Abstract
BTG2 is the prototypical member of the TOB family and is known to be involved in cell growth, differentiation and DNA repair. As a transcriptional co-regulator, BTG2 interacts with CCR4-associated factor 1 (CAF1) and POP2 (CALIF), which are key components of the general CCR4/NOT multi-subunit transcription complex, and which are reported to play distinct roles as nucleases involved in mRNA deadenylation. Here we report the crystal structures of human BTG2 and mouse TIS21 to 2.3 Å and 2.2 Å resolution, respectively. The structures reveal the putative CAF1 binding site. CAF1 deadenylase assays were performed with wild-type BTG2 and mutants that disrupt the interaction with CAF1. The results reveal the suppressive role of BTG2 in the regulation of CAF1 deadenylase activity. Our study provides insights into the formation of the BTG2-CAF1 complex and the potential role of BTG2 in the regulation of CAF1.
INTRODUCTION
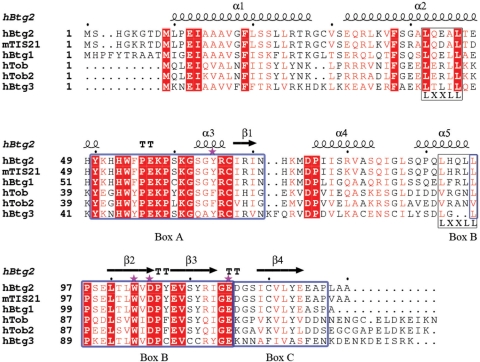
Btg2, the anti-proliferative human homolog of the pc3 (rat) or tis-21 (mouse) genes, belongs to the BTG/TOB family and was originally isolated as an immediate early gene induced by tumor promoters and growth factors in PC12 and Swiss 3T3 cells (1–3). The BTG/TOB family of anti-proliferative proteins is composed of five structurally related members in vertebrates, including BTG1, BTG2, ANA/BTG3, TOB and TOB2 (4–6), which share significant sequence homology (Figure 1) and are reported to be involved in cell growth, differentiation, and survival (7–10).
Figure 1.
Sequence alignment of human BTG2 with homologous proteins. From ‘top’ to ‘bottom’, the sequences are BTG2 from Homo sapiens (NP_006754), TIS21 from Mus musculus (NP_031596), BTG1 from Homo sapiens (NP_001722), Tob1 from Homo sapiens (NP_005740), Tob2 from Homo sapiens (NP_057356) and BTG3 from Homo sapiens (NP_006797). The secondary structure of human BTG2 is indicated in the top line, as well as the conserved domains in the bottom line. Box A, Box B and Box C are shown enclosed by a blue box, while the two LXXLL motifs are shown by a white box. Strictly conserved residues among TOB family members are indicated in red and the most conserved residues are highlighted in red. Residues involved in CAF1 binding are labeled with magenta stars. Sequences were aligned using CLUSTALW(58) and the Figure was produced with ESPript (59).
Studies indicate that BTG2 should have a variety of biological roles (6,11). These include: a transcriptional co-regulator; differentiation; an anti-apoptotic factor in neurogenesis (4,12–16); a key mediator of the stage-specific expansion of thymocyte and a negative regulator of hematopoietic progenitor expansion (17,18); a tumor-suppressor gene in both mouse and human (19–21); a pan-cell cycle regulator (13,22,23); and a regulator of embryo development (24–26).
BTG2 exerts its biological functions by regulating a variety of signaling pathways, which might be achieved by its interaction with different cellular targets via differing mechanisms. As a transcriptional co-regulator, BTG2 has been shown to interact with CAF1 (CCR4-associated factor 1) and POP2 (CALIF) (27,28), and works as a co-activator of ERα-mediated transcription via a CCR4-like complex (29). The main role of the CAF1 homolog in yeast was presumed to be related to its interaction with CCR4, as the CAF1 protein isolated from Saccharomyces cerevisiae did not possess deadenylase activity (30,31). As recombinant yeast CAF1 extracted from Escherichia coli degraded poly(A) in vitro, and CAF1 deletion mutants in yeast show a deadenylation defect, yCAF1 was believed to be required for normal mRNA deadenylation in vivo (31,32). Recently, trypanosome CAF1 was reported to have deadenylation activity, and its depletion led to the delay of deadenylation and degradation of constitutively expressed mRNA (33). Similar results were observed in Drosophila cells, in which depletion of CAF1 brought a marked increase in average poly(A) length and the rate of deadenylation of Hsp70 mRNA was strongly reduced during the recovery from heat shock (34). Mouse CAF1 was identified through its interaction with the CCR4 protein, which can complement the yeast pop2 null mutation in some aspects, and which acts as a processive deadenylase in vitro (35–37). The deadenylase activity of yeast POP2, which is related to RNaseD, is evolutionarily conserved in the CAF1 family including the human homologs CAF1 and POP2 (32,33,38).
There are a number of other known molecular targets of PC3/TIS21/BTG2 in addition to CAF1 and POP2, including PRMT1 (Protein arginine N-methyltransferase 1) (39), Homeoprotein HOXB9 (40), CyclinB1 associated protein kinase Cdc2 (41), Smad1 and Smad8 (24) and Pin-1 (Peptidyl prolyl cis/trans isomerase) (23). Previous studies indicate that BTG2 can modulate the activities of some of its interacting partner proteins, either by stimulation or suppression (24,39–41).
As the prototypical member of an anti-proliferative family, BTG2 contains three highly conserved domains among various species; Box A (Y50–N71), Box B (L96–E115) (4,5) and Box C (D116-P127). Box A, which is also named GR (for growth regulatory, corresponding to residues Y50-I68 in TIS21), and Box B respectively appear to play key roles in anti-proliferative function and in binding to a number of molecular targets (13,27). There are two copies of an LXXLL motif in BTG2 that are involved in the regulation of ERα-mediated activation: L1 (L42-L46) and L2 (L92-L96), referred to as the NR (nuclear receptor) box (28,42). Moreover, Box C is known to interact with PRMT1 in vitro (43).
