Abstract
During eukaryotic translation, mRNAs may form intra-molecular interactions between distant domains. The 5′-cap and the polyA tail were shown to interact through their associated proteins, and this can induce physical compaction of the mRNA in vitro. However, the stability of this intra-molecular association in translating mRNAs and whether additional contacts exist in vivo are largely unknown. To explore this, we applied a novel approach in which several endogenous polysomal mRNAs from Saccharomyces cerevisiae were cleaved near their stop codon and the resulting 3′-UTR fragments were tested either for co-sedimentation or co-immunoprecipitation (co-IP) with their ORFs. In all cases a significant fraction of the 3′-UTR fragments sedimented similarly to their ORF-containing fragments, yet the extent of co-sedimentation differed between mRNAs. Similar observations were obtained by a co-IP assay. Interestingly, various treatments that are expected to interfere with the cap to polyA interactions had no effect on the co-sedimentation pattern. Moreover, the 3′-UTR appeared to co-sediment with different regions from within the ORF. Taken together, these results indicate extensive physical associations between 3′-UTRs and their ORFs that vary between genes. This implies that polyribosomal mRNAs are in a compact configuration in vivo.
INTRODUCTION
Eukaryotic mRNAs are associated with a large array of RNA-binding proteins that facilitate their intracellular processing and function. These proteins contribute to mRNA splicing, export from the nucleus, translation, degradation and more. Two proteins that bind to nearly all eukaryotic mRNAs are the cap-binding protein (eIF4E) and the polyA binding protein (PAB1). These proteins interact with the 5′- and the 3′-ends of the mRNA, respectively. Many lines of evidence, from both in vivo and in vitro systems, indicate that these proteins interact with each other and have a synergistic effect on translation rates (1–8). This intra-molecular interaction was shown in vitro to induce physical compaction of a reporter mRNA into a ‘closed loop’ structure that may facilitate reloading of terminating ribosomes on the 5′-end of the mRNA (9,10). Recently, factors involved in translation termination were also shown to be involved in interactions between the mRNA ends (11). These interactions may lead to further compaction of the mRNA and connection of distant functional regions.
In this study, we explored whether intra-molecular physical interactions occur in translating mRNAs from Saccharomyces cerevisiae. The sedimentation and immunoprecipitation (IP) of 3′-UTRs was tested after the phosphodiester bond linking them to the ORF was cleaved. We observed significant co-sedimentation and co-immunoprecipitation (co-IP) of 3′-UTRs with their ORFs, which differed in extent between mRNAs. Surprisingly, the co-sedimentation was not affected by various treatments that interfere with the known cap-polyA interaction, including removal of the entire 5′-UTR. On the other hand, the 3′-UTR appeared to co-sediment with various domains from within the ORF. This suggests that factors from throughout the ORF (e.g. ribosomes) are involved in this interaction.
MATERIALS AND METHODS
Yeast strains and growth conditions
The strains used in this study are listed in Table 1. Cells were grown in YPD (1% yeast extract, 2% Bacto peptone, 2% glucose) unless they included plasmids that required growth on selection media (SD with the relevant selections).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Genotype | Genetic background | Origin |
|---|---|---|---|
| YA1 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | BY4741 | Euroscarf |
| YA236 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sgn1::kanMX4. | BY4741 | Euroscarf |
| YA586 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tpa1::kanMX4 | BY4741 | Euroscarf |
| YA587 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rpp2a::kanMX4. | BY4741 | Euroscarf |
| YA347 | MATα ade2-144,717 his7-1 lys9-a21 ura3-52 leu2-1 trp1-289. | (12) | |
| YA348 | MATα ade2-144,717 his7-1 lys9-a21 ura3-52 leu2-1 trp1-289 sup35-2 (ts). | (12) | |
| YA127 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 Pep4Δ::HIS3 prbΔ::his3 prc1Δ::hisG rpl25::LEU2 pRPL125FH URA3 CEN | CB012 | (13) |
| YA350 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 fpr1::kanMX4 pRS416 GAL1-FPR1(ORF)-FPR1(3′ UTR) URA3. | BY4742 | This study |
| YA345 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 fpr1::kanMX4 pRS416 GAL1-FPR1(ORF)-SMF3(3′ UTR) URA3. | BY4742 | This study |
| YA421 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 smf3::kanMX4 pRS415 GAL1-SMF3(ORF)- SMF3(3′ UTR) URA3. | BY4741 | This study |
| YA686 | MATα ura3-1 ade2-1 his3-11,15 trp1-1 leu2-3,11 cdc33-42::LEU2 pMDA101 [Cdc33-42/TRP1] | CWO4 | (14) |
Plasmid construction
The plasmids expressing FPR1-FPR1, SMF3-SMF3, SMF3-FPR1 or FPR1-SMF3 were constructed by PCR amplification of the ORFs or the 3′-UTRs from genomic DNA and cloning into pRS416 (FPR1-FPR1 and FPR1-SMF3) or pRS415 (SMF3-SMF3 and SMF3-FPR1) downstream of a GAL1 promoter. All constructs were verified by sequencing. Plasmids were transformed into BY4741 (SMF3-SMF3 or SMF3-FPR1) or BY4742 (FPR1-FPR1 or FPR1-SMF3) strains deleted of their respective ORFs.
Sedimentation analysis of 3′-UTR fragments
The protocol for sedimentation analysis of 3′-UTRs and their ORFs is similar to the ribosome density mapping (RDM) procedure that was previously described (15). Briefly, 50 ml of yeast cells were grown to OD600 0.5–0.8 in YPD (except where indicated), lyzed and separated through sucrose gradient as described before (16). The only modification was that the gradient did not include heparin. The polysomal fraction containing the largest amount of the mRNA of interest was isolated for the cleavage reaction. The standard fraction volume was ∼600 μl, into which DTT and Ribonuclease Inhibitor (Porcine liver, TaKaRa, Otsu, Japan) were added to final concentrations of 0.15 mM and 400 U/ml, respectively. Oligodeoxynucleotides (ODN) complementary to the regions of interest (usually the stop codon region) were added to a final concentration of 0.16 µM and annealed for 20 min with gradual cooling from 37°C to room temperature. Cleavage reactions were initiated by addition of RNase H (10 units per reaction) and RNase H buffer (final concentrations 0.02 M Tris pH 7.4, 0.1 M KCl, 0.02 M MgCl2, 0.1 mM DTT, 0.5 mg/ml CHX). Digestion proceeded for 30 min at 37°C and was terminated by addition of 400 μl of Lysis Minus Detergent (LMD) buffer (20 mM Tris pH 7.4, 140 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.1 mg/ml CHX and 1 mg/ml heparin). Cleavage products were then separated by velocity sedimentation in a 10–50% sucrose gradient, and fractions were collected into 1.5 volumes of 8 M Guanidium HCl and 2.5 volumes of 100% ethanol to precipitate the RNA. Northern blotting was usually performed by separation of half of the RNA from each fraction in 2% agarose/1.1% formaldehayde gels and transfer to a nylon membrane. PCR products that contain either the ORF or the 3′-UTR were labeled radioactively by random priming and hybridized to the membranes. In the analysis presented in Figure 5, separation of the RNA was in 6% acrylamide gel (7 M Urea) and transfer was performed as described (17). Total RNA preparation (Figure 2A) was performed according to the hot phenol protocol (18).
