholar]
- Bristow MR, Hershberger RE, Port JD, Minobe W, Rasmussen R. Beta 1- and beta 2-adrenergic receptor-mediated adenylate cyclase stimulation in nonfailing and failing human ventricular myocardium. Mol Pharmacol. 1989;35:295–303. [PubMed] [Google Scholar]
- Dorn GW, II, Molkentin JD. Manipulating cardiac contractility in heart failure: Data from mice and men. Circulation. 2004;109:150–158. doi: 10.1161/01.CIR.0000111581.15521.F5. [DOI] [PubMed] [Google Scholar]
- van Campen LC, Visser FC, Visser CA. Ejection fraction improvement by beta-blocker treatment in patients with heart failure: An analysis of studies published in the literature. Cardiovasc Pharmacol. 1998;32:S31–S35. doi: 10.1097/00005344-199800003-00006. [DOI] [PubMed] [Google Scholar]
- Poole-Wilson PA, Swedberg K, Cleland JG, Di Lenarda A, Hanrath P, Komajda M, Lubsen J, Lutiger B, Metra M, Remme WJ, Torp-Pedersen C, Scherhag A, Skene A, Carvedilol Or Metoprolol European Trial Investigators Comparison of carvedilol and metoprolol on clinical outcomes in patients with chronic heart failure in the Carvedilol Or Metoprolol European Trial (COMET): Randomised controlled trial. Lancet. 2003;362:7–13. doi: 10.1016/S0140-6736(03)13800-7. [DOI] [PubMed] [Google Scholar]
- Packer M, Fowler MB, Roecker EB, Coats AJS, Katus HA, Krum H, Mohacsi P, Rouleau JL, Tendera M, Staiger C, Holcslaw TL, Amann-Zalan I, DeMets DL, for the Carvedilol Prospective Randomized Cumulative Survival (COPERNICUS) Study Group Effect of carvedilol on the morbidity of patients with severe chronic heart failure: results of the carvedilol prospective randomized cumulative survival (COPERNICUS) study. Circulation. 2002;106:2194–2199. doi: 10.1161/01.CIR.0000035653.72855.BF. [DOI] [PubMed] [Google Scholar]
- Mason DA, Moore JD, Green SA, Liggett SB. A gain-of-function polymorphism in a G-protein coupling domain of the human beta1-adrenergic receptor. J Biol Chem. 1999;274:12670–12674. doi: 10.1074/jbc.274.18.12670. [DOI] [PubMed] [Google Scholar]
- Wagoner LE, Craft LL, Zengel P, McGuire N, Rathz DA, Dorn GW, Liggett SB. Polymorphisms of the beta1-adrenergic receptor predict exercise capacity in heart failure. Am Heart J. 2002;144:840–846. doi: 10.1067/mhj.2002.125325. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74:44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Liggett SB, Mialet-Perez J, Thaneemit-Chen S, Wever SA, Greene SM, Hodne D, Nelson B, Morrison J, Domanski MJ, Wagoner LE, Abraham WT, Anderson JL, Carlquist JF, Krause-Steinrauf HJ, Lazzeroni LC, Port JD, Lavori PW, Bristow MR. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson K, Melander O, Orho-Melander M, Lindblad U, Ranstam J, Rastam L, Groop L. Polymorphism in the beta(1)-adrenergic receptor gene and hypertension. Circulation. 2001;104:187–190. doi: 10.1161/01.cir.104.2.187. [DOI] [PubMed] [Google Scholar]
- Iwai C, Akita H, Kanazawa K, Shiga N, Terashima M, Matsuda Y, Takai E, Miyamoto Y, Shimizu M, Kajiya T, Hayashi T, Yokoyama M. Arg389Gly polymorphism of the human beta1-adrenergic receptor in patients with nonfatal acute myocardial infarction. Am Heart J. 2003;146:106–109. doi: 10.1016/S0002-8703(03)00110-8. [DOI] [PubMed] [Google Scholar]
- Mialet Perez J, Rathz DA, Petrashevskaya NN, Hahn HS, Wagoner LE, Schwartz A, Dorn GW, II, Liggett SB. Beta 1-adrenergic receptor polymorphisms confer differential function and predisposition to heart failure. Nat Med. 2003;9:1300–1305. doi: 10.1038/nm930. [DOI] [PubMed] [Google Scholar]
- Small KM, Forbes SL, Rahman FF, Bridges KM, Liggett SB. A four amino acid deletion polymorphism in the third intracellular loop of the human alpha 2C-adrenergic receptor confers impaired coupling to multiple effectors. J Biol Chem. 2000;275:23059–23064. doi: 10.1074/jbc.M000796200. [DOI] [PubMed] [Google Scholar]
- Neumeister A, Charney DS, Belfer I, Geraci M, Holmes C, Sharabi Y, Alim T, Bonne O, Luckenbaugh DA, Manji H, Goldman D, Goldstein DS. Sympathoneural and adrenomedullary functional effects of alpha2C-adrenoreceptor gene polymorphism in healthy humans. Pharmacogenet Genomics. 2005;15:143–149. doi: 10.1097/01213011-200503000-00002. [DOI] [PubMed] [Google Scholar]
- Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C-adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- Lobmeyer MT, Gong Y, Terra SG, Beitelshees AL, Langaee TY, Pauly DF, Schofield RS, Hamilton KK, Herbert Patterson J, Adams KF, Jr, Hill JA, Aranda JM, Jr, Johnson JA. Synergistic polymorphisms of beta1 and alpha2C-adrenergic receptors and the influence on left ventricular ejection fraction response to beta-blocker therapy in heart failure. Pharmacogenet Genomics. 2007;17:277–282. doi: 10.1097/FPC.0b013e3280105245. [DOI] [PubMed] [Google Scholar]