However, little is known about the precise role of BTG2 in the deadenylase activity of CAF1. Here we report the crystal structures of human BTG2 and the mouse homolog TIS21 to 2.3 Å and 2.2 Å resolution, respectively. The structures reported here reveal features that are important for interaction with their molecular targets. Based on our preliminary structural analysis, wild-type BTG2 and several mutants were constructed for interaction assays and deadenylase assays, the results of which reveal that BTG2 plays a suppressive role in the regulation of CAF1 deadenylase activity. These data provide a structural basis for further analysis of BTG2 in the formation of the BTG2–CAF1 complex and the regulation of CAF1 activity.
MATERIALS AND METHODS
Protein expression and purification
The coding sequences of the human BTG2 and mouse TIS21 genes were inserted into the bacterial expression vector pGEX-6P-1 (GE Healthcare) using EcoRI and XhoI restriction sites. The recombinant plasmids of human BTG2 and mouse TIS21 were transformed into the E. coli strain BL21 (DE3) and overexpressed as glutathione-S-transferase (GST) fusion proteins. The soluble GST-tagged proteins were purified by GST-glutathione affinity chromatography. GST-tagged proteins were employed for in vitro GST ‘pull-down’ experiments, while proteins cleaved with PreScission Protease at 16°C overnight were subjected to further purification.
Recombinant human BTG2 was further purified using Resource S (GE Healthcare) and Superdex-200 size-exclusion columns (GE Healthcare). The purified and concentrated human BTG2 protein (4–6 mg/ml) was stored in 20 mM MES (pH 5.6), 70 mM NaCl at −80°C. Recombinant mouse TIS21 was purified in a similar manner and stored in 20 mM HEPES (pH 7.5), 70 mM NaCl at –80°C.
Mutations of several key residues in the CAF1-binding interface of human BTG2, including Y65A, W103A, D105A, E115A and G64A-W103A were introduced by the PCR method using the human BTG2 expression plasmid as a template, with pairs of primers encoding the mutations at the sites of substitution. All mutants were cloned into the expression plasmid pGEX-6P-1 and overexpressed and purified following a similar protocol to wild-type BTG2. The wild-type and mutants of human BTG2 were also sub-cloned into the vector of pME18S-GST for in vivo interaction assay.
Human CAF1 and POP2 were also cloned into the pGEX-6P-1 vector. The soluble GST-CAF1 and GST-POP2 proteins were purified by GST-glutathione affinity chromatography and cleaved with PreScission Protease at 16°C overnight. Recombinant human CAF1 and POP2 were further purified using Resource Q (GE Healthcare) and Superdex-200 size-exclusion columns (GE Healthcare) using a buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl. The purified and concentrated human CAF1 and POP2 proteins were stored in the deadenylase buffer (50 mM Tris–HCl, 150 mM NaCl, 10% glycerol, 1 mM DTT at pH 7.5) at –80°C.
Crystallization
The human BTG2 protein solution used for crystallization contained 20 mM MES (pH 5.6), 70 mM NaCl and 4 mg/ml protein. Optimal crystals were obtained using the hanging-drop, vapor-diffusion technique with reservoir solutions containing 100 mM Bis–Tris (pH 6.1), 0.2 M sodium chloride, 21% (v/v) PEG3350. Protein solution (1.0 μl) was mixed with reservoir solution (1.0 μl) and equilibrated against 300 μl of reservoir solution at 16°C.
The mouse TIS21 protein solution used for crystallization contained 20 mM HEPES (pH 7.5), 70 mM NaCl and 6 mg/ml protein. The reservoir solution contained 100 mM Bis–Tris (pH 6.5), 23% (v/v) PEG3350. Crystals were obtained following the same method as described for human BTG2.
Data collection and processing
Native crystals of human BTG2 for data collection were flash-cooled in liquid nitrogen in the crystallization reservoir solution and diffracted to 2.3 Å resolution. Data were collected on beamline BL5 of the Photon Factory (Tsukuba, Japan) using an ADSC-Q315 detector. The program HKL2000 was used for data processing, integrating, merging and scaling (44). The crystal belongs to the orthorhombic space group P212121 with unit cell parameters a = 39.39, b = 40.56, c = 67.48 Å, α = β = γ = 90°. There is one BTG2 monomer per asymmetric unit, with a solvent content of 18.6% and a Matthews coefficient of 1.51 Å3 Da−1 (45).
Native crystals of mouse TIS21 diffracted to 2.2-Å resolution. Data were collected in-house on a Mar345dtb image plate with a Rigaku MM007 rotating CuKα anode X-ray source. The crystal belongs to the same space group with unit cell parameters a = 38.02, b = 39.70, c = 83.70 Å, α = β = γ = 90°. There is also one molecule per asymmetric unit, with a solvent content of 30% and a Matthews coefficient of 1.75 Å3 Da−1.
Structure determination and refinement
For structure determination of human BTG2, the phasing problem was solved by the MR method using the program Phaser in the CCP4 program suite (46). The structure of TOB from the TOB-CAF1 complex (PDB entry 2D5R) (47), which shares 40% sequence identity with human BTG2, was employed as a search model. The human BTG2 model was manually adjusted with COOT and refined with Refmac5 (48,49). The final model is composed of one BTG2 monomer, with 120 residues (from residues 7 to 127) and 55 water molecules. The structure of mouse TIS21 was also solved by MR following the same protocol and using the same search model. The final TIS21 model includes 117 residues (from residues 8 to 125) and 82 water molecules. Data collection and structure refinement statistics are summarized in Table 1.
Table 1.
X-ray data collection and structure refinement statistics for human BTG2 and mouse TIS21
| Parameter | Mouse TIS21 | Human BTG2 |
|---|---|---|
| A. Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions a/b/c (Å) | 38.02/39.70/83.70 | 39.39/40.56/67.48 |
| Wavelength (Å) | 1.5418 | 1.0000 |
| Resolution (Å) | 50–2.2 (2.28–2.20) | 50–2.3 (2.34–2.26) |
| Average I/σ (I)a | 16.4 (3.6) | 31.5 (4.2) |
| Total reflections | 47 147 | 26 145 |
| Unique reflections | 6868 | 5309 |
| Completeness (%)a | 100 (100) | 97.6 (95.1) |
| Rmerge (%)a,b | 11.5 (46) | 5.7 (21.3) |
| B. Structure refinement | ||
| Resolution (Å) | 50–2.2 | 50–2.3 |
| Average B-factor (Å)b | 29.7 | 37.5 |
| Rwork/Rfree (%)c | 22.1/25.0 | 19.4/26.3 |
| R.m.s.d. bond lengths (Å)d | 0.006 | 0.014 |
| R.m.s.d. bond angles (°)d | 1.265 | 1.452 |
| C. Ramachandran plot | ||
| Most favored (%) | 91.2 | 89.4 |
| Allowed (%) | 7.8 | 9.6 |
| Generously allowed (%) | 1.0 | 1.0 |
| Disallowed (%) | 0 | 0 |
aNumbers in parentheses correspond to the highest-resolution shell.
bRmerge =Σi |Ii-〈I〉|/Σ〈I〉, where Ii is an individual intensity measurement and 〈I〉 is the average intensity for all the reflections.
cRwork/Rfree = Σ ||Fo|-|Fc||/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
dR.m.s.d. relate to the Engh and Huber parameters.
