Abstract
Although the role of the APOBEC3-dependent retroelement restriction system as an intrinsic immune defense against human immunodeficiency virus type1 (HIV-1) infection is becoming clear, only the rat ortholog of mammalian APOBEC1s (A1) thus far has been shown to possess antiviral activity. Here, we cloned A1 cDNAs from small animal species, and showed that similar to rat A1, both wild-type and Δvif HIV-1 infection was inhibited by mouse and hamster A1 (4- to 10-fold), whereas human A1 had negligible effects. Moreover, rabbit A1 significantly reduced the infectivity of both HIV-1 virions (>300-fold), as well as that of SIVmac, SIVagm, FIV and murine leukemia virus. Immunoblot analysis showed that A1s were efficiently incorporated into the HIV-1 virion, and their packaging is mediated through an interaction with the nucleocapsid Gag domain. Interestingly, there was a clear accumulation of particular C-T changes in the genomic RNAs of HIV-1 produced in their presence, with few G-A changes in the proviral DNA. Together, these data reveal that A1 may function as a defense mechanism, regulating retroelements in a wide range of mammalian species.
INTRODUCTION
It is now clear that the scope of intracellular defense mechanisms against retroviral infections extend beyond the conventional innate and acquired immune responses, involving a series of dominant inhibitory activities that influence retroviral tropism. Two major restriction factors identified thus far are the early block owing to Fv1 and TRIM5α that target incoming retroviral capsids, and cytidine deaminases, such as APOBEC3 (A3) that function at the late phase to hypermutate retroviral genome (1). The A3 proteins have been shown to inhibit the infectivity potential and mobility of a broad and growing number of exogenous retroviruses as well as endogenous retroelements (2,3). A3 edits deoxycytidine (dC) to deoxyuridine (dU) on nascent DNA minus strands during reverse transcription, but the mechanisms underlying the inhibitory effect on retroviruses are not fully understood.
The A3 encoded by mouse genome was found to be about ∼30% identical to the human APOBEC3G (hA3G), initially identified as a critical target for the human immunodeficiency virus type1 (HIV-1) auxiliary protein Vif (4,5). Subsequently, anti-HIV activity of A3 was found to be maintained across diverse mammalian species, such as murine, cat and artiodactyls (cattle, pigs and sheep) in spite of extensive amino acid sequence divergence, and regardless of whether lentiviruses infect the species (5–8). However, the interaction of Vif with A3 molecules is species specific, and this Vif-resistant inhibition of HIV-1 by orthologous A3 proteins appears to contribute to restrict cells from nonprimate mammalian species to support productive HIV-1 replication. Thus, the removal of the A3-mediated block will be required for the development of a small animal model in which HIV-1 replicates efficiently.
Although the role of the A3-dependent retroelement restriction system as an intrinsic resistance mechanism is becoming clear, less well understood is mammalian APOBEC1 (A1), the catalytic component of a complex that deaminates apolipoprotein B mRNA in gastrointestinal tissues (9,10). It has been also shown that A1 exhibit potent DNA mutator activity in an Escherichia coli assay (11). Rodent A1s share ∼70% amino acid sequence identity with human A1, but only rat homolog of A1 was shown to restrict HIV-1 independent of Vif (12,13). To address whether A1 orthologs are involved in an innate pathway of restriction of retrovirus infection, A1 cDNAs from small animal species were cloned, and expressed in order to examine their abilities to influence the infectivities of retroviral virions. Our studies show that several A1s from small animal species were efficiently incorporated into the HIV-1 virion via interaction with the nucleocapsid (NC) Gag domain, and suppressed HIV-1 replication in a cytidine deaminase dependent as well as independent manner. Interestingly, there was a clear accumulation of particular C-T changes in the genomic RNAs produced in the presence of rabbit A1, with few G-A changes in the proviral DNA. Moreover, the local mutational preferences on HIV-1 genomic RNA were found to be similar to those observed in apoB mRNA. Importantly, mutation of the catalytic residue Glu63 significantly reduced antiviral activity, and diminished G-A or C-T changes. Further, these deaminases also inhibited simian immunodeficiency virus (SIV)mac, SIVagm and feline immunodeficiency virus (FIV) infections and to a lesser extent murine leukemia virus (MLV). Together, these data reveal that, unlike their human counterparts, A1 in a wide range of mammalian species may function as a defense mechanism regulating retroelements.
MATERIALS AND METHODS
Molecular cloning of A1s
Primary tissues were prepared from small intestines, which had been removed aseptically from euthanatized ferret (Mustela putorius furo), rabbit (Kbt: NZW), hamster (Slc: Syrian) and mouse (C57BL/6N), respectively. Total RNA was prepared using TRIzol reagent (Invitrogen, Carlsbad, CA) and the synthesis of first strand cDNA was carried out with High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA) using a random primer. cDNA from human small intestine and rat (Sprague-Dawley) liver was purchased from BD Biosciences Clontech, Palo Alto, CA (BDTM Marathon-Ready cDNA, Cat. #639326 and #639413, respectively). The cDNA encoding the entire open reading frame of the A1 were amplified using primer sets designated as seen in Table S1, based on A1 sequences from GeneBank except for ferret. Taq polymerase-amplified PCR products were cloned into pCR-Blunt (Invitrogen) vector and sequenced. The primary PCR product was subsequently reamplified by using oligonucleotides containing EcoRV and NotI cloning sites. Antisense primers encoded the hemagglutinin (HA)-epitope sequence YPYDVPDYA. Amplicons were cleaved at the restriction sites and ligated to similarly cleaved pCAGGS vector (14), yielding HA tagged A1 expression vectors. pCAGGS-based expression plasmids for HA-tagged hA3G has been described elsewhere (15). PCR products generated and digested as described above were also inserted into pcDNA 3.1/Zeo (Invitrogen).
Generation of catalytic site-mutated rabbit A1 expression vectors
Rabbit A1 catalytically inactive mutants E63A and E63Q were constructed with QuickChange® XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) using oligonucleotide primers (Table S1), and inserted into pCAGGS vector.
Amino acid alignments and phylogenetic analysis
Protein sequences of full-length mammalian A1s were aligned with Clustal W software. Phylogenetic trees were reconstructed using the neighbor-joining method with 2000 bootstrap replications. MEGA 3.1 was used for phylogenetic analysis. The GenBank accession numbers of the A1 sequences used in these comparisons were rat (NM012907), mouse (NM031159), human (NM001644), opossum (NM001032982), rabbit (U10695), orangutan (AH013823), chimpanzee (XM001164661), rhesus monkey (XM001112583), cattle (XM594173), dog (XM543826), hamster (AF176577) and horse (XM001493159). The sequence of A1s reported in this article has been deposited in the GenBank data base (accession number AB425821).
