Abstract
Reverse genetics has become pivotal in influenza virus research relying on rapid generation of tailored recombinant influenza viruses. They are rescued from transfected plasmids encoding the eight influenza virus gene segments, which have been cloned using restriction endonucleases and DNA ligation. However, suitable restriction cleavage sites often are not available. Here, we describe a cloning method universal for any influenza A virus strain which is independent of restriction sites. It is based on target-primed plasmid amplification in which the insert provides two megaprimers and contains termini homologous to plasmid regions adjacent to the insertion site. For improved efficiency, a cloning vector was designed containing the negative selection marker ccdB flanked by the highly conserved influenza A virus gene termini. Using this method, we generated complete sets of functional gene segments from seven influenza A strains and three haemagglutinin genes from different serotypes amounting to 59 cloned influenza genes. These results demonstrate that this approach allows rapid and reliable cloning of any segment from any influenza A strain without any information about restriction sites. In case the PCR amplicon ends are homologous to the plasmid annealing sites only, this method is suitable for cloning of any insert with conserved termini.
INTRODUCTION
Many studies in life sciences are based on the efficient generation of multiple plasmid constructs in parallel. Several high-throughput cloning methods are available for generating entire libraries of open reading frames (1). However, for some purposes, it is essential that the inserted cDNA has to be transcribed into RNA of precise length without any additional terminal nucleotides and, therefore, its insertion into an expression cassette is confined to exact nucleotide positions. This constraint also applies to reverse genetics of influenza A viruses. Basic research and vaccine development have been expedited by the generation of recombinant influenza A viruses solely from plasmids (2–6). However, established systems require prior cleavage of amplicons with terminal restriction cleavage sites susceptible to outside cutter enzymes like BsmBI (7) prior to insertion into appropriately cleaved plasmid vectors. Unfortunately, viral genes may contain internal restriction cleavage sites for all available enzymes of this type generating a compatible overhang. At present, this requires subcloning and subsequent removal of unwanted internal restriction cleavage sites before cloning. Such additional time-consuming steps can be different in each case and require prior sequence information of the insert.
Therefore, we sought to establish a cloning method which is independent of restriction enzyme cleavage but still yields high numbers of positive clones. Geiser et al. described a modified Quikchange™ site-directed mutagenesis protocol designed to introduce entire PCR amplicons into plasmids by insertion between two neighboured nucleotides or by exchange of an entire vector region, whereby the two strands of the PCR amplicon serve as megaprimers. Each megaprimer anneals to the plasmid at the complementary site and is then elongated from its 3′-end by the DNA polymerase. The maximum length of successfully cloned inserts in this study was 1117 bp (8), whereas the longest influenza A virus genes (PB2, PB1) each comprise 2341 nt. In order to improve cloning efficiency, we inserted into the plasmid pHW2000 the conserved influenza gene termini (9) and between them the negative selection marker ccdB (10). By use of appropriate primers, this plasmid design provided sufficient homologous annealing sites, but allows shorter PCR primers to be used thus securing efficient amplification. Using this novel assay, we were able to clone the complete genomic sets of eight segments from seven influenza A virus strains and three haemagglutinin genes from strains with subtypes H4, H6 and H8 amounting to a total 59 functional viral genes. These results demonstrate that our approach is suitable for rapid and reliable cloning of basically any insert with conserved termini without any prior information about restriction sites.
MATERIALS AND METHODS
Viruses
Influenza A/Thailand/1(KAN-1)/2004 (H5N1) was propagated in MDCK cells, whereas the strains A/Swan/R65/2006 (H5N1), A/Swine/Belzig/2/2001 (H1N1), A/Denver/57 (H1N1), A/Duck/Ukraine/1/1963 (H3N8), A/Hong Kong/1/1968 (H3N2), A/Chicken/Emirates/R66/2002 (H9N2), A/Mallard/Germany/1740/1/07 (H4N6), A/Turkey/Germany/R617/07 (H6N2) and A/Turkey/Ontario/6118/68 (H8N4) were grown in embryonated hen eggs.