GST ‘pull-down’ experiments in vitro
Following removal of the GST tag, recombinant CAF1 protein purified from E. coli was incubated with GST—and GST-BTG2 WT/mutant—-glutathione-Sepharose beads overnight at 4°C. After washing five times with the binding buffer (PBS with 1% Triton X100), the retained proteins were analyzed on a 12% SDS–PAGE gel and immunoblotted with an anti-CAF1 antibody (50).
Expression vectors and transient transfection
CAF1 and BTG2 cDNAs (wild-type and several mutants) were inserted into pME18S-Flag and pME18S-GST vectors, respectively. Cos 7 cells were transfected with various combinations of the expression vectors by FuGene 6 (Roche) according to manufacturer's protocol. The amount of transfected DNA was adjusted in each experiment by using a control expression vector.
Immunoprecipitation and immunoblotting
In brief, 2 days after transfection, Cos 7 cells were lysed with TNE buffer [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 10 mM NaF, 1 μM phenylmethylsulfonyl fluoride]. Lysates were incubated with glutathione-Sepharose beads for 2 h at 4°C. After washing five times with TNE buffer, the GST-precipitates as well as the lysates were immunoblotted with anti-Flag antibody (M2:Sigma) and anti-GST antibody (Santa Cruz Biotechnology).
Deadenylase assays
The wild-type and mutant human BTG2 proteins, human CAF1 and POP2 were purified as described and dialyzed against the deadenylase buffer (50 mM Tris–HCl, 150 mM NaCl, 10% glycerol, 1 mM DTT, 5 mM MgCl2 at pH 7.5). These proteins were incubated at 37°C for 40 min with a synthesized poly(A) RNA substrate (5′-UCUAAAUAAAAAAAAAAAAAAAAAAAA-3′; final concentration 0.5 μM) labeled with fluorescein isothiocyanate at the 5′ end (51). After addition of formamide, the reaction mixtures were fractionated on a 7 M urea-25% sequence polyacrylamide denaturing gel. The products were analyzed and quantified with a fluorescence imager, FLA-5000 (FUJIFILM).
RESULTS
Overview of the human BTG2 and mouse TIS21 structures
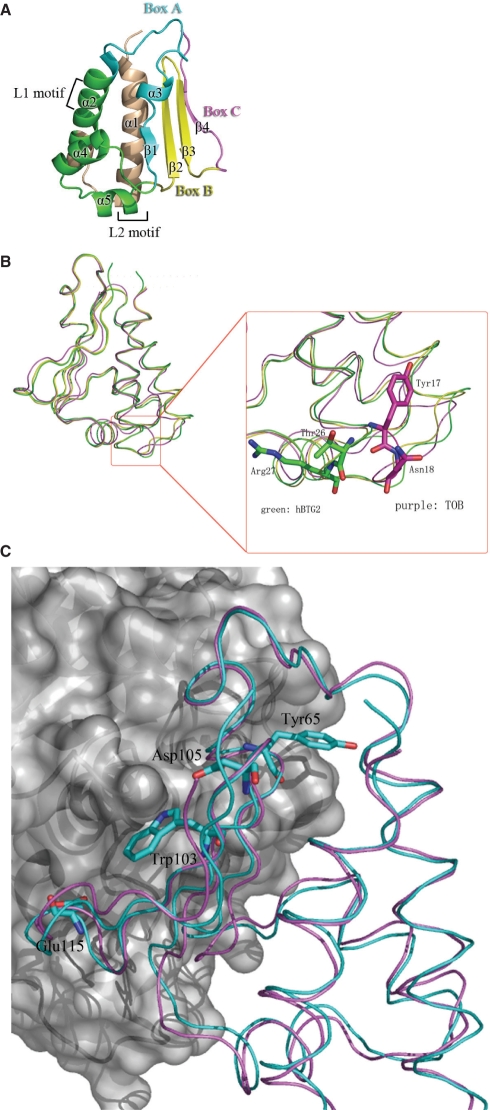
Human BTG2 has approximate dimensions of 49 × 40 × 29 Å3 (Figure 2a). The structure is comprised of five α-helices and four β-strands that form two anti-parallel β-sheets. The N-terminal six residues of human BTG2 are flexible and could not be located from electron density maps. Thr7, the first observable residue, leads immediately into a bundle of three α-helices, followed by four β-strands with two small α-helices inserted between strands β1 and β2. The mouse TIS21 structure is also composed of five α-helices and four β-strands. High structural similarity exists between human BTG2 and mouse TIS21. With a primary sequence identity of 93%, the crystal structures of human BTG2 and mouse TIS21 share an r.m.s.d. of 0.6 Å for the 117 aligned Cα atoms. Superposition of the backbones of human BTG2 and mouse TIS21 is shown in Figure 2b.
Figure 2.
Structures of human BTG2 and mouse TIS21. (A) Ribbon diagram of human BTG2. The conserved domains of Box A, Box B, Box C and the putative HOXB9 interaction region are colored in cyan, yellow, magenta and wheat respectively. The two LXXLL motifs are shown as L1 and L2 motif in black frame. (B) Superposition of human BTG2, mouse TIS21 and TOB (PDB code 2D5R). Human BTG2 is colored in green, mouse TIS21 in yellow and TOB in purple. The regions of the three structures exhibiting main differences are indicated in the red rectangle. The close-up view is shown to the right. The amino acid positions where TOB differs from BTG2/TIS21 are represented as sticks. (C) The conservation of the residues of BTG2/TOB involved in CAF1 binding. CAF1, TOB and BTG2 molecules are colored in gray, violet and blue, while CAF1 is shown in ‘Surface’ diagram. Conserved residues of BTG2 at the interaction surface are represented as sticks.
