Abstract
The tumor suppressor protein p53 is a potent inducer of apoptosis in transformed cells. Hdm2 is an ubiquitin ligase (E3) that acts as a major regulator of p53 by promoting its ubiquitylation and proteasomal degradation. For this reason, inhibiting the E3 activity of Hdm2 has been proposed as a therapeutic approach for cancers expressing wild type p53. We previously identified a family of small molecules (HLI98s, 7-nitro-10-aryl-5-deazaflavins) that inhibit the E3 activity of Hdm2, increase cellular p53 and selectively kill transformed cells expressing wild type p53. However, issues of both potency and solubility in aqueous solution limit the utility of the HLI98s. Here we report that a highly soluble derivative of the HLI98s, which has a 5-dimethylaminopropylamino side chain but lacks the 10-aryl group (HLI373), has greater potency than the HLI98s in stabilizing Hdm2 and p53, activating p53-dependent transcription and in inducing cell death. Furthermore, we show that HLI373 is effective in inducing apoptosis of several tumor cells lines that are sensitive to DNA-damaging agents. These results suggest that HLI373 could serve as a potential lead for developing cancer therapeutics based on inhibition of the ubiquitin ligase activity of Hdm2.
Keywords: ubiquitin, Hdm2/Mdm2, p53, apoptosis, cancer therapeutics
Introduction
Modification of proteins with ubiquitin is catalyzed by a multi-enzyme cascade consisting of E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase). Through proteasomal degradation, ubiquitin plays a critical role in controlling the fate of numerous proteins, including many critical regulators of cell proliferation, differentiation, and apoptosis. Recognition of ubiquitylated proteins by the proteasome is generally believed to require the formation of polyubiquitin chains, usually linked through Lys 48 of ubiquitin (K48). Modification of target proteins through other ubiquitin linkages or by mono-ubiquitylation generally affects proteins in other ways. Specificity in substrate recognition is primarily determined by the E3, which interacts with E2 and recognizes target proteins. RING finger proteins represent the largest class of E3s. These E3s mediate the transfer of ubiquitin from E2s to bound substrates. In addition to targeting specific substrates, they also often mediate their own ubiquitylation and thereby regulate their own levels (1).
The tumor suppressor p53 is believed to primarily function as a transcription factor to control the expression of a variety of target genes, variably leading to growth arrest, senescence or apoptosis (2-5). Recent studies indicate that p53 can also exert a pro-apoptotic role at mitochondria through direct interaction with Bcl-2 family members (6, 7). The intracellular level of p53 is primarily controlled through proteasomal degradation and is kept at very low level in normal cells. At least three RING finger E3s, Hdm2 (Mdm2 in mouse) (8-11), COP-1 (12), and Pirh2 (13) have been shown to ubiquitylate p53 and promote its degradation. The significance of Hdm2/Mdm2 as a major regulator of p53 is underscored by the embryonic lethality of by mdm2−/− mice, which is rescued by loss of p53 (14, 15). Upon various stresses, such as DNA damage and expression of oncogenes, phosphorylation of p53 and Hdm2 prevent their interaction, leading to stabilization and accumulation of p53 (16, 17). These stress stimuli may also promote the auto-ubiquitylation and degradation of Hdm2. Furthermore, cellular proteins such as ARF and several small ribosomal proteins inhibit Hdm2-mediated ubiquitylation, which may contribute to the stabilization of p53 under certain circumstances. There is also evidence that, in addition to promoting proteasomal degradation, the inhibitory action of Hdm2 on p53 may also be related to ubiquitin-mediated translocation of p53 from the nucleus to cytoplasm and by direct inhibition of the transcriptional activity of p53. Moreover, Hdm2 is among the transcriptional targets of p53. This provides a negative feedback mechanism to modulate p53-mediated apoptosis (2-5).
Mutations of p53 are observed in approximately 50% of human cancers. In many tumors retaining wild type p53 there are defects in p53 activation. In particularly, a significant percentage of these tumors exhibit amplification of the Hdm2 gene (18). Further, while expression of p53 often leads to growth arrest in untransformed cells, transformed cells usually died by apoptosis when p53 is expressed (19). It has been shown that blocking the interaction between Hdm2 and p53 results in increased p53 in cells and killing of tumor cells in culture and in athymic nude mice (20, 21). Genetic studies have also demonstrated that restoring p53 in animal models can effectively inhibit tumor growth (22, 23). These results indicate that inhibition of Hdm2-mediated p53 ubiquitylation might upregulate p53 to induce apoptosis in tumor cells. For this reason, we have previously carried out high throughput screening to identify inhibitors of the E3 activity of Hdm2. A family of small molecules named HLI98s (Hdm2 Ligase Inhibitors) that are 7-nitro-10-aryl-5-deazaflavins were identified as cell-permeable inhibitors of the E3 activity of Hdm2 (24). The HLI98s increase the level of p53 in both transformed and non-transformed cells, but preferentially induce apoptosis in transformed cells that express wild type p53. These findings provided proof of principle that inhibition of the ligase activity Hdm2 could be an effective strategy for cancer therapy. A significant limitation of the HLI98s is their lack of solubility in aqueous solution and their relative low potency, which limit their application including in vivo studies. In the present study, we describe a homolog of HLI98s named HLI373, which has a 5-dimethylaminopropylamino side chain but lacks the 10-aryl group (NSC373989). HLI373 is highly soluble in aqueous solution and substantially more potent than the HLI98s in activating p53 and killing transformed cells. Moreover, HLI373 is effective in inducing apoptosis of several tumor cells that are sensitive to DNA-damaging agents. This compound therefore represents a potential “drug-able” lead for the development of therapeutically efficacious inhibitors of Hdm2.
