Abstract
MicroRNAs (miRNAs) are an emerging class of small non-coding RNAs implicated in a wide variety of cellular processes. Research in this field is accelerating, and the growing number of miRNAs emphasizes the need for high-throughput and sensitive detection methods. Here we present the successful evaluation of the Megaplex reverse transcription format of the stem-loop primer-based real-time quantitative polymerase chain reaction (RT-qPCR) approach to quantify miRNA expression. The Megaplex reaction provides simultaneous reverse transcription of 450 mature miRNAs, ensuring high-throughput detection. Further, the introduction of a complementary DNA pre-amplification step significantly reduces the amount of input RNA needed, even down to single-cell level. To evaluate possible pre-amplification bias, we compared the expression of 384 miRNAs in three different cancer cell lines with Megaplex RT, with or without an additional pre-amplification step. The normalized Cq values of all three sample pairs showed a good correlation with maintenance of differential miRNA expression between the cell lines. Moreover, pre-amplification using 10 ng of input RNA enabled the detection of miRNAs that were undetectable when using Megaplex alone with 400 ng of input RNA. The high specificity of RT-qPCR together with a superior sensitivity makes this approach the method of choice for high-throughput miRNA expression profiling.
INTRODUCTION
MicroRNAs (miRNAs) are an emerging class of small non-coding RNAs capable of negatively regulating gene expression. With over 800 human miRNAs reported thus far and many more awaiting experimental validation, these molecules represent one of the largest classes of gene regulators. Recent studies have implicated miRNAs in numerous cellular processes including development, differentiation, proliferation, apoptosis and stress response and thus, not surprising, these same miRNAs are turning out to be important players in cancer development (1). In addition to the dramatic impact on our insight into the fundamental aspects of oncogenesis, the discovery of miRNAs has also potentially great implications for translational research as evidence is emerging that miRNA signatures correlate with diagnosis, tumour classification and prognosis (2). In view of these observations, accurate high-throughput profiling of miRNAs is a major challenge for the field. Various methods, such as microarrays and bead-based flow cytometry, are available enabling the detection of multiple miRNAs in a single experiment, but such approaches generally require significant amounts of input RNA (>1 µg) and preclude the use of very small clinical biopsies or analysis of small subsets of cells or even single cells (2–7).
Real-time quantitative PCR (RT-qPCR) has superior sensitivity, down to the single molecule level. The stem-loop reverse transcription primer method developed by Chen et al. (8) has enabled specific and sensitive PCR-based quantification of small RNA molecules such as miRNAs. This method selectively targets mature miRNAs and easily covers a dynamic range of linear quantification of 7 log10 units. Here we present the evaluation of the Megaplex reverse transcription format of the stem-loop primer-based RT-qPCR approach to quantify miRNA expression.
MATERIALS AND METHODS
Cell culture and RNA samples
Eleven neuroblastoma (NB) cell lines (NGP, IMR-32, SMS-KAN, SK-N-BE(2c), LAN-5, SK-MYC2, SK-N-AS, SK-N-SH, NBL-S, SK-N-FI and CLB-GA) were cultured in RPMI 1640 medium (Invitrogen) supplied with 15% fetal calf serum, 1% penicillin/streptomycin, 1% kanamycin, 1% glutamine, 2% HEPES (1 M), 1% sodiumpyruvate (100 nM) and 0.1% beta-mercapto (50 nM). At 80% confluence, cells were harvested by scraping for total RNA isolation (miRNeasy, Qiagen) or trypsinized for cytospin preparation. Human colon and brain RNA samples were obtained from Stratagene.
Preparation of cells on membrane-coated slides
Cell suspensions were prepared for microdissection by centrifuging the cells for 10 min at 750 g. Pellets were washed twice with phosphate-buffered saline and resuspended to obtain ∼2.5 × 106 cells/ml. In all, 200 µl of the cell suspension was then transferred by centrifugation (120 g for 3 min) onto slides covered with polyethylene naphthalate membrane (PALM Microlaser Technologies, Bernried, Germany).