Figure 5.
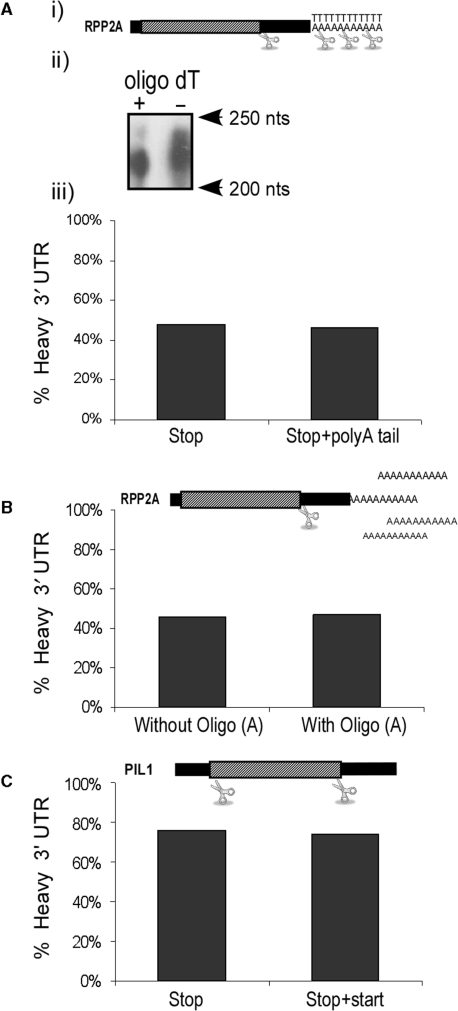
Effect of removal of the polyA or 5′-UTR. (A) Removal of the polyA by oligo dT cleavage. (i) Scheme of the RPP2A mRNA and the cleavage sites. (ii) Control for the oligo dT cleavage efficiency. The polysomal fraction containing most of the RPP2A mRNA was subjected to RNAse H cleavage with ODN complementary to the stop codon region with (+) or without (−) oligo dT20. Samples were resolved by 6% PAGE and subjected to northern analysis. (iii) Quantification of the percentage of the 3′-UTR that sediments in heavy fractions following cleavage at the stop codon and the polyA (‘Stop + PolyA’) or without cleavage at the polyA (‘Stop’). (B) Addition of excess of polyA. During the RNase H cleavage reaction an excess of A20 oligomer (103-fold, assuming 1012 mRNA molecules in the sample) was added to the cleavage reaction to titrate Pab1 from the polyA tail. The histogram presents the percentage of 3′-UTR that sedimented in the heavy fractions without or with addition of oligo (A). (C) Removal of the entire 5′-UTR. The histogram presents the percentage of PIL1 3′-UTR that sedimented in the heavy fractions after cleavage only at the stop codon region (‘Stop’) or also at the start codon region (‘Stop + Start’). Results are representative of two experimental repeats.
Figure 2.
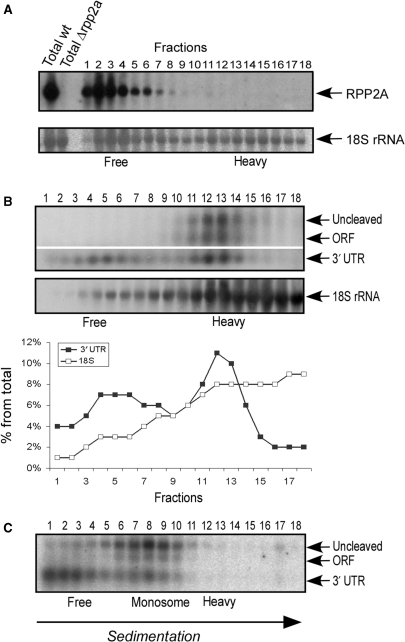
The 3′-UTR is associated with its ORF. (A) RNA isolated from a wild-type strain (wt) by the hot-phenol method was added to an extract from cells deleted of the RPP2A (rpp2aΔ) gene, and subjected to separation in sucrose gradient. The sedimentation position of RPP2A mRNA (top) and the 18S rRNA (bottom) was determined by northern analysis. Note that the 18S rRNA signals in fractions 1–6 are from the added wt RNA. (B) The fraction containing mRNAs associated with three ribosomes was isolated and mixed with a two volumes of heavier fractions containing mRNAs associated with 10 or more ribosomes. RPP2A mRNA was cleaved at the stop codon region and subjected to sedimentation analysis in a sucrose gradient. The sedimentation position of RPP2A fragments (top) and of the 18S rRNA (bottom) was determined by northern analysis. Graphs present the quantification of the northern signals. (C) Cleavage reaction of RPP2A was performed in the presence of 0.8 M KCl. Under these conditions the ORF fragment sediments as associated with a single ribosome (‘monosome’). The sedimentation position of some of the 3′-UTR fragments also appears in this position.
IP of ribosomes with their associated mRNA fragments
A strain expressing Flag-tagged ribosomal protein (Rpl25) (13) was utilized for this assay. Cells’ growth, polysomal mRNA separation and isolation, and cleavage reactions were as described above. Samples were then immunoprecipitated by adding one-fifth volume of anti-Flag beads (Sigma St. Louis, Mi, USA Cat. #A2220) and mixing for 3 h at 4°C. Bound material was precipitated by centrifugation at 800g and washed three times in LMD buffer. Bound RNA was eluted by adding LMD buffer supplemented with 20 mM EDTA or 150 ng/μl Flag peptide and the eluted sample was subjected to phenol extraction followed by precipitation with ethanol and sodium acetate.
Antisense ODNs
The antisense ODNs (Sigma-Genosys, Cambridge, UK) that were used in the RNAse H cleavage reactions are listed in Table 2. The numbers in the ODN names indicate the distance from the start codon. Underlined bases were added for cloning purposes.