Viral preparation and infectivity assay
293T, GP293, HeLa, Caco-2 and Mus dunni tail fibroblasts (MDTF) cells were maintained in DMEM supplemented with 10% fetal calf serum (Gibco, Grand Island, NY). VSV-G pseudotyped HIV-1-based luciferase reporter virus stocks were produced in 293T cells by cotransfection of wild-type or Δvif pNL4-3 Luc E−R− (5), together with pVSV-G and one of several expression vectors encoding APOBEC proteins, which are HA-epitope tagged or a control empty vector using Effectene® (Qiagen, Hilden, Germany). Culture supernatants were harvested, filtered and frozen in aliquots. The p24 content of the viruses was determined in ELISA kits (ZeptoMetrix, Buffalo, NY). Target fresh 293T cells were infected with 0.5–1.5 ng equivalent of luciferase reporter viruses and cultured for 48 h. Infected cells were lyzed, and each lysate was assayed for luciferase activity as previously described (16). Single-round SIVmac and SIVagm luciferase reporter virus stocks with or without Vif were produced as VSV-G pseudotypes in 293T cells by cotransfection of pSIV Luc E−R− or pSIV Luc E−R− Δvif (5) and an expression vectors for APOBECs or a mock vector. The p27 content of the viruses was determined in ELISA kits (ZeptoMetrix). Target fresh 293T cells were infected for with 4.0–80 ng equivalent of luciferase reporter viruses and cultured for 48 h. Infected cells were lyzed and each lysate was assayed for luciferase activity.
To generate FIV-GFP virus, 293T cells were transfected with pFIV-H1/copGFP and pFIV-34N (System Bioscience, Mountain View, CA), pVSV-G and APOBEC expression vector or an empty vector. Culture supernatants were filtered and centrifuged at 40 000g for 1 h. Target cells were infected with FIV-GFP viruses, equivalent to 8 ng of reverse transcriptase estimated by using Reverse Transcriptase Assay (Roche, Basel, Switzerland) and the infectivity was measured by flow cytometry at 48 h postinfection. Single-round MLV reporter virus stocks were produced as VSV-G pseudotypes in GP293 cells expressing Moloney MLV gag and pol genes (17) by cotransfection of pFB-Luc or pFB-hrGFP (Stratagene) together with expression vectors for APOBECs or a mock vector. Virus-containing supernatants were normalized for equal MLV p30 CA content estimated by Western analysis. Target MDTF cells were infected with equivalent amounts of MLV reporter viruses and cultured for 48 h. Values are presented as the percent infectivity relative to the value of the wild-type virus without the expression of APOBECs.
Editing of HIV-1 proviral DNA and genomic RNA
The cells were harvested 48 h postinfection, and total DNA was isolated using the QIAamp DNA Blood Mini Kit (Qiagen). A 408 bp of the HIV-1 pol region was amplified with the high-fidelity DNA polymerase (Takara, Ohtsu, Japan) subsequent to the digestion with DpnI. Amplified fragments were subsequently gel purified, then cloned into pCR-Blunt vector (Invitrogen) and sequenced. The viral genomic RNA in cell-free virions was purified using QIAamp viral RNA Mini Kit (Qiagen) and converted to cDNA in vitro using a High Capacity cDNA Archive kit with random primers, sebsequent to the treatment with DNase. The pol region was amplified by PCR, cloned and sequenced as described above.
Western blot analysis
The encapsidation of APOBEC proteins into HIV-1 or MLV virions were detected by pelleting the supernatant of 293T cells transfected with viral DNA and HA-epitope tagged forms of the APOBEC expression vector through a 20% sucrose cushion. The pellets were solubilized in 50 μl of 1% Triton-containing buffer and the equivalent p24 of each solubilized virions was subjected to immunoblot analysis. Cell lysates, virion lysates and immunoprecipitates were subjected to SDS–PAGE, and then transferred to a PVDF membrane (Millipore, Bedford, MA). The membranes were probed with the anti-HA epitope (HA.11; Covance, Princeton, NJ) or anti-β-actin (AC-74; Sigma, Saint Louis, MO) antibody. A monoclonal antibody recognizing the p24 CA (18) and a goat anti-MLV p30 serum (ViroMed Biosafety Labs, Camden, NJ) was used for detection of HIV-1 and MLV CA, respectively. Reactive proteins were detected using biotin-conjugated rabbit immunoglobulin (Sigma), streptoavidin-conjugated peroxidase (Sigma) and developed using Chemi-Lumi One (Nacalai Tesque, Kyoto, Japan). The plasmids expressing HIV-1 Gag or the deletion mutants were described previously (22). Bands in western blots were quantified on a VersaDoc 5000 imager (BIO-RAD, Hercules, CA).
Nucleotide sequence accession numbers
The sequences determined in this study have been submitted to GenBank and assigned accession no. AB425821.
RESULTS
Molecular cloning and phylogenetic analysis of mammalian A1s
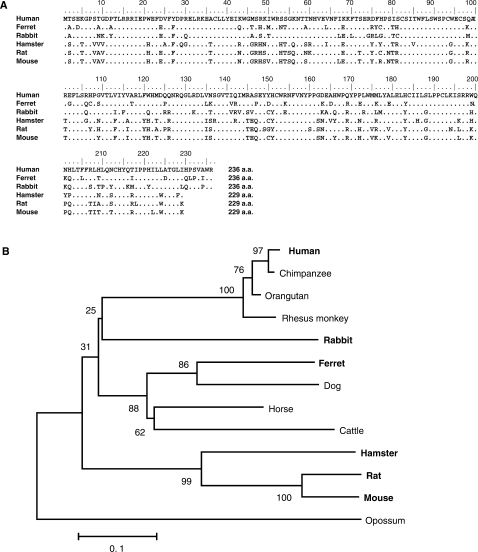
We cloned, sequenced and compared the predicted amino acid sequences of A1 cDNAs from primate (human), carnivora (ferret), lagomorphs (rabbit) and rodents (hamster, rat, mouse). Sequence analysis revealed that the clones obtained encoded genes that were identical to the GenBank sequences of A1s (rabbit;U10695, hamster; AF176577, rat;NM012907 and mouse;NM031159), except for one amino acid residue (M80I) substitution in human A1 (NM001644). Results showed that human A1 cDNA encodes an open reading frame of 236 amino acids that has a 78.0% amino acid sequence identity with ferret, 75.8% with rabbit and 70–72% with rodent orthologs. A short C-terminal extension was found in human and rabbit as previously reported (19), and also in ferrets but not in rodents. In particular, the active site motif, designated as His-X-Glu-(X)23-28-Pro-Cys-X2-Cys (X can be any amino acid) (20), was well conserved (Figure 1A). Phlyogenetic tree analyses revealed that the rabbit A1 gene is related to primate A1 genes, while A1s from rodents form a single separate cluster (Figure 1B).