Isolation of cells by laser microdissection and pressure catapulting
Single cells were isolated using the PALM MicroBeam system (PALM Microlaser Technologies) as described previously (9) and collected in a 200 µl Eppendorf tube cap containing 3.94 µl RT master mix as detailed below. Following collection of cells, all tubes were centrifuged for 1 min at 16 000 g. Cells were lysed by heating at 95°C for 5 min, after which the entire lysate was used in a Megaplex RT reaction followed by pre-amplification of the miRNA complementary DNA (cDNA) (see further).
miRNA reverse transcription
For miRNA cDNA synthesis, RNA was reverse transcribed using the miRNA reverse transcription kit (Applied Biosystems) in combination with the stem-loop Megaplex primer pool (Applied Biosystems), allowing simultaneous reverse transcription of 450 miRNAs and endogenous controls. Briefly, 8 µl of total RNA (50 ng/µl) was supplemented with RT primer mix (10×), RT buffer (10×), MultiScribe Reverse Transcriptase (10 U/µl), dNTPs with dTTP (0.5 mM each), MgCl2 (3 mM) and AB RNase inhibitor (0.25 U/µl) in a total reaction volume of 80 µl. For RT reactions with subsequent pre-amplification, the reaction volume is proportionally reduced to 5 µl. Concentration of each stem-loop primer in the RT reaction mix was 1 nM, a 50-fold dilution compared with a singleplex RT reaction, ensuring minimal non-specific interactions between the different stem-loop primers. To increase reverse transcription efficiency, a pulsed RT reaction was used (40 cycles of 16°C for 2 min, 42°C for 1 min and 50°C for 1 s, followed by a final reverse transcriptase inactivation at 85°C for 5 min).
Pre-amplification of cDNA
Megaplex RT product (5 µl) was pre-amplified using Applied Biosystems’ TaqMan PreAmp Master Mix (2×) and PreAmp Primer Mix (5×) in a 25-µl PCR reaction. The primer pool consisted of forward primers (50 nM) specific for each of the 450 miRNAs and a universal reverse primer (50 nM) (Applied Biosystems, early access). The pre-amplification cycling conditions were as follows: 95°C for 10 min, 55°C for 2 min and 75°C for 2 min followed by 14 cycles of 95°C for 15 s and 60°C for 4 min.
Real-time qPCR
For each cDNA sample, 384 small RNAs were profiled using a gene maximization PCR plate setup in a 384-well plate. As instrument and liquid handling variations were shown to be minimal, no PCR replicates were measured. This approach allowed us to profile one sample per 384-well plate. Without pre-amplification, RT product was diluted 400-fold; when pre-amplification was applied, the dilution factor was 1600. PCR amplification reactions were carried out in a total volume of 8 µl, containing 4 µl of TaqMan Master Mix (Applied Biosystems), 1 µl of cDNA and 3 µl of miRNA TaqMan probe and primers (Applied Biosystems). Cycling conditions were as follows: 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. All PCR reactions were performed on the 7900HT RT-qPCR system (Applied Biosystems). Raw Cq values were calculated using the SDS software v.2.1 using automatic baseline settings and a threshold of 0.2. The crossing point between the baseline corrected amplification curve and threshold line is called the quantification cycle (Cq) (according to RDML guidelines, http://www.rdml.org) (10).
Assessment of pre-amplification bias
Potential pre-amplification bias was addressed by analyzing the preservation of differential expression. Briefly, differential miRNA expression between different sample pairs (ΔCq) was determined for each approach (with or without pre-amplification). Subsequently, the difference in differential miRNA expression (ΔΔCq) was calculated. Minimal bias should result in low ΔΔCq values.
LNA microarrays
In total, 5 µg of total RNA was hybridized to immobilized locked nucleic acid (LNA)-modified capture probes according to Castoldi et al. (11). Background- and flag-corrected median intensities were log transformed and normalized according to the average signal of each array.