The structure clearly shows the three highly conserved domains. Box A is composed of strand β1, the short helix α3, part of the α2 helix and a connecting loop between them. Two antiparallel β-strands (β2 and β3) form Box B, while Box C is composed of strand β4 and the extended C-terminal loop. Helix α1, part of the α2 helix and the connecting loop between them constitute the putative HOXB9 interaction region. Previous studies have shown that BTG2 can associate with various molecular targets via different regions (27,40,43). Consistent with our structure, these interfaces are located on different surfaces of BTG2 and may not interfere with each other, which raises the possibility that BTG2 may bind two or more molecular targets simultaneously in order to fulfill different regulatory requirements.
The two LXXLL motifs known as the nuclear receptor box (NR box) are located on helices α2 and α5, respectively. Interestingly, these two motifs are located on opposing faces of BTG2 and provide a hydrophilic surface, which might facilitate contact with nuclear receptors, while the hydrophobic residues are buried inside the core of the protein. The leucine residues of the NR box are located in the middle of their respective α-helices.
Location of the putative CAF1 binding site
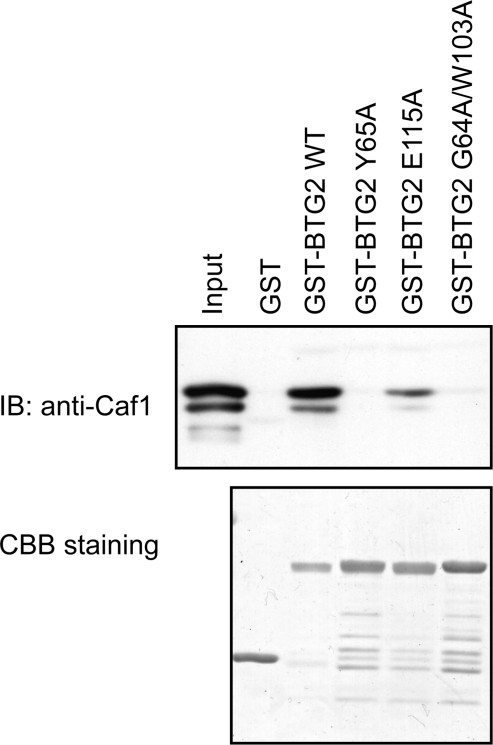
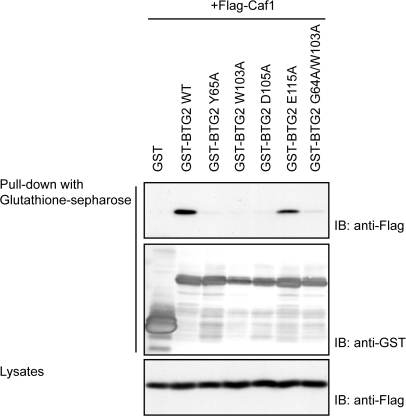
The primary sequence identity between human BTG2 and human TOB is 40%, and their structures share an r.m.s.d of 1.2Å for 114 aligned residues. Superposition of the backbones of human BTG2 and TOB (PDB code 2D5R, unpublished data) is shown in Figure 2b. Previous studies have highlighted the importance of Box B of the TOB family proteins for interaction with CAF1 (27), while the TOB–CAF1 complex structure shows that both Box A and Box B participate in association with CAF1. Structural comparison of the interaction surfaces of BTG2 and TOB reveals significant conservation of the CAF1 interface (Figure 2c), as predicted by the conservation of sequence in these domains. In our crystal structure of human BTG2, the Tyr65 residue in Box A orients its polar NH2 group against the outside of the hydrophilic cluster, while its aromatic side chain is buried inside and may form hydrogen bonds with CAF1; the Trp103 residue projects out its aromatic side chain which may participate in strong hydrophobic interactions with CAF1; the Asp105 residue is located in a cavity that may form the entrance to CAF1; and the Glu115 residue forms a protuberant head which may bind into a groove on the CAF1 molecule but with a weaker association. As a result, we speculate that Tyr65 in Box A and Trp103 and Asp105 in Box B may be important for the interaction of BTG2 with CAF1, while Glu115 alone may not play a predominant role. In order to test this hypothesis, several conserved residues (Y65, W103, D105 and E115) were individually mutated to alanine and the resulting mutant proteins were assayed for their ability to bind to CAF1. Due to the poor stability of the BTG2 W103A mutant in E. coli, a G64A-W103A double mutant was constructed for interaction assays and subsequent deadenylase assays. First, the in vitro interaction assays of BTG2 wild-type and mutants with CAF1 were performed by GST ‘pull-down’ (Figure 3). As mentioned before, in respect to the poor stability of BTG2 W103A and D105A mutants in E. coli, only wild-type BTG2 and Y65A, E115A and G64A-W103A mutants were applied for in vitro interaction assays. The results show that both the Y65A and G64A-W103A mutants were unable to interact with CAF1 in vitro. In contrast, the E115A mutant appeared to retain some binding to CAF1. For further confirmation, we performed co-immunoprecipitation experiments of CAF1 with wild-type BTG2 and the mutants Y65A, W103A, D105A, E115A and G64A-W103A. A vector expressing Flag-tagged CAF1 was transfected into Cos 7 cells, together with a GST-tagged expression vector encoding GST-tagged BTG2 wild type or mutants. Lysates of transfectants were applied for pull-down with glutathione-Sepharose beads, followed by immunoblotting with anti-Flag antibody (Figure 4). Only the wild-type BTG2 and E115A mutant were observed to interact with Flag-CAF1, while no interaction was seen for the Y65A, W103A, D105A and G64A-W103A mutants. The in vivo and in vitro interaction assays consistently demonstrated that the missense mutations Y65A, W103A or D105A resulted in abrogation of the interaction between BTG2 and CAF1. In agreement with our hypothesis, the residues Y65 in Box A and W103, D105 in Box B of BTG2 are critical for interaction with CAF1. Only one single point mutation will abolish the interaction between BTG2 and CAF1.