Generation of the plasmid pHWSccdB
For amplification of the ccdB gene from the Gateway™ conversion cassette (Invitrogen GmbH) (nt from 897 to 1583 of reading frame cassette A), we used primers with elongated 5′-ends containing the conserved influenza A gene termini and adjacent BsmBI restriction cleavage sites. The primer sequences were:
5′-ATATCGTCTCAGGGAGCAAAAGCAGGACGCGTGGATCCGGC-3′ and
5′-ATATCGTCTCTTATTAGTAGAAACAAGGGTCGACCTGCAGACTG-3′.
The forward primer contains 12 nt from the 5′-end of influenza A cRNA (underlined) and the reverse primer 13 nt from 5′-end of influenza A vRNA (underlined). After digestion of the PCR amplicon and recipient plasmid pHW2000 with BsmBI, the resulting fragments were ligated and an aliquot was transformed into ccdB-resistant DB 3.1™ bacteria (Invitrogen).
Primers
For amplification of influenza A virus genes, we used primers modified from ref. (11) (Table 1). The forward primer PHW-PB1-17F was newly designed by extending its 3′-end with additional conserved nucleotides (results from Blast query not shown), because the primer with the original 3′-end caused erroneous amplification and subsequent integration of the PA gene instead of PB1 into pHWSccdB. For the neuraminidase gene, four primer pairs should be designed according to the subtype (separately for N1, N2, N4, N5, N8; for N3; for N6, N7; and for N9) (11).
Table 1.
Primer set used for PCR amplification of influenza A virus genes
| Gene | Forward primer | Reverse primer | Expected size (bp) |
|---|---|---|---|
| PB2 | PHW-PB2f: | PHW-PB2-2341r: | 2341 + 26 |
| gaagttggggggg agcgaaagcaggTC | ccgccgggttatt agtagaaacaaggTCGTTT | ||
| PB1 | PHW-PB1-17f: | PHW-PB1-2341r: | 2341 + 26 |
| gaagttggggggg agcgaaagcaggCAAAC | ccgccgggttatt agtagaaacaaggCATTT | ||
| PA | PHW-PAf: | PHW-PA-2233r: | 2233 + 26 |
| gaagttggggggg agcgaaagcaggTAC | ccgccgggttatt agtagaaacaaggTACTT | ||
| HA | PHW-HAf: | identical with PHW-NSr | 1779 + 26 |
| gaagttggggggg agcaaaagcaggGG | |||
| NP | PHW-NPf: | PHW-NPr: | 1565 + 26 |
| gaagttggggggg agcaaaagcaggGTA | ccgccgggttatt agtagaaacaaggGTATTTTT | ||
| NA | PHW-N12458f: | PHW-N12458r: | 1399 + 26 |
| gaagttggggggg agcaaaagcaggAGT | ccgccgggttatt agtagaaacaaggAGT | ||
| M | PHW-Mf: | PHW-Mr: | 1027 + 26 |
| gaagttggggggg agcaaaagcaggTAG | ccgccgggttatt agtagaaacaaggTAG | ||
| NS | PHW-NSf: | PHW-NSr: | 875 + 26 |
| gaagttggggggg agcaaaagcaggGTG | ccgccgggttatt agtagaaacaaggGTG |
Primers are modified from universal influenza A primers11. Their 5′ends (italic) are derived from pHW2000 regions preceding the viral gene termini followed by the conserved influenza A virus gene termini (lowercase), and conserved gene-specific nucleotides at the 3′ ends (uppercase bold). The expected size of the PCR products is based on the length of the genes of strain A/Thailand/1(KAN-1)/2004 (H5N1) plus the noninfluenza sequences and may differ slightly in case of HA, NA and the NS genes from those of other influenza A isolates.