RESULTS
Minimal pre-amplification bias and sensitivity
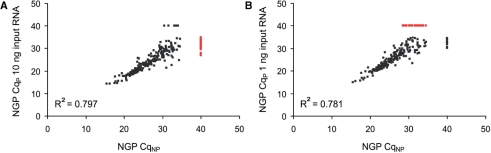
A major concern for introducing a pre-amplification step is the possibility that the relative miRNA expression levels in the original cell population are not maintained. In order to obtain an unbiased pre-amplification of the miRNA cDNA, equal amplification efficiency and a high degree of amplification specificity is required. Previous studies already reported the use of pre-amplification in combination with the stem-loop procedure, but no study thus far evaluated the effects of the pre-amplification step on the fidelity of miRNA expression measurement (12,13). Here we performed an in-depth analysis of the potential bias of a pre-amplification step through direct comparison of miRNA expression profiles obtained with and without pre-amplification. To this purpose, we profiled 384 miRNAs in three different NB cancer cell lines using the Megaplex reverse transcription either with or without pre-amplification of the miRNA cDNA. Because each sample was profiled in a separate PCR run, raw Cq values were corrected for inter-run variation. Calibration and normalization of raw Cq values was done by equalizing the average Cq value across all detectable miRNAs between the different sample runs. For further data analysis, only those miRNAs with a Cq value equal to or below 35 were taken into account. It is generally accepted that a Cq value of 35 represents single molecule template detection; Cq values above 35 are, therefore, considered noise. Low-copy template detection is also subjected to a higher degree of variability, mainly due to Poisson distribution sampling effects. For Megaplex RT without pre-amplification, 400 ng of total RNA was used, whereas for pre-amplified samples, 1 and 10 ng of total RNA were used. Calibrated Cq values obtained with pre-amplification (CqP) were plotted against those obtained without pre-amplification (CqNP) (Figure 1). MiRNAs that were either undetectable or had a Cq value above 35 were assigned a Cq of 40. The Cq–Cq plots for the NGP cells reveal a suboptimal correlation between both data sets (Megaplex RT alone versus Megaplex with pre-amplification), especially for those miRNAs with a high Cq value. Nevertheless, the slope of the linear trendline fitted along the correlation plot nearly equals 1, indicating unbiased and efficient pre-amplification efficiency irrespective of miRNA expression level. Therefore, miRNA quantification using the pre-amplification procedure should result in relative miRNA expression levels that represent the actual situation in the cell population.
Figure 1.
Correlation plot between Cq values obtained through Megaplex RT alone (CqNP) and Megaplex RT with pre-amplification (CqP) using 10 ng of total RNA (A) and 1 ng of total RNA (B) from NGP neuroblastoma cells. (A) Data points highlighted in red indicate miRNAs that are only detectable if pre-amplification is applied. (B) Data points highlighted in red indicate miRNAs that are only detectable if no pre-amplification is applied.
Interestingly, a higher number of miRNAs were detected with the pre-amplification procedure, despite the fact that 40-times less input RNA was used (Figure 1A). Assuming 100% amplification efficiency, a 14-cycle pre-amplification of 10 ng total RNA would result in 200 ng of miRNA cDNA (total RNA equivalents) in the final PCR reaction. When no pre-amplification is applied, each RT-qPCR assay contains only 1 ng of miRNA cDNA (total RNA equivalents). The predicted 200-fold difference in input with or without a pre-amplification step clearly contributes to the superior sensitivity of the pre-amplification approach. When the amount of input RNA is further reduced from 10 to 1 ng, the increased sensitivity is less pronounced as some of the miRNAs become undetectable again (Figure 1B). When using 1 ng of input, both Poisson distribution sampling effects and reduced RT efficiency for low copy numbers come into play. Similar results were obtained with the other cell lines (data not shown).