Figure 1.
Alignment of the amino acid sequence and phylogenetic analysis of mammalian A1 proteins. (A) Amino acid sequence alignment of A1 from human, ferret, rabbit, hamstar, rat and mouse. The predicted amino acid sequences of these cloned A1 molecules are aligned with Clustal W software to the previously identified sequences of mammalian A1s. The numbers are amino acid residue positions. (B) Phylogenetic analysis of the protein sequences of full-length mammalian A1s. The tree was reconstructed by neighbor-joining method with protein p-distances using MEGA 3.1. Shown interior nodes are bootstrap percentages derived from 2000 replications. Branch lengths represent the number of substitution per site. Opossum is a separate group. The species used for further experiments in this study are indicated in bold letters.
A1s from small animal species inhibit HIV-1 virion infectivity
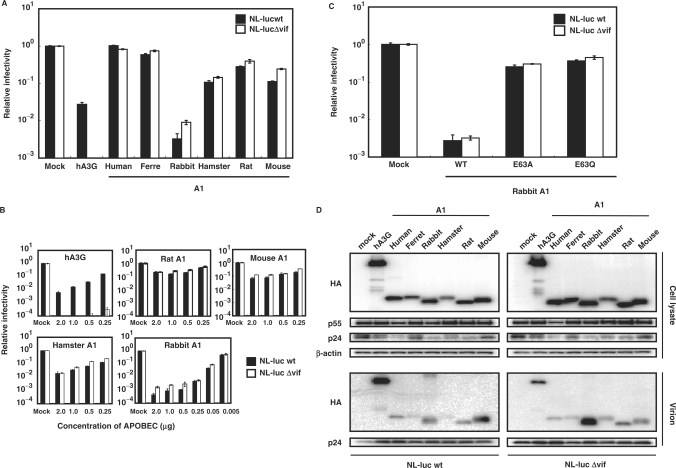
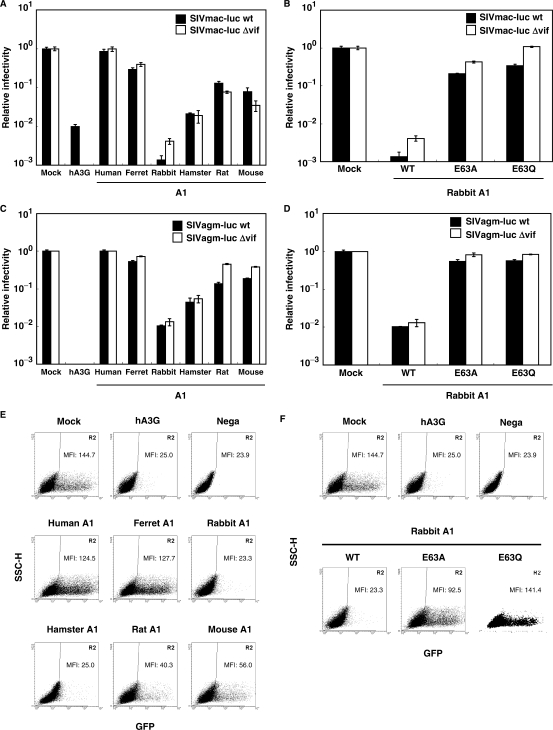
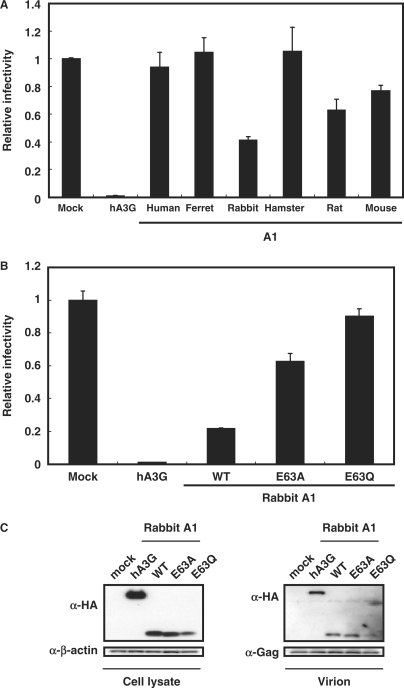
To examine the anti-HIV activities of A1s from these small animal species, single-round infectivity assay with VSV-G pseudotyped wild-type and Δvif HIV-1 luciferase reporter viruses (5) produced in the presence of the influenza HA epitope-tagged A1 proteins was performed. hA3G was used as a control in these experiments. To examine the effect on HIV-1 infectivity quantitatively, virions were normalized based on p24 content. Virus-induced intracellular luciferase activity, which is directly proportional to the infectivity of the virus, was measured 48 h after infection and calculated relative to the APOBEC-negative control virus (Figure 2A, Mock). As expected, hA3G caused a modest decrease in infectivity of the wild-type and a more pronounced decrease in infectivity of the Δvif HIV-1virus (Figure 2A). A1 from human intestine did not showed any antiviral activity, while, in agreement with previous observations (12,13), A1 from rat caused a relatively small (2- to 3-fold) decrease in infectivity of the Δvif HIV-1virus and a >5-fold decrease in infectivity of the wild-type virus (Figure 2A). In contrast with previous findings however (5,12,13), the A1s from rodents, such as hamster and mouse were equally active against wild-type and Δvif HIV-1 viruses, reducing their infectivity >10-fold in some experiments. A modest reduction in infectivity of wild-type and Δvif viruses was also found with A1 from ferrets, but of more interest are the findings with A1 from rabbits. Over 100-fold decrease in the Δvif HIV-1 virus and a more pronounced 300-fold decrease in infectivity of the wild-type virus was seen with this cytidine deaminase from rabbits. Identical results were obtained when rabbit A1 was expressed with an alternate expression vector, pcDNA3.1 (Figure S1), ruling out an effect specific to the pCAGGS construct used in these experiments. Dose titration studies showed that as little as 0.05 μg of rabbit A1 was sufficient to achieve significant inhibition against HIV-1 (Figure 2B), 10- to 20-fold more potent than APOBECs from other small animal species. For further confirmation, the anti-HIV activity of A1s from small animal species was examined with the use of a HIV-1-based the green fluorescent protein (GFP)-expressing reporter virus. As shown in Figure S2, the results obtained with the HIV-GFP vector are consistent with those seen using the HIV-Luc vector. These multiple lines of investigations indicate that, similar to hA3G, A1s from small animal species can function on HIV-1.
Figure 2.