RT– PCR from isolated RNA
Viral RNA was isolated with the QIAamp Viral RNA Kit (Qiagen, Hilden, Germany) according to manufacturer's instructions and reverse-transcribed using Omniscript RT Kit (Qiagen) along with a primer complementary to the conserved 12 nt of 3′-end of the viral RNA (primer Uni12) (11). Using gene-specific primers two microlitres of each RT reaction were subjected to PCR (total reaction volume 100 µl) by an initial denaturation step (98°C 30 s), followed by 35 cycles each consisting of 98°C 10 s, 60°C 30 s, 72°C 6 min and final elongation (72°C 5 min) utilizing 2 U Phusion High-Fidelity DNA Polymerase (New England BioLabs, Frankfurt am Main, Germany) according to the manufacturer's instructions. For determination of fragment sizes and quantification of amplicons, we used the MassRuler™ DNA ladder (Mix) (Fermentas, St Leon-Rot, Germany).
Target-primed plasmid amplification and transformation
Target-primed plasmid amplification was performed with 100 ng pHWSccdB plasmid and PCR amplicon, which had been agarose-gel purified and pooled from 1 to 6 PCR reactions, using 2 U Phusion High-Fidelity DNA Polymerase according to manufacturer's instructions (total reaction volume 50 µl). The amount of PCR amplicon was determined using ImageJ 1.38X (12) from a calibrated image of an agarose gel electrophoresis run including quantitative DNA standards. An initial denaturation step (98°C 30 s) was followed by 35 cycles each consisting of 98°C 10 s, 48°C 1 min and 72°C 5:30 min. Prior to transformation into bacteria, which are not completely sensitive to ccdB [such as the SURE2™, Xl1-Blue™ or XL-10 Gold™ strains (Stratagene Europe, Amsterdam, The Netherlands)], cleavage with DpnI (New England Biolabs (UK) Ltd.) was performed in order to remove the methylated parent plasmid pHWSccdB and any hemimethylated hybrid molecules. In case of the TOP10™ strain (Invitrogen Ltd, UK) which is fully sensitive to ccdB, this step is not required. Following target-primed plasmid amplification, 1, 5 or 10 µl of the reaction mixture were transformed into TOP10™, SURE2™, Xl1-Blue™ or XL-10 Gold™ bacteria according to the manufacturer's instructions.
Virus rescue by transfection of plasmids
Cloned genes from all virus strains except the haemagglutinins of the highly pathogenic H5N1 isolates A/Thailand/1(KAN-1)/2004 and A/Swan/R65/2006 were tested by cotransfection (13) with seven plasmids encoding the genes of A/WSN/33 (2) or the hvPR8 strain (14). A plasmid clone was demonstrated to be functional by the appearance of a cytopathic effect on MDCK cells and positive haemagglutination test from the supernatant.