Table 2.
ODNs used for RNAse H cleavage
| ODN | Sequence |
|---|---|
| RPL41A - 70–88 | CATAATCCGCTTATTTGGA |
| HYP2 - 473–492 | GCCATGATGTTAACCGGTTT |
| RPP2A - 293–312 | TAAACCGAAACCCATGTCGT |
| ARO7 - 748–771 | TTACTCTTCCAACCTTCTTAGCAA |
| YHB1 - 1177–1200 | CTAAACTTGCACGGTTGACATCTT |
| NOP15 - 640–663 | TCACCATTTGAATTCGATACCTGA |
| SHM2 - 1387–1410 | TTACACAGCCAATGGGTATTCGCC |
| PIL1 - 1063–1085 | GGTAACTTGTTCTTTTCTGCTGG |
| PIL1 - 721–740 | GAGGCTTCATACCCATCGTA |
| PIL1 - 280–300 | GATGTCGGAAACATCGTCAT |
| PIL1 - 19–29 | GAGAGGCGGTAGGTGCCCTG |
| PIL1 - 1010–1030 | GTTTGTTGGGGAAGAGACTC |
| FPR1 - 322–345 | GGATCC TTAGTTGACCTTCAACAATTCGA |
| SMF3 - 1402–1422 | GGATCCTTAAAAATGGATGTCGGCACCT |
Western blot analysis
Proteins from sucrose gradient fractions were precipitated by addition of two sample volumes of ethanol. Samples were separated on 10% denaturing polyacrylamide gels and subjected to western analysis with the antibodies indicated in Table 3.
Table 3.
Antibodies used for western analysis
| Antibody | Dilution | Source |
|---|---|---|
| Mouse anti-Pab1 | 1:5000–10 000 | Dr Motti Choder |
| Rabbit anti-eIF4E | 1:500 | Dr Motti Choder |
| Mouse anti-Flag | 1:10 000 | Sigma F3165 |
RESULTS
Co-sedimentation of 3′-UTRs with their 5′-ends
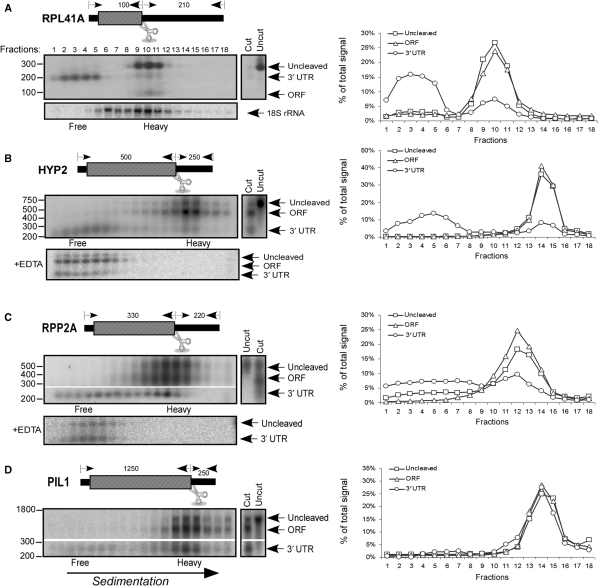
To investigate the physical interactions between the ends of translating mRNAs, we used an experimental procedure similar to RDM (15). Polysomal fractions containing the majority of the mRNA of interest were isolated, and mRNAs were cleaved at the stop codon region by adding RNase H and ODNs complementary to this region. Following cleavage, samples were separated on a sucrose gradient, and the sedimentation position of the 3′-UTR fragment or the 5′ fragment (which contains the 5′-UTR and ORF) was determined by northern analysis [Figure 1 presents representative blots and Table 4 summarize the results of multiple experimental repeats (n)]. Note that in every such cleavage reaction there are residual uncut mRNA molecules (indicated as ‘uncleaved’) that serve as a convenient marker for the sedimentation position of the full-length transcript. As expected, the 5′ fragment, which contains the ORF, sediment as associated with ribosomes in all cases, and its sedimentation is identical to the sedimentation of the uncleaved transcript. The sedimentation of the ORF differs between mRNAs because they are normally associated with different numbers of ribosomes [i.e. RPL41 is usually associated with one ribosome, HYP2 with 4–5 ribosomes, RPP2A with 2–3 ribosomes and PIL1 with 7 ribosomes (19,20)].
Figure 1.
Sedimentation analysis for 3′-UTRs. The sedimentation patterns of the 3′-UTRs of RPL41A (A), HYP2 (B), RPP2A (C) and PIL1 (D) were determined by cleavage and separation by velocity sedimentation in sucrose gradients. A schematic illustration of the cleavage position and approximate lengths of the resulting fragments is presented in each panel. The polysomal fraction containing the majority of each mRNA was isolated and the mRNA was cleaved at the indicated position. Cleavage products were separated on a sucrose gradient into 18 fractions, and the sedimentation position of the cleavage products (3′-UTR or ORF) and the remaining uncut full-length mRNA (Uncleaved) was determined by northern analysis. Note that 3′-UTRs were usually (e.g. C and D) detected by specific probes and required different exposure times. Control lanes with uncut and cut mRNA that were not subjected to separation in a sucrose gradient are included in each panel. In (B) and (C), the panels designated ‘+EDTA’ present the results of a similar cleavage and sedimentation analysis, but in the presence of 20 mM EDTA. Graphs present the quantification of the signals for the different bands. ‘Free’ indicates the fractions that contain RNA free of any ribosomal subunits and ‘Heavy’ indicates the fractions that contain RNA associated with one or more ribosomes.
Table 4.