Reverse transcription variability for low-copy RNA molecules
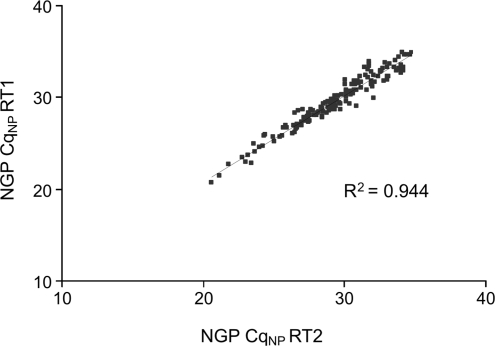
To further investigate the suboptimal correlation in the Cq–Cq plot from a sample analyzed with or without pre-amplification, we characterized the variation introduced by the RT reaction. By performing two independent RT reactions on the same RNA sample, we evaluated the variation induced by the RT step. The Cq–Cq plot for the two RT reactions indeed shows that a certain degree of variation is introduced by the RT reaction (R2 = 0.944) (Figure 2). A similar experiment with the two other cell lines resulted in a correlation coefficient of 0.943 and 0.928. To determine which miRNAs display the highest degree of variation following two subsequent RT reactions, we divided the data set into three subsets according to Cq value: highly expressed miRNAs (Cq below 25), moderately expressed miRNAs (Cq between 25 and 30) and low abundant miRNAs (Cq above 30). Individual correlation coefficients for each subset suggest a much higher RT variation for the low abundant miRNAs (R2 = 0.685) compared to the moderately (R2 = 0.806) and highly (R2 = 0.872) expressed miRNAs. This partly explains the observed variation for the low abundant miRNAs when comparing the data sets obtained with and without pre-amplification (Figure 1) as both were generated with a different RT reaction. Another source of variation could be liquid handling and instrumentation. However, when performing repeated RT-qPCR runs for the same sample, almost no variation was observed (R2 = 0.983).
Figure 2.
Cq–Cq correlation plot for two independent reverse transcription reactions without pre-amplification.
To further evaluate and compare RT variation for the procedures with and without pre-amplification, we determined the variability of individual miRNAs. Triplicate RNA samples from human brain and human colon were reverse transcribed using the method with pre-amplification and the method without pre-amplification. In case of pre-amplification, 10 ng of total RNA was used whereas 400 ng of RNA was used when no pre-amplification was applied. Sixteen individual miRNAs were profiled in triplicate RT-qPCR experiments for each RT sample. Raw Cq values were transformed according to 2−Cq and averaged across the triplicate RT-qPCR experiments for each RT sample. Per miRNA, the averaged expression values were used to calculate the coefficient of variation (CV) across the triplicate RT samples. CV values for each RNA sample (human colon and human brain) profiled with each of the two methods (Megaplex with and without pre-amplification) were then plotted in function of the average Cq value for the sample under consideration (Supplementary Figure S1). Results clearly indicate low CV values for high and moderately expressed miRNAs (average Cq < 30) in both colon and brain samples, independently of the quantification method used. For the low abundant miRNAs (Cq > 30) CV values drastically increase (up to 50%) in colon and brain samples, again independently of the quantification method. This confirms the increased RT variation for low abundant miRNAs.
Preservation of differential expression
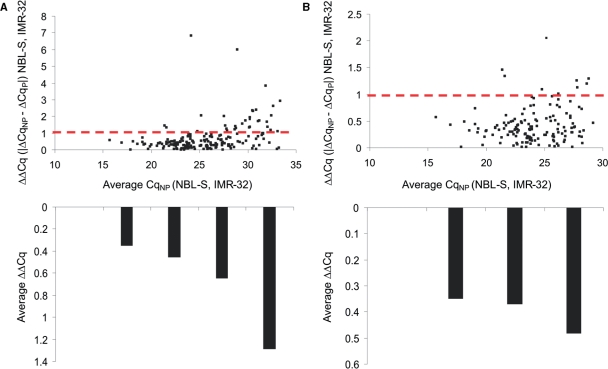
To assess the potential bias introduced through a pre-amplification step, differential miRNA expression levels (ΔCq) between the three different cell lines were determined for each procedure, i.e. with (ΔCqP) or without pre-amplification (ΔCqNP). The difference in differential miRNA expression between two cell lines, measured both with and without pre-amplification (calculated by the ΔΔCq = |ΔCqNP − ΔCqP|), was then plotted against the average Cq value of the miRNA in the two cell lines under consideration, as measured by the procedure with no pre-amplification step (Figure 3A). In all, 80% of all miRNAs with a Cq value <35 display a ΔΔCq < 1. Of these miRNAs, 75% have a ΔΔCq < 0.5. The plot clearly indicates that the ΔΔCq value increases for low abundant miRNAs. While for the most abundant miRNAs (average Cq value between 15 and 20) there is a near-perfect correlation in differential miRNA expression (average ΔΔCq = 0.35), the lowest expressed miRNAs (average Cq value between 30 and 35) display an average ΔΔCq value of 1.3 (Figure 3A). To some degree, this intensity-dependent variation is attributable to the fact that each data set was generated using a different RT reaction as low abundant miRNAs are more susceptible to variation during reverse transcription (see higher). By lowering the detection Cq cutoff from 35 to 30, thereby excluding the lowest expressed miRNAs, 94% of all miRNAs display a ΔΔCq < 1 (Figure 3B). Similar results were obtained in other cell types (Supplementary Figure S2). Interestingly, over half the miRNAs with a ΔΔCq > 1 were differentially expressed (ΔCq > 1), irrespective of the quantification method (with or without pre-amplification). For example, miR-299-5p has a ΔΔCq value > 1, meaning a more than 2-fold bias in differential expression when comparing two samples with and without pre-amplification. However, the individual ΔCq values indicate that this miRNA shows a highly differential expression in the two cell lines (ΔCqP = −6.5, ΔCqNP = −8.3). Large-scale screening studies often apply a ΔCq value of 1 (2-fold difference) to select differentially expressed miRNAs so these miRNAs would definitely be selected, regardless of the use of pre-amplification or not.