RESULTS
Interdependent design of cloning vector pHWSccdB and PCR primers
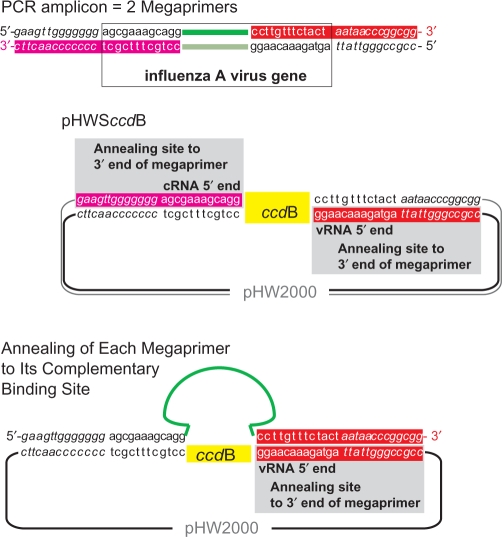
For the introduction of entire PCR amplicons into plasmids by target-primed plasmid amplification, homologous termini are required (8). Therefore, we extended the 5′-ends of universal influenza A primers (11) to achieve homology to the cloning vector pHW2000 (2). However, these primers with a resulting size of 40 nt performed only poorly in the PCR (data not shown). This result could be attributed to the high GC-content of the adjacent vector regions. Therefore, in order to secure high PCR yields in combination with the required homology with vector sequences of at least 20 nt and for high cloning efficiency, we introduced into pHW2000 the conserved influenza A gene termini (9) and between them the negative selection marker ccdB resulting in plasmid pHWSccdB (Figure 1). A set of eight primer pairs is required for full-length amplification of all influenza virus genomic segments. These primers are based on the universal influenza A primers (11) with modifications (Table 1). Their 5′-ends consist of 13 nt homologous to vector sequences preceding the influenza A gene termini in pHWSccdB. This interdependent design of primers and plasmid vector resulted in terminal homologous regions of 25 and 26 nt between amplicon and vector (Figure 1 and Table 1) allowing the use of shorter PCR primers, which ensured efficient amplification. In addition, the introduced ccdB gene enables selection against parental pHWSccdB after transformation into ccdB-sensitive bacteria such as the TOP10™ strain.
Figure 1.
Schematic representation of PCR amplicon and plasmid pHWSccdB. The two strands of the PCR amplicon serve as megaprimers. Each 3′-end of the megaprimer anneals to the complementary annealing site within the plasmid pHWSccdB and is elongated during target-primed plasmid amplification leading to replacement of the ccdB marker by the viral gene. The two newly synthesized strands hybridize to nicked circular molecules.
Universal influenza A PCR and target-primed plasmid amplification
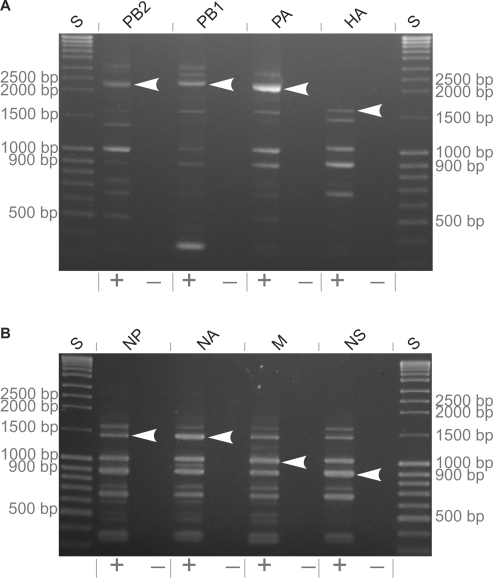
To clone the complete set of all eight genomic segments of influenza A/Thailand/1(KAN-1)/2004 (H5N1) (15), viral RNA was reverse-transcribed and amplified by the modified universal influenza A virus PCR (11) (Figure 1). For amplification of the neuraminidase gene of strain A/Thailand/1(KAN-1)/2004 (H5N1) (15), the appropriate primer pair was used. Since the gene-specific primers differ at their 3′-ends by only a few nucleotides (Table 1), other viral genes may be co-amplified. To assure insertion of the correct amplicon, for each of the eight viral genes, three PCR reactions were pooled (for PB1 only one reaction was used) and amplicons of correct size (Figure 2) were isolated from an agarose gel and used for target-primed plasmid amplification.
Figure 2.
Full-length PCR amplicons from genes of the strain A/Thailand/1(KAN-1)/2004 (H5N1). Lanes: S DNA size marker (3 µl), PCR from H2O control (−) and from cDNA transcribed from viral RNA (+) (each 5 µl from 100 µl total reaction volume).