Inhibition of HIV-1 infection by A1s. (A) VSV-G pseudotyped wild-type and Δvif HIV-1 luciferase viruses were produced in 293T cells transfected with 1.5 μg of luciferase reporter viruses, 1.0 μg of pVSV-G and 0.5 μg of HA-tagged APOBEC expression vector or empty vector. Virus-containing supernatants were normalized for equal p24 content and used for the infection of the fresh 293T cells. Virus-induced intracellular luciferase activity was measured. Data are presented as a percentage of the level of luciferase activity detected in cells infected with virions derived from cells that did not express an exogenous APOBEC protein. The average of three experiments with standard deviation is indicated. (B) Wild-type and Δvif HIV-1 luciferase reporter viruses pseudotyped were produced in 293T cells transfected with decreasing amounts of HA-tagged APOBEC expression vector. The amount of expression vector plasmid transfected is shown in micro gram on the X-axis. (C) The catalysis domain of A1 is required for the efficient anti-HIV-1 potency of the rabbit A1. The HA-tagged rabbit A1 (WT), A1 mutated in the active site motif (E63A and E63Q) were used. (D) A1 proteins from small animal species are encapsidated into HIV-1 virions. The producer cells were collected and lyzed, while the released virus in the supernatants were collected by ultracentrifugation. The cells and virion lysates were then subjected to Western analysis using an antibody specific for the HA tag and HIV-1 Gag CA. The immunoblot probed with anti-β-actin antibody of the proteins present in the cell lysates is shown.
The cytidine deaminase domains of APOBEC proteins contains an active site with conserved consensus motifs in which the His–Cys–Cys residues coordinate a Zn2+ ion, and the Glu residue serving an essential role in catalysis as a proton shuttle (21). Therefore, we generated the catalytic site mutant forms of rabbit A1 in which the critical Glu-63 of active site was changed to Ala or Gln (E63A, E63Q), and examined their antiviral activity. Results showed that the ability of the mutant proteins to restrict the infectivity of wild-type and Δvif viruses were severely, but not completely impaired (Figure 2C), suggesting the existence of an albeit weak deaminase-independent restriction mechanism by A1.
A1s are packaged into HIV-1 virions
It is now well established that human and murine A3s inhibit HIV-1 infectivity by being packaged into progeny virions. Therefore, we verified whether A1 proteins (which are epitope tagged) would indeed be selectively packaged into HIV-1 virions. For these studies, transfected cells were harvested, and proteins in whole-cell lysates were analyzed by western blotting with anti-HA, anti-HIV-1 CA p24 and anti-β-actin monoclonal antibodies with the β-actin blot serving as a loading control. Data showed comparable amounts of A1 proteins and HIV-1 Gag precursor protein (p55) expression, but minor effects on the processing efficiency into p24 CA with some A1 proteins (e.g. human and rabbit, Figure 2D). Cell-free virus was concentrated by pelleting through a 20% sucrose cushion, and then equivalent amount of solubilized virions was subjected into the western blotting. A1 proteins from small animal species were found to be packaged into HIV-1 virions, but variations in the efficiency of the incorporation of each A1 protein were seen. Rabbit A1 was incorporated efficiently, which could explain for its potent antiviral activity. The incorporations into HIV-1 virions of A1s from small animal species were not affected by expression of the Vif protein (Figure 2D). We therefore conclude that A1s like hA3G are specifically packaged into HIV-1 virions. However, unlike hA3G, this incorporation is not inhibited with coexpression of HIV-1 Vif. The mutant rabbit A1 E63A E63Q proteins were correctly expressed within the producer cells and also incorporated into the HIV-1 virions efficiently, as judged by western blot analysis (data not shown).
The HIV-1 Gag NC domain is required for A1 packaging
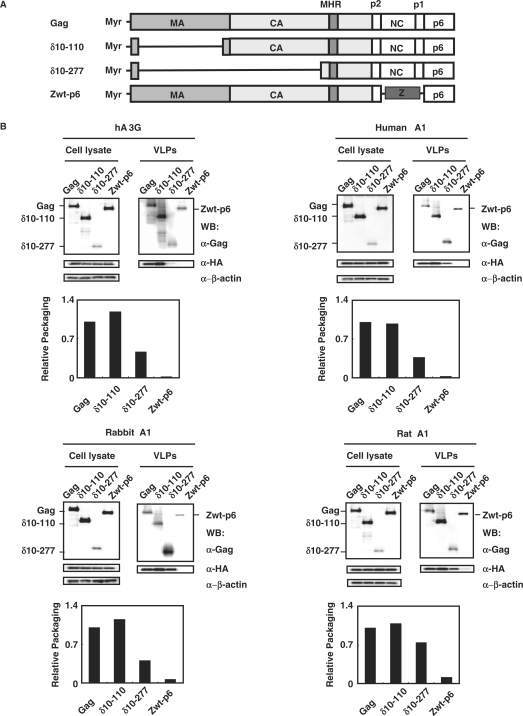
We next determined whether parts of Gag were dispensable for A1 packaging into virions, by use of previously described Gag mutants (22). Two Gag deletion mutants, δ10-110 mutant lacking residues 10 through 110, δ10-277 mutant lacking residues 10 through 277 or the Zwt-p6 mutant, in which NC is replaced by a leucine zipper from GCN4 were tested (Figure 3A). A1s from human, rabbit and rat as well as hA3G were efficiently incorporated into virus like particles (VLPs) formed by both Gag deletion δ10-110 and δ10-277 mutants (Figure 3B). Consistent with previous observations (22), hA3G was not incorporated into VLP formed by Zwt-p6 mutant. A1 from human and rabbit did not exhibit any detectable incorporation, while, A1 from rat caused a relatively small amount of incorporation into VLP formed by Zwt-p6 mutant (Figure 3B). Thus, almost all matrix and the amino-terminal two-thirds of capsid appear to be largely dispensable for A1 packaging, similar to those observed in hA3G packaging.
Figure 3.
A1 proteins are encapsidated into HIV-1 virions via through the interaction with NC, but not the majority of Gag. (A) Schematic representation of the intact HIV-1 Gag precursor and its derivatives used in this study. The Gag are myristoylated (Myr-) on the N-tereminus of MA. The positions of the major homology region (MHR) and the various Gag cleavage sites are shown. (B) Western blot analysis of cell lysates and extracellular VLPs generated following transfection of 293T cells with plasmids expressing HIV-1 Gag or its derivatives shown in (A) and the HA-epitope tagged forms of the indicated APOBEC proteins. The producer cells were collected and lyzed, while the supernatants were harvested and released virus were collected by ultracentrifugation. The cells and virion lysates were then subjected to Western analysis using antibodies specific for the HIV-1 Gag CA and HA tag. The immunoblot probed with anti-β-actin antibody of the proteins present in the cell lysates is shown. Results were normalized to those obtained from the transfection of cells with the full-length Gag and APOBEC constructs, and were graphically shown as relative packaging.