Materials and Methods
Reagents and antibodies
Camptothecin and the proteasome inhibitors ALLN and MG132 were purchased from Calbiochem (La Jolla, CA). Adriamycin and β-actin antibody were from Sigma (St. Louis, MO). Antibodies recognizing Hdm2 (Ab-1, Ab-2 and Ab-4) were from Oncogene (Boston, MA). Antibodies recognizing p53, p21WAF1/CIP1, GFP, and PARP were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody recognizing gp78 was described previously (25). Antibody recognizing the HA epitope tag was from Roche (Indianapolis, IN). HLI373 (5-(3-dimethylamino-propylamino)-3,10-dimethyl-10H-pyrimido[4,5-b]quinoline-2,4-dione) was provided by the Developmental Therapeutic Program, National Cancer Institute, NIH (Bethesda, MD).
Cell lines and plasmids
RPE, RPE-E1A, U2OS, U2OS-pG13, C8 and A9 cells and p53−/−mdm2−/− MEFs were cultured as described (24). HT1080 (26) and HCT116-p53+ and HCT116-p53− cells (27) were grown in DMEM media supplemented with 10% FBS. LOX-IMVI, A549, MDA-MB-231, SW-620, PC-3 and U251 cells (28) were cultured in RPMI1640 media supplemented with 10% FBS. All media were supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin and 2.5 mM L-glutamine. Plasmid encoding pEGFP-N1 was from Clontech (Mountain View, CA). Plasmids pCMV-Hdm2, pCB6-p53, pCI-gp78, and pcDNA3-HA-AO7 were used to express Hdm2, p53, gp78, and HA-AO7 in mammalian cells respectively. pG13-Luc plasmid contains 13 copies of p53-binding site (5′-CCTGCCTGGACTTGCCTGG-3′) upstream of the luciferase coding region, while pG13mut-Luc plasmid has 15 copies of mutated p53-binding site (5′-CCTTAATGGACTTTAATGG-3′) (24, 25, 29-31). Plasmid encoding HA-ubiquitin (pMT123) has been described (32).
Immunoblotting
Cells were lysed in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM iodoacetamide, 1 μg/ml aprotinin, 100 μg/ml PMSF and 5 μg/ml leupeptin) and centrifuged at 12,000 rpm (∼14,000 g) for 20 min at 4°C. Equal amount of post-nuclear supernatants were resolved by SDS-PAGE, transferred to PVDF membranes (Millipore, Billerica, MA) and immunoblotted with specific antibodies using standard procedures (24). Bands corresponding to specific proteins were visualized with horseradish peroxidase-labeled secondary antibodies and chemiluminescence agents (Pierce, Rockford, IL). Immunoprecipitation of Hdm2 was carried out as described (33).
In vitro auto-ubiquitylation assay
Assays were carried out using GST-Hdm2 immobilized to glutathione sepharose, E1, E2 and 32P-ubiquitin as described (24). Ubiquitylated Hdm2 was visualized by Storm PhosphoImager (Molecular Dynamics, Sunnyvale, CA).
Assessment of DNA damage
DNA damage was assessed using alkaline elution of DNA as described previously (34, 35). Briefly, U2OS cells were labeled with [3H] thymidine (1 μCi/ml) for ∼72 h. After an overnight chase with fresh medium, cells were treated with 3 Gy or incubated with the indicated compounds for 4 h. Single strand breaks (SSB) were assessed by their failure to be retained on a 2 μM polycarbonate filter. To assess DNA-protein cross-links (DPC) and protein-associated strand breaks cells were treated with compounds for 4 hr as indicated followed by irradiation of all samples with 30 Gy to induce strand breaks. Samples were lysed and evaluated for binding of protein-DNA crosslinks as measured by retention on a 0.8 μM polyvinyl chloride/acrylic copolymer.
Luciferase assay
U2OS stably transfected with pG13-Luc and U2OS transiently transfected with pG13-Luc or pG13mut-Luc were lysed with reporter lysis buffer (Promega, Madison, WI). The resulting lysates were centrifuged at 12,000 rpm (∼14,000 g) for 20 min. Luciferase activity in the supernatant was determined using the luciferase assay substrates (Promega) and Microlumat Plus LB 96V luminometer (EG&G Berthold, Germany).
Results
HLI373 increases p53 and Hdm2 protein levels in cells
To identify more effective small molecules that inhibit Hdm2 and activate p53, a search of the National Cancer Institute Developmental Therapeutics Program chemical database was carried out. Among six identified 5-deazaflavins related to the HLI98s, a single compound that we designated HLI373, 5-(3-dimethylaminopropylamino)-3,10-dimethyl-10H-pyrimido[4,5-b]quinoline-2,4-dione (C18H24ClN5O2, MW: 377.868) (NSC373989) (Figure. 1A), was determined to be soluble in aqueous solutions at concentrations up to 200 mM and found to increase p53 and Hdm2 in cells (data not shown and Figure 1B). Notably, HLI373 differs from HLI98s by lacking the 10-aryl group and by having N-methyl groups at position 3 and 10, and a 3-dimethylamino-propylamino substituent at position 5.
Figure 1. HLI373, a highly soluble derivative of HLI98, is more potent than HLI98s.