Percentage of sedimentation in heavy fractions and other characteristics of the tested mRNAs
| Gene name | Average % in heavy fractions (n)a | Standard Deviation | ORF length (nts) | 3′ UTR length (nts)b | 5′ UTR length (nts)b | Number of ribosomes on mRNA | mRNA half life (min)c | mRNA copy/cellc | % G/C in 3′ UTR |
|---|---|---|---|---|---|---|---|---|---|
| FPR1 | 12 (3) | 2 | 345 | 223 | 17 | 3 | 16 | 6 | 30 |
| RPL41A | 21 (3) | 11 | 69 | ∼210 | 22 | 1 | 23 | 49 | 30 |
| HYP2 | 27 (3) | 3 | 474 | ∼250 | 53 | 4–5 | 17 | 22 | 30 |
| PMA1 | 33 (3) | 11 | 2754 | ∼500 | ∼50 | >10 | 22 | 16 | 27 |
| ARO7 | 39 (3) | 1 | 771 | ∼200 | ∼50 | 4–5 | 10 | 2 | 36 |
| YHB1 | 40 (3) | 9 | 1200 | ∼300 | 20 | 7 | 14 | 11 | 32 |
| NOP15 | 45 (2) | 663 | ∼200 | 38 | 4–5 | 9 | 4 | 39 | |
| RPP2A | 44 (6) | 3 | 321 | ∼250 | 30 | 3 | 19 | 21 | 26 |
| SHM2 | 52 (3) | 7 | 1410 | ∼250 | 92 | >10 | 42 | 17 | 33 |
| PIL1 | 61 (6) | 9 | 1020 | ∼200 | 108 | 7 | 19 | 7 | 28 |
| SMF3 | 56 (3) | 10 | 1422 | ∼200 | 200 | 7 | 8 | 1 | 30 |
The sedimentation of the 3′-UTR fragments appears to be bi-modal for all tested mRNAs, with a clear signal in the light fractions (1–6) and another signal in heavier fractions (‘heavy UTRs’). The two populations of UTRs appeared to be of similar length (Figure 1). Finer comparison of the lengths of 3′-UTRs of RPP2A by northern analysis in 6% acrylamide gel, or by analysis of their elution profile from polyU beads at different temperatures (21) did not reveal any differences either (data not shown). This suggests similar lengths of the polyA tails among the light- and heavy-sedimenting 3′-UTRs.
Fractions 1–6 contain complexes <40S, as determined from the OD254 profile of the gradient (data not shown) and from the sedimentation position of the 18S rRNA, the main constituent of the 40S ribosomal subunit (Figure 1A). Thus, the signal in these fractions is from 3′-UTRs not associated with ribosomal subunits. To the best of our knowledge, this is the first experimental indication that neither ribosomes nor ribosomal subunits are associated with eukaryotic 3′-UTRs in vivo. The simplest explanation for this observation is that ribosomes fully dissociate from the mRNA at the translation termination site, consistent with recent in vitro findings (22).However, other models, such as rapid scan of the 3′-UTR by CHX-resistant ribosomes cannot be excluded.
The signals obtained from the heavy UTRs appear in the same fractions as the signals obtained from the 5′ fragment containing both the ORF and the 5′-UTR. For example, about 21 ± 11% of the RPL41A 3′-UTR appears in fractions 7–9 along with the corresponding 5′ fragment (Figure 1A). It is unlikely that the sedimentation of the 3′-UTR in this position is due to its association with postterminating ribosomes that continue to scan the 3′-UTR. In such a case, the full-length transcript (‘uncleaved’) would have sedimentated as heavier than both the ORF and the 3′-UTR (i.e. as the sum of their associated ribosomes). The similar sedimentation position of the 3′-UTR and the ORF suggests that 3′-UTRs are associated with their ORF even after they are cleaved by the RNase H, and this leads to their heavy sedimentation. Interestingly, the percentage of 3′-UTRs that co-sediment with their 5′ fragments differs between genes (from 12 ± 3% for FPR1 to 61 ± 9% for PIL1, Table 4), hence suggesting different extent of association for different mRNAs.
To verify that complexes sedimenting in the heavy region of the gradients contain ribosomes, we performed similar experiments in the presence of 20 mM EDTA, which is known to disrupt polyribosomal complexes. As can be seen in Figure 1B and C (‘+EDTA’), both the full length and the 3′-UTR changed their sedimentation position to a lighter fractions. Thus, the heavy sedimentation of the 3′-UTR necessitates interaction with polyribosomal complexes.
The 3′-UTR co-sediments with its own 5′ fragment and not with unrelated polysomal mRNAs
The heavy sedimentation of the 3′-UTR may be due to association with its own mRNA or with other polysomal complexes. To discriminate between these options, we mixed a sample of mRNA isolated by the hot phenol method (i.e. cleaned of any associated protein) with lysate from cells deleted of the RPP2A gene. This sample was subjected to sedimentation analysis and the position of the phenol-purified RPP2A was determined by northern analysis (Figure 2A). These transcripts appeared in fractions 1–5, free of any ribosomal complexes. This demonstrates that the RPP2A mRNA sequence does not interact with unrelated polysomal complexes.
To further establish that there is no general interaction with any large polysomal complexes, we performed a mixing experiment in which the polysomal fraction that contains RPP2A mRNA (associated with three ribosomes) was mixed with a 2-fold excess of a much heavier polysomal fraction (mRNAs associated with more than 10 ribosomes) and then subjected to a cleavage reaction. If the 3′-UTR of RPP2A can interact with other polysomal mRNAs after the cells’ lysis, then its sedimentation is expected to shift towards the heavier fractions. As shown in Figure 2B, the 3′-UTR of RPP2A co-sediments exactly with its 5′ fragment (fractions 9–12) even in the presence of much heavier polysomal complexes. This sedimentation position differs from that of the excess of added heavy polysomal mRNAs, as revealed by the signal of the 18S rRNA (Figure 2B).
Another indication for the association of the 3′-UTR with its own polysomal ORF came from an experiment in which we added high amounts of KCl (0.8 M) to the cleavage reaction. This led to extensive release of ribosomes from the mRNAs and a shift in the ORF sedimentation to a single ribosome position. Importantly, the cleaved 3′-UTR also shift its sedimentation position to this position (Figure 2C). Thus, the shift in sedimentation of a 3′-UTR correlates with the change in sedimentation of its ORF.
