Abstract
Expression of amyloid precursor protein (APP) is critical to the etiology of Alzheimer's disease (AD). Consequently, regulating APP expression is one approach to block disease progression. To this end, APP can be targeted at the levels of transcription, translation, and protein stability. Little is currently known about the translation of APP mRNA. Here, we report that endogenous APP mRNA is translated in neural cell lines via an internal ribosome entry site (IRES) located in the 5′-untranslated leader. The functional unit of the APP IRES is located within the 5′ 50 nucleotides of the 5′-leader. In addition, we found that the APP IRES is positively regulated by two conditions correlated with AD, increased intracellular iron concentration and ischemia. Interestingly, the enhancement of APP IRES activity is dependent upon de novo transcription. Taken together, our data suggest that internal initiation of translation of the APP mRNA is an important mode for synthesis of APP, a mechanism which is regulated by conditions that also contribute to AD.
INTRODUCTION
Alzheimer's disease (AD) is a neurodegenerative disorder characterized by the presence of intracellular neurofibrillary tangles as well as extracellular plaques composed of aggregates of β-amyloid peptide (Aβ) in the brain. Aβ is central to the pathophysiology of AD and may initiate the disorder (1). The peptide is derived from the sequential cleavage of amyloid precursor protein (APP) by two proteases, β- and γ-secretase. A β-secretase (Bace1) (2) and the catalytic subunits of the γ-secretase, presenilin 1 and 2 (PS1 and PS2) (3,4) are now accepted as genes that play an essential role in the onset and progression of the disease (5).
If expression of the Aβ peptide is a major contributor to the etiology of AD, then a logical target for amelioration of the disease is to regulate the cleavage of its precursor protein, APP, into the Aβ peptide products. An alternative approach would be to regulate availability of APP itself, which should also decrease levels of the Aβ peptide. Multiple avenues may be amenable to alter APP expression. For example, transcription of the APP gene has been well studied with identification of cell type-specific promoter regions and different promoter alleles that may contribute to AD (6–9). Once APP has been translated, reducing APP protein stability could decrease the likelihood of its cleavage by secretases. However, nascent APP already exhibits a short half-life (ranging from 20 min to 90 min) (10–12) and once it is cleaved, the Aβ peptide is very stable and insoluble, likely making it difficult to further affect protein half-life or increase peptide solubility.
Translation initiation is an additional process to regulate protein expression (13). The main form of initiation in eukaryotes is cap-dependent translation, which relies upon the rate-limiting protein eukaryotic initiation factor (eIF) 4E to bind the 7-methyl-guanosine (m7G) cap structure at the 5′-end of the mRNA. eIF-4E subsequently recruits the scaffolding protein eIF-4G, to which the remainder of the translational machinery (including the 40S ribosomal subunit and methionine-loaded tRNA) is attached, forming the 43S pre-initiation complex [for review, see Ref. (14)]. The 43S complex is proposed to then migrate or scan along the mRNA 5′-leader until it encounters the first initiator codon (AUG). Upon recognition of the initiator codon by the 43S complex, GTP hydrolysis occurs, the 60S ribosomal subunit joins the 40S subunit to form the 80S ribosome, translation initiation factors are liberated and peptide synthesis ensues (15,16).
Cap-dependent translation is diminished during mitosis (17) as well as in reaction to cellular stressors such as decreased oxygen supply resulting from ischemia (18). Cap-dependent translation is also inhibited in response to particular viral infections, such as poliovirus (19). In response to these stressors, a subset of eukaryotic mRNAs continue to be translated via initiation at an internal ribosome entry site (IRES) generally situated in their 5′-leader (20–23). IRESes directly recruit translational machinery, independently of the 5′-m7G cap structure and enable a select pool of mRNAs to be translated in response to conditions when global or cap-dependent translation is inhibited (24). Moreover, recent studies have found that cells containing mutations in the dyskerin gene exhibit a decrease in the expression of a selective group of proteins via a disruption in the internal initiation of translation of their mRNAs (25). This result indicates that IRES-dependent translation may be a primary translational mechanism for a subset of mRNAs.
Recent evidence indicates that the APP mRNA may be translated through an IRES. Qin and Sarnow (17) found that APP mRNA is one of several mRNAs which remain associated with polyribosomes during mitosis, when cap-dependent translation is greatly reduced. In addition, translation of a second cistron from a dicistronic DNA construct increased when the APP 5′-leader was placed into its intercistronic region (17). However, many questions remain and some reports have challenged the use of dicistronic DNA constructs to assay IRES activity. The presence of a cryptic promoter or cryptic splice acceptor site in the DNA sequence of a 5′-leader may generate RNA species which only contain the open reading frame (ORF) for the second cistron (26). The resulting monocistronic RNA would lead to an artifactual increase in the expression of the second cistron (27). Perhaps more importantly, it is not known whether IRES-dependent translation is a physiologically significant mechanism for APP expression in vivo and whether cap- and/or IRES-dependent translation is utilized under conditions in which APP synthesis is upregulated, thus promoting progression of AD.
Several studies suggest a role for elevated intracellular iron in the etiology of AD (28). During normative aging, there is an increase in both iron and the iron-sequestering protein, ferritin, in the central nervous system. However, in AD patients, there is a further enrichment of iron in the brain without the concomitant increase in ferritin (29,30). This increased level of labile iron is available to participate in the formation of reactive oxygen species and contributes to multiple cellular reactions which promote AD; the synthesis of APP, the rate of Aβ oligomerization, aggregation and precipitation are all enhanced through its interaction with iron (31,32). Once in an extracellular plaque, iron can enhance Aβ toxicity through the generation of hydroxyl radicals and the resulting oxidative stress (33).
Perturbations in the cerebral vasculature which induce ischemic conditions in the brain are also proposed to be a causal factor for AD [for reviews, see Refs (34,35)]. Indeed, at autopsy most AD patient brains exhibit pathology of prior cerebrovascular damage (36), including vascular irregularities as well as a decrease in capillary density (37). Recent data also suggest that ischemia and reperfusion lead to the sequestration of canonical cap-dependent translation factors into mRNA storage bodies in rat hippocampus, leading to a global translational arrest (38). Despite this finding, ischemia has also been shown to lead to an increase in APP expression (39) and cleavage (40).
Interestingly, both elevated intracellular iron levels and ischemia individually appear to promote AD at the level of translation. Despite the global inhibition of cap-dependent protein synthesis imparted by these cellular stressors (41,42), APP expression is enhanced (32,43). These results support the hypothesis that APP synthesis takes place via an alternate translation initiation mechanism allowing for it to be synthesized even under these conditions. Indeed, in the present report we validate the initial report that the APP 5′-leader contains an IRES and show that IRES-dependent translation is a mechanism by which endogenous APP mRNA is translated. Moreover, increased intracellular iron levels and chemical ischemia both augment APP IRES activity and endogenous APP synthesis in neural cell lines. These results indicate that the APP IRES may be a target for regulating APP expression in vivo.
MATERIALS AND METHODS
Cloning, reporter constructs and RNA transcription
PCR primers were designed to amplify the 5′-leader of the human APP and β-globin (GenBank accession numbers: APP, NM_000484; β-globin, V00497) mRNAs from human adult brain cDNA. A 4-nt deletion (−64 AGAG −61) in the APP 5′-leader (APPΔ) was generated by Gene Tailor Site Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA). The 5′-leaders were inserted into an EcoRI and NcoI restriction site between the Renilla luciferase and the Photinus luciferase cistrons of a dicistronic construct (44) and subcloned into the PBluescript II SK (+) vector (Stratagene, La Jolla, CA, USA). Monocistronic luciferase constructs (45) containing the 5′-leader upstream of the Photinus luciferase ORF were also subcloned into PBluescript II SK (+). Dicistronic and monocistronic RNA were in vitro transcribed from linearized plasmids and poly A tailed, all according to the manufacturer's protocol, using mMessage mMachine T7 and mMessage mMachine T7 Ultra (Ambion, Austin, TX, USA), respectively. For studies comparing translation from untailed m7G-capped mRNAs to that from ApppG-capped mRNAs, Megascript T7 (Ambion) was used to in vitro transcribe the transcripts with ApppG cap analog (New England Biolabs, Ipswich, MA) used in place of m7G cap analog in the reaction. All RNAs were heat-denatured, run on nondenaturing 3-(N-morpholino) propanesul fonic acid (MOPS) agarose gels (Lonza Rockland Inc., Rockland, ME, USA), and visualized with SYBR gold to check their size and integrity prior to transfection in cells.