Editing of HIV-1 proviral DNA and genomic RNA by A1 proteins
A3-mediated antiretroviral activity has been shown to be associated with cytidine deamination of nascently transcribed viral cDNAs following infection of target cells. Such dC-to-dU deamination in minus-strand DNA is detected as replacement of dG-to-dA in integrated proviral genomes. To address whether A1 functions in a similar manner, accumulation of dG-to-dA changes in the proviral DNA during reversetranscription was examined. Viral DNA was prepared from cells infected with wild-type or Δvif HIV-1 viruses produced in the presence of A1s from small animal species, and the 3′-end of the pol gene was analyzed for evidence of hypermutation. Consistent with previous reports, significant dG-to-dA hypermutation was observed for hA3G serving as a control for these studies (Table 1) and no dG-to-dA mutation was observed when the virus was prepared in the absence of an APOBEC protein (data not shown). In contrast, the predominant mutations induced by rabbit A1 that exhibited the most prominent antiviral activity were dC-to-dT changes (22 and 14 events in 23 664 and 24 480 bases in wild-type and Δvif viruses, respectively), which could have arisen through deamination of unpaired plus-stranded cDNA or virion RNA. To address the latter possibility, cell-free virions were purified and the genomic RNA was converted to cDNA for pol gene analyses. As can be seen in Table 2, there was a clear accumulation of dC-to-dT (U in the template RNA) changes in the RNAs of HIV-1 produced in the presence of rabbit A1 (39 and 51 events in 9792 and 8976 bases in wild-type and Δvif viruses, respectively), a frequency that is 4- to 5-fold higher than those observed in DNA sequencing. Thus, HIV-1 genome RNA as well as reversetranscribed proviral DNA could be a substrate for A1-mediated deamination.
Table 1.
Sequence analysis of reverse transcribed second-strand proviral DNA in the presence of mammalian A1s
| A1 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hA3G |
Human |
Rabbit |
Rabbit E63A |
Rabbit E63Q |
Hamster |
Mouse |
Rat |
|||||||||
| wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | |
| Clones sequenced | 34 | 36 | 24 | 24 | 58 | 60 | 24 | 23 | 24 | 23 | 34 | 36 | 24 | 24 | 24 | 24 |
| Total base pair sequenced | 13 872 | 14 688 | 9792 | 9792 | 23 664 | 24 480 | 9792 | 9384 | 9792 | 9384 | 13 872 | 14 688 | 9792 | 9792 | 9792 | 9792 |
| Clones with G to A | 15 | 22 | 2 | 1 | 13 | 24 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 6 | 0 | 0 |
| Number of G to A mutations | 71 | 318 | 5 | 7 | 69 | 118 | 0 | 0 | 0 | 2 | 1 | 1 | 0 | 24 | 0 | 0 |
| Number of G to A mutations per 1 kb | 5.12 | 21.65 | 0.51 | 0.71 | 2.92 | 4.82 | 0 | 0 | 0 | 0.21 | 0.07 | 0.07 | 0 | 2.45 | 0 | 0 |
| Clones with C to T | 0 | 0 | 0 | 0 | 20 | 10 | 1 | 0 | 1 | 0 | 3 | 3 | 0 | 5 | 6 | 6 |
| Number of C to T mutations | 0 | 0 | 0 | 0 | 22 | 14 | 2 | 0 | 1 | 0 | 3 | 4 | 0 | 5 | 7 | 7 |
| Number of C to T mutations per 1 kb | 0 | 0 | 0 | 0 | 0.93 | 0.57 | 0.20 | 0 | 0.10 | 0 | 0.22 | 0.27 | 0 | 0.51 | 0.71 | 0.71 |
| Number of Other mutations | 0 | 1 | 0 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
A 408 bp fragment of HIV-1 pol region was amplified from reverse transcripts infected with wt or Δvif NL-Luc viruses produced in the presence of hA3G or mammalian A1s. The number in the clones of each group are shown (wt, wild-type). All mutations are designated using the conventional plus-strand nomenclature.
Table 2.
Sequence analysis of HIV-1 genomic RNA in the presence of mammalian A1s
| A1 |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mock |
hA3G |
Rabbit |
Rabbit E63A |
Rabbit E63Q |
Rat |
|||||||
| wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | wt | Δvif | |
| Clones sequenced | 23 | 23 | 21 | 24 | 24 | 22 | 21 | 23 | 24 | 24 | 22 | 24 |
| Total base pair sequenced | 9384 | 9384 | 8568 | 9792 | 9792 | 8976 | 8568 | 9384 | 9792 | 9792 | 8976 | 9792 |
| Clones with C to T | 0 | 0 | 0 | 0 | 15 | 17 | 0 | 0 | 0 | 0 | 6 | 13 |
| Number of C to T mutations | 0 | 0 | 0 | 0 | 39 | 51 | 0 | 0 | 0 | 0 | 13 | 17 |
| Number of C to T mutations per 1 kb | 0 | 0 | 0 | 0 | 3.98 | 5.68 | 0 | 0 | 0 | 0 | 1.45 | 1.74 |
| Number of Other mutations | 0 | 2 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 |
This experiment was performed as described in the legend for Table 1, except that the genomic RNA of HIV-1 was amplified (wt, wild-type).
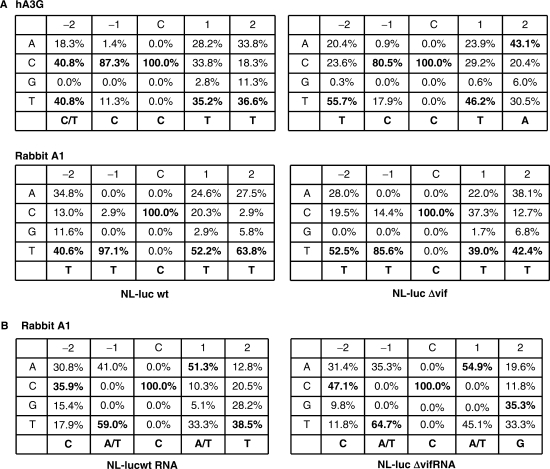
The preferential sites of hypermutations induced by rabbit A1 were also examined (Figure 4). Consistent with previous observations (23–26), hA3G prefers to edit dC (marked by an asterisk) in the viral target DNA sequence C/TCC*. In constrast, rabbit A1, similar to mouse A3 (27) and human A3F (13,28,29), show preference for TTC* (Figure 4A). These results suggest that rabbit A1 inhibits HIV-1 infection by a deamination-dependent mechanism through targeting of proviral DNA sequences that are different from hA3G. Sequence context of the cytidine in viral RNA sequences mutated by rabbit A1 showed a clear WCW (W is A or T) trinucleotide preference on both the wild-type and Δvif HIV-1 genomic RNA (Figure 4B). The cytosine residue within WCW sequences was reported to be highly conserved as the apoB mRNA-editing site sequence by A1 protein among divergent of mammalian species (30). Rabbit A1 deaminated the cytosines in the genomic RNA but with different frequencies; some cytosines were changed at high frequencies, whereas others were not changed in any of the clones (Figure S4). Taken together, analysis of these mutations confirms that the inhibitory effects observed in rabbit A1s were based on, at least in part, cytosine-deaminating activity on viral genomic RNA, and the analysis of mutational hot spots indicated that the molecular mechanisms for editing of HIV-1 genome RNA and apoB mRNA overlap.