Figure 3.
Difference in differential miRNA expression (ΔΔCq) between two neuroblastoma cell lines (NBL-S and IMR-32) as measured using Megaplex RT alone (ΔCqNP) and Megaplex RT with pre-amplification (ΔCqP). ΔΔCq values are plotted in function of the average expression of each miRNA in the two cell lines as quantified by Megaplex RT alone (average CqNP). Bar plots indicate the mean ΔΔCq value for miRNAs with an average CqNP value ranging between 15–20, 20–25, 25–30 and 30–35. (A) Results when a Cq detection cutoff of 35 is applied. (B) Results when Cq detection cutoff is lowered to 30.
To assess whether the high degree of variation observed for the low abundant miRNAs could also be due to miRNA sequence characteristics, six different sequence parameters were evaluated for each mature miRNA (number of A, U, C and G bases, GC percentage and sequence length). For each individual miRNA, the average ΔΔCq value was used as a measure of variation. All 384 miRNAs analyzed were divided into four subgroups according to the average ΔΔCq value (Group 1, ΔΔCq ⩽ 0.5; Group 2, 0.5 < ΔΔCq ⩽ 1; Group 3, 1 < ΔΔCq ⩽ 2; Group 4, ΔΔCq > 2). The number of A bases was the only parameter that significantly differed between the different subgroups (Kruskal–Wallis, P < 0.05). Paired analysis of all subgroups revealed that the number of A bases is significantly higher for miRNAs belonging to Group 4 compared to Groups 1 and 2 (Mann–Whitney, P = 0.009 and P = 0.002, respectively). In general, miRNAs with a ΔΔCq value ⩽1 have a higher number of A bases compared to miRNAs with a ΔΔCq value >1 (Mann–Whitney, P = 0.006). To assess whether these A bases appear randomly across the miRNA sequence or in stretches of consecutive bases, the number of miRNAs containing one or more stretches of at least three A bases was determined for each subgroup. Interestingly, miRNAs with a ΔΔCq value ⩽1 were more likely to contain one or more stretches of at least three A bases as compared to miRNAs with a ΔΔCq value >1 (Fisher exact, P = 0.0049). It remains to be determined how exactly the number of A bases influences the ΔΔCq value.