Cell culture
C6 and SH-SY5Y cells were obtained from American Type Culture Collection (ATCC) (Manassas, VA) and grown in complete culture medium: Dubelco's Modified Eagle Medium (DMEM; Sigma Aldrich, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; Sigma Aldrich) and 1% penicillin (10 000 U/ml)/streptomycin (10 000 μg/ml)/glutamine (29.2 mg/ml) (Cellgro, Herdon, VA, USA).
Dicistronic DNA transfection
C6 cells were plated at a density of 1.0 × 105, 17 h prior to transfection. Immediately prior to transfection, complete growth media described above was refreshed. Cells were transfected using FuGene6 transfection reagent (Roche, Indianapolis, IA, USA) with 1 µg of dicistronic DNA construct. Cells were lysed with 400 µl 1× Passive Lysis Buffer (Promega, Madison, WI, USA) for 15 min at room temperature with rocking. Fifty microliters of the lysate was assayed for luciferase activity using the dual reporter assay system (Promega) in a Luminoskan Ascent luminometer (Thermo Labsystems, Franklin, MA).
Monocistronic and dicistronic RNA transfection
C6 cells were plated at a density of 1 × 106 in 35-mm well plates for 17 h. Cells were rinsed with PBS and complete culture media (described above) was replaced with serum-free media (DMEM containing Glutamine, Sigma Aldrich). Cells were then transfected with 4 μg of dicistronic m7G-capped and tailed reporter RNAs with 16 μl of TransMessenger transfection reagent (Qiagen, Valencia, CA, USA). After 3 h, the cells were rinsed with PBS and complete culture media was added for an additional 4 h. Cells were rinsed with PBS and lysed with 400 μl of 1× Passive Lysis Buffer (Promega) for 15 min at room temperature with rocking. Fifty microliters of lysate was assayed for luciferase activity using the dual reporter assay system (Promega) in a Luminoskan Ascent luminometer (Thermo Labsystems). For monocistronic RNA transfections, the same transfection protocol was followed, except that C6 cells were co-transfected with 2 μg of either m7G-capped or ApppG-capped monocistronic Photinus luciferase RNA along with 2 μg of an m7G-capped Renilla luciferase RNA to control for transfection efficiency.
Cellular treatments
To study the effect of iron on IRES activity, cells were exposed to 50 μM of ferric ammonium citrate (FAC; Sigma Aldrich) and/or 2.5 μg/ml of Actinomycin D (Calbiochem, San Diego, CA, USA). Cells were treated for a total of 24 h: 17 h before the RNA transfection, 3 h during the transient RNA transfection and 4 h after the RNA transfection. To study the effect of ischemia on IRES activity, C6 cells were exposed to 10 mM sodium azide (Acros organics, Morris Plains, NJ, USA) and 10 mM dideoxyglucose (Calbiochem) for 10 min. The cells were then rinsed with PBS and transfected with RNA. For ex vivo studies using rapamycin (Sigma Aldrich), C6 cells were treated with 40 nM rapamycin (Sigma Aldrich) for 16 h. In studies using cycloheximide (Sigma Aldrich), C6 cells were treated with 300 µg/ml cycloheximide in complete cell culture media for 0 min, 30 min, 60 min, 90 min, 120 min or 180 min prior to harvest using 200 µl of 1× cell culture lysis buffer (Promega) containing phosphatase inhibitors (Pierce, Rockford, IL, USA) and protease inhibitors (Roche) as per manufacturer's specifications. Lysates were either immediately used for western blotting or immediately stored at −80°C. GraphPad Prism software was used to calculate half-life using the nonlinear one-phase decay equation.
Cellular siRNA treatments
Nonsense siRNA of 10 μM (Dharmacon, Thermo Scientific, Lafayette, CO, USA, D-001206-13-20) or rat eIF-4E siRNA (Dharmacon, L-088826-01-0010) were incubated in 35-mm plate wells with 12 μl of Interferin transfection reagent (PolyPlus-transfection, NY, USA, 40950) at 37°C for 10 min. During this time, cultured C6 cells were plated at 1.0 × 106 in wells already containing siRNA complexes and serum-free growth media. Complete growth media was then added to a final volume of 2.2 ml. Cells were harvested after 72 h with 200 µl of 1× cell culture lysis buffer (Promega) containing phosphatase inhibitors (Pierce) and protease inhibitors (Roche) as per manufacturer's specifications. Lysates were either immediately stored at −80°C or used for western blotting.
In vitro translation
Capped and tailed monocistronic RNAs were in vitro translated in micrococcal nuclease-treated rabbit reticulocyte lysate (SpeedRead Lysates, Novagen, Madison, WI, USA). The 0.5 μg of RNA, 100 mM of KCl, 0.25 mM of Mg acetate, 312.5 μM methionine and increasing concentrations of m7G cap analog (New England Biolabs) were added to the reaction and incubated for 1 h at 30°C. Luciferase activity was then measured as detailed above. To verify stability of the transcripts following reactions, RNA was isolated from each rabbit reticulocyte lysate (RRL) reaction using MegaClear (Ambion). RNA (1 µg of each) was then analyzed by northern blot analysis and probed with DIG-labeled RNA probe against Photinus luciferase.
Western blot analysis
Cells were harvested with 1× cell culture lysis buffer (Promega) with protease and phosphatase inhibitors (Roche). The cell lysate was analyzed via western blotting by separating the proteins on a 10% SDS–polyacrylamide gel with subsequent transfer onto nitrocellulose. The membranes were blocked for 1 h at room temperature in 3% milk and TBST (Tris pH 7.9, NaCl, Tween 20). Nitrocellulose blots were then probed with: primary monoclonal antibody to APP (Millipore, Billerica, MA, USA) 1:1000 overnight at 4°C; primary monoclonal antibody to eIF-4E (BD Biosciences, Boston, MA, USA) 1:1000 overnight at 4°C; primary polyclonal antibody to CX37/GJA4 (LifeSpan Biosciences, Seattle, WA, USA) 1:500 overnight at 4°C; primary polyclonal antibody to GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA) 1:500 for 1 h at room temperature; primary polyclonal antibody to phospho-p70s6Kinase (Cell Signaling Technology, Danvers, MA, USA) 1:1000 overnight at 4°C; primary polyclonal antibody to total p70s6Kinase (Cell Signaling Technology) 1:1000 overnight at 4°C; or JunB (P169) (Cell Signaling Technology) 1:500 for 4 h at room temperature, all diluted in TBST. The blots were then rinsed briefly with TBST and incubated with anti-mouse (1:10 000) or anti-rabbit (1:50 000) horseradish peroxidase-conjugated secondary antibody (Promega) for 1 h at room temperature. Immunoreactive bands were detected by developing the blots with ECL plus chemiluminescence reagents (GE Healthcare, Piscataway, NJ, USA) and scanned with a Storm phosphorimager. Protein expression levels were quantified using ImageQuant software (GE Healthcare) and each lane normalized for protein loading using GAPDH levels.
RNA Isolation
For RT–PCR experiment, TRI-Reagent (Sigma Aldrich) was used to isolate total RNA from cells in culture. Briefly, RNA was isolated from the aqueous phase of the TRI-reagent preparation, purified with phenol/chloroform extraction and precipitated with isopropanol followed by an ethanol wash. Dried and pelleted RNA was resuspended in RNAse-free water (Ambion) and incubated at 65°C for 10–15 min. For isolation of exogenous RNA from RRL experiments, MegaClear (Ambion) was used. For northern analysis of endogenous mRNA following cellular treatments in C6 cells, total RNA was isolated using RNAqueous (Ambion) as per manufacturer's specifications.
Northern blot analysis
Total RNA was isolated from samples as detailed above and heat-denatured for 15 min at 65°C. For northerns of endogenous RNA, 1 µg was run on a 1.25% agarose gel and transferred to a positive nylon membrane. RNA was UV cross-linked to the membrane. Hybridization of the DIG-labeled in vitro transcribed Photinus luciferase gene, APP gene or GAPDH gene-specific probe was done overnight at 68°C. Anti-DIG-AP (Roche) identified the bound probes and the addition of CDP-star substrate was used to visualize RNA using X-ray film exposure.