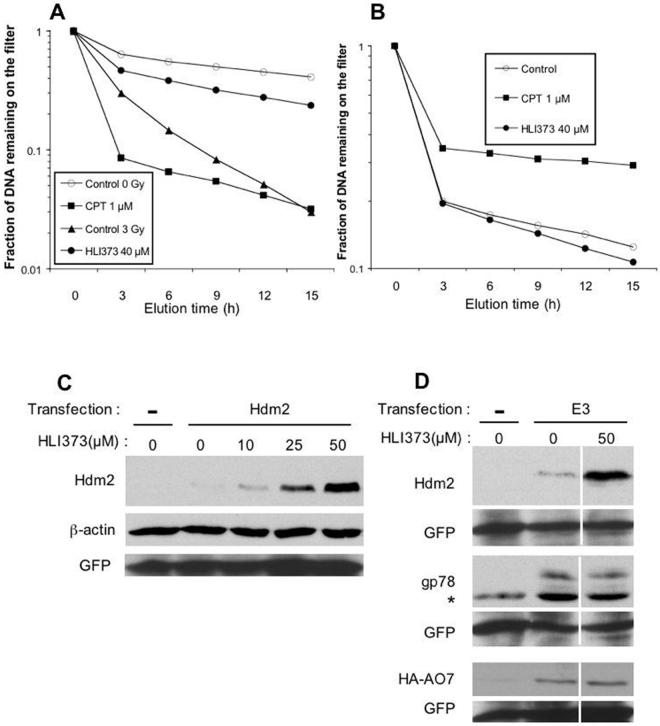
A, Structure of HLI98 family and HLI373. HLI98C: R2 = Cl, HLI98D: R1 = Cl, HLI98E: R1 = CH3. B, RPE cells were treated with 1 μg/ml Adriamycin (Adr), 50 μM MG132 (MG), or HLI373 dissolved in PBS or DMSO and added to cells at the indicated final concentrations for 8 h. Levels of p53, Hdm2 and β-actin, used as a loading control, were assessed by immunoblotting with antibodies directed against the indicated proteins. C, RPE cells were incubated with 1 μg/ml Adriamycin, 50 μM ALLN, or the indicated concentrations of HLI373 for 8 h followed by assessment as in B. D, Immobilized bacterially-expressed GST-Hdm2 was treated as indicated prior to carrying out an in vitro auto-ubiquitylation reaction using 32P-labeled ubiquitin. Lack of E1 in the reaction mixture served as a negative control.
Soluble HLI373 exhibits higher potency than HLI98s
To further characterize the ability of HLI373 to increase p53 and Hdm2, Tert-immortalized human retinal pigment epithelial (RPE) cells were incubated with HLI373 for 8 h. HLI373 increased p53 and Hdm2 to a similar level whether it was dissolved in PBS or in DMSO. The increase observed was similar to that with a peptide aldehyde proteasome inhibitor MG132 (50 μM) (Figure 1B). Using RPE cells, HLI373 maximally increased cellular p53 and Hdm2 at 5 μM. The water solubility of HLI373 is significant as almost half of potential therapeutics have not been developed because of poor solubility in aqueous solutions (36). Adriamycin (doxorubicin) is a DNA-damaging chemotherapeutic agent known to induce stabilization of p53 through phosphorylation. Notably HLI373 was as effective at 5 μM as Adriamycin at its optimal concentration (1 μg/ml) in increasing p53. We next evaluated the relative potency of HLI373 and HLI98s in stabilizing p53 and Hdm2 in RPE cells following an 8 h treatment. Strikingly, HLI373 at 3 μM was superior to the HLI98s at concentrations from 5 to 50 μM and similar in effect to another peptide aldehyde proteasome inhibitor, ALLN (Figure 1C). Consistent with its effect in cells, HLI373 also inhibited auto-ubiquitylation of bacterially-expressed Hdm2 in a cell-free ubiquitylation assay system (Figure 1D).
Since it is well known that p53 is activated and its level increases after exposure of cells to DNA-damaging agents, we asked whether HLI373 causes DNA damages directly. Alkaline elutions were performed to assess the formation of single strand breaks (SSB) and DNA-protein crosslinks (DPC) in U2OS (human osteosarcoma) cells. Compared with cells treated with 3 Gy of radiation and with the topoisomerase I inhibitor camptothecin (1 μM), HLI373-treated cells had significantly more DNA retained on the filter, indicating that it does not induce SSB in U2OS cells under these conditions (Figure 2A). Alkaline elution assay under non-deproteinizing condition also indicated that HLI373 did not contribute to the production of DPC in irradiated cells (Figure 2B). As a positive control, camptothecin induced DNA filter retention, which is indicative of DPC resulting from topoisomerase I cleavage complexes (34). These results are consistent with HLI373 increasing p53 in cells through inhibiting Hdm2-mediated ubiquitylation and not by inducing a DNA damage response.
Figure 2. HLI373 does not cause DNA damage and inhibits Hdm2 auto-ubiquitylation.
A, U2OS cells labeled with [3H] thymidine were treated with HLI373 (40 μM) for 4 h. Ionizing radiation (3 Gy) and camptothecin (1 μM) (CPT) were used as positive control. Untreated cells are also shown. The level of single strand breakage (SSB) is expressed as the fraction of labeled DNA remaining on the filter after alkaline elution. The data are representative results from two independent experiments. B, U2OS cells labeled with [3H] thymidine were treated with HLI373 (40 μM) for 4 h followed by irradiation with 30 Gy. Alkaline elution was used to determine DNA-protein cross-links (DPC) as described in Materials and Methods.. The data are representative of two independent experiments. Cells treated without drug (30 Gy only) and with 30 Gy plus camptothecin (1 μM) served as negative and positive controls, respectively. C, Fibroblasts from p53−/−mdm2−/− mice were transfected with Hdm2 cDNA under the control of a CMV promoter. Forty-eight h after transfection, cells were treated with HLI373 for 8 h. Cellular Hdm2, β-actin and GFP, used as a transfection efficiency control, were determined by immunoblotting. Transfection of plasmid without insert is indicated by (−). D, Fibroblasts from p53−/−mdm2−/− mice were transfected with plasmids encoding Hdm2, gp78 or HA-AO7. After 48 h, cells were treated with 50 μM HLI373 for 8 h. Hdm2, gp78 and HA-AO7 were evaluated by immunoblotting. GFP was used as a transfection efficiency control. Asterisk indicates non-specific band.
HLI373 selectively inhibits auto-ubiquitylation of Hdm2
To further ensure that the increase in p53 and Hdm2 was not the result of stress-induced increased p53 accompanied by the p53-dependent transcription of Hdm2, p53−/−mdm2−/− mouse embryo fibroblasts (MEFs) were transiently transfected with plasmid encoding Hdm2 under the control of a p53-independent CMV promoter. HLI373 stabilized cellular Hdm2 in a dose-dependent manner (Figure 2C). This result indicates that, like the HLI98s, HLI373 does not act primarily by initiating a p53 response.
An area of great interest is whether small molecules can be developed that can target ubiquitin ligases with some degree of specificity, as there are more than 500 substrate-specific E3s encoded in the human genome. While the HLI98s show some selectivity, it is clear that they are not totally specific (24). To begin to evaluate the selectivity of HLI373, we compared its effect on Hdm2 to effects on two other RING finger ubiquitin ligases that are known to mediate self-ubiquitylation and that are degraded in a proteasome-dependent manner when overexpressed (25, 31) (Kitagaki, J., Yang, Y., and Weissman A.M. unpublished observations). gp78/RNF45/AMFR is an E3 implicated in endoplasmic-reticulum associated degradation (25), while AO7/RNF25 is an E3 that has been suggested to be involved in NF-κB signaling (31, 37). p53−/−mdm2−/− MEFs were transiently transfected with plasmids encoding Hdm2, gp78 or AO7 bearing an N terminal HA epitope tag (HA-AO7) prior to incubation with HLI373. While Hdm2 was stabilized, HLI373 had no discernable effect on gp78 or AO7 (Figure 2D). These results suggest that HLI373 may preferentially inhibit the ubiquitin ligase activity of Hdm2.
HLI373 inhibits ubiquitylation and degradation of p53
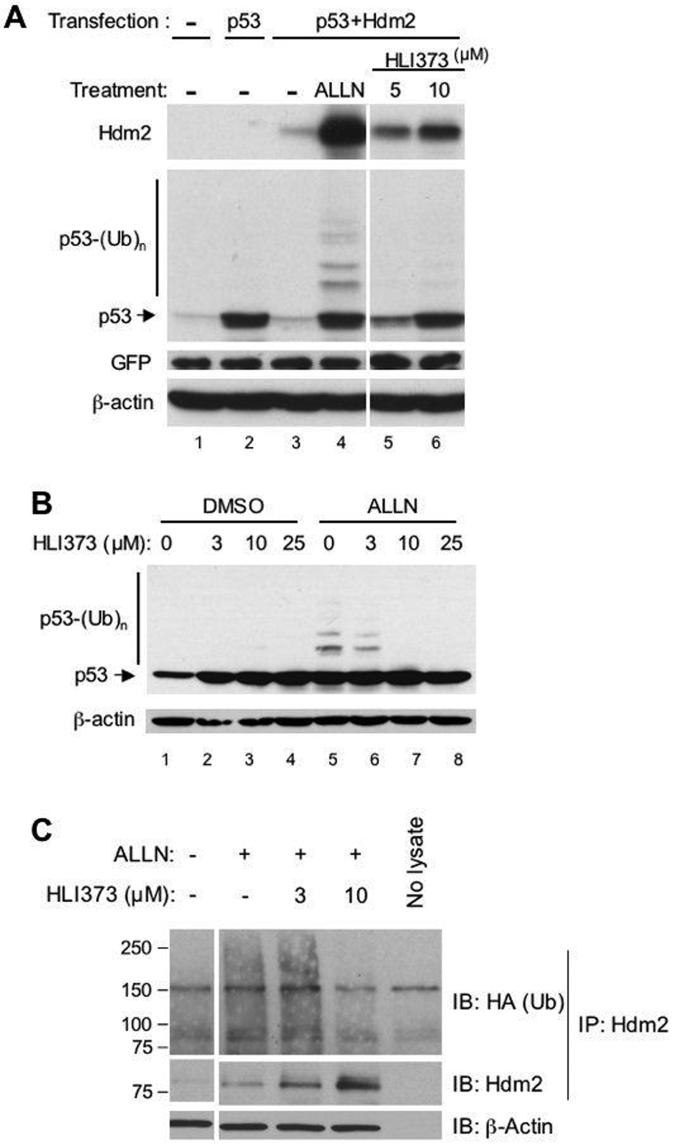
To determine whether HLI373 is functioning by blocking Hdm2-mediated p53 ubiquitylation, U2OS cells, which express low levels of endogenous p53 and Hdm2, were co-transfected with plasmids encoding p53 and Hdm2 and then treated with the proteasome inhibitor ALLN or with HLI373. Co-transfection with plasmids encoding p53 and Hdm2 resulted in degradation of p53 (Figure 3A compare lanes 2 and 3). ALLN inhibited Hdm2-mediated p53 degradation and also resulted in a discernable ladder of higher molecular weight immunoreactive species, indicative of ubiquitylation (Figure 3A, lane 4); this is consistent with stabilizing p53 by inhibition of the proteasome. Incubation with HLI373 also blocked p53 degradation. However, despite similar levels of p53 to that seen with ALLN, ubiquitylated p53 did not accumulate to a significant extent in HLI373-treated cells (Figure 3A, compare lanes 4 to 6), this is consistent with the idea that HLI373 acts by preventing p53 ubiquitylation. To confirm this effect with endogenous p53, the human colon carcinoma cell line HCT116, which expresses higher levels of wild type p53 (HCT116-p53+), were treated with HLI373 for 1 h prior to incubation with or without ALLN for 7 h. When cells were exposed to HLI373 together with ALLN, the ALLN-induced accumulation of ubiquitylated p53 was markedly reduced in a dose-dependent manner (Figure 3B lanes 5-8). These results definitively establish that HLI373 prevents Hdm2-dependent ubiquitylation of p53. Consistent with these findings, treatment of U2OS cells with HLI373 at 10 μM also resulted in a marked decrease in ubiquitylated species immunoprecipitated with anti-Hdm2 (Figure 3C upper panel), while the level of immunoprecipitated Hdm2 increased (Figure 3C middle panel).
Figure 3. HLI373 inhibits degradation and ubiquitylation of p53 and Hdm2.
A, U2OS cells were transfected with plasmids encoding p53 and Hdm2. After 16 h, cells were treated with 50 μM ALLN or with HLI373 as indicated for 8 h. Proteins were detected by immunoblotting. B, HCT116-p53+ cells were treated as indicated with HLI373 for 1 h before treatment either with 50 μM ALLN or vehicle alone for 7 h followed by assessment of cell lysates by immunoblotting. C, U2OS cells were transfected with plasmid encoding HA-tagged ubiquitin. Twenty-four hours after transfection, the cells were treated with HLI373 as indicated prior to addition of 50 M proteasome inhibitor ALLN for an additional 4 h. β-actin corresponding to 5 % of the lysate used for IP serves as a loading control.
Stabilization of p53 by HLI373 results in transcriptionally active p53
One of the principal activities of p53 is as a transcription factor that induces the expression of numerous genes that contribute to either cell cycle arrest or apoptosis. To examine whether HLI373 increases transcriptional activity of p53, we utilized U2OS cells expressing endogenous wild type p53 and stably transfected with a p53-responsive luciferase reporter (pG13-Luc) (24, 29). HLI373 induced a dose-dependent increase in luciferase activity and demonstrated substantially greater potency than the HLI98s (Figure 4A). To ensure that this response was specific and dependent on the p53 response element, U2OS cells were transfected with either pG13-Luc or pG13 that is mutated in its p53-binding sites (pG13mut-Luc). While HLI373 activated transcription from pG13-Luc, HLI373 had no significant effect on transactivation from pG13mut-Luc (Figure 4B). Equal transfection efficiency with the two plasmids was confirmed by GFP co-transfection (data not shown). These results indicate that the stabilization of p53 by HLI373 results in activation of p53-dependent transcription.
Figure 4. HLI373 activates p53 transcription.
A, pG13 stably transfected U2OS cells (U2OS-pG13) were incubated with 1 μg/ml Adriamycin, or the indicated concentration of HLI373, HLI98C, HLI98D, or HLI98E for 22 h and luciferase activity assessed. Data represents average and standard deviation of three independent experiments. B, U2OS cells were transiently transfected with plasmids encoding pG13-Luc or pG13mut-Luc. Twenty-four h after transfection, cells were treated with 1 μg/ml Adriamycin, or the indicated concentrations of HLI373 for 20 h, followed by assessment of luciferase activity. Data represents average and standard deviation of three independent experiments. C, RPE cells were treated with Adriamycin, ALLN or HLI373 for 24 h. Cell lysates were immunoblotted as indicated. D, RPE cells were treated for 4 h with the indicated additions as in C and cell lysates evaluated.
We next examined the effect of HLI373 on the level of p21WAF1/CIP1, which is a p53 target gene. When RPE cells were treated with 3 μM of HLI373 for 24 h, the level of p21 increased to a level similar to that seen with Adriamycin or ALLN (Figure 4C). This provides further evidence that the p53 that accumulates with HLI373 is transcriptionally active. However, p21 is itself a target for ubiquitin-mediated proteasomal degradation by either SCFSkp2 or APC/CCdc20, depending on the phase of the cell cycle (38, 39). When cells were treated for only 4 h HLI373 resulted in only a low level of p21, compared to the marked stabilization with ALLN, while stabilization of p53 and p21 were found to be similar with either ALLN or HLI373 (Figure 4D). Whether this slight increase seen with HLI373 represents the initiation of a p53 response, or indicates some small effect on E3s involved in p21 degradation will require further evaluation. These results with p21, which are similar to those obtained previously with the HLI98s (24), provide further evidence both for the stabilization of p53 by HLI373 and for its relative specificity.
Induction of selective apoptosis of transformed cells and MEFs harboring wild type p53 by HLI373
To examine whether HLI373, like the HLI98s, induces differential killing of transformed cells, we compared RPE cells and RPE cells transformed with adenovirus E1A (RPE-E1A), which interacts with the retinoblastoma gene product but apparently not p53 (40, 41). As predicted, treatment with the proteasome inhibitor MG132 killed both RPE and RPE-E1A cells. However, HLI373 resulted in cell death in RPE-E1A cells, but had no effect on parental RPE cells. Similar differential killing was observed with Adriamycin (Figure 5A). In accord with the relative stabilization and activation of p53, HLI373 was substantially more active in inducing death in transformed cells at 10 μM than the HLI98s (Figure 5B). Consistent with apoptotic cell death, cleavage of the caspase substrate poly (ADP-ribose) polymerase (PARP) was selectively observed in transformed (RPE-E1A) HLI373-treated cells (Figure 5C). These results indicate that HLI373 preferentially induces apoptosis in transformed cells.
Figure 5. HLI373 selectively kills transformed cells.
A, Parental RPE cells and E1A transformed RPE (RPE-E1A) cells were incubated with 1 μg/ml Adriamycin, 50 μM MG132 or the indicated concentrations of HLI373 for 26 h. Cell death was assessed by trypan blue exclusion. B, RPE and RPE-E1A cells were treated with 1 μg/ml Adriamycin or 10 μM of HLI373, HLI98C, HLI98D, or HLI98E for 22 h. Cell death was determined by trypan blue exclusion. C, RPE and RPE-E1A cells were treated with 1 μg/ml Adriamycin, 50 μM MG132, or indicated concentrations of HLI373 for 26 h. PARP was assessed by immunoblot.
HLI373 selectively kills tumor cells harboring wild type p53
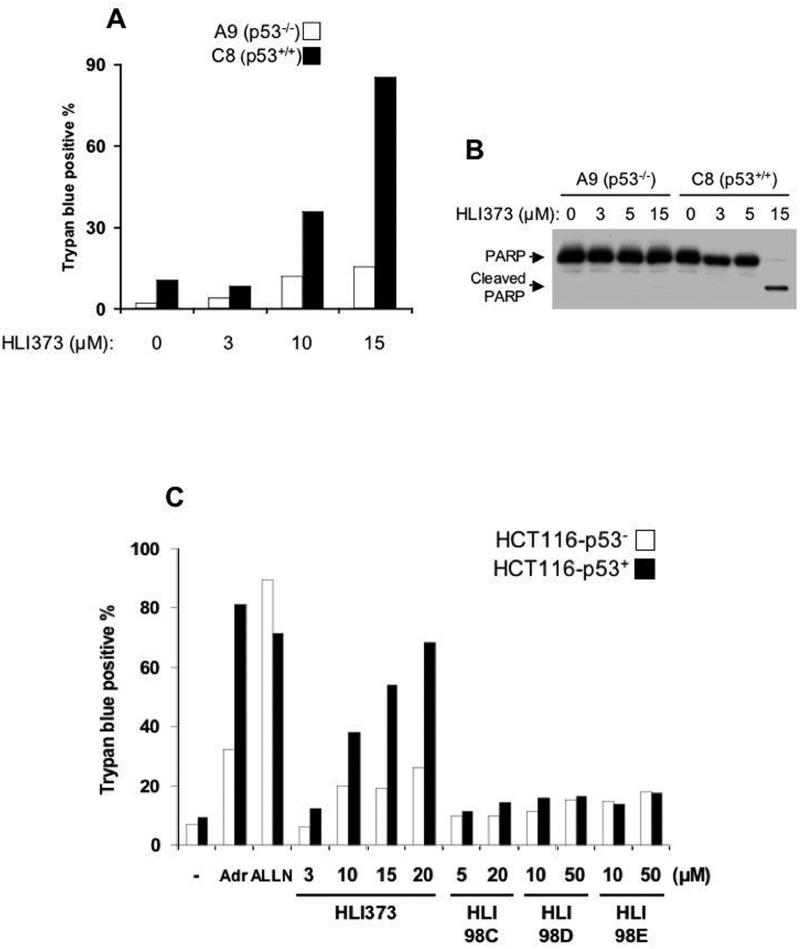
A premise behind this work is that p53-expressing cells will be differentially susceptible to apoptosis in response to HLI373. A well-established system for measuring p53-dependent cell death is MEFs transformed with the adenovirus E1A protein and activated Ha-ras (19). To evaluate whether HLI373 induces apoptosis in a p53-dependent manner, we compared wild type p53 MEFs (C8) to p53-deficient MEFs (A9) (Figure 6A). After incubation with HLI373 for 15 h, cell viability was assessed by trypan blue exclusion. While HLI373 increased cell death in C8 cells in a dose-dependent manner, A9 cells were relatively resistant. HLI373 also increased PARP cleavage in C8 cells but not A9 cells after 15 h (Figure 6B), consistent with HLI373 inducing apoptotic cell death as a consequence of p53 activation.
Figure 6. HLI373 selectively kills cultured cells expressing wild type p53.
A, p53-deficient (A9) or wild type (C8) MEFs was treated with HLI373 for 15 h. Cell death was assessed by trypan blue exclusion. B, A9 and C8 cells were incubated with HLI373 for 15 h and PARP assessed by immunoblotting. C, Human colon carcinoma HCT116-p53+ and HCT116-p53− cells were treated with 1 μg/ml Adriamycin, 50 μM ALLN, or HLI373 for 46 h. D, The indicated cells were incubated with 25 μM HLI373 (upper) or 1 μg/ml Adriamycin (lower) for 30 h, and cell viability assessed. Results calculated based on cells specifically killed by the indicated treatment and represents average and standard error of three independent experiments [100 × (total cell death with drug treatment – background cell death) / (total viable cells without treatment)].
To confirm that HLI373 selectively kills tumor cells in a p53-dependent manner, HCT116 cells that either do or do not express 53 were compared (HCT116-p53+, HCT116-p53−) (27). HLI373 increased cell death in HCT116-p53+ in a dose dependent manner, while HCT116-p53− cells were substantially more resistant (Figure 6C). In contrast, proteasome inhibitor (ALLN), which affects the levels of many different target proteins, was indiscriminant in causing death. Furthermore, the effects on cell death observed with HLI373 were substantially greater and more specific to p53 expressing cells than those seen with HLI98s. Consistent with the apoptotic nature of the induced-cell death, HLI373 increased cleaved caspase 3 in HCT116-p53+ cells but not in HCT116-p53− cells (data not shown).
To further evaluate p53 dependence of tumor cell killing induced by HLI373 multiple different human tumor cell lines were assessed by trypan blue exclusion. Four cell lines [LOX-IMVI (melanoma), A549 (lung carcinoma), HT1080 (fibrosarcoma), and U2OS (osteosarcoma)] that express wild type p53 and that are sensitive to Adriamycin (Figure 6D lower graph) were found to also be substantially more sensitive to HLI373 than four cell lines [MDA-MB-231 (breast cancer), SW-620 (colon cancer), PC-3 (prostate cancer), and U251 (glioblastoma)] in which p53 is inactive (Figure 6D, upper) (28). These results further support the notion that HLI373 kills tumor cells in a p53-dependent manner and that this compound represents a potential water-soluble lead for the development of cancer therapeutics.
Discussion
The ubiquitin system represents a rich source of molecular targets for cancer, prominent among these is the p53 ubiquitin ligase, Hdm2. Our initial attempt to identify inhibitors of this E3 yielded three related 5-deazaflavins, the HLI98s, which were limited in utility by solubility and overall potency (42). We have now identified a related compound, referred to as HLI373, as a water soluble and more potent inhibitor of Hdm2. This small molecule preferentially induces apoptosis in transformed cells and a number of tumor cell lines that express wild type p53. Maximal stabilization of p53 is observed in the low micromolar range when endogenous p53 and Hdm2 were assessed (IC50 ∼ 3 μM). Therefore, HLI373 represents a potential lead for development of anti-cancer therapeutics.
Due to its essential role in controlling cell growth, its frequent inactivation in human tumors and its propensity to induce apoptosis in tumor cells, activation of p53 is well recognized as being a highly desirable goal in the treatment of cancer. For example, p53 delivered by adenovirus effectively prevents the growth of certain tumors in culture and nude mice (42). More recently, it has been demonstrated that restoration of expression of p53 in animal models using genetic method blocks tumor growth (22, 23). Therefore, developing small molecules that increase p53 is of great clinical importance. Many studies have shown that the level of p53 in cells is tightly controlled through ubiquitylation and proteasomal degradation (2-5).
As a consequence of its central role in regulation of p53, Hdm2 represents a particularly attractive molecular target for inactivation so as to increase p53 activity in cancers that express active p53. One approach to this is to prevent the binding of p53 to Hdm2. Accordingly, small peptides derived from p53 have been shown to inhibit the interaction of Hdm2 with p53, resulting in accumulation of p53 in cells (43). At least three non-peptidic small molecules, inhibit the interaction of p53 and Hdm2, resulting in p53-dependent apoptosis (20, 21, 44). In particularly, Nutlin-3 has shown potential promise in tumors that express wild type p53 (45). We and others (24, 46) have taken an alternative approach, by screening for small molecules that inhibits Hdm2's E3 activity so as to prevent the ubiquitylation and subsequent proteasomal degradation of p53. Our data first with the HLI98s and now with HLI373 demonstrate that inhibition of Hdm2-mediated ubiquitylation in cells can result in stabilization of both p53 and Hdm2 and preferential killing of tumor cells expressing wild type p53.
Although the HLI98s can stabilize p53 and Hdm2, they have limited solubility in aqueous solution and relative low potency, which prevent their further application in animal models of human tumors. Recently the 7-nitro group present in HLI98s was found to be dispensable for p53 activation (47). Similarly, HLI373 lacks the 7-nitro group and unlike previously described members of this family, it has the added benefit of water solubility. This property makes it potentially suitable for administration through a variety of routes in vivo and makes if more amenable to cellular studies than the HLI98s.
There are a number of issues that remain regarding the HLI compounds. One relates to mechanism of action. The HLI compounds inhibit Hdm2 ubiquitin ligase activity in cells, but whether this is a consequence of blocking E2-Hdm2 interactions or altering the RING finger structure remains to be determined. A second issue relates to specificity, it is unlikely that without substantial medicinal chemistry surrounding these leads that a highly specific compound could be generated. However, as we have previously shown, the HLI98s are relatively specific, and the limited analysis we have undertaken with HLI373 also suggests a degree of specificity for Hdm2/Mdm2. While a high degree of specificity is generally considered desirable, it is evident that drugs possessing a certain level of off-target activity can be acceptable (48, 49) and, in some cases, might even contribute to therapeutic efficacy. A third important issue relates to potency. In some cells we observe stabilization of p53 at 1 μM of HLI373 and activation of a discernable p53 response at 3 to 5 μM. While it is common wisdom that highly potent reagents with activity in the nanomolar levels are essential in therapeutics, this may not necessarily the case when targeting p53. Animal studies have demonstrated that minimal increases in p53 activity are capable of resulting in a substantial p53 response (22). When p53 levels are acutely increased massive toxicity, particularly in proliferating tissues, can occur (50). Thus, it may be that agents such as HLI373 that may have an appropriate level of bioavailability are of sufficient activity to be of clinical utility.
In conclusion, HLI373 is a potentially drug-able compound that may serve as a potential lead for therapeutics targeting the E3 activity of Hdm2.
Acknowledgements
We thank Dr. Susan Holbeck (Developmental Therapeutics Program, National Cancer Institute) for performing the homology search for compounds with similarity to HLI98s, Dr. Andrew B. Fotia (LPDS, National Cancer Institute) for HA-AO7, Dr. Karen H. Vousden and Robert L. Ludwig (The Beatson Institute for Cancer Research) for plasmids and cell lines. We are also grateful to Dr. John Beutler (Molecular Targets Discovery Program, NCI) for helpful discussions and advice, to the NIH Fellows Editorial Board for revisions on this manuscript and to Dr. Lee J. Helman for critical review of this manuscript.
Support: J.K. is a fellow of Japanese Society for the Promotion of Science. This work was supported by the Center for Cancer Research, National Cancer Institute.
Footnotes
The authors have no conflicting interests related to the work described in this study.
References
- 1.Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–61. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096–106. doi: 10.1038/sj.onc.1207411. [DOI] [PubMed] [Google Scholar]
- 3.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–36. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 4.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 6.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–12. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 7.Moll UM, Wolff S, Speidel D, Deppert W. Transcription-independent pro-apoptotic functions of p53. Curr Opin Cell Biol. 2005;17:631–6. doi: 10.1016/j.ceb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 8.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–9. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 9.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 10.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–7. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 11.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275:8945–51. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 12.Dornan D, Wertz I, Shimizu H, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 13.Leng RP, Lin Y, Ma W, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–91. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 14.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 15.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 16.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–34. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 17.Prives C. Signaling to p53: breaking the MDM2-p53 circuit [In Process Citation] Cell. 1998;95:5–8. doi: 10.1016/s0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 18.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–9. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe SW, Ruley HE, Jacks T, Housman DE. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–67. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 20.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 21.Issaeva N, Bozko P, Enge M, et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–8. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 22.Mendrysa SM, O'Leary KA, McElwee MK, et al. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–34. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, Ludwig RL, Jensen JP, et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–59. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci U S A. 2006;103:341–6. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–33. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 27.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 28.O'Connor PM, Jackman J, Bae I, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–300. [PubMed] [Google Scholar]
- 29.el-Deiry WS, Tokino T, Velculescu VE, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–25. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Lin J, Levine AJ. Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med. 1995;1:142–52. [PMC free article] [PubMed] [Google Scholar]
- 31.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96:11364–9. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–98. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 33.Tang J, Qu LK, Zhang J, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–62. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- 34.Rao VA, Agama K, Holbeck S, Pommier Y. Batracylin (NSC 320846), a dual inhibitor of DNA topoisomerases I and II induces histone gamma-H2AX as a biomarker of DNA damage. Cancer Res. 2007;67:9971–9. doi: 10.1158/0008-5472.CAN-07-0804. [DOI] [PubMed] [Google Scholar]
- 35.Kohn KW. DNA filter elution: a window on DNA damage in mammalian cells. Bioessays. 1996;18:505–13. doi: 10.1002/bies.950180613. [DOI] [PubMed] [Google Scholar]
- 36.Dimond PF. Using nanotechnologies In biotech and medicine: nanomaterials are more than the sum of tiny parts. Gen Eng News. 2005;25:21–8. [Google Scholar]
- 37.Asamitsu K, Tetsuka T, Kanazawa S, Okamoto T. RING finger protein AO7 supports NFkappaB-mediated transcription by interacting with the transactivation domain of the p65 subunit. J Biol Chem. 2003;278:26879–87. doi: 10.1074/jbc.M211831200. [DOI] [PubMed] [Google Scholar]
- 38.Yu ZK, Gervais JL, Zhang H. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21(CIP1/WAF1) and cyclin D proteins. Proc Natl Acad Sci U S A. 1998;95:11324–9. doi: 10.1073/pnas.95.19.11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amador V, Ge S, Santamaria PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27:462–73. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blint E, Phillips AC, Kozlov S, Stewart CL, Vousden KH. Induction of p57(KIP2) expression by p73beta. Proc Natl Acad Sci U S A. 2002;99:3529–34. doi: 10.1073/pnas.062491899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bandara LR, La Thangue NB. Adenovirus E1a prevents the retinoblastoma gene product from complexing with a cellular transcription factor. Nature. 1991;351:494–7. doi: 10.1038/351494a0. [DOI] [PubMed] [Google Scholar]
- 42.Yang C, Cirielli C, Capogrossi MC, Passaniti A. Adenovirus-mediated wild-type p53 expression induces apoptosis and suppresses tumorigenesis of prostatic tumor cells. Cancer Res. 1995;55:4210–3. [PubMed] [Google Scholar]
- 43.Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–9. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 44.Li WD, Wang MJ, Ding F, Yin DL, Liu ZH. Cytotoxic effect of a non-peptidic small molecular inhibitor of the p53-HDM2 interaction on tumor cells. World J Gastroenterol. 2005;11:2927–31. doi: 10.3748/wjg.v11.i19.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vassilev LT. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Lai Z, Yang T, Kim YB, et al. Differentiation of Hdm2-mediated p53 ubiquitination and Hdm2 autoubiquitination activity by small molecular weight inhibitors. Proc Natl Acad Sci U S A. 2002;99:14734–9. doi: 10.1073/pnas.212428599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson JM, Henderson G, Black F, et al. Synthesis of 5-deazaflavin derivatives and their activation of p53 in cells. Bioorg Med Chem. 2007;15:77–86. doi: 10.1016/j.bmc.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 48.Adams J, Kauffman M. Development of the proteasome inhibitor Velcade (Bortezomib) Cancer Invest. 2004;22:304–11. doi: 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 49.Sharp S, Workman P. Inhibitors of the HSP90 molecular chaperone: current status. Adv Cancer Res. 2006;95:323–48. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 50.Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–14. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]