Confirmation of MYCN-regulated miRNAs
To further evaluate the performance of the Megaplex and pre-amplification method outlined above, we set up a miRNA profiling screen to identify miRNAs that are regulated by the oncogenic MYCN transcription factor, known to play a role in an aggressive type of childhood NB. Recently, a subset of miRNAs regulated by MYCN has been identified (14). Among these miRNAs were four members of the miR-17-92 cluster on chromosome 13 that were identified in at least one of three studied model systems (miR-17-5p, miR-18, miR-20a and miR-92). The miR-17-92 cluster contains a total of seven different miRNAs (miR-17-5p, miR-17-3p, miR-18a, miR-19a, miR-20a, miR-19b and miR-92) and has recently been shown to be a direct target of MYC, the best studied and founding member of the MYC family of bHLH/LZ transcription factors (15). To see whether we could confirm previously published results on MYCN-regulated miRNAs, we profiled all seven miRNAs from the miR-17-92 cluster in a panel of 11 NB cell lines using the Megaplex reverse transcription with pre-amplification starting with 20 ng of input RNA. The panel of cell lines consisted of five MYCN-amplified cell lines, one cell line with stable overexpression of MYCN and five cell lines with normal MYCN copy number. If the miR-17-92 cluster is activated by MYCN, these miRNAs should display a higher expression in the cell lines with MYCN amplification and overexpression. Five miRNAs displayed a significant differential expression between both groups of cell lines (miR-17-3p, miR-19a, miR-19b, miR-20a and miR-92) (Mann–Whitney test, P < 0.05) (Supplementary Figure S3). For miR-17-5p and miR-18a*, there was a trend for higher expression in MYCN-activated cells (P = 0.052). We also quantified the expression of the miR-17-92 cluster in the same 11 cell lines without the pre-amplification step. Mann–Whitney analysis again revealed four differentially expressed miRNAs (miR-17-5p, miR-19b, miR-20a and miR-92). Apparently, both the Megaplex RT with and without pre-amplification are capable of identifying the majority of the miRNAs in the cluster as being differentially expressed with respect to the MYCN status. Moreover, the introduction of the pre-amplification step slightly increases the sensitivity of the detection, identifying significant differential expression for five miRNAs and nearly significant differential expression for the remaining two miRNAs. The entire miR-17-92 cluster is transcribed into a single primary miRNA transcript (pri-miR-17-92). If this cluster is activated by MYCN activity, most likely all miRNAs from the cluster should be activated. The results obtained with pre-amplification most closely resemble this situation.
To further validate our PCR-based approach, we also compared our PCR-based results with an independent miRNA expression profiling study using a microarray platform (3,11). Again, the same subset of cell lines was used, and miRNAs with a significantly differential expression were selected with the Mann–Whitney test (P < 0.05). Three miRNAs from the miR-17-92 cluster displayed a significant differential expression (miR-17-3p, miR-19b and miR-92). For miR-19a, the P-value was very close to being significant (P = 0.052).
Profiling of single cells
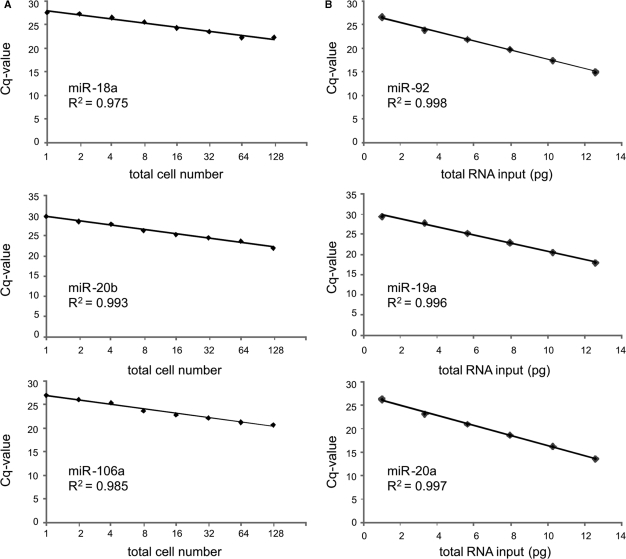
Finally we investigated whether our method would be suited for single-cell analysis. Single-cell RNA levels are cell type dependent and vary between 10 and 30 pg of total RNA. We, therefore, serially diluted human brain total RNA to obtain a dilution series ranging from 6250 to 2 pg of total RNA. All samples from the dilution series underwent Megaplex reverse transcription and pre-amplification prior to RT-qPCR. Expression of both high and low abundant miRNAs was determined, and Cq values were plotted in function of total input RNA (Figure 4B). These plots display perfect linearity, with slopes equaling the theoretical slope of −1 and suggest that the method is suited to profile single cells. However, when analyzing a single cell or few cells, it is desirable that the expression profiles can be readily obtained from total cell lysate. This way, RNA yield is unaffected as no downstream manipulations such as RNA purification or deoxyribonuclease (DNase) treatment is required. The stem-loop reverse transcription primers selectively target mature miRNAs thus ruling out the necessity of a DNase treatment. Second, it is important to verify that the method works for individually isolated cells. To test the possibility of obtaining accurate miRNA expression data from total cell lysate of individual cells, we prepared cytospins of NGP cells on membrane-coated slides. Individual cells were microdissected to obtain a dilution series of 1, 2, 4, 8, 16, 32, 64 or 128 cells that were lysed by heating, followed by Megaplex reverse transcription, pre-amplification and subsequent RT-qPCR. The expression level of three miRNAs, miR-18a, miR-20b and miR-106a, was determined for each point in the dilution series and Cq values were plotted in function of cell numbers (Figure 4A). The resulting dilution series presents with perfect linearity with a slope of −0.856, −1.07 and −0.935, respectively, approaching the theoretical slope of −1.