Reverse transcription and PCR
ImPROMII Reverse Transcription System (Promega) was performed on total RNA. Nonspecific RNA primers [Oligo(dT)] provided in the kit were used in the reverse transcription reaction to amplify all mRNA. The positive control was prepared as suggested in the product manual. The first negative control contained no RNA template but did contain reverse transcriptase (RT) enzyme. The second negative control did not contain RT enzyme but did contain the RNA template. The RT reaction was carried out as described in the product manual. PCR was performed using Proofstart (Qiagen). For PCR, RT reaction products were amplified with 5′-end primers to amplify upstream of the Renilla codon (GGC CTA GGC TTT TGC AAA) and 3′-primers to amplify the Photinus codon (ATC TTC CAG CGG ATA GAA TGG CGC CGG). PCR cycles and temperature settings were applied as per manufacturer's instructions, except that the number of amplification cycles was increased to 50.
RESULTS
APP is endogenously synthesized in an mTOR-independent manner in neural cells
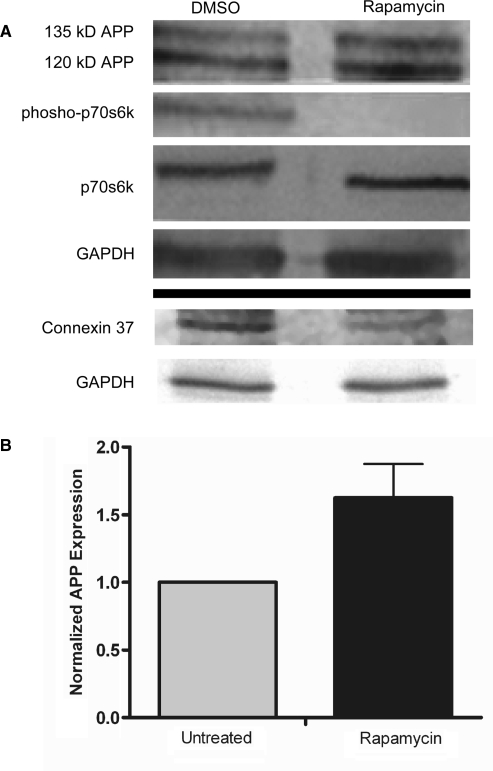
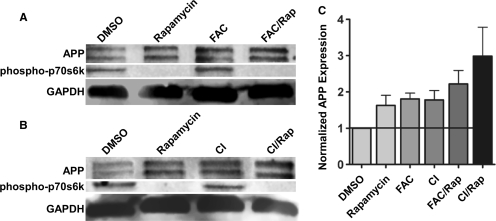
To determine if APP can be synthesized by a cap-independent mechanism, rat neural C6 cells in culture were treated for 16 h with rapamycin to inhibit mammalian target of rapamycin (mTOR) and consequently cap-dependent translation (46). In the rapamycin-treated cells, phosphorylation of p70s6 kinase (p70s6K), a protein which is a substrate of mTOR, was dramatically diminished indicating that the drug treatment was effective. In the presence of rapamycin, the amount of full length APP (mature ∼135 kDa and immature ∼120 kDa) (47) actually increased by ∼50% demonstrating that APP continues to be synthesized when cap-dependent translation is inhibited (Figure 1). We also observed similar results in human neuroblastoma SH-SY5Y cells (Figure S1). Previous studies using transfected plasmids encoding for APP have found a half-life between 20 min and 90 min (10–12). In C6 cells, we found a half-life of 45 min for the mature form of APP (135 kDa) and a half-life of 19 min for the immature form (120 kDa) (Figure S2). The rapid turnover of APP indicates that protein stability did not confound interpretation of the results. Alternatively, expression of connexin 37 protein was reduced (Figure 1). This result is in agreement with the previous observation of a loss of connexin 37 mRNA on polyribosomes following poliovirus infection (48). Taken together, these results indicate that connexin 37 is translated in a cap-dependent manner, while translation of APP can be maintained or even increased when cap-dependent translation is diminished by inhibiting mTOR.
Figure 1.
APP is endogenously synthesized in a cap-independent manner in rat C6 glioma cells. C6 cells in culture were treated for 16 h with either DMSO or rapamycin. Cell lysates were analyzed via western blotting for (A) APP, phosphorylated p70s6 kinase and total p70s6 kinase, (B) connexin37 and GAPDH as a loading control. Imagequant Software was used to quantify APP expression from six separate experiments.
Knockdown of the cap-binding protein eIF-4E does not affect endogenous APP expression
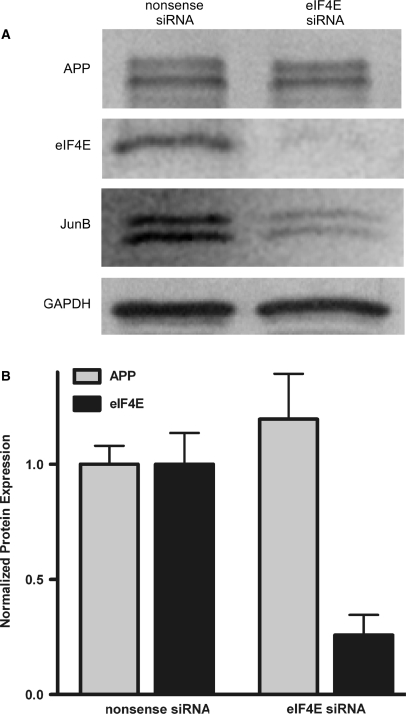
To confirm that APP mRNA can be translated in a cap-independent manner, we knocked down expression of the cap-binding protein eIF-4E (Figure 2). Rat C6 neural cells were treated with siRNA directed against eIF-4E or with control nonsense siRNA (Dharmacon). After 72 h, APP expression was equivalent in the eIF-4E siRNA and control siRNA cells despite the observation that eIF-4E protein was knocked down by >80%. JunB, whose mRNA is translated by a cap-dependent mechanism (49), was reduced in expression in cells treated with eIF-4E siRNA. GAPDH was used as a loading control because it has a very stable half-life, ranging from 90 h to 120 h (50).
Figure 2.
Knockdown of the cap-binding protein eIF-4E in rat C6 cells does not affect APP expression. (A) C6 cells were treated with either nonsense siRNA or siRNA directed against eIF-4E and harvested after 72 h. Western blotting was used to detect levels of APP, eIF-4E, JunB and GAPDH. (B) Imagequant software was used to quantify protein levels from five separate experiments.
The APP 5′-leader exhibits IRES activity
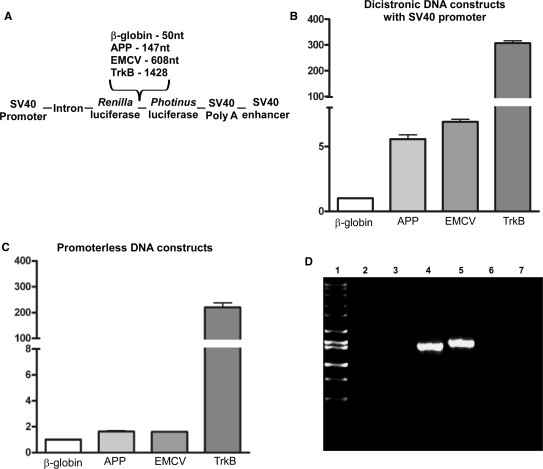
To determine whether the APP mRNA is translated via an IRES in its 5′-leader, we assayed the ability of the APP 5′-leader and other 5′-leaders to initiate translation when placed in the intercistronic region of a dicistronic construct (Figure 3) [dicistronic DNA constructs generously provided by Dr Anne Willis (44)]. The constructs contained the ORFs for Renilla luciferase and Photinus luciferase and were transfected into C6 neural cells for 24 h. Cell lysates were then obtained and assayed for luciferase activity. Ratios of Photinus luciferase to Renilla luciferase (P:R) obtained from each 5′-leader construct were normalized to the P:R ratio obtained from the control β-globin 5′-leader construct. The β-globin mRNA is translated exclusively in a cap-dependent manner (51). P:R ratios above a value of 1 therefore indicate the presence of an IRES. The dicistronic construct containing the encephalomyocarditis virus (EMCV) IRES was used as a positive control and exhibited a P:R ratio of ∼8-fold over the negative control (Figure 3A and B). The P:R ratio from the dicistronic DNA containing the APP 5′-leader was ∼6-fold higher than the β-globin construct. A similar finding was shown by Qin and Sarnow (17) using a comparable dicistronic luciferase DNA construct and suggests that the APP 5′-leader has an IRES.