Figure 3.
Interaction of CAF1 with wild-type Btg2 and mutants in vitro. CAF1 protein was incubated with GST and GST-BTG2 wild-type and mutants. The retained proteins were analyzed by immunoblotting with an anti-CAF1 antibody. The aliquots of GST-fusion proteins used in the upper panel were subjected to gel electrophoresis and CBB staining which were shown in the bottom.
Figure 4.
Interaction of CAF1 with BTG2 wild-type and the mutants in vivo. Cos 7 cells were transfected with the plasmids coding for Flag-tagged CAF1 and GST-tagged BTG2 wild-type or mutants. Cell lysates were prepared 48 h after transfection. Interactions were examined by pull-down with glutathione-Sepharose beads, followed by immunoblotting (IB) with anti-Flag antibody and anti-GST antibody. Expression of Flag-tagged CAF1 is shown in the bottom.
Although the main chain amide group of Tyr65 in BTG2, rather than the side chain, may mediate its involvement in the interaction with CAF1, the side chain does play important roles as an anchor. Compared with the Phe55 residue of TOB, the Tyr65 residue of BTG2 has an additional hydroxyl group which can form an intramolecular hydrogen bond with the His49 residue. The hydrogen bond between Tyr65 (on helix α3) and His49 (on helix α2) helps to anchor Tyr65 and facilitates the α-helical bundles, which lie behind, to provide support and stability to the CAF1 interface. Thus if the side chain of Tyr65 residue is lost, the residue will become too flexible and may no longer be strong enough to exert a force on CAF1. This may explain why the Y65A mutant causes a significant loss-of-interface interactions compared with the wild-type BTG2. Glu115 is further away from CAF1 than other main residues in the BTG2-CAF1 interface. In agreement with our prediction, the role of Glu115, in interaction with CAF1 is not as crucial as Trp103 since there is no observed reduction in the CAF1 binding affinity of the E115A mutant. The rapid degradation of the W103A mutant suggests that Trp103 not only plays important roles in the formation of the BTG2-CAF1 complex, but also contributes to the stability of BTG2. From a careful analysis of the crystal structure of human BTG2, it is evident that the residues discussed above at the interface all fit the electron density map very well and their mean temperature factors are not high, implying that the orientation of these residues in our structure is correct and credible.
BTG2 suppresses the deadenylase activity of CAF1
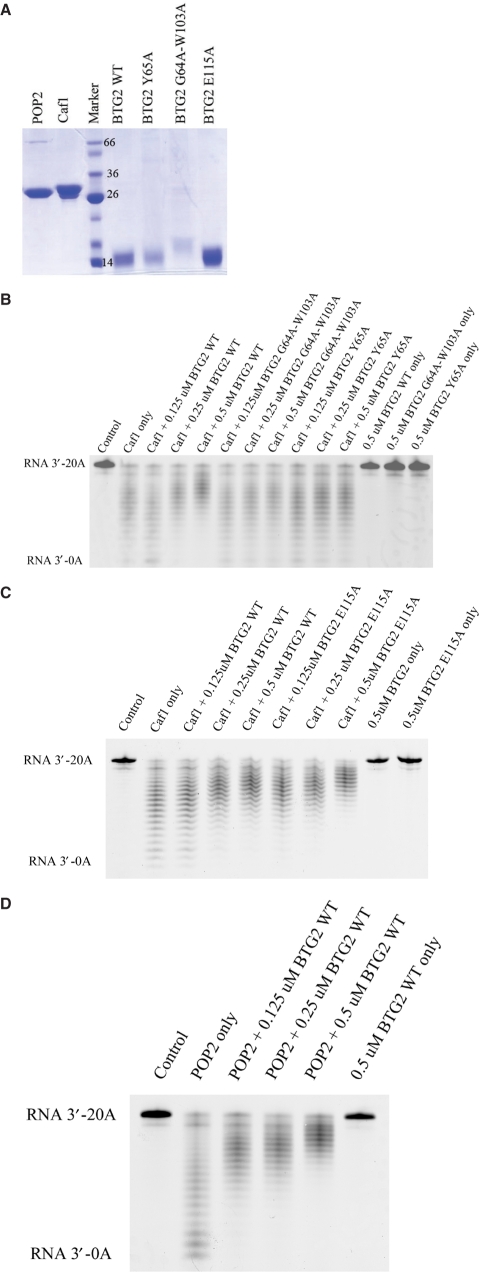
To examine whether BTG2 affects the mRNA deadenylation mediated by CAF1, we carried out in vitro deadenylase assays with single-strand poly(A) RNA as a substrate. We first examined the deadenylase activity of CAF1. GST-tagged CAF1 was over-expressed in E. coli cells, and the soluble GST-tagged proteins were purified by GST-glutathione affinity chromatography and cleaved with PreScission Protease at 16°C overnight. The CAF1 proteins were further purified using a Resource Q ion exchange column and a Superdex-200 size-exclusion column. The purified and concentrated CAF1 proteins (Figure 5a) were diluted in the deadenylase buffer and incubated with a poly(A) RNA substrate at 37°C for 40 min. Analysis of the reaction products on the denaturing sequencing gel revealed cleavage of the poly(A) RNA substrate (Figure 5b). The generation of an RNA ladder, which indicated trimming of the substrate from the 3′-end due to the 5′-end label, confirmed that CAF1 had deadenylase activity in vitro.
Figure 5.
The deadenylase assay of CAF1/POP2 with BTG2 wild-type and mutants in vitro. (A) Protein profile of the CAF1, POP2, BTG2 wild-type and mutants Y65A, G64A-W103A and E115A. The migration of a molecular weight size marker is indicated. According to the result of CBB staining and UV spectrophotometer, proteins were diluted to the same concentration for deadenylase assay. (B) CAF1 only or CAF1 with BTG2 wild-type or G64A-W103A or Y65A mutant proteins were incubated with 5′-fluorescein isothiocyanate-labeled RNA substrate (RNA 3′-7N+20 As) for 40 min. (C) CAF1 only or CAF1 with BTG2 E115A mutant were employed for deadenylase assays as described. (D) POP2 alone or POP2 with wild-type BTG2 were employed for deadenylase assays as described. The labeled RNA was then analyzed on a denaturing sequencing gel. The control lane shows the starting substrate; the concentrations of CAF1 and POP2 applied in this assay were all 0.5 μM.