Figure 4.
(A) Standard curve analysis depicting perfect linearity and correlation between input cell number (1–128) and measured Cq value for three different miRNAs (miR-18a, miR-20b and miR-106a). (B) Standard curve analysis depicting perfect linearity and correlation between total RNA input (2–6250 pg) and measured Cq value for three different miRNAs (miR-92, miR-19a and miR-20a). Total input RNA values on X-axis are log2 based.
DISCUSSION
In this study, we have evaluated the sensitivity and reliability of a pre-amplification step for high-throughput stem-loop primer-based RT-qPCR measurement of miRNA expression.
Our result convincingly demonstrate that such quantification is feasible starting from minute amounts of RNA (10 ng) or even single-cell RNA, thus opening the way for profiling the miRNAome from small cell populations or individual cells. It is important, however, to address the potential bias of the pre-amplification reaction. By evaluating the preservation of differential miRNA expression through comparison of sample pairs with and without pre-amplification, we used a relevant approach to evaluate possible pre-amplification bias. In contrast to previous studies on the use of a pre-amplification step in a stem-loop PCR reaction setup, we have made a direct comparison between results generated with and without pre-amplification and also analyzed the effect on low abundant miRNAs. Further, our data set contains expression data for almost 400 miRNAs, allowing for a more robust analysis. For the most abundant miRNAs, the difference in differential expression, as measured by Megaplex alone and Megaplex followed by pre-amplification, did not exceed 0.8 PCR cycles for 90% of the genes. For the low abundant miRNAs the differential expression difference exceeded 1 more frequently. We identified the RT reaction itself as a major factor contributing to the observed variation. RT-induced variation was particularly high for the low abundant miRNAs, confirming previous studies on mRNA templates (16). In these analyses, variation in RT efficiency plays a major role as paired sample comparison required four independent RT reactions. Most likely, a certain degree of variation will be attributable to the pre-amplification reaction itself. By further expanding the stem-loop primer pool, we not only cover the majority of all human miRNAs but also avoid the need for multiple separate RT reactions per sample.
Our analysis reveals that expression data for low abundant miRNAs obtained through Megaplex reverse transcription followed by pre-amplification should be interpreted with caution. In view of the variability in reverse transcription combined with Poisson distribution effects, results should be confirmed in independent experiments or biological replicates. Important to note, however, is that the direction of differential gene expression is almost always preserved.
We demonstrated the sensitivity of our approach by confirming (and extending) differential expression of the miR-17-92 cluster. For five out of seven miRNAs residing within this cluster we could confirm significant MYCN-dependent expression while MYCN-dependent expression for the other two was borderline significant. MYCN-dependent expression was already reported for four miRNAs within the cluster. Furthermore, this cluster is directly activated by MYC, a transcription factor with high homology to MYCN. Finally, when repeating the same analysis using LNA-based microarray technology, three of seven miRNAs displayed significant MYCN-dependent expression, confirming literature and stem-loop RT-qPCR profiling results obtained in this study. The above results underscore the high sensitivity and accuracy of Megaplex reverse transcription followed by limited-cycle pre-amplification. The fact that more miRNAs are detected when applying the pre-amplification procedure further illustrates its impact on detection sensitivity.
To address the possibility of single-cell miRNA expression analysis, we evaluated preservation of detection linearity in a 2-fold dilution series ranging from 1 to 128 individually isolated cells. For the miRNAs analyzed, detection linearity was maintained down to the single-cell level. Moreover, miRNA profiling could be performed directly on total cell lysate avoiding the need for sample manipulations that affect the RNA yield. Tang and colleagues (13) have shown that using a 220-plex stem-loop primer pool, accurate miRNA expression profiling from single handpicked cells is possible. Here we show that an increased complexity of the stem-loop primer pool still allows for highly accurate miRNA quantification on whole-cell lysate from individually picked cells. Important to note, however, is the discrepancy between the expression level in a single cell and the average expression in a population of cells belonging to the same cell type, due to the lognormal distribution of gene expression (17). This can explain the slightly larger variation that was observed for the single-cell dilution series compared to the RNA dilution series. Fundamental biological questions on early tumour development or stem cell differentiation are best tested at the single-cell level. Often, specific cells are microdissected from a heterogeneous population, thereby ensuring that the target population is free of contamination with non-target cells (18,19). Also, studies on embryonic development are typically performed on the single-cell level (20). We strongly believe that the method presented here will aid in the unraveling of miRNA function in single-cell studies or pure cell populations isolated by microdissection, flow sorting or bead-based selection.
In this study, we have shown that the Megaplex stem-loop PCR procedure in combination with limited-cycle pre-amplification is a powerful method for miRNA expression profiling in both large and small cell populations capable of covering the majority of human miRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Scientific Research (FWO) Flanders (FWO postdoc to J.V.); Kinderkankerfonds (a non-profit childhood cancer foundation under Belgian law); Ghent University Research Fund (BOF; 01D31406 to P.M.). Funding for open access charge: EU STREP EET-pipeline n° 037260.
Conflict of interest statement. Nathalie Bernard, Caifu Chen and Simone Guenther are employees of Applied Biosystems.
Supplementary Material
References
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru CD, Shimizu M, Zupo S, Dono M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods. 2004;1:155–161. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
- 6.Sioud M, Rosok O. Profiling microRNA expression using sensitive cDNA probes and filter arrays. Biotechniques. 2004;37:574–576. doi: 10.2144/04374ST01. 578–580. [DOI] [PubMed] [Google Scholar]
- 7.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 8.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schutze K, Lahr G. Identification of expressed genes by laser-mediated manipulation of single cells. Nat. Biotechnol. 1998;16:737–742. doi: 10.1038/nbt0898-737. [DOI] [PubMed] [Google Scholar]
- 10.Taylor CF, Field D, Sansone SA, Aerts J, Apweiler R, Ashburner M, Ball CA, Binz PA, Bogue M, Booth T, et al. Promoting coherent minimum reporting guidelines for biological and biomedical investigations: the MIBBI project. Nat. Biotechnol. 2008;26:889–896. doi: 10.1038/nbt.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castoldi M, Schmidt S, Benes V, Hentze MW, Muckenthaler MU. miChip: an array-based method for microRNA expression profiling using locked nucleic acid capture probes. Nat. Protoc. 2008;3:321–329. doi: 10.1038/nprot.2008.4. [DOI] [PubMed] [Google Scholar]
- 12.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 2008;14:27–41. doi: 10.3233/jad-2008-14103. [DOI] [PubMed] [Google Scholar]
- 13.Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, Krause M, Astrahantseff K, Klein-Hitpass L, Buettner R, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int. J. Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 16.Stahlberg A, Hakansson J, Xian X, Semb H, Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004;50:509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 17.Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res. 2005;15:1388–1392. doi: 10.1101/gr.3820805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin MS, Kim HS, Kang CS, Park WS, Kim SY, Lee SN, Lee JH, Park JY, Jang JJ, Kim CW, et al. Inactivating mutations of CASP10 gene in non-Hodgkin lymphomas. Blood. 2002;99:4094–4099. doi: 10.1182/blood.v99.11.4094. [DOI] [PubMed] [Google Scholar]
- 19.De Preter K, Vandesompele J, Heimann P, Yigit N, Beckman S, Schramm A, Eggert A, Stallings RL, Benoit Y, Renard M, et al. Human fetal neuroblast and neuroblastoma transcriptome analysis confirms neuroblast origin and highlights neuroblastoma candidate genes. Genome Biol. 2006;7:R84. doi: 10.1186/gb-2006-7-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.