The 3′-UTR co-immunoprecipitates with polyribosomal mRNA
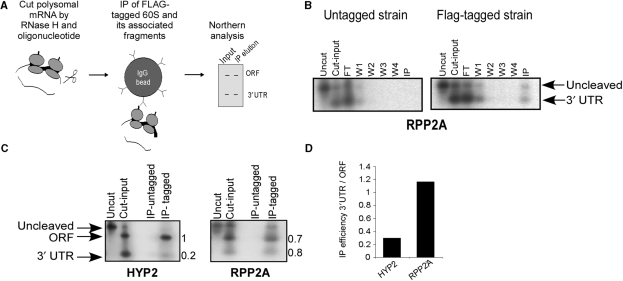
To support the indication that the 3′-UTR interacts stably with the 5′ fragment by an alternative experimental approach, we devised a co-IP assay of ribosomes and their associated mRNA fragments (Figure 3A). In this assay, polysomes were isolated from a strain carrying a Flag-tagged ribosomal protein that is part of the large ribosomal subunit (Rpl25, kindly provided by Dr T. Inada). Cleavage reactions by RNase H and ODN were performed as described, but instead of separating the reaction products on a sucrose gradient, the samples were subjected to co-IP using anti-Flag antibodies coupled to agarose beads. Following co-IP, the complexes were eluted from the beads and the mRNA fragments were detected by northern blot analysis. Methylene blue staining of the blots revealed in the immunoprecipitate lane bands corresponding to rRNAs of both the large and small subunits (data not shown). Rpl25p is located only in the large subunit, therefore the appearance of rRNA of the small subunit (18S) indicates the precipitation of an assembled ribosome. Figure 3B presents the results of the northern analysis following co-IP from the wild-type and RPL25-Flag strains after cleavage of the RPP2A mRNA. Only the IP from the tagged strain recovered a substantial amount of the RPP2A 3′-UTR (note that in Figure 3B a probe specific to the 3′-UTR was used, therefore the ORF was not observed).
Figure 3.
Co-IP of ribosomes with their associated mRNA fragments. (A) Scheme of the procedure. Polysomal mRNA from a strain-expressing Flag-tagged Rpl25 was isolated and subjected to cleavage with specific ODN. Samples were then subjected to IP with beads coated with anti-Flag antibodies. Finally, the co-IP of the 3′-UTR with the ORF (and ribosomes) is determined by northern analysis. (B) Co-IP of RPP2A fragments in untagged (left) and Flag-tagged (right) strains. Samples of mRNA from each step of the IP procedure were collected and analyzed by northern analysis with a probe recognizing the RPP2A 3′-UTR. Lane labels: Uncut—isolated polysomal sample before addition of antisense ODN. Cut input-sample after cleavage reaction, before mixing with the anti-Flag beads. FT (Flow-through)—material not bound to the anti-Flag beads during incubation. W1–W4—material eluted during washes in binding buffer. IP—eluted material from the beads after addition of binding buffer supplemented with a 150 ng/μl Flag peptide. (C) Comparison of the co-IP of HYP2 (left) and RPP2A (right) fragments. Lane labels are as in (B) (in this experiment the elution was done with EDTA). Numbers to the right of each panel indicate the ratio of the signals between the IP and the input of the 3′-UTR or ORF. (D) Efficiency of the IP of HYP2 and RPP2A 3′-UTRs, normalized to the efficiency of their ORF.
We compared the extent of co-precipitation between HYP2 and RPP2A, which appeared to have a relatively low and high level of co-sedimentation, respectively (Table 4). In both cases, signals corresponding to the 3′-UTR appeared only in the IP from the tagged strain (Figure 3C). Furthermore, there were notable differences in the extent of co-IP between HYP2 and RPP2A. HYP2 has a relatively low level of IP efficiency (ratio of input to IP of ∼0.2) and RPP2A has a relatively high efficiency (ratio of ∼0.8) (Figure 3C). Considering the efficiency of IP of the ORF fragments (Figure 3D), these results imply weaker interactions between the 3′-UTR and the 5′ fragment in HYP2 than in RPP2A. Thus, the co-IP results are consistent with the results from the co-sedimentation assay with respect to the general association of the 3′-UTR with the 5′ region and with respect to the variation of the strength of this association between mRNAs.
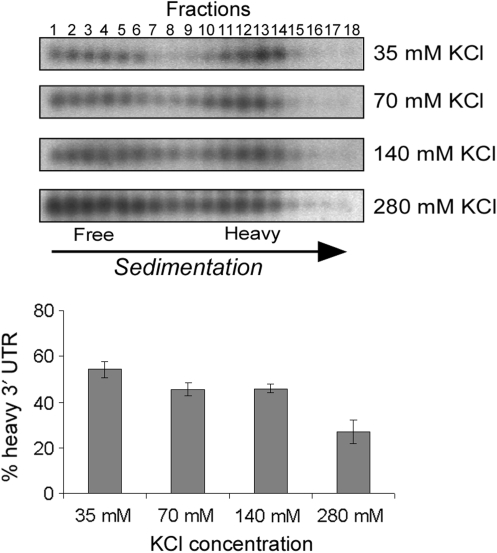
The interaction is stable at various KCl concentrations
The co-sedimentation of the 3′-UTR with its 5′ fragment indicates a stable interaction that withstands the experimental procedure. To further explore the stability of this apparent interaction, we performed the experimental procedure for one of the mRNAs (RPP2A) with KCl present in all solutions (including the lysis buffer, LMD and gradients) at the indicated concentrations (Figure 4). A simple expectation is that higher salt concentrations will interrupt protein–protein interactions, yet will stabilize interactions that are based on nucleotide base-pairing. The KCl concentrations used herein did not affect the number of ribosomes associated with the entire RPP2A transcript (data not shown).
Figure 4.
Analysis of the effect of KCl on the co-sedimentation of RPP2A 3′-UTR. The indicated concentrations of KCl were added to all solutions of the cleavage procedure (including the lysis buffer, RNase H buffer, LMD and sucrose gradients) and RPP2A mRNA was subjected to analysis as in Figure 1. The histogram presents the results of three independent repeats.
We found a significant co-sedimentation in salt concentrations ranging from 35 mM to 280 mM with a decrease in association towards the higher concentrations. Since base-pairing become more stable at higher salt concentrations, the decrease in association may suggest the presence of a more complex RNA structure or involvement of proteins in the interactions.
The basis for the co-sedimentation is not the cap-polyA interaction
The results presented thus far demonstrate that even after cutting the phosphodiester bond that links the 3′-UTR to the ORF region of the transcript, these two fragments co-sediment and co-precipitate. To test if this is due to interactions between the 3′-UTR and the cap structure, we performed the experiments presented in Figures 5–7.
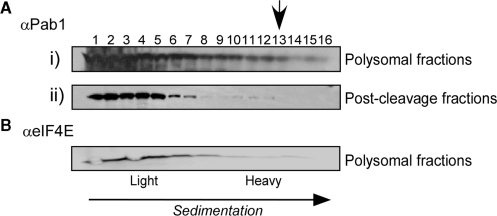
Figure 6.
Western analysis for Pab1 and eIF4E sedimentation in sucrose gradients. Cell extracts were separated by velocity sedimentation into 16 fractions and the sedimentation of Pab1 [A(i)] or eIF4E (B) was determined by western analysis. [A(ii)] A polysomal fraction [indicated by an arrow in A(i)] was isolated and incubated under the same conditions as in a cleavage reaction. The sample was then separated in a second sucrose gradient, and the sedimentation of Pab1 was determined by western analysis.