Recombinant human BTG2 was overexpressed and purified in a similar manner, albeit using a Resource S ion exchange column for purification in place of the Resource Q ion exchange column. When purified wild-type BTG2 protein was added to the deadenylase reaction system of CAF1 in increasing amounts, the deadenylase activity of CAF1 was suppressed in a dose-dependent manner, while the Y65A and G64A-W103A mutants had no effect on the deadenylase activity of CAF1 (Figure 5b). Figure 5b also shows that the wild-type BTG2 and mutants alone had no deadenylase activity. Deadenylase assays were also performed with another BTG2 mutant, E115A, which can still interact with CAF1, with similar suppression of CAF1 activity as for the wild-type BTG2 (Figure 5c). The above data suggest that BTG2 both qualitatively and quantitatively suppresses CAF1 activity, and that the suppressive role of BTG2 may be achieved via the direct interaction of these two proteins. Consistent with previous data, BTG2 mutants shown not to interact with CAF1 also failed to suppress CAF1 activity. The BTG2 E115A mutant, which could still associate with CAF1, also suppressed CAF1 activity similar to the wild-type protein.
Similar results were obtained to show that BTG2 also suppressed the deadenylase activity of CALIF/Pop2 (Figure 5d). The data from Figure 5d suggest that this suppressive role of BTG2 towards the CAF1 family of proteins is conserved.
DISCUSSION
Comparison with related structures
A search for similar structures with the DALI server (52) did not yield any other matches for BTG2 besides the TOB structure (PDB entry 2D5R), which belongs to the same family. This suggests that structures of the TOB family are highly conserved but quite distinct from other proteins. Interestingly, in contrast to the tight loop between helices α1 and α2 of TOB, BTG2 contains a more flexible loop (T26-G28) that may be important for its interaction with molecular targets such as HOXB9. It has been reported that the N-terminal region of BTG1 (1–38), whose corresponding residues are 1–36 in BTG2, is sufficient for interaction with HOXB9 using the yeast two-hybrid system (40). However, there are no reports of a direct interaction between HOXB9 and TOB to date. Thus, we postulate that the flexible loop between helices α1 and α2 of BTG2 is likely to be important for contact with HOXB9.
Despite belonging to the same family, BTG2 and TOB have different molecular targets and are involved in diverse signaling pathways. Therefore, the subtle differences between these two protein structures may provide a basis to explain their different functions. Nevertheless, we cannot rule out the possibility that differences in expression may also contribute to these effects.
A model of BTG2–CAF1 complex
Combined with all the data above, we built a three-dimensional model of the BTG2-CAF1 complex by FTDOCK (53). Our model suggests a total of ∼1400 Å2 of solvent accessible surface area on the two proteins would be buried as a result of complex formation. The BTG2–CAF1 interface would involve hydrophobic, charged and polar residues together with an elaborate network of hydrogen bonding, constituting two main patches of interactions. For clarity, the CAF1 and BTG2 interface residues are subscripted with A and B, respectively. In the first patch, the amide group of TyrB65 would form a hydrogen bond with the O atom of PheA245. The second patch would consist of the TrpB103 and AspB105 residues and the Ser201A and Lys203A residues. Of these, the TrpB103 residue would be highlighted as the core of the interface that would make extensive contacts both through the hydrophobic interaction and hydrogen bonds with CAF1. Specifically, the aromatic ring of the TrpB103 residue would form a typical π–cation interaction with the Lys203A residue, and the Nɛ1 atom of TrpB103 would form a hydrogen bond with the O atom of Ser201A. Moreover, the AspB105 residue would form a salt bridge with the Lys203A residue. Taken together, our model indicates that there would be two invariant residues (Trp103 and Asp105) and one conserved residue (Tyr65) of BTG2 in close contact with CAF1. Although our model fits the two protein molecules reasonably well and is consistent with all of the observations to date, other modes of CAF1 binding cannot be excluded.
Interestingly, our experiments reveal that BTG2 suppresses the deadenylase activity of CAF1 via direct interaction. However, the interface of BTG2 and CAF1 is not close to the active site of CAF1. One possibility would be that the binding of BTG2 is enough to induce local conformational changes that influence the activity of CAF1 or even distort the active site. Some previously published examples have highlighted the conformational rearrangements of enzymes upon complex formation, such as the Trm8–Trm82 complex and HP1451–HP0525 complex (54,55). Trm8, the catalytic subunit of a yeast m7G46 tRNA methyltransferase, undergoes subtle conformational changes upon binding the noncatalytic subunit Trm82, which results in the regulation of Trm8 activity (54). HP1451 binds to HP0525, which is an inner membrane associated ATPase, inducing notable changes in the HP0525 structure; it therefore acts as an inhibitory factor (55). However, the proteins in our complex model are regarded as rigid molecules in docking and we cannot derive any information about their conformational flexibility. BTG2 lacks any known RNA-binding domains and may not directly interact with RNA. Nevertheless, it is tempting to speculate that BTG2 may block access of CAF1 to RNA for cleavage, particularly for longer RNA substrates, and this may provide an alternative potential mechanism for the suppression of CAF1 deadenylase activity. It will be of interest to determine whether BTG2 blocks the substrate binding of CAF1 and thus explain how BTG2 contributes to the suppression of the deadenylase activity of CAF1.
BTG2 was recently shown to be a general activator of mRNA degradation, for which the deadenylase activities of both CAF1 and CCR4 are involved (56). However, the precise mechanism by which BTG2 enhances mRNA deadenylation remains to be determined, as BTG2 was unable to trigger the deadenylase activity of CAF1 in vitro (56). We have demonstrated that BTG2 could suppress the deadenylase activity of CAF1 in vitro. We recently showed that TOB interacts with the CCR4–NOT complex and inhibits its deadenylase activity in vitro (57). Therefore, at least in vitro, the TOB/BTG family proteins inhibit deadenylase activity associated with the CAF1-containing CCR4–NOT complex. It is apparent that CAF1 by itself is not the decisive factor in mRNA deadenylation, and alternative pathways of cellular mRNA decay may unusually operate when its deadenylase activity is suppressed by BTG2. One possibility would be that CAF1 may release the mRNA substrate and transfer it to other deadenylases such as Pan2-Pan3. Further studies would help understand the contradictory roles of BTG2 on mRNA decay between in vivo and in vitro, and clarify the complicated pathways and network of cellular mRNA turnover.
CONCLUSIONS
In summary, our structural analysis of BTG2 provides useful insights into the putative CAF1-binding interface and indicates the main difference with TOB, which may explain the different roles of these two pleiotropic proteins, and suggest how the BTG2–CAF1 complex may form. The deadenylase assay of CAF1 with BTG2 provides an unexpected insight into the suppressive role of BTG2, which is the first to reveal the precise role of BTG2 to CAF1.
FUNDING
The Ministry of Science and Technology International Cooperation Project (grant number 2006DFB32420); the Ministry of Science and Technology 973 Project (grant number 2007CB914301); the National Natural Science Foundation of China (grant numbers 30221003 and 30770438); the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms partial), MEXT, Japan. Funding for open access charge: Ministry of Science and Technology of China.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Sheng Fu, Xue Li, Mingxiao He for their technical assistance. We are grateful to Dr Zhiyong Lou for his kind help with data collection and processing. We also thank Dr Yiwei Liu, Dr Xuehui Chen and Yunqian Guo for useful discussion.
REFERENCES
- 1.Bradbury A, Possenti R, Shooter EM, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc. Natl Acad. Sci. USA. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fletcher BS, Lim RW, Varnum BC, Kujubu DA, Koski RA, Herschman HR. Structure and expression of TIS21, a primary response gene induced by growth factors and tumor promoters. J. Biol. Chem. 1991;266:14511–14518. [PubMed] [Google Scholar]
- 3.Altin JG, Kujubu DA, Raffioni S, Eveleth DD, Herschman HR, Bradshaw RA. Differential induction of primary-response (TIS) genes in PC12 pheochromocytoma cells and the unresponsive variant PC12nnr5. J. Biol. Chem. 1991;266:5401–5406. [PubMed] [Google Scholar]
- 4.Tirone F. The gene PC3(TIS21/BTG2), prototype member of the PC3/BTG/TOB family: regulator in control of cell growth, differentiation, and DNA repair? J. Cell Physiol. 2001;187:155–165. doi: 10.1002/jcp.1062. [DOI] [PubMed] [Google Scholar]
- 5.Matsuda S, Rouault J, Magaud J, Berthet C. In search of a function for the TIS21/PC3/BTG1/TOB family. FEBS Lett. 2001;497:67–72. doi: 10.1016/s0014-5793(01)02436-x. [DOI] [PubMed] [Google Scholar]
- 6.Duriez C, Moyret-Lalle C, Falette N, El-Ghissassi F, Puisieux A. BTG2, its family and its tutor. Bull. Cancer. 2004;91:E242–E253. [PubMed] [Google Scholar]
- 7.Iacopetti P, Barsacchi G, Tirone F, Maffei L, Cremisi F. Developmental expression of PC3 gene is correlated with neuronal cell birthday. Mech. Dev. 1994;47:127–137. doi: 10.1016/0925-4773(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 8.Mesner PW, Epting CL, Hegarty JL, Green SH. A timetable of events during programmed cell death induced by trophic factor withdrawal from neuronal PC12 cells. J. Neurosci. 1995;15:7357–7366. doi: 10.1523/JNEUROSCI.15-11-07357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rouault JP, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, et al. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Dibenedetto AJ, Pittman RN. Genes induced in programmed cell death of neuronal PC12 cells and developing sympathetic neurons in vivo. Dev. Biol. 1997;188:322–336. doi: 10.1006/dbio.1997.8655. [DOI] [PubMed] [Google Scholar]
- 11.Lim IK. TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J. Cancer Res. Clin. Oncol. 2006;132:417–426. doi: 10.1007/s00432-006-0080-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortes U, Moyret-Lalle C, Falette N, Duriez C, Ghissassi FE, Barnas C, Morel AP, Hainaut P, Magaud JP, Puisieux A. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol. Carcinog. 2000;27:57–64. [PubMed] [Google Scholar]
- 13.Guardavaccaro D, Corrente G, Covone F, Micheli L, D’Agnano I, Starace G, Caruso M, Tirone F. Arrest of G(1)-S progression by the p53-inducible gene PC3 is Rb dependent and relies on the inhibition of cyclin D1 transcription. Mol. Cell Biol. 2000;20:1797–1815. doi: 10.1128/mcb.20.5.1797-1815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J. Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti MT, Tata AM, Augusti-Tocco G, Mattei E, Lakshmana MK, Krizhanovsky V, Reeves SA, et al. Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1. J. Neurosci. 2004;24:3355–3369. doi: 10.1523/JNEUROSCI.3860-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl Acad. Sci. USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konrad MA, Zuniga-Pflucker JC. The BTG/TOB family protein TIS21 regulates stage-specific proliferation of developing thymocytes. Eur. J. Immunol. 2005;35:3030–3042. doi: 10.1002/eji.200526345. [DOI] [PubMed] [Google Scholar]
- 18.Oswald J, Steudel C, Salchert K, Joergensen B, Thiede C, Ehninger G, Werner C, Bornhauser M. Gene-expression profiling of CD34+ hematopoietic cells expanded in a collagen I matrix. Stem Cells. 2006;24:494–500. doi: 10.1634/stemcells.2005-0276. [DOI] [PubMed] [Google Scholar]
- 19.Lim IK, Lee MS, Lee SH, Kim NK, Jou I, Seo JS, Park SC. Differential expression of TIS21 and TIS1 genes in the various organs of Balb/c mice, thymic carcinoma tissues and human cancer cell lines. J. Cancer Res. Clin. Oncol. 1995;121:279–284. doi: 10.1007/BF01209594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K, Kubota Y, Lu KP, Aoki I. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin. Cancer Res. 2005;11:7523–7531. doi: 10.1158/1078-0432.CCR-05-0457. [DOI] [PubMed] [Google Scholar]
- 21.Elmore LW, Di X, Dumur C, Holt SE, Gewirtz DA. Evasion of a single-step, chemotherapy-induced senescence in breast cancer cells: implications for treatment response. Clin. Cancer Res. 2005;11:2637–2643. doi: 10.1158/1078-0432.CCR-04-1462. [DOI] [PubMed] [Google Scholar]