Figure 4.
Comparison of the preferred sequence context for cytidine deamination by rabbit A1 in first strand cDNA and the genomic RNA of wild-type orΔvif viruses. (A) TTC*T in the HIV-1 minus-strand cDNA was the preferred tetranucleotide target of rabbit A1. Shown are percentages of each nucleotide found at the −2, −1, +1 and +2 positions relative to the dC residue targeted for deamination (position zero). The consensus DNA sequence is shown at the bottom of each minitable by bold. (B) WC*W sequence in the HIV-1 genome was the preferred RNA target of rabbit A1. Comparison of the preferred sequence observed in the viral genomic RNA produced in the presence of rabbit A1. All of the mutations in HIV-1 genomic RNA aligned with respect to the cytidine (C) targeted for deamination (position zero). The frequency with which each of the four bases found at positions adjacent to the deaminated C is indicated. The consensus RNA sequence is shown at the bottom of each minitable by bold.
A1s from small animal species exhibit broad antilentiviral activity
To assess the breath of A1-mediated antiviral activity, the activities of these proteins on SIV infectivity were examined. A single-round assay was used to measure the infectivity of wild-type and Δvif SIVmac, and SIVagm luciferase reporter viruses (5), in the presence of A1s. VSV-G pseudotyped viral supernatants were collected, normalized for SIV CA p27 and used to infect 293T cells. As expected, hA3G caused a profound decrease in infectivity of both wild-type and Δvif SIV viruses (Figure 5A and C). A1 from rodents were found to be moderately active, but rabbit A1 significantly (>100-fold) reduced the infectivity of wild-type and Δvif SIV viruses. In contrast to A1 from small animal species, A1 from human intestine showed no antiviral activity. Expression of catalytic site mutant forms of rabbit A1 (E63A, E63Q) has little effect on the infectivity of SIV (Figure 5B and D), suggesting that the deaminase activity is also important for SIV repressive activities.
Figure 5.
Inhibition of SIV and FIV infection by A1s. Wild-type and Δvif SIVmac (A and B), wild-type and Δvif SIVagm (C and D) luciferase reporter viruses pseudotyped were prepared. 293T cells were transfected with 1.5 μg of luciferase reporter viruses, 1.0 μg of pVSV-G and 0.5 μg of HA-tagged APOBECs (A and C) or rabbit A1 with catalytic site mutations (B and D). Virus-containing supernatants were normalized for equal p27 content and used for the infection. At 48 h postinoculation, virus-induced luciferase activity was measured and presented as described. VSV-G pseudotyped FIV GFP reporter viruses were produced in 293T cells transfected with 1.5 μg of pFIV-H1/copGFP and pFIV-34N, 1.0μg of pVSV-G and 0.5 μg of HA-tagged APOBECs (E) or rabbit A1 with catalytic active site mutations (F). At 48 h postinoculation with FIV-GFP virions normalized for equal RT activity, cells exhibiting GFP fluorescence of target 293T cells were analyzed on flow cytometry. The level of GFP MFI detected within the GFP-positive windows are indicated. Comparable results were obtained in three additional experiments.
To measure the activities of A1 proteins on FIV infectivity, plasmids expressing APOBECs were cotransfected with an FIV (pFIV-H1/U6-copGFP) genome along with the packaging pFIV-34N plasmid and pVSV-G. Viral supernatants of pseudotyped virions were collected, normalized for RT activity and used to infect 293T cells. The GFP expression was analyzed by flow cytometry at 48 h postinfection. Large proportion of of 293T cells were positive for GFP expression, with a MFI of 144.7, while in the cells transduced with hA3G containing FIV virions, only a few green cells were present, with 5.7-fold lower MFI of 25.0 (Figure 5E). Interestingly, similar to primate lentiviruses, the infectivity of FIV was significantly reduced in the presence of rabbit A1, with a 6.2-fold lower MFI of 23.3. We also detected reduced GFP signals in the cells transduced with rodent A1-containing virions, while human and ferret A1s caused minimal reduction in the number of GFP positive cells (Figure 5E). As seen in Figure 5F, rabbit A1 with catalytic site mutation E63A caused a slight decrease in FIV infectivity, but the E63Q mutation had no effect. Thus, the results using HIV-1, SIVmac, SIVagm and FIV reporter viruses combined suggest that the A1s from small animal species have a relatively broad lentivirus restriction potential that is mainly mediated through deaminase-dependent mechanism.
A1s affect MLV virion infectivity
Evidence is mounting that some APOBEC proteins can target a variety of retroviral substrates, such as various oncovirus and spumavirus. The A3 orthologs from artiodactyls; cattle, pigs and sheep, as well as hAB and hA3G have been reported to exert antiviral activity on MLV (6,13,23,24). These findings suggest that A3s in artiodactyls could function as barriers of cross-subspecies transmission of MLV from mice. Interestingly, this simple oncovirus is resistant to the mouse A3, explaining the absence of a Vif-like activity in MLV (27,31), but the underlying mechanism is currently unknown (32). Therefore, we examined whether the A1s expressed in small animal species affect MLV infection. MLV-based reporter viruses were produced by transient transfection of pFB-Luc, encoding the luciferase gene, into the MLV packaging cell line GP293 with pVSV-G, in the presence of APOBEC proteins. As shown in Figure 6A, hA3G was able to restrict the infectivity of MLV, consistent with previous reports (13,23,24). Interestingly, MLV infectivity was inhibited ∼7-fold by rabbit A1, while A1s from other mammalian species had none or only moderate effect. Similar results were obtained with another murine retroviral vector, pFB-hrGFP (data not shown). Deaminase-defective rabbit A1s retained only partial antiviral activity (Figure 6B) despite comparable levels of MLV virion incorporation (Figure 6C). These data suggest that A1 from small animal species functions as potential barriers of cross-species transmission of this gammmaretroviruses from mice.
Figure 6.