Forty to 700 ng of each isolated amplicon (Table 2 and Figure 3) were used together with the uncleaved pHWSccdB for the target-primed plasmid amplification. Then, an aliquot was transformed into competent Xl1-Blue™ bacteria. For cloning of the PB1 gene, SURE2™ bacteria had to be used for transformation. The amount of DNA transformed was ∼1 ng. If the bacteria are not completely sensitive to ccdB selection, the DNA should be digested with DpnI prior to transformation in order to select against unmodified methylated pHWSccdB and hemimethylated hybrid molecules. The numbers of bacterial colonies after transformation ranged from 5 to 150 per plate. Insertion of the respective fragment was confirmed by restriction enzyme cleavage with NheI and sequencing (data not shown). From the analyzed clones, 75–100% carried an insert of the expected size (Table 2). These data demonstrate that our cloning method does not require prior knowledge of restriction sites of the insert and yields a high rate of positive clones. Using this method, we generated plasmid sets of all eight gene segments representing the entire genomes of the seven influenza A virus strains A/Thailand/1(KAN-1)/2004 (H5N1), A/Swan/R65/2006 (H5N1), A/Swine/Belzig/2/2001 (H1N1), A/Duck/Ukraine/1/1963 (H3N8), A/HongKong/1/1968 (H3N2), A/Denver/57 (H1N1) and A/Chicken/Emirates/R66/2002 (H9N2) as well as of three haemagglutinin genes of strains A/Mallard/Germany/1740/1/07 (H4N6), A/Turkey/Germany/R617/07 (H6N2) and A/Turkey/Ontario/6118/68 (H8N4). For transformation, we used TOP10™, XL1-Blue™ or XL10-Gold™ and in case of some PB1 genes SURE2™ bacteria. No sequence information relevant for cloning had been available from 18 of those viral genes [all eight genes from both A/Swine/Belzig/2/2001 (H1N1) and A/Chicken/Emirates/R66/2002 (H9N2), and haemagglutinin genes of A/Mallard/Germany/1740/1/07 (H4N6) and A/Turkey/Germany/R617/07 (H6N2)]. Taken together, 59 influenza virus genes were cloned using target-primed plasmid amplification. Except for the haemagglutinin genes of the highly pathogenic H5N1 strains A/Thailand/1(KAN-1)/2004 and A/Swan/R65/2006, which require enhanced security precautions (BSL3+ conditions), all cloned genes were tested in virus rescue and found to be functional. From strains A/Swine/Belzig/2/2001 (H1N1), A/Duck/Ukraine/1/1963 (H3N8), A/HongKong/1/1968 (H3N2) and A/Denver/57 (H1N1) fully homologous recombinant viruses could be rescued after transfection of the complete plasmid sets.
Table 2.
Cloning efficiency: numbers of colonies and of clones with correct length
| Viral Gene | Length (nt) | PCR amplicon (ng) | Transformed (µl) | Bacteria colonies | Correct clones/ Number analyzed |
|---|---|---|---|---|---|
| PB2 | 2341 | 120 | 1 | 0 | 11/12 |
| 120 | 10 | 34 | |||
| PB1 | 2341 | 40 | 1 | 60 | 14/16 |
| 40 | 10 | 20 | |||
| PA | 2233 | 600 | 1 | 5 | 6/8 |
| 600 | 10 | 92 | |||
| HA | 1779 | 490 | 1 | 26 | 8/8 |
| 490 | 10 | 65 | |||
| NP | 1565 | 500 | 1 | 23 | 8/8 |
| 500 | 10 | 48 | |||
| NA | 1399 | 430 | 1 | 18 | 4/5 |
| 430 | 10 | 39 | |||
| M | 1027 | 150 | 1 | 12 | 8/8 |
| 150 | 10 | 140 | |||
| NS | 875 | 690 | 1 | 19 | 12/12 |
| 690 | 10 | 150 |
Numbers of bacteria colonies (XL1-Blue™ bacteria, in case of the PB1 gene SURE2™ bacteria) and of positive plasmid clones carrying the cDNA of viral genes from strain A/Thailand/1(KAN-1)/2004 (H5N1) with correct length in relation to viral gene length, approximate amounts of PCR amplicons used for target-primed plasmid amplification and amounts of it used for transformation.