Figure 3.
The APP 5′-leader exhibits IRES activity which cannot be attributed to the presence of a cryptic promoter or cryptic splice acceptor activity. (A) C6 cells were transfected for 24 h with dicistronic luciferase DNA constructs containing an SV40 promoter and the β-globin, APP, EMCV or full-length mouse TrkB 5′-leader inserted into the intercistronic region. (B) Luciferase activity is shown as the ratio of Photinus luciferase to Renilla luciferase (P:R) and is normalized to the activity obtained from the construct containing the β-globin 5′-leader. (C) C6 cells were transfected for 24 h with promoterless dicistronic luciferase DNA constructs containing the β-globin, APP, EMCV or mouse TrkB 5′-leaders and assayed for Photinus and Renilla luciferase activity. (D) C6 cells were transfected with promoter-containing dicistronic DNA constructs for 24 h. Total RNA was harvested and nonspecific RT was performed. cDNA products were then amplified with 5′-primers annealing upstream of the Renilla and 3′-primers annealing to the Photinus cistron. PCR products were analyzed on a 1% agarose gel. Lane 1, Hi-Lo ladder (Bionexus, Oakland, CA, USA); lane 2, no template; lane 3, mock transfection; lane 4, β-globin construct transfection; lane 5, APP construct transfection; lane 6, no RT enzyme; and lane 7, no PCR reaction.
The use of dicistronic DNA constructs has two major limitations. Cryptic promoter or cryptic splice acceptor activity may be present in the 5′-leader leading to an abundance of capped monocistronic Photinus luciferase transcripts and thereby presenting an artificially augmented P:R ratio. To test for the presence of a cryptic promoter in the APP 5′-leader, the leader was inserted into the intercistronic region of a promoterless dicistronic DNA construct [graciously provided by Dr Anne Willis (52)]. We used the β-globin 5′-leader as a negative control and the full-length mouse neurotrophin receptor TrkB 5′-leader as a positive control, which we showed previously to contain a cryptic promoter (53). When C6 cells were transfected with the promoterless DNA constructs for 24 h, the APP 5′-leader and EMCV IRES constructs produced P:R ratios similar to that of the negative control β-globin; while, the TrkB 5′-leader exhibited a P:R ratio of >200. This result indicates that the APP 5′-leader does not contain a cryptic promoter.
The dicistronic DNA construct contains an upstream intron and a cryptic splice acceptor site in a 5′-leader situated in the intercistronic region would result in the splicing of the Renilla luciferase ORF. To determine whether the APP 5′-leader exhibits cryptic splicing activity, we transfected C6 cells with the promoter-containing dicistronic DNA constructs for 24 h and amplified the resulting RNA species using reverse transcriptase (RT) RT–PCR. The 5′-PCR primer was designed to lie upstream of the Renilla ORF, while the 3′-primer was designed to anneal in the middle of the Photinus ORF as previously demonstrated (54). With these primer designs, both mono- and dicistronic RNA species would be amplified regardless of the presence of the Renilla luciferase ORF in the RNA transcript, as shown previously (54). Only single mRNA species were observed from the transfections of the dicistronic DNA constructs containing the β-globin or APP 5′-leader at ∼1.5 KB, which is the size expected for full-length dicistronic mRNA. Negative controls including an RT reaction in absence of a template followed by PCR, RT–PCR on total RNA from mock-transfected cells, no RT enzyme and no PCR reaction (Figure 3D) did not show any bands. This data indicate that the APP 5′-leader does not contain a cryptic splice acceptor site. Moreover, the data support the conclusion that the enhanced P:R ratio from the APP 5′-leader in the DNA dicistronic constructs is due to the presence of an IRES.
Transfection of dicistronic RNA confirms that the APP mRNA 5′-leader contains an IRES
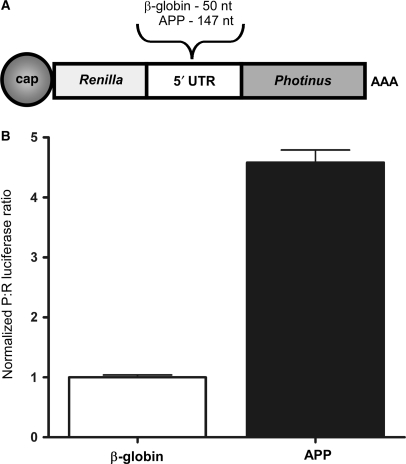
To overcome any possible limitations of the dicistronic DNA constructs and validate the presence of an IRES in the APP 5′-leader, we in vitro transcribed dicistronic m7GpppG (m7G)-capped and polyA-tailed mRNA with intercistronically inserted 5′-leader sequences and transfected rat neural C6 cells with these individual mRNAs. Seven hours later, the cell lysates were harvested and assayed for luciferase activity. From these transfections, the dicistronic mRNA containing the APP 5′-leader RNA revealed a P:R ratio of nearly 5-fold over the negative control β-globin 5′-leader (Figure 4), similar to the P:R ratios obtained using dicistronic DNA construct transfections (Figure 3B). This result further supports the presence of an IRES in the APP 5′-leader.
Figure 4.
Ex vivo transfection of dicistronic RNA confirms that the APP mRNA 5′-leader contains an IRES. (A) Dicistronic luciferase RNAs containing the 5′-leader from the β-globin or APP mRNA inserted into the intercistronic region of a dicistronic luciferase construct were individually transfected into C6 cells. (B) Luciferase activity is shown as the ratio of Photinus luciferase to Renilla luciferase (P:R) and is normalized to the activity obtained from the mRNA containing the β-globin 5′-leader.
The APP 5′-leader can initiate translation independent of the 5′-m7G cap structure in vitro
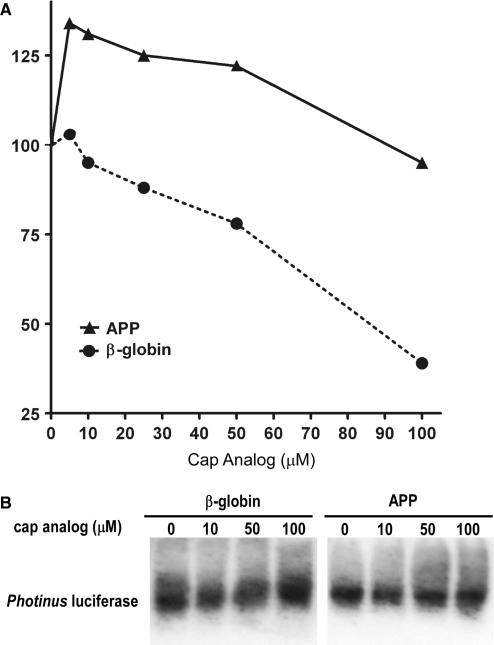
While dicistronic constructs are useful tools to identify IRESs, most eukaryotic mRNAs are monocistronic and positioning of the IRES within the mRNA and in particular its distance from the 5′-end of the mRNA may alter its activity. Consequently, we examined the ability of the APP 5′-leader to initiate translation of a monocistronic mRNA. In vitro transcribed monocistronic capped and tailed mRNA containing the β-globin or APP 5′-leaders placed upstream of the Photinus luciferase ORF were added to RRL. An increasing concentration of m7G cap analog (New England Biolabs) was also added to compete with the cap structure for eIF-4E and inhibit cap-dependent translation. In the presence of 100 µM of cap analog, translation of the β-globin 5′-leader-containing RNA was reduced by >60% (Figure 5A). In contrast, translation of the mRNA containing the APP 5′-leader was remarkably resistant to the effects of cap analog addition. In fact, low concentrations of cap analog actually led to an increase in Photinus luciferase activity, suggesting there may be a competition between the cap and IRES for the translation initiation machinery. At the highest concentrations of cap analog, Photinus luciferase activity from the APP 5′-leader mRNA was equivalent to that obtained from the untreated lysate. A 2-way ANOVA confirmed a significant difference in protein expression from the mRNAs containing the APP and β-globin 5′-leaders (P < 0.05). Moreover, northern blot analysis did not show any differential change in the levels of the two mRNAs after addition of cap analog indicating that RNA stability was not confounding interpretation of the results (Figure 5B). Consequently, this in vitro experiment demonstrates that the APP 5′-leader can mediate cap-independent translation.
Figure 5.