Inhibition of MLV infection by A1s. (A and B) MLV packaging cell line GP293 were transfected with 1.5 μg of luciferase pFB-Luc reporter plasmids, 1.0 μg of pVSV-G and 0.5 μg of HA-tagged APOBECs or rabbit A1 with catalytic site mutations. Virus-containing supernatants were normalized for equal MLV p30 CA content and used for the infection of the MDTF cells. Virus-induced intracellular luciferase activity was measured and presented as described. (C) Rabbit A1 proteins are encapsidated into MLV virions. After transfection, released virion was collected by ultracentrifugation, while the producer cells were collected and lyzed. The cells and virion lysates were then subjected to Western analysis using antibodies specific for the HA tag and MLV Gag CA. An immunoblot probed with anti-β-actin antibody of the proteins present in the cell lysates is also shown. While only the immunoblot of p30 CA performed with the disrupted virions is presented, closely similar results were also obtained using the cell lysates (data not shown).
DISCUSSION
In this study, we showed that single domain cytidine deaminase A1 from rodents (mouse, rat and hamster) and largomorphs (rabbit) are capable of inhibiting the infectivity of various lentiviruses in tissue culture models. A rank order in anti-HIV potency was seen, with rabbit A1 showing the greatest activity. The finding of more efficient virion incorporation of rabbit as compared to other small animal species A1 proteins may be a contributing factor. Catalytic site mutant analysis suggested a deaminase-dependent restriction mechanism, with genomic RNA as well as reversetranscribed proviral DNA serving as substrates for A1-mediated deamination.
A clear accumulation of C-T changes in the genomic RNAs of HIV-1 produced in the presence of rabbit A1 was observed, with G-A changes in the proviral DNA. Furthermore, expression of catalytic site mutant forms of rabbit A1 has little effect on the viral infectivity, supporting the importance of the deaminase activity for these repressive activities. Cytidine deaminase-defective A3 mutants have been shown to exhibit significant antiviral activity (33), implying that antiviral and deaminase activities can be uncoupled. Further evidence in support of editing-independent antiviral mechanism comes from studies on the enzymatically inactive, high-molecular mass complex of hA3G (34) as well as on the antiviral activity against hepatitis B virus (HBV) (35). Nevertheless, more recent studies using deaminase-defective A3 mutants show that efficient inhibition of HIV-1 or retroelements requires catalytically active A3 (36,37). In this regard, although deaminase-defective rabbit A1 mutants were shown to inhibit various lentivirus and MLV, these antiviral activities were significantly lower than those seen with wild-type A1. The suppressive activity of A1, therefore, is principally associated with its cytidine deaminase activity.
We demonstrated that the molecular mechanism for A1 editing of the HIV-1 genomic RNA and apoB mRNA overlapped. The C to U editing of apoB mRNA is shown to be a nuclear event (38), mediated by a complex composed of A1 homodimer and an A1 complementation factor (ACF) (39–41). Expression of ACF mRNA in cells such as Caco-2, a human colon cancer-derived cell line demonstrated to edit endogenous apoB mRNA has been documented (42). However, mRNA for ACF could not be detected by RT-PCR in 293T cells used for retorovirus production in this study (Supplementary Figure S5). Thus, the site(s) in virus-producing cells (e.g. nucleus, cytoplasm or both) where the deamination of HIV-1 genomic RNA takes place remains to be identified, and further experiments are needed to fully understand the role of ACF in the editing of retroviral genome observed in the present study.
In our study, A1s were expressed using both a chick β-actin (CAG) and a cytomegalovirus promoter-driven expression vector, and the concentration of A1s required to mediate antiviral activity was titrated carefully. We found that only 0.05 μg of rabbit A1 was required to achieve significant inhibition against HIV-1 (Figure 2B). Moreover, the antiviral activity of rabbit A1 appeared to be more potent than hA3G (Supplementary Figure S1), and this is unlikely to be explained by differences in virion incorporation of the two enzymes (Figure 2D). Taken together, the data suggest that antiretroviral activities of A1s observed in this study were not solely due to using overexpressed protein systems.
We found that human A1 exhibited no antiretroviral activity, consistent with reports of others (12,13,29,43). A1 from hominoids, therefore, appears to exclusively mediate the C to U editing of apoB mRNA, giving rise to two proteins with different sizes in the gastrointesitinal tissues that function in lipid transport and metabolism. As there have been no reports thus far of lentivirus infection in rodents, the antiviral activity of A1 proteins in rodents seen against HIV, SIV and FIV are also unlikely to have evolved originally to restrict infection of these lentiviruses. Nevertheless, the finding that rabbit A1 can restrict MLV raises the possibility that A1s from small animals species evolved to restrict cross-subspecies transmission of oncoviruses from mice. Furthermore, endogenous lentivirus of rabbits has recently been described (44), lending biological significance to the antilentiviral activity in lagomorphs reported here. Further studies of the antiviral activities of A1 proteins from other members of the placentalia super-order of Laurasiatheria; cetartiodactyla (cow, pig, sheep) and carnivore (cat) will be required to fully understand the complex evolutionary history of APOBEC genes as an intrinsic resistance mechanism against retroelements.
HIV-1 exhibits a highly restricted host cell tropism. The identification of chemokine receptors as entry cofactors with human CD4 raises the possibility that small animal species, in particular, rodents could be engineered to express these molecules, thereby rendering them able to support a productive HIV-1 infection. However, HIV-1 replication in rodents (45–47) and rabbits (48,49) expressing human versions of the HIV-1 receptors appeared to be limited and variable. Our findings with A1 suggest that this enzyme may be partly responsible for the inefficient replication of HIV-1 observed in rabbit as well as rodent cells. Blocking of antiviral A1 function by RNA interference in cells from small animal species should verify whether A1 acts as an intrinsic resistance factor. Further understanding of these species-nonspecific repressive activities to HIV-1 replication at the late phase, in conjugation with the early block owing to the different classes of activities, such as TRIM5α (50,51), may suggest approaches to the development of small animal models of HIV-1 infection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Higo Bank; Grants from the Japan Health Science foundation, Ministry of Health and Welfare of Japan; The Cooperative Research Project on Clinical and Epidemiological Studies of Emerging and Re-emerging Infectious Diseases; Research and Education Program for Development of Therapy of Emerging and Reemerging Infectious Diseases Including AIDS. Funding for open access charge: Higo Bank.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank C. Cheng-Mayer for critical reviewing the article. We also thank P. D. Bieniasz, N. Landau, D. Trono, M. Hayami and H. Amanuma for providing reagents; J. Shibata, K. Monde and K. Yusa for discussions.