Figure 3.
Purified PCR amplicons used as megaprimers for target-primed plasmid amplification. For each lane, three PCR assays (each 100 µl) were pooled and agarose-gel purified (total volume 50 µl), except for PB1 for which only one assay was used. Lanes: S DNA size marker (3 µl) and purified amplicons (each 5 µl).
DISCUSSION
Reverse genetics of influenza A viruses requires the cloned cDNA of the eight viral genomic segments to be transcribed into RNA of precise length without any additional terminal nucleotides. In this study, we established a method for rapid and reliable uniform cloning of influenza A genes without any information about restriction sites. Using this approach, we generated complete genomic sets of eight plasmids from seven viral strains of different serotypes.
In the previously described modified Quikchange™ method (8), amplicons with a size of up to 1171 nt had been inserted between two adjacent nucleotides. However, the two largest influenza virus genes, PB2 and PB1, comprise 2341 nt each. Earlier attempts to insert them into unmodified pHW2000 were not successful (data not shown). The results obtained in this study suggest that for longer inserts the exchange of a stretch, in this case the ccdB gene within pHWSccdB as selectable marker, by the PCR amplicon yields significantly higher success rates than direct insertion between two neighboured positions. The cloning of several viral genes was performed using the ccdB-sensitive TOP10 bacteria or ccdB-resistant bacteria such as XL1-Blue™ or SURE2™ and the highly processive Phusion™ polymerase. In case of the strain A/Thailand/1(KAN-1)/2004 (H5N1), the strain XL1-Blue™ was used except for the PB1 gene for which the strain SURE2™ had to be used. This observation demonstrates that the negative selection against the parent plasmid by using the ccdB marker can be replaced by DpnI digestion commonly used in the Quikchange™ protocol. Earlier attempts to use either the original pHW2000 plasmid or a modified version of this vector containing the conserved influenza gene ends only or the pHWSccdB plasmid together with Turbo-Pfu™ polymerase for cloning of the lengthy PB2 gene were not successful (data not shown). However, when using the highly processive Phusion polymerase instead, the usage of vector pHWSccdB led to positive clones. Therefore, a homology of at least 20 nt between amplicon ends and annealing sites of the plasmid, a spacer between annealing sites, and the high processivity of the used polymerase together are essential for cloning of lengthier inserts.
The PCR primers used for amplification of the full-length influenza gene segments take advantage of the conserved gene termini. These termini are identical for all eight segments of all influenza A virus strains. Adjacent to these termini are a few nucleotides identical in all virus strains but gene-specific. Therefore, the universal influenza primers, used in this study, modified from Hoffmann et al. (11) differ only by a few nucleotides at their 3′-ends allowing misamplification of other virus genes. These additional amplicons contain mutations at these sites or may be fragmented and are therefore not suitable for cloning.
The 5′-terminus of the cRNAs from the influenza A virus polymerase genes PB2, PB1 and PA differs from that of the other viral genes by the nucleotide at position 4. Such minor differences can be addressed by appropriate design of the PCR primers (Figure 1 and Table 1) and are corrected during target-primed plasmid amplification demonstrating that complete homology between the annealing sites of vector and amplicon is not required.
Sufficient amounts of insert (Table 2) are critical for cloning efficiency via target-primed plasmid amplification. Otherwise, the rate of positive clones can decline considerably. The Quikchange™ method (developed by Stratagene, La Jolla, CA, USA) yields circular nicked molecules because the primers are not phosphorylated and the primer annealing sites are situated at corresponding positions on the two opposite strands of the plasmid template. Therefore, the newly synthesized strands cannot serve as template during following cycles because the primer would anneal close to the 5′-end of the template in contrast to the PCR in which the primer annealing site is situated at the 3′-end. The products are two strands each with one megaprimer strand as 5′-end; these two strands hybridize to nicked circular molecules. There are two cyclic linear primer extensions from the two plasmid template strands, but not an approximately exponential amplification as in the PCR. This linear amplification explains why such big amounts of PCR amplicon and plasmid are necessary. Accordingly, attempts to use 20 cycles instead of 35 yielded no bacterial colonies after transformation.