The APP 5′-leader can initiate cap-independent translation in vitro. (A) Monocistronic luciferase RNAs containing the 5′-leader from the β-globin and APP mRNAs inserted upstream of the Photinus luciferase ORF were individually translated in micrococcal nuclease-treated RRL in the presence of increasing concentrations of cap analog. The initial level of Photinus luciferase activity from each RNA was normalized to 100. (B) Northern blot analysis was performed on monocistronic RNA isolated from the RRL in vitro translation assays in presence of increasing amounts of cap analog to demonstrate transcript integrity. RNA was probed with DIG-labeled in vitro transcribed probe against Photinus luciferase.
The 5′ 50 nt of the APP 5′-leader are sufficient to internally initiate translation
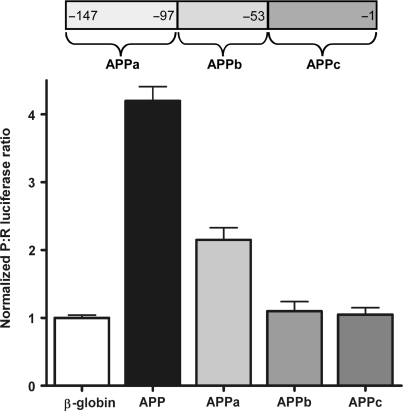
To locate the region within the APP 5′-leader that is required for basal IRES activity, the 5′-leader was divided into three nonoverlapping regions of 44–53 nt (APPa, APPb and APPc; Figure 6). When C6 cells were transfected with dicistronic RNAs containing APPa, APPb or APPc 5′-leader regions, only APPa exhibited IRES activity above control levels (Figure 6), although total IRES activity from the APPa leader was reduced by approximately half of the full-length APP leader. This result indicates that the minimal sequence to initiate translation is ⩽50 nt. However, the full-length 5′-leader is required for optimum IRES activity.
Figure 6.
The 5′ 50 nt of the APP 5′-leader are sufficient to internally initiate translation. The APP 5′-leader was divided into three nonoverlapping regions (see schematic) consisting of the 5′ 50 nt (APPa), the internal 44 nt containing the IRE (APPb) and the 3′ 53 nt (APPc). The three APP segments along with the full-length APP 5′-leader and β-globin 5′-leader were inserted into the intercistronic region of the dicistronic luciferase mRNA and transfected into C6 cells. The P:R ratio is normalized to the activity obtained from the mRNA containing the β-globin 5′-leader.
The APP 5′-leader initiates translation cap-independently ex-vivo
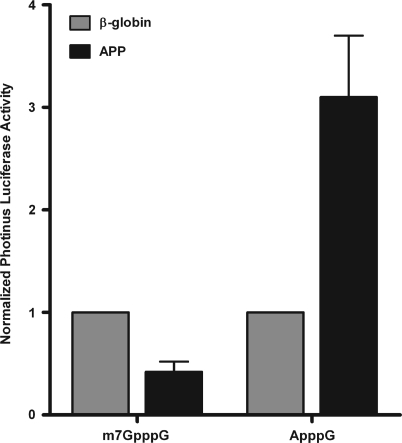
To assess the effect of the APP 5′ leader on translation, the β-globin and APP 5′-leaders were individually placed upstream of the Photinus luciferase ORF. The β-globin 5′-leader is short and unstructured and should be an excellent substrate for cap-dependent translation; while the APP 5′-leader is longer, G/C rich and presumably more structured than the β-globin 5′-leader. Monocistronic RNA was in vitro transcribed and capped with an m7G cap or an ApppG cap. Inclusion of the ApppG cap inhibits 5′- to 3′-exonuclease activity, but it does not bind to eIF-4E and therefore cannot initiate cap-dependent translation. The mRNA was co-transfected with an m7G-capped mRNA coding for humanized Renilla luciferase (phRL, Promega) to normalize for transfection efficiency. Following a 7 h transfection into C6 cells, Photinus and Renilla luciferase activity was assayed. The m7G-capped RNA containing the β-globin 5′-leader was translated very efficiently at a level of ∼2.5-fold over the m7G-capped RNA containing the APP 5′-leader (Figure 7). However, these translational differences were reversed when ApppG capped RNA was examined. In this situation, translation of the RNA containing the APP 5′-leader was >3-fold higher than the ApppG capped β-globin reporter RNA (Figure 7). These results indicate that basal APP translation may be lower than mRNAs with less structured and G/C rich 5′-leaders, but more importantly it demonstrates that the APP 5′-leader can initiate cap-independent translation in cells. Additionally, it supports the hypothesis that during events when eIF-4E is not utilized (when the N-terminus of eIF-4G is cleaved or during an upregulation of the homolog of the eIF-4G c-terminus—DAP5/p97), synthesis of APP can be maintained in a cap-independent manner.
Figure 7.
The APP 5′-leader demonstrates IRES activity in a monocistronic context. Monocistronic Photinus reporter mRNA with the β-globin or APP 5′-leader downstream of either an m7G cap or ApppG cap were co-transfected with monocistronic m7G-capped humanized Renilla luciferase mRNA, to normalize for transfection efficiency, into C6 cells. Following a 7 h transfection into C6 cells, Photinus and Renilla luciferase activities were assayed.
Increased intracellular iron concentration and chemical ischemia increase cap-independent translation of APP
In postmortem AD brain, an increase in intracellular iron concentration is often evident and coincident with presence of Aβ plaques (28,29). In addition, there is often evidence in postmortem brain of acute ischemic injury. Iron affects protein synthesis at multiple levels (33); while ischemia and reperfusion inhibits cap-dependent translation (41). Therefore, we examined whether ex vivo treatments which mimic these conditions correlated with AD, namely excess intracellular iron and ischemia, could augment endogenous levels of APP.
To determine if translation of the endogenous APP mRNA is affected by iron or ischemic conditions, C6 cells were treated either with FAC for 24 h or acutely with chemical ischemia reagents for 10 min. Both treatments were performed in the absence or presence of rapamycin to inhibit mTOR and diminish cap-dependent translation (Figure 8). FAC is a soluble ferric iron complex which is a good source for increasing intracellular iron in neural cells (55). To mimic ischemia and reperfusion, C6 cells were treated for 10 min with 10 mM sodium azide (an inhibitor of oxidative phosphorylation) and 10 mM dideoxyglucose (an inhibitor of glycolysis) (56), and then placed back into complete growth media for 7 h. Regardless of the presence or absence of rapamycin, both FAC and chemical ischemia increased APP expression, ranging from 50% to 80%. Moreover, rapamycin by itself also increased APP expression by 50% (Figure 8; also see Figure 1). This result suggests that iron and chemical ischemia stimulate cap-independent translation of endogenous APP. Similar results were observed in the undifferentiated human dopaminergic neuroblastoma cell line, SH-SY5Y (Figure S1B). Interestingly, FAC and chemical ischemia did not decrease phosphorylation of p70s6kinase, suggesting that stimulating cap-independent translation of APP by these reagents does not require inhibition of the mTOR pathway.
Figure 8.
ex vivo conditions which mimic the AD brain state lead to an increase in cap-independent translation of APP. Rat neural C6 cells were either treated with FAC for 24 h or acutely with chemical ischemia reagents (sodium azide and dideoxyglucose), for 10 min, 7 h prior to harvest both treatments carried out in the absence or presence of rapamycin for 16 h. Cell lysates were probed for (A & B) APP, phospho-p70s6 kinase and GAPDH levels using Western blotting. (C) APP levels were quantified using Molecular Dynamics Imagequant software and normalized for protein loading. APP protein levels were normalized to DMSO (untreated) controls. Data graphed represent five separate experiments.
To determine whether protein or RNA stability influenced these results, we performed a western and northern blot analysis, respectively. For protein expression, C6 cells exposed to the different treatments were treated with cycloheximide for the last 30 or 60 min of the experiment. The results show that APP protein stability was unaffected (Figure S3A and S3B). A northern blot analysis of RNA obtained from C6 cells exposed to the different stimuli showed that RNA stability was also unaffected (Figure S3C). Taken together, these results indicate that cap-independent translation is utilized for the translation of the endogenous APP mRNA both in a basal state and in response to the stressful cellular conditions induced by excess iron and ischemia.