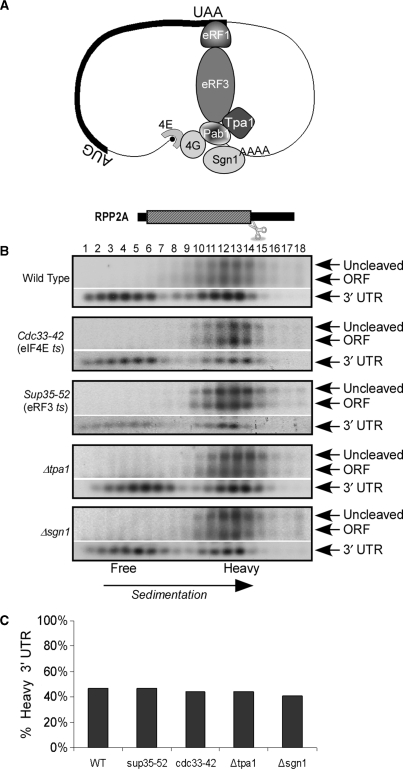
Figure 7.
Effect of mutations on the sedimentation of RPP2A 3′-UTR. (A) Schematic representation of an mRNA in a closed loop configuration with the factors tested in section B depicted. (B) Cleavage and sedimentation analysis of the various strains; eIF4E ts and eRF3 ts were grown at 25°C to logarithmic phase and then shifted to 37°C for 60 min; tpa1Δ and sgn1Δ were grown at 30°C in YPD. (C) Quantification of the percentage of 3′-UTR signals that appear in the heavy fractions (fractions 13–18).
We performed a cleavage assay for RPP2A with addition of oligo dT, which is expected to cleave the polyA tail (Figure 5A). The addition of oligo dT indeed led to shortening of the 3′-UTR (indicative of cleavage of the polyA tail), yet in two experimental repeat only a marginal effect (<5%) was observed on the 3′-UTR that sediment in the heavy fractions of the gradient. No significant effect was observed also when another mRNA (PMA1) was tested (data not shown).
No effect on RPP2A (Figure 5B), PIL1 or PMA1 (data not shown) sedimentation was observed also when the reaction was performed in the presence of 103 excess of oligo (A) to compete for Pab1 binding, further demonstrating that the polyA tail is unnecessary for this interaction.
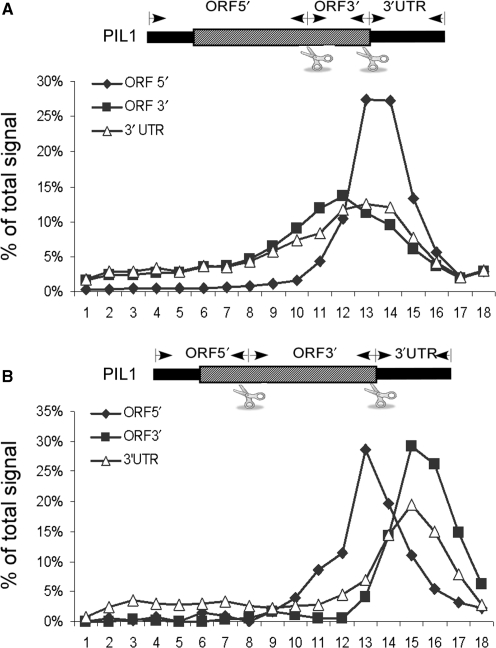
The possible involvement of the entire 5′-UTR in co-sedimentation was tested by cleaving the PIL1 mRNA at two positions simultaneously; immediately downstream of the start codon and downstream of the stop codon (PIL1 has a relatively long 5′-UTR therefore the cleaved ORF can be distinguished from the uncleaved). We performed two experimental repeats and in both the 3′-UTR sedimented at the same position as the ORF fragment and with similar percentages whether or not the 5′-UTR was cleaved (Figure 5C and data not shown). Another cleavage scheme for PIL1 (Figure 8), further downstream to the start codon, also indicted that the 5′ fragment does not control the sedimentation of the 3′-UTR. Similar negligible effect for the 5′-UTR (<5% change) was observed when RPP2A mRNA was tested (data not shown). This indicates that the observed heavy sedimentation of the 3′-UTR is not due to association with elements within the 5′-UTR (e.g. the cap structure) but due to elements within, or associated with, the ORF.
Figure 8.
Cleavage at sites within the ORF. PIL1 mRNA was cleaved at the stop codon and the last third (A) or the stop codon and the first third (B) according to the scheme in each section. Samples were then subjected to velocity sedimentation in sucrose gradients and the sedimentation profile of each fragment was determined by northern analysis using specific probes. Results are representative of two experimental repeats.
We also tested whether proteins involved in these interactions co-sediment with the cleaved 3′-UTR. We used antibodies recognizing the polyA binding protein and cap binding protein (αPab1 and αeIF4E, respectively, kindly provided by Dr M. Choder) to perform western analysis with fractions collected after the cleavage reaction (Figure 6). Most of the signal for these proteins appeared in the fractions containing mRNAs free of ribosomes, and the levels at the heavy fractions where the 3′-UTRs co-sediment were almost undetectable. Pab1 and eIF4E were expected to be associated with polysomal mRNAs; therefore this sedimentation position was probably due to dissociation during our experimental procedure (23). In any case, the sedimentation of Pab1 and eIF4E did not correlate with the sedimentation pattern of the 3′-UTR. It should be noted, however, that the possibility that small, undetectable fraction of these proteins do sediment in these fractions cannot be excluded.
To further examine the dispensability of the cap-polyA interaction for the 3′-UTR heavy sedimentation, we performed sedimentation analyses for RPP2A 3′-UTR in yeast strains lacking various proteins that are involved in the cap-polyA interaction (Figure 7). We used a temperature sensitive strain of the cap-binding protein (cdc33-42) in which eIF4E levels were undetectable after 60 min at 37°C (data not shown). Lower yet significant amounts of polysomal complexes were detected after the temperature shift; this may be due to a decrease in ribosomal dissociation rates and/or cdc33-independent initiation. The sedimentation of the 3′-UTR of RPP2A was determined in this strain after the temperature shift, and was found to be similar to its sedimentation in the parental strain. Similar results were obtained in a temperature-sensitive mutant of a translation termination factor (sup35-52), or in strains deleted of other proteins involved in translation initiation (sgn1Δ) and termination and polyA tail length control (tpa1Δ) (24,25). A PAB1-mutated strain (pab1-16), which carries two amino acid exchanges (in RRMs 1 and 2) was also tested. These point mutations are known to significantly reduce the affinity of Pab1 to oligo (A) (26). However, even these specific mutations led to a strong decrease in ribosomal association and in mRNA stability, therefore the northern data was inconclusive (data not shown).