- 22.Lim IK, Lee MS, Ryu MS, Park TJ, Fujiki H, Eguchi H, Paik WK. Induction of growth inhibition of 293 cells by downregulation of the cyclin E and cyclin-dependent kinase 4 proteins due to overexpression of TIS21. Mol. Carcinog. 1998;23:25–35. doi: 10.1002/(sici)1098-2744(199809)23:1<25::aid-mc4>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 23.Hong JW, Ryu MS, Lim IK. Phosphorylation of serine 147 of tis21/BTG2/pc3 by p-Erk1/2 induces Pin-1 binding in cytoplasm and cell death. J. Biol. Chem. 2005;280:21256–21263. doi: 10.1074/jbc.M500318200. [DOI] [PubMed] [Google Scholar]
- 24.Park S, Lee YJ, Lee HJ, Seki T, Hong KH, Park J, Beppu H, Lim IK, Yoon JW, Li E, et al. B-cell translocation gene 2 (Btg2) regulates vertebral patterning by modulating bone morphogenetic protein/smad signaling. Mol. Cell Biol. 2004;24:10256–10262. doi: 10.1128/MCB.24.23.10256-10262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakaguchi T, Kuroiwa A, Takeda H. Expression of zebrafish btg-b, an anti-proliferative cofactor, during early embryogenesis. Mech. Dev. 2001;104:113–115. doi: 10.1016/s0925-4773(01)00374-4. [DOI] [PubMed] [Google Scholar]
- 26.Sugimoto K, Hayata T, Asashima M. XBtg2 is required for notochord differentiation during early Xenopus development. Dev. Growth Differ. 2005;47:435–443. doi: 10.1111/j.1440-169X.2005.00819.x. [DOI] [PubMed] [Google Scholar]
- 27.Rouault JP, Prevot D, Berthet C, Birot AM, Billaud M, Magaud JP, Corbo L. Interaction of BTG1 and p53-regulated BTG2 gene products with mCaf1, the murine homolog of a component of the yeast CCR4 transcriptional regulatory complex. J. Biol. Chem. 1998;273:22563–22569. doi: 10.1074/jbc.273.35.22563. [DOI] [PubMed] [Google Scholar]
- 28.Prevot D, Morel AP, Voeltzel T, Rostan MC, Rimokh R, Magaud JP, Corbo L. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J. Biol. Chem. 2001;276:9640–9648. doi: 10.1074/jbc.M008201200. [DOI] [PubMed] [Google Scholar]
- 29.Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J. Cell Sci. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- 30.Ohn T, Chiang YC, Lee DJ, Yao G, Zhang C, Denis CL. CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucleic Acids Res. 2007;35:3002–3015. doi: 10.1093/nar/gkm196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 32.Daugeron MC, Mauxion F, Seraphin B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res. 2008;36:3374–3388. doi: 10.1093/nar/gkn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 2004;23:2862–2871. doi: 10.1038/sj.emboj.7600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Draper MP, Salvadore C, Denis CL. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell Biol. 1995;15:3487–3495. doi: 10.1128/mcb.15.7.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu-Yoshida Y, Sasamoto M, Yoshida A, Yoshioka T, Matsumoto A, Sakai A. Mouse CAF1, a mouse homologue of the yeast POP2 gene, complements the yeast pop2 null mutation. Yeast. 1999;15:1357–1364. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1357::AID-YEA465>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 37.Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/3′-5′-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J. Biol. Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- 38.Bianchin C, Mauxion F, Sentis S, Seraphin B, Corbo L. Conservation of the deadenylase activity of proteins of the Caf1 family in human. RNA. 2005;11:487–494. doi: 10.1261/rna.7135305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- 40.Prevot D, Voeltzel T, Birot AM, Morel AP, Rostan MC, Magaud JP, Corbo L. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J. Biol. Chem. 2000;275:147–153. doi: 10.1074/jbc.275.1.147. [DOI] [PubMed] [Google Scholar]
- 41.Ryu MS, Lee MS, Hong JW, Hahn TR, Moon E, Lim IK. TIS21/BTG2/PC3 is expressed through PKC-delta pathway and inhibits binding of cyclin B1-Cdc2 and its activity, independent of p53 expression. Exp. Cell Res. 2004;299:159–170. doi: 10.1016/j.yexcr.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 43.Berthet C, Guehenneux F, Revol V, Samarut C, Lukaszewicz A, Dehay C, Dumontet C, Magaud JP, Rouault JP. Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells. 2002;7:29–39. doi: 10.1046/j.1356-9597.2001.00497.x. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Methods in Enzymology. New York: Elsevier; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 45.Matthews BW. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Nishida K, Horiuchi M, Noda NN, Takahasi K, Iwasaki N, Minami A, Inagaki F. Crystallization and preliminary crystallographic analysis of the Tob-hCaf1 complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;63:1061–1063. doi: 10.1107/S1744309107057466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura T, Yao R, Ogawa T, Suzuki T, Ito C, Tsunekawa N, Inoue K, Ajima R, Miyasaka T, Yoshida Y, et al. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 2004;36:528–533. doi: 10.1038/ng1344. [DOI] [PubMed] [Google Scholar]
- 51.Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol. Cell Biol. 2007;27:4980–4990. doi: 10.1128/MCB.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 53.Gabb HA, Jackson RM, Sternberg MJ. Modelling protein docking using shape complementarity, electrostatics and biochemical information. J. Mol. Biol. 1997;272:106–120. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 54.Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure. 2008;16:52–61. doi: 10.1016/j.str.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 55.Hare S, Fischer W, Williams R, Terradot L, Bayliss R, Haas R, Waksman G. Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase. EMBO J. 2007;26:4926–4934. doi: 10.1038/sj.emboj.7601904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauxion F, Faux C, Seraphin B. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J. 2008;27:1039–1048. doi: 10.1038/emboj.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyasaka T, Morita M, Ito K, Suzuki T, Fukuda H, Takeda S, Inoue J, Semba K, Yamamoto T. Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Sci. 2008;99:755–761. doi: 10.1111/j.1349-7006.2008.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]