APP IRES activity is augmented by iron treatment
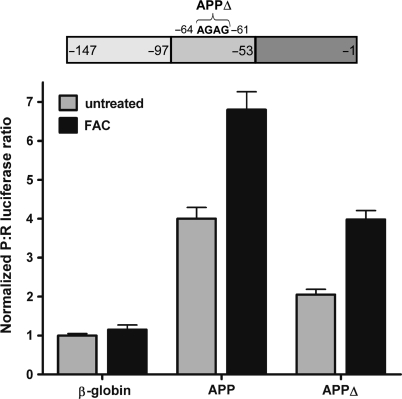
To determine whether iron treatment augments APP IRES activity, we exposed C6 cells to FAC for 24 h. During the final 7 h of FAC treatment, the cells were transfected with dicistronic RNA containing the β-globin or APP 5′-leaders (Figure 9). Iron had no significant effect on the P:R ratio obtained from the β-globin 5′-leader. However, the P:R ratio obtained from the RNA containing the APP 5′-leader was enhanced by 1.7-fold, from 4.0 to 6.8. This result indicates that activity of the APP IRES is positively regulated by iron.
Figure 9.
Iron exposure increases APP 5′-leader IRES activity independently of the IRE. Dicistronic luciferase mRNA containing the β-globin, APP 5′-leader, or APP 5′-leader with the IRE mutated (APPΔ) in the intercistronic region were transfected individually into C6 cells which had been treated for a total of 24 h with FAC-supplemented media or with complete culture media. Luciferase activity is shown as the ratio of Photinus luciferase to Renilla luciferase (P:R) and is normalized to the activity obtained from the mRNA containing the β-globin 5′-leader.
The APP 5′-leader has previously been reported to contain an iron-responsive element [IRE; (32)]. IREs modulate the stability and translation of a number of mRNAs coding for proteins involved in iron metabolism through their binding to iron regulatory proteins [IRPs; (57)]. To determine if the APP 5′-leader IRE is required for the iron-induced increase in IRES activity, 4 nt of the IRE were deleted (APPΔ, see schematic in Figure 6). This deletion inhibits binding of IRPs to the APP IRE, as well as the IREs in the ferritin light- and heavy-chain mRNAs (32,58). Transfection of dicistronic RNA containing the APPΔ 5′-leader showed that the ability of iron to enhance APP IRES activity still persisted (Figure 9). The mutation resulted in a decrease of the basal level of APP IRES activity by approximately half, but the enhancement of the basal IRES activity remained as iron increased the P:R ratio by almost 2-fold (Figure 9). This result indicates that the putative IRE in the APP 5′-leader affects internal initiation mediated by the APPa region (Figure 6), but it is not required for the iron-induced increase in APP IRES activity.
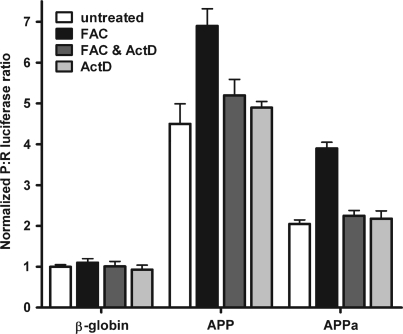
The mechanism and regulation of IRES-dependent translation of eukaryotic mRNAs is not well understood. However, the utilization of internal initiation of translation often coincides with periods when global changes in transcription occur, including during mitosis and apoptosis (24). This observation suggests that protein products of genes which are preferentially transcribed during these periods may regulate IRES-dependent translation. To determine whether de novo transcription is required for the iron-induced increase in IRES activity, C6 cells were either untreated or treated with FAC in the presence or absence of actinomycin D (an inhibitor of de novo transcription) and then transfected with dicistronic RNAs containing the β-globin, APP and APPa 5′-leaders (Figure 10). Exposure to iron not only increased the P:R ratio obtained from the APP dicistronic RNA, but the P:R ratio from the APPa RNA was also enhanced by almost 2-fold with iron. Interestingly, the iron-induced increase in IRES activity mediated by both the APP and APPa 5′-leaders was abolished in the presence of actinomycin D. Cells treated only with actinomycin D did not exhibit any change from the basal P:R ratio. These results indicate that the 5′ 50 nt of the APP 5′-leader (APPa) is sufficient to exhibit the iron-enhanced increase in IRES activity and furthermore this augmentation requires de novo transcription.
Figure 10.
The 5′ 50 nt of the APP 5′-leader are sufficient to mediate the iron-enhanced increase in APP IRES activity, a process which requires de novo transcription. Dicistronic luciferase mRNA containing the β-globin, APP or APPa 5′-leader were transfected into C6 cells and treated with complete growth medium, FAC, FAC and actinomycin D or actinomycin D alone. The P:R ratio is normalized to the activity obtained from the mRNA containing the β-globin 5′-leader under each individual treatment.
APP IRES activity is stimulated by chemical ischemia
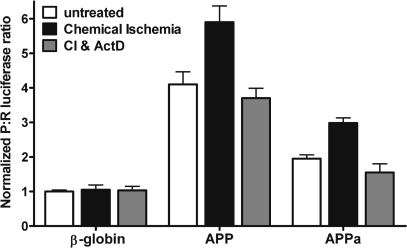
To determine if APP IRES activity is specifically enhanced by ischemia, we transfected dicistronic RNAs into C6 cells and acutely (10 min) treated the cells with the chemical ischemia reagents sodium azide and dideoxyglucose as previously described (56). After 7 h, we assayed for luciferase activity. The P:R ratio obtained from the β-globin 5′-leader RNA was unaltered in response to chemically induced ischemia (Figure 11). However, the P:R ratio from both the APP and APPa 5′-leaders were increased by 1.4- and 1.5-fold, respectively. This ischemia-induced enhancement of IRES activity was abolished in the presence of actinomycin D, a result similar to that observed with the iron-enhanced increase in APP IRES activity (Figure 10). These results indicate that de novo transcription is required for the enhancement of APP IRES activity by chemical ischemia and that the 5′ 50 nt of the APP 5′-leader are sufficient to mediate this augmentation of IRES activity.
Figure 11.
Chemical ischemia enhances APP IRES activity in a transcription-dependent manner. Dicistronic luciferase RNAs containing the β-globin, APP or APPa 5′-leaders were transfected into C6 cells and treated with either complete medium or complete medium with a 10 min exposure to sodium azide and dideoxyglucose in the absence or presence of actinomycin D. The P:R ratio is normalized to the activity obtained from the mRNA containing the β-globin 5′-leader.
DISCUSSION
In the present report, we show that endogenous APP can be translated in a cap-independent manner in neural cells in the presence of rapamycin or in the absence of the cap-binding protein eIF-4E. Additionally, endogenous APP translation is enhanced by iron and chemical ischemia treatment even when cap-dependent translation is inhibited using rapamycin. These observations led us to confirm that the APP 5′-leader contains an IRES using a combination of RNA and DNA reporter constructs. Moreover, the IRES was localized to the 5′ 50 nt of the 5′-leader. APP IRES activity was also enhanced by increased intracellular iron concentration and chemical ischemia, both effects which were dependent upon de novo transcription. Taken together, these results indicate that IRES-dependent translation of the APP mRNA may be physiologically relevant.
Mechanism of in vivo translation of APP mRNA
In the present studies, we provide support for the biological relevance of the APP IRES. APP synthesis was maintained and even enhanced when cap-dependent translation was reduced. Moreover, even under these translation-repressing conditions, iron and chemical ischemia treatments led to an increase in APP expression without affecting protein or RNA stability.
It is possible that rapamycin treatment and knockdown of eIF-4E may also be perceived as a stressful event by the cell and consequently upregulate IRES-dependent translation. However, methods to selectively reduce IRES-dependent translation without affecting cap-dependent translation are relatively unknown. To date, only hypomorphic alleles of the gene dyskerin have been shown to repress IRES activity (25). Nonetheless, in vitro and ex vivo assays demonstrated that the APP 5′-leader is capable of initiating translation at levels similar to cap-dependent translation. These results support the premise that IRES-dependent translation can be as efficient as cap-dependent translation.
Regulating APP IRES activity
Two perturbations, an increase in intracellular iron and a decrease in oxygen are proposed to exacerbate AD (28,35). In the current studies, both conditions led to increased endogenous APP expression and APP IRES activity. Interestingly, the increase in APP IRES activity required de novo transcription. IRES trans-acting factors (ITAFs) are proposed to be one of the rate-limiting factors for eukaryotic IRESs as well as for some viral IRESes [for review, see Ref. (59)]. ITAFs are thought to be molecular chaperones which bind the 5′-leader and either maintain the secondary structure or alter it in such a way that the ribosomal subunits and/or canonical eukaryotic translation initiation factors can bind and facilitate initiation of translation (20). Consequently, it is tempting to speculate that increased intracellular iron and chemical ischemia treatments enhance transcription of a particular ITAF. However, multiple scenarios could be occurring, including synthesis of ITAFs which compete with already-present inhibitory ITAFs (60,61) for binding sites in the APP mRNA 5′-leader, thus alleviating repression of IRES activity.
Dissecting the APP IRES
The minimal functional unit of the APP IRES is located in the 5′ 50 nt of the APP 5′ leader and this 5′ region is sufficient for both iron- and ischemia-enhanced IRES activity. Post-transcriptional regulation of mammalian genes involved in iron metabolism is often mediated by an IRE (57). Rogers and colleagues (32) have proposed that APP synthesis is regulated through a unique type II IRE in the APP 5′-leader and found that chelating iron inhibits IRP binding to the APP IRE as well as APP synthesis. The type II APP IRE is unusual; traditionally, binding of an IRP to an IRE occurs in the presence of iron [as previously observed for IRP binding to the ferritin and transferrin receptor IREs (62)], while IRPs proposed to bind the type II APP IRE occur in the absence of iron (32). Interestingly, abrogating IRP binding by deleting four nucleotides within the IRE (32) also led to a decrease in basal APP IRES activity. This result suggests that IRP binding to the APP IRE may act as an ITAF and enhance IRES activity, since deletion of this entire region is dispensable for IRES activity (Figure 6).
The intracellular pathway which links iron and ischemia to an enhancement in APP IRES activity is unknown. However, oxidative stress is a common downstream affect of excess intracellular iron [for review see Ref. (63) and ischemia (64)]. Iron contributes to the production of hydroxyl radicals through the Fenton reaction (65). Ischemia results in the production of peroxynitrite also leading to hydroxyl radical formation (66). Intracellular signaling pathways elicited by free radicals can alter transcription and may link iron and ischemia to enhanced APP IRES activity. Alternatively, distinct mechanisms could mediate the iron and ischemia response. For example, increased levels of labile iron enhances synthesis of ferritin, which along with a ferritin-like protein K562, can bind DNA and alter transcription (67,68).
Regulating APP synthesis as a therapeutic target in AD
Inhibiting APP expression is a viable target for the treatment of AD. Expression of APP can be regulated at transcriptional, translational or protein levels. Our present results indicate that the APP mRNA is translated in an IRES-dependent mechanism. This work further elucidates the mechanism for the translation of APP and provides a new avenue for exploration. If ITAFs are indeed a rate-limiting step for eukaryotic IRES translation, then studies directed toward identifying ITAFs that bind the APP 5′-leader and designing mimetics to interfere with the binding may be a useful approach toward the treatment of AD.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (AG028156). Funding for open access charge: National Institutes of Health AG028156.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Tara Dobson, Amandine Bastide, Anne-Marie Fernandes-Romao and Olga Hartman for excellent technical assistance and to Brian Kempf, Jessica Tyler and members of the Krushel lab for critical reading of the article.
REFERENCES
- 1.Farlow MR. Etiology and pathogenesis of Alzheimer's disease. Am. J. Health Syst. Pharm. 1998;55(Suppl. 2):S5–S10. doi: 10.1093/ajhp/55.suppl_2.S5. [DOI] [PubMed] [Google Scholar]
- 2.Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, et al. Membrane-anchored aspartyl protease with Alzheimer's disease beta-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 3.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 4.Brunkan AL, Goate AM. Presenilin function and gamma-secretase activity. J. Neurochem. 2005;93:769–792. doi: 10.1111/j.1471-4159.2005.03099.x. [DOI] [PubMed] [Google Scholar]
- 5.Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Lahiri DK. Functional characterization of amyloid beta precursor protein regulatory elements: rationale for the identification of genetic polymorphism. Ann. N. Y. Acad. Sci. 2004;1030:282–288. doi: 10.1196/annals.1329.035. [DOI] [PubMed] [Google Scholar]
- 7.Lahiri DK, Ge YW. Role of the APP promoter in Alzheimer's disease: cell type-specific expression of the beta-amyloid precursor protein. Ann. N. Y. Acad. Sci. 2004;1030:310–316. doi: 10.1196/annals.1329.039. [DOI] [PubMed] [Google Scholar]
- 8.Lahiri DK, Ge YW, Maloney B. Characterization of the APP proximal promoter and 5′-untranslated regions: identification of cell type-specific domains and implications in APP gene expression and Alzheimer's disease. FASEB J. 2005;19:653–655. doi: 10.1096/fj.04-2900fje. [DOI] [PubMed] [Google Scholar]
- 9.Lahiri DK, Ge YW, Maloney B, Wavrant-De Vrieze F, Hardy J. Characterization of two APP gene promoter polymorphisms that appear to influence risk of late-onset Alzheimer's disease. Neurobiol. Aging. 2005;26:1329–1341. doi: 10.1016/j.neurobiolaging.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Allinquant B, Hantraye P, Mailleux P, Moya K, Bouillot C, Prochiantz A. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J. Cell Biol. 1995;128:919–927. doi: 10.1083/jcb.128.5.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso GL, Gandy SE, Buxbaum JD, Greengard P. Chloroquine inhibits intracellular degradation but not secretion of Alzheimer beta/A4 amyloid precursor protein. Proc. Natl Acad. Sci. USA. 1992;89:2252–2256. doi: 10.1073/pnas.89.6.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weidemann A, Konig G, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K. Identification, biogenesis, and localization of precursors of Alzheimer's disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 13.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 14.Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- 15.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray NK, Wickens M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 17.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 18.Bornes S, Prado-Lourenco L, Bastide A, Zanibellato C, Iacovoni JS, Lacazette E, Prats AC, Touriol C, Prats H. Translational induction of VEGF internal ribosome entry site elements during the early response to ischemic stress. Circ. Res. 2007;100:305–308. doi: 10.1161/01.RES.0000258873.08041.c9. [DOI] [PubMed] [Google Scholar]
- 19.Sonenberg N, Pelletier J. Poliovirus translation: a paradigm for a novel initiation mechanism. Bioessays. 1989;11:128–132. doi: 10.1002/bies.950110504. [DOI] [PubMed] [Google Scholar]
- 20.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 21.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 22.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 23.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol. Cell. 2008;100:27–38. doi: 10.1042/BC20070098. [DOI] [PubMed] [Google Scholar]
- 25.Yoon A, Peng G, Brandenburger Y, Zollo O, Xu W, Rego E, Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. Alternative ways to think about mRNA sequences and proteins that appear to promote internal initiation of translation. Gene. 2003;318:1–23. doi: 10.1016/s0378-1119(03)00774-1. [DOI] [PubMed] [Google Scholar]
- 27.Grundhoff A, Ganem D. Mechanisms governing expression of the v-FLIP gene of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2001;75:1857–1863. doi: 10.1128/JVI.75.4.1857-1863.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Connor JR, Menzies SL, St Martin SM, Mufson EJ. A histochemical study of iron, transferrin, and ferritin in Alzheimer's diseased brains. J. Neurosci. Res. 1992;31:75–83. doi: 10.1002/jnr.490310111. [DOI] [PubMed] [Google Scholar]
- 29.Connor JR, Snyder BS, Arosio P, Loeffler DA, LeWitt P. A quantitative analysis of isoferritins in select regions of aged, parkinsonian, and Alzheimer's diseased brains. J. Neurochem. 1995;65:717–724. doi: 10.1046/j.1471-4159.1995.65020717.x. [DOI] [PubMed] [Google Scholar]
- 30.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. Copper, iron and zinc in Alzheimer's disease senile plaques. J. Neurol. Sci. 1998;158:47–52. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, Moir RD, Tanzi RE, Bush AI, Rogers JT. Redox-active metals, oxidative stress, and Alzheimer's disease pathology. Ann. N. Y. Acad. Sci. 2004;1012:153–163. doi: 10.1196/annals.1306.012. [DOI] [PubMed] [Google Scholar]