- Small KM, Mialet-Perez J, Seman CA, Theiss CT, Brown KM, Liggett SB. Polymorphisms of cardiac presynaptic alpha2C adrenergic receptors: Diverse intragenic variability with haplotype-specific functional effects. Proc Natl Acad Sci USA. 2004;101:13020–13025. doi: 10.1073/pnas.0405074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small KM, Mialet-Perez J, Liggett SB. Genetic variation within the β1-adrenergic receptor gene results in haplotype-specific expression phenotypes. J Cardiovasc Pharmacol. 2008;51:106–110. doi: 10.1097/FJC.0b013e31815a958f. [DOI] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II: Methods for discrete population genetic data. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- Parmar M, Machin D. Survival Analysis:A practical approach. Chichester: John Wiley & Sons; 1995. [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genome-wide experiments. Proc Natl Acad Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JMG, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Lee , Wen-Chung Searching for Disease-Susceptibility Loci by Testing for Hardy-Weinberg Disequilibrium in a Gene Bank of Affected Individuals. Am J Epimdeiol. 2003;158:397–400. doi: 10.1093/aje/kwg150. [DOI] [PubMed] [Google Scholar]
- Gilsbach R, Brede M, Beetz N, Moura E, Muthig V, Gerstner C, Barreto F, Neubauer S, Vieira-Coelho MA, Hein L. Heterozygous alpha 2C-adrenoceptor-deficient mice develop heart failure after transverse aortic constriction. Cardiovasc Res. 2007;75:728–737. doi: 10.1016/j.cardiores.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Manly KF. Reliability of statistical associations between genes and disease. Immunogenetics. 2005;57:549–558. doi: 10.1007/s00251-005-0025-x. [DOI] [PubMed] [Google Scholar]
- Stone M. Cross-validatory choice and assessment of statistical predictions. J Royal Statist Soc Series B. 1974;36:111–147. [Google Scholar]
- Zhu J, Hastie T. Classification of gene microarrays by penalized logistic regression. Biostatistics. 2004;5:427–443. doi: 10.1093/biostatistics/kxg046. [DOI] [PubMed] [Google Scholar]
- Ritchie MD, Motsinger AA. Multifactor dimensionality reduction for detecting gene-gene and gene-environment interactions in pharmacogenomics studies. Pharmacogenomics. 2005;6:823–834. doi: 10.2217/14622416.6.8.823. [DOI] [PubMed] [Google Scholar]
- Gong R, Liu Z, Li L. Epistatic effect of plasminogen activator inhibitor 1 and beta-fibrinogen genes on risk of glomerular microthrombosis in lupus nephritis: interaction with environmental/clinical factors. Arthritis Rheum. 2007;56:1608–1617. doi: 10.1002/art.22598. [DOI] [PubMed] [Google Scholar]
- Larsen JE, Pavey SJ, Passmore LH, Bowman RV, Hayward NK, Fong KM. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13:2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- Pohjanen E, Thysell E, Jonsson P, Eklund C, Silfver A, Carlsson IB, Lundgren K, Moritz T, Svensson MB, Antti H. A multivariate screening strategy for investigating metabolic effects of strenuous physical exercise in human serum. J Proteome Res. 2007;6:2113–2120. doi: 10.1021/pr070007g. [DOI] [PubMed] [Google Scholar]
- Agranoff D, Fernandez-Reyes D, Papadopoulos MC, Rojas SA, Herbster M, Loosemore A, Tarelli E, Sheldon J, Schwenk A, Pollok R, Rayner CFJ, Krishna S. Identification of diagnostic markers for tuberculosis by proteomic fingerprinting of serum. Lancet. 2006;368:1012–1021. doi: 10.1016/S0140-6736(06)69342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood IA, Visscher PM, Mengersen KL. Classification based upon gene expression data: Bias and precision of error rates. Bioinformatics. 2007;23:1363–1370. doi: 10.1093/bioinformatics/btm117. [DOI] [PubMed] [Google Scholar]
- Mertens BJ, De Noo ME, Tollenaar RA, Deelder AM. Mass spectrometry proteomic diagnosis: enacting the double cross-validatory paradigm. J Comput Biol. 2006;13:1591–1605. doi: 10.1089/cmb.2006.13.1591. [DOI] [PubMed] [Google Scholar]
- Rathz DA, Brown KM, Kramer LA, Liggett SB. Amino acid 49 polymorphisms of the human beta1-adrenergic receptor affect agonist-promoted trafficking. J Cardiovasc Pharmacol. 2002;39:155–160. doi: 10.1097/00005344-200202000-00001. [DOI] [PubMed] [Google Scholar]