REFERENCES
- 1.Bieniasz PD. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. [DOI] [PubMed] [Google Scholar]
- 2.Holmes RK, Malim MH, Bishop KN. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 2007;32:118–128. doi: 10.1016/j.tibs.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Chiu YL, Green WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 4.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 5.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 6.Jonsson SR, Hache G, Stenglein MD, Fahrenkrug SC, Andresdottir V, Harris RS. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006;34:5683–5694. doi: 10.1093/nar/gkl721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munk C, Zielonka J, Constabel H, Kloke BP, Rengstl B, Battenberg M, Bonci F, Pistello M, Lochelt M, Cichutek K. Multiple restrictions of human immunodeficiency virus in feline cells. J. Virol. 2007;81:7048–7060. doi: 10.1128/JVI.02714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O’Brien SJ, et al. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol. 2008;9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chester A, Scott J, Anant S, Navaratnam N. RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochem. Biophys. Acta. 2000;1494:1–13. doi: 10.1016/s0167-4781(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 10.Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 11.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 12.Bishop KN, Holmes RK, Sheehy AM, Malim MH. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. [DOI] [PubMed] [Google Scholar]
- 13.Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho S-J, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 14.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 15.Ohsugi T, Koito A. Human T cell leukemia virus type I is resistant to the antiviral effects of APOBEC3. J. Virol. Methods. 2007;139:93–96. doi: 10.1016/j.jviromet.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Koito A, Kameyama Y, Cheng-Mayer C, Matsushita S. Susceptibility of mink (Mustera vision)-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 2003;77:5109–5117. doi: 10.1128/JVI.77.9.5109-5117.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns JC, Friedmann T, Driever W, Burrascano M, Yee J.-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koito A, Hattori T, Matsushita S, Maeda Y, Nozaki C, Sagawa K, Takatsuki K. Conserved immunogenic region of a major core protein (p24) of human and simian immunodeficiency viruses. AIDS Res. Hum. Retroviruses. 1988;4:409–417. doi: 10.1089/aid.1988.4.409. [DOI] [PubMed] [Google Scholar]
- 19.Yamanaka S, Poksay KS, Balestra ME, Zeng G.-Q, Innerarity TL. Cloning and mutagenesis of the rabbit apoB mRNA editing protein. A zinc motif is essential for catalytic activity, and noncatalytic auxiliary factor(s) of the editing complex are widely distributed. J. Biol. Chem. 1994;269:21725–21734. [PubMed] [Google Scholar]
- 20.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 21.Betts L, Xiang S, Short SA, Wolfenden R, Carter CWJ. Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- 22.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 24.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcript. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi M, Takaori-Kondo A, Shindo K, Abudu A, Fukunaga K, Uchiyama T. APOBEC3G targets specific virus species. J. Virol. 2004;78:8238–8244. doi: 10.1128/JVI.78.15.8238-8244.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 29.Wiegand HL, Doehle BP, Bogerd HP, Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujino T, Navaratnam N, Jarmuz A, Von Haeseler A, Scott J. C – U editing of apolipoprotein B mRNA in marsupials: identification and characterization of APOBEC-1 from the American opossum Monodelphus domestica. Nucleic Acids Res. 1999;27:2662–2671. doi: 10.1093/nar/27.13.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doehle BP, Schafer A, Wiegand HL, Bogerd HP, Cullen BR. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 2005;79:8201–8207. doi: 10.1128/JVI.79.13.8201-8207.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browne EP, Littman DR. Species specific restriction of Apobec3, mediated hypermutation. J. Virol. 2008;82:1305–1313. doi: 10.1128/JVI.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newman EN, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 34.Chiu Y-L, Soros VB, Kreisberg JF, Stopak K, Yonemoto W, Greene WC. Cellular APOBEC3G restricts HIV-1 infection in restricting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- 35.Turelli P, Mangeat B, Jost SS, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 36.Miyagi E, Opi S, Takeuchi H, Khan M, Goila-Gaur R, Kao S, Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumacher AJ, Hache G, MacDuff DA, Brown WL, Harris RS. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD and human immunodeficiency virus type 1 restriction. J. Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lau PP, Xiong WJ, Zhu HJ, Chen SH, Chan L. Apolipoprotein B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation. J. Biol. Chem. 1991;266:20550–20554. [PubMed] [Google Scholar]
- 39.Lellek H, Kirsten R, Diehl L, Apostel F, Buck F, Greeve J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J. Biol. Chem. 2000;275:19848–19856. doi: 10.1074/jbc.M001786200. [DOI] [PubMed] [Google Scholar]
- 40.Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol. Cell. Biol. 2000;20:1846–1854. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O’Keefe R, Scott J, Navaratnam N. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao S, Moberly JB, Schonfeld G. Editing of apolipoprotein B messenger RNA in differentiated Caco-2 cells. J. Lipid Res. 1990;31:695–700. [PubMed] [Google Scholar]
- 43.Zheng Y-H, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katzourakis A, Tristem M, Pybus OG, Gifford RJ. Discovery and analysis of the first endogenous lentivirus. Proc. Natl Acad. Sci. USA. 2007;104:6261–6265. doi: 10.1073/pnas.0700471104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Browning J, Horner JW, Pettoello-Mantovani M, Raker C, Yurasov S, DePinho RA, Goldstein H. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc. Natl Acad. Sci. USA. 1997;94:14637–14641. doi: 10.1073/pnas.94.26.14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawada S, Gowrishankar K, Kitamura R, Suzuki M, Suzuki G, Tahara S, Koito A. Disturbed CD4+ T cell homeostasis and in vitro HIV-1 susceptibility in transgenic mice expressing T cell line-tropic HIV-1 receptors. J. Exp. Med. 1998;187:1439–1449. doi: 10.1084/jem.187.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keppler OT, Welte FJ, Ngo TA, Chin PA, Patton KS, Tsou C.-L, Abbey NW, Sharkey ME, Grant RM, You Y, et al. Progress toward a human CD4/CCR5 transgenic rat model for de novo infection by human immunodeficiency virus type 1. J. Exp. Med. 2002;195:719–736. doi: 10.1084/jem.20011549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn CS, Mehtali M, Houdebine LM, Gut J.-P, Kirn A, Aubertin A.-M. Human immunodeficiency virus type 1 infection of human CD4-transgenic rabbits. J. Gen. Virol. 1995;76:1327–1336. doi: 10.1099/0022-1317-76-6-1327. [DOI] [PubMed] [Google Scholar]
- 49.Speck RF, Penn ML, Wimmer J, Esser U, Hague BF, Kindt TJ, Atchison RE, Goldsmith MA. Rabbit cells expressing human CD4 and human CCR5 are highly permissive for human immunodeficiency virus type 1 infection. J. Virol. 1998;72:5728–5734. doi: 10.1128/jvi.72.7.5728-5734.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in old world monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 51.Schaller T, Hue S, Towers GJ. An active TRIM5 in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 2007;81:11713–11721. doi: 10.1128/JVI.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.