The co-sedimentation is due to interactions throughout the ORF
The apparent association of the 3′-UTR with the ORF may be confined to a specific region of the ORF or distributed throughout the coding region. To explore these possibilities, we cleaved the PIL1 mRNA at the stop codon region and at the second third (Figure 8A) or at the first third of the coding region (Figure 8B). These cleavages generated ORF fragments of different lengths from either end of the ORF. Analysis of the 3′-UTR sedimentation revealed that in both cases it co-sedimented with the larger ORF fragment, whether it was from the beginning or the end of the coding region. Similar results, yet of lower ribosomal separation (due to the shorter lengths of ORF fragments), were obtained when RPP2A mRNA was cleaved at two positions simultaneously (data not shown). While these cleavage schemes cannot exclude the possibility of interaction with a site in the center of the ORF (that is present in the center of all tested ORFs), we find it more likely that the association of the 3′-UTR is due to interactions throughout the ORF, and not specific to a unique site. This is also consistent with the small amount of 3′-UTRs that co-sediment with the shorter ORF fragment (e.g. Figure 8A).
The association is dependent on features from the 3′-UTR and the ORF
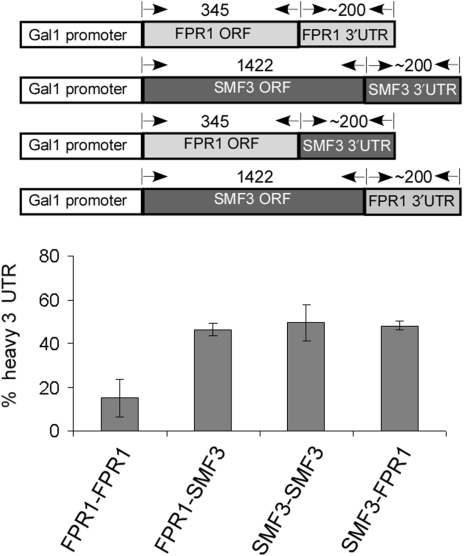
We created a series of constructs in which we replaced the 3′-UTR of one mRNA by another, and expressed them from a strong heterologous promoter (GAL1) (Figure 9). These constructs were used to determine if the extent of co-sedimentation of a 3′-UTR is maintained outside the context of its native ORF. Analysis of the co-sedimentation values of the plasmid-expressed FPR1 and SMF3 3′-UTRs linked to their native ORFs produced values of 15 ± 9% and 50 ± 8%, respectively. Similar values were obtained for the endogenous, chromosomally expressed mRNAs (Table 4), indicating that the genomic context is not critical for the apparent association. When SMF3 3′-UTR was linked to FPR1 ORF, about 46 ± 3% of the 3′-UTR fragments co-sedimented with the ORF fragment (Figure 9). This value differs from the value of FPR1 3′-UTR (15%), indicating that FPR1 ORF is not sufficient to control the extent of co-sedimentation with its associated 3′-UTR. On the other hand, merging the 3′-UTR of FPR1 to the ORF of SMF3 (SMF-FPR) resulted in similar heavy sedimentation as that of SMF3 3′-UTR. Thus, the tendency of a 3′-UTR to associate with its ORF is not dominated by one of the two regions, but may be controlled by factors from either the ORF or the 3′-UTR.
Figure 9.
Analysis of chimeric mRNAs. Plasmids expressing the FPR1 ORF fused to the FPR1 3′-UTR (FPR1-FPR1) or to SMF3 3′-UTR (FPR1-SMF3) or the SMF3 ORF fused to the SMF3 3′-UTR (SMF3-SMF3) or FPR1 3′-UTR (SMF3-FPR1) were introduced to strains deleted of the respective ORF. Cells were grown to mid-logarithmic phase and the mRNAs were subjected to cleavage at the indicated sites and separation in sucrose gradients. The percentage of the 3′-UTR that sediments in the heavy fractions (average of at least three experimental repeats) is indicated in the histogram.
DISCUSSION
Eukaryotic mRNA compaction is known to occur through interactions between the polyA and cap-binding complex. Recently, the translation termination factor eRF3 was also implicated in this interaction. These interactions were shown to induce translation initiation, and one model suggests that terminating ribosomes are recycled more efficiently to the initiation site following translation termination. In this study, we examined the physical association of 3′-UTRs with the upstream portion of the mRNA in the context of polyribosomes isolated from yeast cells. This association was tested by both co-sedimentation in sucrose gradients (Figure 1) and by co-IP (Figure 3). Both methods indicated stable and extensive interactions between the 3′-UTR and the ORF that withstand our experimental procedure, and that these interactions are of different strengths in different mRNAs (Table 4).
Interactions between the ends of an mRNA were identified previously. The polyA binding protein (PAB1) is associated with the translation initiation factor eIF4G that interacts with eIF4E (10). These interactions are known to enhance translation (1,2,5,7,8,27) and to induce mRNA circularization (9). We therefore suspected that these interactions are the basis for the apparent co-sedimentation of the 3′-UTR. Several assays designed to interfere with this interaction, however, had minimal effect on the observed co-sedimentation. These assays included depletion of the cap-binding protein in a temperature sensitive strain, removal of the polyA from the mRNA by cleavage with oligo dT, and removal of the entire 5′-UTR from the mRNA. The lack of effect on co-sedimentation following perturbations to the cap–polyA interaction indicate that its contribution to the overall phenomenon is minimal (Figures 5–8). We hypothesize that the cap–polyA interactions are not strong enough to withstand our experimental conditions. Milder experimental conditions could potentially maintain the cap–polyA interaction and therefore yield higher percentages of co-sedimentation.