Another critical aspect for high efficiency is the protection of the PCR amplicon from short-waved UV light during excision from the agarose gel. If no precautions are taken, clones with repetitions of vector–insert junctions may be isolated (data not shown). Moreover, PCR amplicons have to be purified from agarose gels in order to eliminate the cloning of polymerase gene segments with large internal deletions. These aberrant products which occur in defective interfering particles carry the same termini as the intact full-length genes (16,17). If present in high frequency, they can interfere with efficient amplification of intact polymerase gene segments. Moreover, they can be efficiently integrated into the cloning vector by target-primed plasmid amplification even if they appear only in trace amounts following PCR.
In summary, we established a novel method which can be used for cloning of any influenza A virus strain independent of restriction cleavage. This approach is based on target-primed plasmid amplification using the amplicons of the influenza A gene segments as megaprimers. For improvement, the plasmid vector and primer pairs were adapted to each other: the conserved termini of the influenza A gene segments were integrated into the cloning vector flanking the negative selection marker ccdB. These modifications could also be introduced into the vectors of other reverse genetics systems for influenza A (3–6) and B viruses (18,19). Provided that the termini of the insert are only homologous to the annealing sites within the plasmid vector and not to any other region within vector or insert, this approach is also suitable for standardized cloning of any insert with conserved termini without the need for prior sequence information.
FUNDING
Forschungssofortprogramm Influenza (FSI) of the Government of the Federal Republic of Germany, the European Commission [SPSB-CT-2006-044263 (FLUPOL) and SSPE-CT-2006-44372 (Innflu)]; Deutsche Forschungsgemeinschaft (DFG-Kl238/9-1). Funding for open access charge: Forschungssofortprogramm Influenza (FSI) of the Government of the Federal Republic of Germany.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank E. Hoffmann and R. G. Webster for the pHW2000 plasmid. We are very grateful to Pilaipan Puthavathana for providing us with the influenza A virus A/Thailand/1(KAN-1)/2004 (H5N1), to Elke Lange for strain A/Swine/Belzig/2/2001 (H1N1) and to Timm Harder for all other strains used in this study.
REFERENCES
- 1.Marsischky G, LaBaer J. Many paths to many clones: a comparative look at high-throughput cloning methods. Genome Res. 2004;14:2020–2028. doi: 10.1101/gr.2528804. [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl Acad. Sci. USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, et al. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl Acad. Sci. USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J. Virol. 1999;73:9679–9682. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massin P, Rodrigues P, Marasescu M, van der Werf S, Naffakh N. Cloning of the chicken RNA polymerase I promoter and use for reverse genetics of influenza A viruses in avian cells. J. Virol. 2005;79:13811–13816. doi: 10.1128/JVI.79.21.13811-13816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit E, Spronken MI, Vervaet G, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. A reverse-genetics system for influenza A virus using T7 RNA polymerase. J. Gen. Virol. 2007;88:1281–1287. doi: 10.1099/vir.0.82452-0. [DOI] [PubMed] [Google Scholar]
- 7.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geister M, Cebe R, Drewello D, Schmitz R. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 2001;31:88–90, 92. doi: 10.2144/01311st05. [DOI] [PubMed] [Google Scholar]
- 9.Skehel JJ, Hay AJ. Nucleotide sequences at the 5′ termini of influenza virus RNAs and their transcripts. Nucleic Acids Res. 1978;5:1207–1219. doi: 10.1093/nar/5.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couturier M, Bahassi el M, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 12.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- 13.Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl Acad. Sci. USA. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimm D, Staeheli P, