Abstract
The plant hormone ethylene is involved in many developmental processes, including fruit ripening, abscission, senescence, and leaf epinasty. Tomato contains a family of ethylene receptors, designated LeETR1, LeETR2, NR, LeETR4, and LeETR5, with homology to the Arabidopsis ETR1 ethylene receptor. Transgenic plants with reduced LeETR4 gene expression display multiple symptoms of extreme ethylene sensitivity, including severe epinasty, enhanced flower senescence, and accelerated fruit ripening. Therefore, LeETR4 is a negative regulator of ethylene responses. Reduced expression of this single gene affects multiple developmental processes in tomato, whereas in Arabidopsis multiple ethylene receptors must be inactivated to increase ethylene response. Transgenic lines with reduced NR mRNA levels exhibit normal ethylene sensitivity but elevated levels of LeETR4 mRNA, indicating a functional compensation of LeETR4 for reduced NR expression. Overexpression of NR in lines with lowered LeETR4 gene expression eliminates the ethylene-sensitive phenotype, indicating that despite marked differences in structure these ethylene receptors are functionally redundant.
The plant hormone ethylene regulates many processes, including fruit ripening, seed germination, senescence, and abscission of leaves and flowers (1). Ethylene produced in one part of a plant to regulate one of these processes may be deleterious to another tissue or organ, requiring the plant to minimize the response in some tissues while amplifying it in others. To achieve this aim, a plant must differentially regulate ethylene perception and signal transduction. Recent research has focused on Arabidopsis mutants defective in components of the ethylene signal transduction pathway (2–4). Several mutations in the Arabidopsis ETR1 gene result in dominant ethylene insensitivity (5, 6). By analogy to bacterial two-component systems, the Arabidopsis ETR1 protein can be divided into three domains (7, 8). The amino-terminal sensor domain contains three putative transmembrane segments and has been shown to bind ethylene when expressed in yeast. The etr1–1 mutation in this domain abolishes ethylene binding (8). The protein functions as a dimer in the membrane, and the transition metal cofactor copper is necessary for ethylene binding (8, 9). The second domain exhibits homology to histidine kinases that, in bacterial two-component sensing systems, are autophosphorylated. This portion of the ETR1 protein has been shown to have histidine kinase activity in vitro (10). By analogy to bacterial two-component systems, the third domain, the response regulator, may receive the phosphate from the histidine of the histidine kinase domain at an aspartate residue (7). A region of the ETR1 protein between the membrane spanning and histidine kinase domains also has homology to GAF domains that have been associated with cGMP binding in other proteins. (6, 11).
ETR1 is part of a gene family consisting of five members: ETR1, ERS1, ETR2, EIN4, and ERS2 (12–14). Loss-of-function mutants of the Arabidopsis ETR1, ETR2, EIN4, and ERS2 genes do not have defects in ethylene responses, although etr1 mutants do exhibit an ethylene-independent defect in cell elongation (15). However, triple and quadruple mutants for these genes have constitutive ethylene response phenotypes in the absence of ethylene, indicating that coordinately these genes negatively regulate ethylene responses (15). The constitutive ethylene phenotype of the multiple mutants suggests that the receptor is in an active state of suppression of the signal transduction pathway in the absence of ethylene and that ethylene binding inactivates this suppression (15). The lack of an ethylene-related phenotype in single and double loss-of-function mutants also suggests that the family members are, at least partially, functionally redundant.
Tomato also contains a family of putative ethylene receptor genes, designated LeETR1, LeETR2, NR, LeETR4, and LeETR5 (16–18). A semidominant mutation in the NR gene results in the phenotype of the tomato Never ripe (Nr) mutant. Nr originally was identified by the inability of its fruit to undergo ripening (19). The Nr mutant also exhibits delayed flower and leaf senescence and delayed flower pedicel abscission (20). LeETR1, LeETR2, LeETR4, and LeETR5 were cloned by homology to the Arabidopsis ETR1, ETR2, and tomato NR genes. All of the deduced proteins encoded by these genes contain carboxyl-terminal response regulator domains except for NR (17, 18). LeETR4 and LeETR5 are structurally divergent from the other tomato ETR1 homologs. LeETR5 is predicted to have a fourth N-terminal hydrophobic region and also lacks the histidine that has been shown to be autophosphorylated in ETR1 (18). Introduction of a Cys to Ser mutation in the membrane spanning domain of LeETR4 or LeETR5 at a position analogous to the Arabidopsis etr1–1 mutation site results in ethylene insensitivity in transgenic Arabidopsis (18). We have further shown that all five genes encode functional ethylene receptors by demonstrating ethylene binding of yeast-expressed proteins (H.J.K. and A. Bleecker, unpublished work). RNA expression patterns vary among the tomato ETR1 homologs. LeETR1 is expressed constitutively in all tissues examined. LeETR2 is expressed at low levels in all tissues with induction in seeds before germination and down-regulation in elongating seedlings and senescing leaf petioles. NR mRNA is up-regulated in ovaries and ripening fruit (16, 17). LeETR4 is present at high levels in fruit but is low in vegetative tissues. The LeETR5 expression pattern is similar to LeETR4, but absolute mRNA levels are lower (18). Here we show that reduction of LeETR4 mRNA results in increased sensitivity to ethylene in multiple plant tissues. However, plants with reduced NR mRNA have a phenotype similar to wild-type plants, apparently as a result of functional compensation by increased levels of LeETR4 gene expression. Finally, we show that the ethylene-sensitive phenotype associated with LeETR4 depletion can be complemented with a NR-overexpressing transgene, indicating functional redundancy in these two structurally divergent ethylene receptors.
Methods
Plant Material.
Tomato (Lycopersicon esculentum cv. Floradade) plants were grown under standard greenhouse conditions. Flowers were tagged at anthesis, and fruits were harvested at the indicated days after anthesis or at the breaker stage (when the first red color was observed at the blossom end). Breaker stage fruits were allowed to ripen at room temperature until harvest, frozen in liquid nitrogen, and stored at −80°C. To obtain fruit set, flowers of LeETR4 antisense and control plants were treated with a solution of 1 mM silver thiosulfate, or individual inflorescences were treated with 4 mg/liter of 1-methylcyclopropene (MCP) (EthylBloc, Floralife, Walterboro, SC) overnight.
Production of Transgenic Plants.
Transgenic tomato plants were produced by using standard Agrobacterium-mediated plant transformation methods with either neomycin phosphotransferase for kanamycin resistance (LeETR4 antisense constructs) or 5-enolpyruvylshikimate-3-phosphate synthase for glyphosate resistance (NR antisense constructs) as a selectable marker (21). Full-length LeETR4 or NR cDNAs were introduced in their antisense orientations behind the figwort mosaic virus 35S promoter (22) and followed by the nopaline synthase 3′ terminator. Introduction and inheritance of the transgenes were confirmed by PCR using primers specific for the selectable marker genes.
RNA Isolation and Analysis.
RNA was extracted with SDS/phenol followed by LiCl precipitations (25). RNase protection assays were performed by using a manufacturer's kit (Ambion, Austin, TX) as described (17, 18).
Seedling Triple Response Assays.
Assays were performed on homozygous NR and LeETR4 antisense and control lines by using the designated concentrations of 1-aminocyclopropane-1-carboxylate (ACC) as described in ref. 20 or ethylene gas as described in ref. 23. Solid KMnO4 was added to the culture vessels to remove ethylene emitted by the seedlings.
Ripening Analysis of Fruits.
Fruit ethylene production was determined by gas chromatography. Color analysis was performed by using a Minolta color meter on a red to green scale with five measurements taken per fruit as described (24).
Crosses.
Homozygous lines of the NR overexpressing lines described earlier (25) were crossed to LeETR4 antisense lines LeETR4AS-1 (homozygous for the transgene) or LeETR4AS-2 (heterozygous for the transgene).
Results
To understand the roles of the tomato ethylene receptor homologs in plant development, we have produced transgenic tomato lines expressing antisense genes of LeETR4 and NR. We examined the effects of reduced NR or LeETR4 gene expression on several developmental processes that are regulated by ethylene. Ethylene treatment induces leaf epinasty, a downward curvature of the leaves resulting from selective cell expansion on the upper side of the petiole (1). Transgenic plants expressing an LeETR4 antisense gene were severely epinastic in the absence of ethylene (Fig. 1A). Leaf ethylene production by the LeETR4 antisense plants was not significantly different from that of wild type (Table 1). Application of the ethylene action inhibitors MCP or silver thiosulfate reversed the epinasty, indicating that it was a result of altered ethylene response. NR antisense lines did not exhibit epinasty under normal growth conditions.
Figure 1.
Phenotypes of the LeETR4 or NR antisense lines and crosses between NR overexpressing lines and LeETR4 antisense lines. (A) Epinasty of petioles and leaves of LeETR4 antisense plants. (B) Flowers from NR antisense line NRAS-1. (C) Prematurely senescing flowers from LeETR4 antisense line, LeETR4AS-1. (D) Flowers from the NR overexpressing line (NROE-2) used as a parent for the cross shown in E. (E) Flowers from a cross between LeETR4 antisense line LeETR4AS-1 and NR overexpressing line NROE-2. Magnification: ×0.5.
Table 1.
LeETR expression and ethylene synthesis in leaves of NR and LeETR4 antisense lines
| Line | LeETR1 mRNA, % | LeETR2 mRNA, % | LeETR5 mRNA, % | Ethylene, nl/gfw/hr |
|---|---|---|---|---|
| Wild type | 1.3 ± 0.1 × 10−3 | 3.2 ± 1.8 × 10−4 | 4.1 ± 0.1 × 10−4 | 3.3 ± 0.3 |
| NRAS-1 | 1.5 ± 0.2 × 10−3 | 5.3 ± 1.5 × 10−4 | 6.9 ± 0.2 × 10−4** | 3.1 ± 0.4 |
| NRAS-2 | 1.4 ± 0.4 × 10−3 | 5.0 ± 1.2 × 10−4 | 6.8 ± 1.8 × 10−4 | 2.9 ± 0.5 |
| NRAS-3 | 1.2 ± 0.1 × 10−3 | 4.5 ± 1.7 × 10−4 | 5.2 ± 1.7 × 10−4 | 3.1 ± 0.5 |
| Wild type | 7.5 ± 2.9 × 10−4 | 3.9 ± 0.3 × 10−4 | 1.6 ± 0.4 × 10−3 | 4.6 ± 0.7 |
| LeETR4AS-1 | 7.3 ± 3.2 × 10−4 | 4.0 ± 1.4 × 10−4 | 1.9 ± 0.6 × 10−3 | 4.8 ± 0.7 |
| LeETR4AS-2 | 9.0 ± 2.1 × 10−4 | 4.5 ± 0.6 × 10−4 | 1.2 ± 0.3 × 10−3 | 5.4 ± 0.6 |
| LeETR4AS-3 | 5.5 ± 1.0 × 10−4 | 2.6 ± 0.3 × 10−4* | 1.3 ± 0.4 × 10−3 | 6.0 ± 0.7 |
LeETR mRNA levels expressed as % of total mRNA were determined as described in Methods. Numbers represent means ± SE.
*Significantly different from wild type (P ≤ 0.05).
**Significantly different from wild type (P ≤ 0.01).
Ethylene application stimulates floral senescence in many plant species, including tomato (1). NR antisense flowers were indistinguishable from wild-type flowers, and fruit set was normal in those plants (Fig. 1B). Three LeETR4 antisense lines produced flowers that underwent senescence before fully opening (Fig. 1C), and fruit set did not occur. Floral senescence occurred in all flowers in LeETR4 antisense plants from these three lines, but could be reversed by the application of silver thiosulfate or MCP.
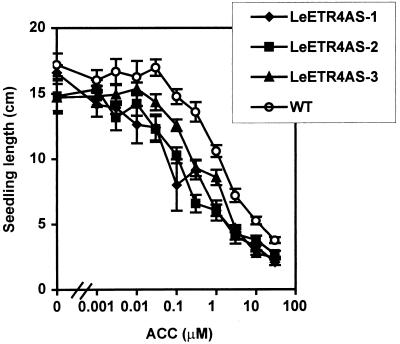
To determine the effects of antisense gene expression on ethylene sensitivity, we generated dose–response curves with etiolated NR and LeETR4 antisense seedling length grown in increasing concentrations of the ethylene precursor ACC (Fig. 2). Etiolated wild-type seedlings responded to ACC levels in a dose-dependent manner with little reduction in growth at ACC levels up to 0.03 μM followed by increasing reductions in seedling length from 0.1 to 30 μM. In contrast, LeETR4 antisense (LeETR4AS) seedlings were significantly shorter than wild type at all ACC concentrations. Etiolated LeETR4AS and wild-type seedlings grown in the presence of the ethylene action inhibitor MCP were not significantly different in length (data not shown), suggesting that the difference in length at 0 μM ACC was the result of differences in sensitivity to endogenous ethylene. Results of ethylene dose–response curves were similar to ACC dose–response curves. LeETR4AS seedlings grown in the presence of 1 ppm ethylene were 35–50% shorter than wild-type seedlings (data not shown). No significant differences in length of NRAS and wild-type seedlings were observed.
Figure 2.
Ethylene sensitivity of etiolated seedlings in LeETR4 (LeETR4AS-1–3) antisense lines. Seedlings were grown in the dark for 2 weeks in the presence of increasing concentrations of the ethylene precursor ACC as described in Methods.
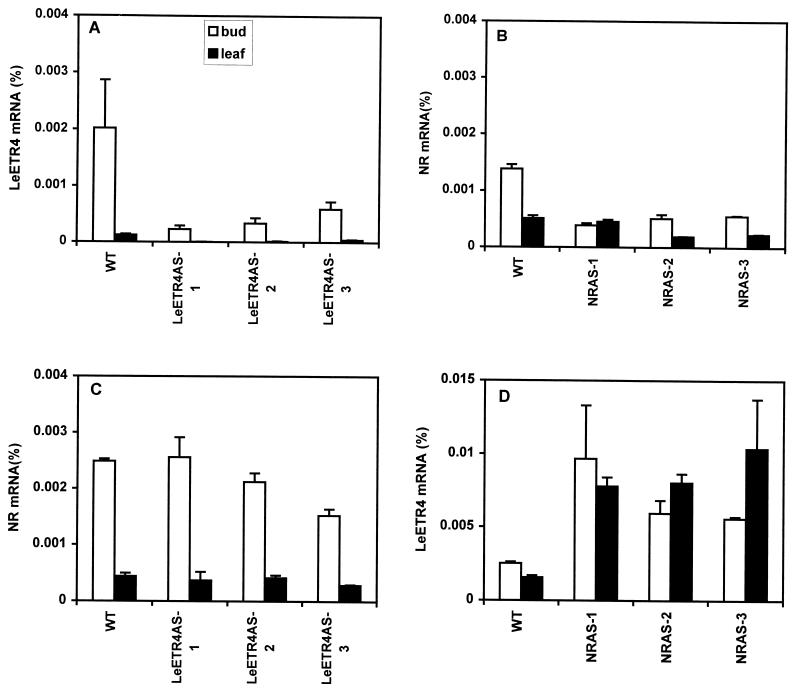
To determine the basis of the ethylene sensitivity of the LeETR4 antisense lines and the wild-type phenotype of the NR antisense lines, we examined the LeETR mRNA levels in the transgenic plants. Three lines containing the LeETR4 antisense gene (LeETR4AS-1, LeETR4AS-2, and LeETR4AS-3) exhibited reductions of 65–97% in LeETR4 mRNA in leaves and flower buds (Fig. 3A). These lines exhibited the most pronounced ethylene-response phenotype. Levels of NR mRNA were reduced by 60–72% in flower buds in the NR antisense lines, whereas levels in leaves were reduced by only 11–62% (Fig. 3B). Antisense expression of LeETR4 did not affect expression of NR (Fig. 3C) or any other LeETR (Table 1). However, antisense lines with even modest reductions of NR exhibited 2- to 3-fold higher levels of LeETR4 mRNA than wild-type flower buds or leaves (Fig. 3D). Levels of LeETR1, LeETR2, or LeETR5 mRNAs in flower buds or leaves did not differ significantly among the control and NR antisense lines (Table 1). Elevation of LeETR4 mRNA levels, but not levels of other LeETRs, in NR antisense buds and leaves suggests a unique mode of regulation of the LeETR4 gene.
Figure 3.
Expression of NR and LeETR4 in LeETR4 and NR antisense lines at the mRNA level. (A) LeETR4 mRNA in LeETR4 antisense lines. (B) NR mRNA in NR antisense lines. (C) NR mRNA in LeETR4 antisense lines. (D) LeETR4 mRNA in NR antisense lines. Note difference in scale of y axis in D as compared with A–C. RNase protection assays were performed on RNAs from flower buds and leaves of wild-type (WT) plants and several independent transgenic lines for both the NR (lines NRAS-1–3) and LeETR4 (lines LeETR4AS-1–3) antisense constructs.
In wild-type fruits, NR mRNA levels are low at the immature green stage and increase several-fold with the onset of ripening (16, 17); however, in ripening fruits from NR antisense plants, NR mRNA levels were reduced by 78–90% (Table 2). Levels of NR protein were also significantly reduced in fruits from the three NR antisense lines (data not shown). LeETR4 mRNA levels in fruits of NR antisense plants were somewhat reduced compared with control plants (Table 2). LeETR4 antisense lines were at least 90% reduced in levels of LeETR4 mRNA and exhibited somewhat lower levels of NR mRNA (Table 2).
Table 2.
Fruit ripening in the LeETR4 and NR antisense lines
| Line | Ethylene, nl⋅g−1FW⋅hr−1 | Color (a) | Days to breaker | NR mRNA, % | LeETR4 mRNA, % |
|---|---|---|---|---|---|
| Wild type | 7.9 ± 0.7 | 15.7 ± 1.2 | 47.0 ± 0.4 | 1.5 ± 0.1 × 10−2 | 5.6 ± 0.4 × 10−3 |
| NRAS-1 | 4.4 ± 0.4** | 11.3 ± 1.7* | 49.6 ± 0.6** | 1.5 ± 0.5 × 10−3** | 2.7 ± 0.3 × 10−3** |
| NRAS-2 | 3.5 ± 0.2** | 10.5 ± 2.4 | 47.6 ± 0.4 | 1.3 ± 0.4 × 10−3** | 4.0 ± 1.5 × 10−3 |
| NRAS-3 | 3.9 ± 0.2** | 8.0 ± 2.2** | 47.4 ± 0.3 | 3.4 ± 1.2 × 10−3** | 2.2 ± 0.1 × 10−3** |
| Wild type | 9.0 ± 0.7 | 15.4 ± 1.1 | 52.9 ± 0.4 | 1.1 ± 0.1 × 10−2 | 7.0 ± 1.0 × 10−3 |
| LeETR4AS-1 | 7.4 ± 0.4 | 21.9 ± 0.7** | 49.8 ± 0.4** | 5.7 ± 0.2 × 10−3** | 2.2 ± 0.3 × 10−4** |
| LeETR4AS-2 | 8.4 ± 0.5 | 19.6 ± 0.7** | 50.8 ± 0.4** | 8.2 ± 0.5 × 10−3* | 7.6 ± 0.6 × 10−4** |
| LeETR4AS-3 | 18.6 ± 1.6** | 26.9 ± 0.5** | 41.7 ± 0.3** | 1.0 ± 0.1 × 10−2 | 3.9 ± 0.1 × 10−4** |
Ethylene, color, and mRNA level determinations were taken from fruit 4 days after the first appearance of red color. Color measurements are on a green to red scale with higher “a” values corresponding to more red pigmentation. Days to breaker represent the number of days from anthesis to the onset of ripening. NR and LeETR4 mRNA levels expressed as % of total mRNA were determined as described in Methods. Numbers represent means ± SE.
*Significantly different from wild type (P ≤ 0.05).
**Significantly different from wild type (P ≤ 0.01).
Although immature fruits do not respond to ethylene by ripening, ethylene exposure accelerates the time to the onset of ripening; fruits in some way measure the cumulative exposure to ethylene and use this as a developmental clock (26). If reduction in LeETR4 gene expression increased ethylene sensitivity, fruit ripening should be accelerated. In several antisense LeETR4 lines we observed decreased time from flowering to appearance of the first red color on the fruit (days to breaker) as well as accelerated accumulation of red pigmentation after the onset of ripening (Table 2). Fruits from the line with the most extreme phenotype (LeETR4AS-3) initiated ripening 11 days earlier than wild-type fruits. These fruits also developed external red color after the onset of ripening more quickly than control fruits, indicating that the increase in ethylene sensitivity was occurring during both fruit growth and ripening (Table 2). The LeETR4AS-3 line also produced more ethylene during ripening, although the other LeETR4 antisense lines did not. In contrast, reduction in NR gene expression by antisense mRNA did not have obvious effects on fruit development, and timing to onset of ripening was not altered. However, color development in ripening fruit was delayed, and levels of ethylene were approximately half those produced by wild-type fruits.
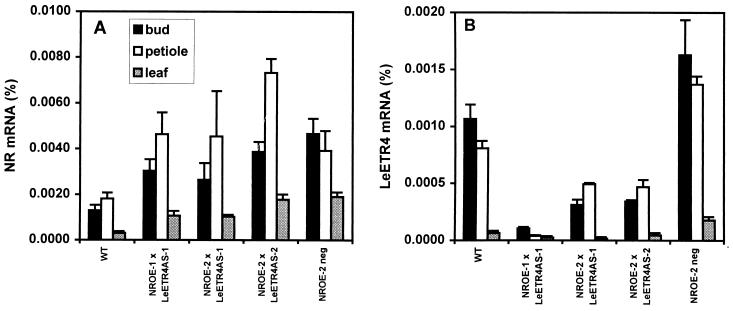
The wild-type phenotype of NR antisense lines suggested that the increase in LeETR4 gene expression in buds and leaves of NR antisense lines (Fig. 3D) may be compensating for the reduction in NR gene expression. To determine whether the elevated expression of one LeETR can compensate for reduced levels of another, we crossed ethylene-sensitive lines LeETR4AS-1 and LeETR4AS-2 with NR-overexpressing lines NROE-1 and NROE-2 (25). These NR-overexpressing lines exhibited characteristics of decreased ethylene sensitivity in some tissues, including a reduced triple response of etiolated seedlings, increased stem elongation, and increased tolerance to the virulent pathogen Xanthomonas campestris pv. vesicatoria. Plants containing both transgenes exhibited increased levels of NR mRNA in leaves, petioles, and flower buds, whereas the same tissues had reduced levels of LeETR4 mRNA as compared with wild-type tissues (Fig. 4). These F1 plants were phenotypically indistinguishable from wild type (Fig. 1 C–E), whereas the original LeETR4 antisense plants, either heterozygous or homozygous for the transgene, exhibited the severe epinasty and flower senescence phenotypes. Flowers and fruits of the F1 plants developed normally and leaves were not epinastic under normal growth conditions. Progeny from the cross carrying the gene for NR overexpression, but not the LeETR4 antisense gene, had elevated levels of NR gene expression, and normal levels of LeETR4 gene expression in leaves and buds (Fig. 4).
Figure 4.
Expression levels of NR and LeETR4 mRNA in crosses of NR overexpressors and LeETR4 antisense lines in flower buds, leaves, and leaf petioles. (A) NR mRNA. (B) LeETR4 mRNA. RNase protection assays were performed by using specific probes for the NR and LeETR4 genes in tissues from wild-type (WT) plants, progeny from crosses of the LeETR4 antisense and NR overexpressing lines (NROE × LeETR4AS), and progeny from the cross overexpressing NR but not expressing the LeETR4 antisense gene (NROE-2 neg).
Discussion
Why would a tomato plant have at least five ethylene receptors with such structural divergence and different patterns of expression? Transgene-mediated alterations in tomato LeETR4 and NR gene expression have led to phenotypes that begin to address these questions. Transgenic plants with reduced LeETR4 expression have multiple phenotypes indicative of increased ethylene sensitivity. Leaves and stems are epinastic in the absence of ethylene (Fig. 1A). Flowers senesce prematurely and fruit does not set (Fig. 1C). Epinasty and flower senescence can be reversed by application of silver thiosulfate or MCP, further suggesting that these phenotypes are the result of increased ethylene sensitivity. Ethylene levels in leaves of the LeETR4 antisense plants did not differ significantly from wild type, indicating that the phenotypes were not the result of increased ethylene production. Ethylene receptors appear to function as negative regulators of the signal transduction pathway. It has been suggested that when ethylene is bound by the receptor this negative regulation is suppressed, resulting in an ethylene response. By reducing the levels of receptors the repression of the signal transduction pathway is further diminished, resulting in enhanced ethylene responsiveness (15). Our results also indicate that lowering ethylene receptor levels increases sensitivity to ethylene, and that LeETR4 is a negative regulator of the ethylene signal transduction pathway.
Although the LeETR4 antisense plants have an increased ethylene-sensitive phenotype, the NR antisense plants appear normal. Our results suggest the lack of developmental abnormalities in NR antisense lines is the result of functional compensation of LeETR4 mRNA levels for reduced NR gene expression. Functional compensation for reduced expression of one member of a gene family by increased expression of another member of the same family has been demonstrated in several mammalian multigene families, including retinoic acid receptor, retinoblastoma, and connexin gene families (27–29). Why reduction in NR gene expression results in functional compensation, while reduction in LeETR4 does not, is unknown. Why levels of LeETR4, but not the other LeETRs, are increased in NR antisense plants is also unknown. The different behavior of LeETR4 could be explained by a differential regulation by ethylene. The LeETR4 gene does have a lower threshold for ethylene induction than the other four receptor genes (ref. 25; D.M.T., unpublished work). At this point, however, we cannot rule out regulation of receptor levels by a mechanism that does not directly involve ethylene. Nor can we eliminate the possibility of an as yet unidentified member of the gene family although extensive screens of genomic and cDNA libraries as well as expressed sequence tag databases makes this unlikely.
Alterations in expression of other ethylene receptor genes is a possible explanation for the wild-type phenotype in the single loss-of-function mutants of Arabidopsis ETR1, ERS2, EIN4, or ETR2. Only one double loss-of-function mutant (etr1;ein4) exhibits increased ethylene sensitivity, whereas all triple and quadruple mutants show increased ethylene sensitivity (15). It is possible that there is functional compensation in Arabidopsis as well. Only by reducing expression of several ETR1 homologs can functional compensation be removed. Three of the five ETR family members do show ethylene inducibility. Whether levels of the other ETR1 homologs are altered in the loss-of-function mutants remains to be determined.
NR and LeETR4 are quite different structurally. They are only 51% similar and 41% identical at the amino acid level. NR lacks the response regulator domain found in the other LeETRs. LeETR4 lacks several of the conserved domains suggested to be necessary for histidine kinase activity. LeETR4 also contains a 24-aa amino terminal extension that is not present in NR (18). Despite these differences in structure, the LeETR4 antisense phenotype can be ameliorated by overexpressing NR under the control of a constitutive promoter. Plants from crosses of NR overexpressors and LeETR4 antisense lines are normal in phenotype with respect to lack of epinasty, flower senescence, and normal fruit set. Although we cannot yet address the stoichiometry of the complementation at the protein level, the results suggest that LeETR4 and NR are functionally equivalent ethylene receptors. There is a rough proportionality between NR gene expression and NR protein, as quantitated by Western blotting of transgenic lines (data not shown), but we cannot rule out quantitative differences among receptors in their abilities to transduce the ethylene signal. Further examination of these plants may reveal differences in ethylene responsiveness as compared with wild-type plants, particularly under biotic or abiotic stress.
Fruit ripening is somewhat paradoxical in that receptor levels increase in parallel with ethylene sensitivity (16, 17). The key to understanding this apparent contradiction is that ethylene synthesis also is increasing rapidly during this time. At the onset of fruit ripening ethylene production becomes autocatalytic, resulting in a rapid increase in ethylene levels. Ripening may proceed in the presence of high levels of negatively regulating receptors as a result of saturating levels of ethylene being produced by the fruits. Ethylene appears to be saturating in ripening fruit, because transgenic lines with up to 80% reduced ethylene production do not display differences in ripening rate (H.J.K., unpublished results). The time to onset of ripening is reduced in several LeETR4 antisense lines (Table 1). It has been suggested that fruit must reach competency to ripen, and that the cumulative ethylene exposure of immature fruits signals the beginning of the ripening process (26). LeETR4 antisense fruit may need less overall ethylene exposure to initiate ripening as a result of lowered receptor levels early in development. One possible role of LeETR4 in immature fruits is to delay the onset of ripening until the seed have matured. Seed germination is reduced in several antisense lines, perhaps as a result of a lack of maturation (unpublished results).
The presence of at least five ethylene receptors in tomato with very different structures, expression patterns, and modes of regulation indicates the necessity for the plant to tightly control the response to ethylene and the environmental and developmental signals that result in ethylene production. Receptors that are negative regulators of the signal transduction pathway and whose expression is up-regulated in response to ethylene may reduce and delay the response. After a long period of ethylene exposure the plant becomes less sensitive to increased ethylene levels and resumes normal functioning. Generally, plants respond to rapid increases in hormone levels by attempting to reduce the effect in multiple ways. Mechanisms to maintain homeostasis can include conjugation, degradation, sequestration, and alteration of sensitivity. Increasing ethylene receptor levels is an effective method of regulating the ethylene responsiveness of tissues. Although clear evidence has been seen for regulation of ethylene levels by conjugation of the ethylene precursor ACC, the plant appears to remove ethylene solely by dissipation of the gas (1, 30). In attached tomato fruits the skin acts as a barrier to diffusion, resulting in an accumulation of ethylene (31). By regulating receptor levels the plant can modulate the ethylene response even when high levels of ethylene are present. The regulation of both ethylene synthesis and perception allows the plant to initiate an ethylene response in one tissue while suppressing the response in others.