We suggest two general models for interactions between the 3′-UTR and the ORF. The first model poses extensive intra-molecular base-pairing between the ORF and the 3′-UTR and stems from the tendency of RNA molecules to form secondary and tertiary structures. The proposed interactions may be stable enough to maintain the association of the 3′-UTR with the ORF during our experimental procedure. In this model, translating ribosomes would occasionally interrupt these interactions, which may explain the appearance of 3′-UTRs in the light fractions. This model implies that mRNAs are in extensively structured conformations, which are maintained even when ribosomes translate the coding region. The second model suggests interactions between the 3′-UTR and factors that are associated with the ORF, most likely ribosomes. According to this model, the 3′-UTR interacts with ribosomal proteins and remains associated with them during translation. This interaction is maintained during translation (and during the experimental procedure), thereby leading to the apparent heavy sedimentation of 3′-UTRs. To distinguish between the two models it is necessary to compare the association of the 3′-UTR with the ORF either in the presence or absence of ribosomes. The methods utilized herein are not suitable for this purpose because both the sedimentation and the co-IP assays require ribosomes as a handle for separation and therefore it is impossible to test the physical association when no ribosomes are present.
Interestingly, the amount of 3′-UTRs that co-sedimented with their 5′-fragments differed significantly between genes (Table 4); some rarely sedimented in the heavy fractions (i.e. their majority appeared as free 3′-UTRs) while for others, the majority of the 3′-UTR co-sedimented with the ORF. This implies that the affinity of the interactions between the 3′-UTR and the rest of the mRNA vary from one mRNA to another. We compiled a list of parameters that might be related to such association (Table 4). None of these parameters appeared to be strongly correlated with the extent of co-sedimentation (though a larger sample is probably necessary for a more thorough comparison). This suggests that the extent of interaction is a function of features beyond these simple parameters. Assuming that the 3′-UTRs interact with ribosomes, features such as elongation rates or specific ribosome-associated proteins might be involved. Alternatively, if the association is through extensive base-pairing that generates large mRNA structures that include both the ORF and the 3′-UTR, then the nucleotide sequence will be an important feature. Treatment of cells with low concentrations of cycloheximide (2.5–5 μg/ml) to lower translation activity (as measured by 35S incorporation into proteins) or use of elongation factor mutants (tef5-7 and tef5-1, kindly provides by Dr T. Kinzy) (28) had no effect on the extent of co-sedimentation of RPP2A mRNA (data not shown). This suggests that reducing elongation rates does not affect the extent of interaction between the 3′-UTR and the ORF.
What may be the implications of the interactions between the 3′-UTR and the ORF? We consider two likely consequences of the interactions between the 3′-UTR and ORF: (i) mRNA stability—compaction of the mRNA through extensive intra-molecular interactions may maintain it in a form that is less accessible to various degradation factors. Upon relaxation of this compact form, the 3′-UTR would become exposed and decay processes could start. One prediction of this model is that mRNAs with more associations between the 3′-UTR and the ORF (i.e. higher percentage of sedimentation in heavy fractions) will have higher stability. (ii) Translation rates—another possibility is that compaction of the mRNA allows more efficient recycling of terminating ribosomes to the translation initiation site. This hypothesis, which stems from the ‘closed loop’ model for interactions between the ends of the mRNA, predicts that mRNAs with stronger intra-molecular associations will have higher translation rates. Analyses of the parameters in Table 4 that are related to the above hypotheses (i.e. mRNA half life and ribosomal association of the ORF) do not reveal a clear correlation between those parameters and the percentage of association. These options cannot be dismissed, however, because other gene-specific processes may obscure the underlying correlation.
FUNDING
Israel Science Foundation (1096/05); the Ministry of Lower Saxony; J. and A. Taub biological research fund. Funding for Open Access Publication Charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
ACKNOWLEDGMENTS
We thank Dr T. Inada for the tagged Rpl25 strain, Dr Motti Choder for antibodies and Dr Lilach Pnueli for technical assistance and members of the Arava lab for fruitful discussions.
REFERENCES
- 1.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 2.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarun SZ, Jr, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- 4.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 5.Borman AM, Michel YM, Kean KM. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 2000;28:4068–4075. doi: 10.1093/nar/28.21.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michel YM, Poncet D, Piron M, Kean KM, Borman AM. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 7.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 8.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 10.Sachs AB. Physical and functional interactions between the mRNA cap structure and the polyA tail. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor: CSHL press; 2000. [Google Scholar]
- 11.Uchida N, Hoshino S, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 2002;277:50286–50292. doi: 10.1074/jbc.M203029200. [DOI] [PubMed] [Google Scholar]
- 12.Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol. Cell Biol. 2002;22:3301–3315. doi: 10.1128/MCB.22.10.3301-3315.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inada T, Winstall E, Tarun SZ, Jr, Yates J.R., 3rd, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. Rna. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altmann M, Sonenberg N, Trachsel H. Translation in Saccharomyces cerevisiae: initiation factor 4E-dependent cell-free system. Mol. Cell Biol. 1989;9:4467–4472. doi: 10.1128/mcb.9.10.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eldad N, Arava Y. Detecting ribosomal association with the 5′ leader of mRNAs by ribosome density mapping (RDM) Methods Enzymol. 2007;431:163–175. doi: 10.1016/S0076-6879(07)31009-4. [DOI] [PubMed] [Google Scholar]
- 16.Arava Y. Isolation of polysomal RNA for microarray analysis. Methods Mol. Biol. 2003;224:79–87. doi: 10.1385/1-59259-364-X:79. [DOI] [PubMed] [Google Scholar]
- 17.Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M. The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev. 2005;19:3004–3016. doi: 10.1101/gad.353205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melamed D, Arava Y. Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol. 2007;431:177–201. doi: 10.1016/S0076-6879(07)31010-0. [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Warner JR. Expression of a micro-protein. J. Biol. Chem. 2001;276:33821–33825. doi: 10.1074/jbc.M103772200. [DOI] [PubMed] [Google Scholar]
- 20.Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. Rna. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valasek L, Szamecz B, Hinnebusch AG, Nielsen KH. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- 24.Winstall E, Sadowski M, Kuhn U, Wahle E, Sachs AB. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 2000;275:21817–21826. doi: 10.1074/jbc.M002412200. [DOI] [PubMed] [Google Scholar]
- 25.Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM. Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol. Cell Biol. 2006;26:5237–5248. doi: 10.1128/MCB.02448-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deardorff JA, Sachs AB. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol. 1997;269:67–81. doi: 10.1006/jmbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 27.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carr-Schmid A, Durko N, Cavallius J, Merrick WC, Kinzy TG. Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange. J. Biol. Chem. 1999;274:30297–30302. doi: 10.1074/jbc.274.42.30297. [DOI] [PubMed] [Google Scholar]
- 29.Hurowitz EH, Brown PO. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z, Dietrich FS. Mapping of transcription start sites in Saccharomyces cerevisiae using 5′ SAGE. Nucleic Acids Res. 2005;33:2838–2851. doi: 10.1093/nar/gki583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc. Natl Acad. Sci. USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]