- 32.Rogers JT, Randall JD, Cahill CM, Eder PS, Huang X, Gunshin H, Leiter L, McPhee J, Sarang SS, Utsuki T, et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer's amyloid precursor protein transcript. J. Biol. Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- 33.Richardson DR. Novel chelators for central nervous system disorders that involve alterations in the metabolism of iron and other metal ions. Ann. N. Y. Acad. Sci. 2004;1012:326–341. doi: 10.1196/annals.1306.026. [DOI] [PubMed] [Google Scholar]
- 34.de la Torre JC. Pathophysiology of neuronal energy crisis in Alzheimer's disease. Neurodegener. Dis. 2008;5:126–132. doi: 10.1159/000113681. [DOI] [PubMed] [Google Scholar]
- 35.Bazan NG, Palacios-Pelaez R, Lukiw WJ. Hypoxia signaling to genes: significance in Alzheimer's disease. Mol. Neurobiol. 2002;26:283–298. doi: 10.1385/MN:26:2-3:283. [DOI] [PubMed] [Google Scholar]
- 36.Kalaria RN. The role of cerebral ischemia in Alzheimer's disease. Neurobiol. Aging. 2000;21:321–330. doi: 10.1016/s0197-4580(00)00125-1. [DOI] [PubMed] [Google Scholar]
- 37.Buee L, Hof PR, Delacourte A. Brain microvascular changes in Alzheimer's disease and other dementias. Ann. N. Y. Acad. Sci. 1997;826:7–24. doi: 10.1111/j.1749-6632.1997.tb48457.x. [DOI] [PubMed] [Google Scholar]
- 38.Jamison JT, Kayali F, Rudolph J, Marshall M, Kimball SR, Degracia DJ. Persistent redistribution of poly-adenylated mRNAs correlates with translation arrest and cell death following global brain ischemia and reperfusion. Neuroscience. 2008;154(2):504–20. doi: 10.1016/j.neuroscience.2008.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluta R, Kida E, Lossinsky AS, Golabek AA, Mossakowski MJ, Wisniewski HM. Complete cerebral ischemia with short-term survival in rats induced by cardiac arrest. I. Extracellular accumulation of Alzheimer's beta-amyloid protein precursor in the brain. Brain Res. 1994;649:323–328. doi: 10.1016/0006-8993(94)91081-2. [DOI] [PubMed] [Google Scholar]
- 40.Bennett SA, Pappas BA, Stevens WD, Davidson CM, Fortin T, Chen J. Cleavage of amyloid precursor protein elicited by chronic cerebral hypoperfusion. Neurobiol. Aging. 2000;21:207–214. doi: 10.1016/s0197-4580(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 41.DeGracia DJ, Kumar R, Owen CR, Krause GS, White BC. Molecular pathways of protein synthesis inhibition during brain reperfusion: implications for neuronal survival or death. J. Cereb. Blood Flow Metab. 2002;22:127–141. doi: 10.1097/00004647-200202000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Pantopoulos K. Conditional derepression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol. Cell Biol. 2002;22:4638–4651. doi: 10.1128/MCB.22.13.4638-4651.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tohda M, Suwanakitch P, Jeenapongsa R, Hayashi H, Watanabe H, Matsumoto K. Expression changes of the mRNA of Alzheimer's disease related factors in the permanent ischemic rat brain. Biol. Pharm. Bull. 2004;27:2021–2023. doi: 10.1248/bpb.27.2021. [DOI] [PubMed] [Google Scholar]
- 44.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 45.Pinkstaff JK, Chappell SA, Mauro VP, Edelman GM, Krushel LA. Internal initiation of translation of five dendritically localized neuronal mRNAs. Proc. Natl Acad. Sci. USA. 2001;98:2770–2775. doi: 10.1073/pnas.051623398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 47.Su Y, Ryder J, Ni B. Inhibition of Abeta production and APP maturation by a specific PKA inhibitor. FEBS Lett. 2003;546:407–410. doi: 10.1016/s0014-5793(03)00645-8. [DOI] [PubMed] [Google Scholar]
- 48.Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staber PB, Vesely P, Haq N, Ott RG, Funato K, Bambach I, Fuchs C, Schauer S, Linkesch W, Hrzenjak A, et al. The oncoprotein NPM-ALK of anaplastic large-cell lymphoma induces JUNB transcription via ERK1/2 and JunB translation via mTOR signaling. Blood. 2007;110:3374–3383. doi: 10.1182/blood-2007-02-071258. [DOI] [PubMed] [Google Scholar]
- 50.Voet D, Voet JG. Biochemistry. John Wiley & Sons: New York:; 1995. [DOI] [PubMed] [Google Scholar]
- 51.Lockard RE, Lane C. Requirement for 7-methylguanosine in translation of globin mRNA in vivo. Nucleic Acids Res. 1978;5:3237–3247. doi: 10.1093/nar/5.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 53.Li PW, Li J, Timmerman SL, Krushel LA, Martin SL. The dicistronic RNA from the mouse LINE-1 retrotransposon contains an internal ribosome entry site upstream of each ORF: implications for retrotransposition. Nucleic Acids Res. 2006;34:853–864. doi: 10.1093/nar/gkj490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hoepken HH, Korten T, Robinson SR, Dringen R. Iron accumulation, iron-mediated toxicity and altered levels of ferritin and transferrin receptor in cultured astrocytes during incubation with ferric ammonium citrate. J. Neurochem. 2004;88:1194–1202. doi: 10.1046/j.1471-4159.2003.02236.x. [DOI] [PubMed] [Google Scholar]
- 56.Kume T, Nishikawa H, Taguchi R, Hashino A, Katsuki H, Kaneko S, Minami M, Satoh M, Akaike A. Antagonism of NMDA receptors by sigma receptor ligands attenuates chemical ischemia-induced neuronal death in vitro. Eur. J. Pharmacol. 2002;455:91–100. doi: 10.1016/s0014-2999(02)02582-7. [DOI] [PubMed] [Google Scholar]
- 57.Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- 58.Ke Y, Sierzputowska-Gracz H, Gdaniec Z, Theil EC. Internal loop/bulge and hairpin loop of the iron-responsive element of ferritin mRNA contribute to maximal iron regulatory protein 2 binding and translational regulation in the iso-iron-responsive element/iso-iron regulatory protein family. Biochemistry. 2000;39:6235–6242. doi: 10.1021/bi9924765. [DOI] [PubMed] [Google Scholar]
- 59.Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Cornelis S, Tinton SA, Schepens B, Bruynooghe Y, Beyaert R. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res. 2005;33:3095–3108. doi: 10.1093/nar/gki611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobson T, Minic A, Nielsen K, Amiott E, Krushel L. Internal initiation of translation of the TrkB mRNA is mediated by multiple regions within the 5′ leader. Nucleic Acids Res. 2005;33:2929–2941. doi: 10.1093/nar/gki605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levenson CW, Tassabehji NM. Iron and ageing: an introduction to iron regulatory mechanisms. Ageing Res. Rev. 2004;3:251–263. doi: 10.1016/j.arr.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Honda K, Casadesus G, Petersen RB, Perry G, Smith MA. Oxidative stress and redox-active iron in Alzheimer's disease. Ann. N. Y. Acad. Sci. 2004;1012:179–182. doi: 10.1196/annals.1306.015. [DOI] [PubMed] [Google Scholar]
- 64.Sugawara T, Fujimura M, Noshita N, Kim GW, Saito A, Hayashi T, Narasimhan P, Maier CM, Chan PH. Neuronal death/survival signaling pathways in cerebral ischemia. NeuroRx. 2004;1:17–25. doi: 10.1602/neurorx.1.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warner DS, Sheng H, Batinic-Haberle I. Oxidants, antioxidants and the ischemic brain. J. Exp. Biol. 2004;207:3221–3231. doi: 10.1242/jeb.01022. [DOI] [PubMed] [Google Scholar]
- 66.Eliasson MJ, Huang Z, Ferrante RJ, Sasamata M, Molliver ME, Snyder SH, Moskowitz MA. Neuronal nitric oxide synthase activation and peroxynitrite formation in ischemic stroke linked to neural damage. J. Neurosci. 1999;19:5910–5918. doi: 10.1523/JNEUROSCI.19-14-05910.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Broyles RH, Belegu V, DeWitt CR, Shah SN, Stewart CA, Pye QN, Floyd RA. Specific repression of beta-globin promoter activity by nuclear ferritin. Proc. Natl Acad. Sci. USA. 2001;98:9145–9150. doi: 10.1073/pnas.151147098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Surguladze N, Thompson KM, Beard JL, Connor JR, Fried MG. Interactions and reactions of ferritin with DNA. J. Biol. Chem. 2004;279:14694–14702. doi: 10.1074/jbc.M313348200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.