Acknowledgments
We thank Jack Wilkinson and Jeanne Layton at Monsanto for producing the NR antisense plants. This work was supported in part by U.S. Department of Agriculture Grant No. 95–37304-2326 to H.J.K. This is Florida Agricultural Experiment Station Journal Series No. R07445.
Abbreviations
- ACC
1-aminocyclopropane-1-carboxylate
- NR
Never Ripe
- MCP
1-methylcyclopropene
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090550597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090550597
References
- 1.Abeles F, Morgan P, Saltveit M. Ethylene in Plant Biology. 2nd Ed. San Diego: Academic; 1992. [Google Scholar]
- 2.Guzman P, Ecker J. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roman G, Lubarsky B, Kieber J, Rothenberg M, Ecker J. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woeste K, Kieber J. Philos Trans R Soc London B. 1998;353:1431–1438. doi: 10.1098/rstb.1998.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleecker A, Estelle M, Somerville C, Kende H. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- 6.Bleecker A, Esch J, Hall A, Rodriguez F, Binder B. Philos Trans R Soc London B. 1998;353:1405–1412. doi: 10.1098/rstb.1998.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C, Kwok S, Bleecker A, Meyerowitz E. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 8.Schaller G E, Bleecker A. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez F, Esch J, Hall A, Binder B, Schaller G E, Bleecker A. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- 10.Gamble R, Coonfield M, Schaller G E. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L, Ponting C. Trends Biochem Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 12.Hua J, Chang C, Sun Q, Meyerowitz E. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- 13.Hua J, Sakai H, Nourizadeh S, Chen Q, Bleecker A, Ecker J, Meyerowitz E. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai H, Hua J, Chen Q, Chang C, Medrano L, Bleecker A, Meyerowitz E. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua J, Meyerowitz E. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson J, Lanahan M, Yen H-C, Giovannoni J, Klee H. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 17.Lashbrook C, Tieman D, Klee H. Plant J. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- 18.Tieman D, Klee H. Plant Physiol. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rick C, Butler L. Adv Genet. 1956;8:267–382. [Google Scholar]
- 20.Lanahan M, Yen H-C, Giovannoni J, Klee H. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick S, Neidermeyer J, Fry J, Barnason A, Horsch R, Fraley R. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- 22.Richins R, Scholthof H, Shepard R. Nucleic Acid Res. 1987;15:8451–8466. doi: 10.1093/nar/15.20.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yen H-C, Lee S, Tanksley S, Lanahan M, Klee H, Giovannoni J. Plant Physiol. 1995;107:1343–1353. doi: 10.1104/pp.107.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klee H. Plant Physiol. 1993;102:911–916. doi: 10.1104/pp.102.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciardi, J., Tieman, D., Lund, S., Jones, J., Stall, R. & Klee, H. (2000) Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- 26.Yang S F. In: Plant Senescence: Its Biochemistry and Physiology. Thompson W, Nothnagel E, Huffaker R, editors. Rockville, MD: Am. Soc. Plant Physiol.; 1987. pp. 156–165. [Google Scholar]
- 27.Bérard J, Luo H, Chen H, Mukuna M, Bradley W E C, Wu J. J Immunol. 1997;159:2586–2598. [PubMed] [Google Scholar]
- 28.Mulligan G J, Wong J, Jacks T. Mol Cell Biol. 1998;18:206–220. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minkoff R, Bales E, Kerr C, Struss W. Devel Genet. 1999;24:43–56. doi: 10.1002/(SICI)1520-6408(1999)24:1/2<43::AID-DVG6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 30.Peiser G, Yang S F. Plant Physiol. 1998;116:1527–1532. doi: 10.1104/pp.116.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cameron A, Yang S F. Plant Physiol. 1982;70:21–23. doi: 10.1104/pp.70.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]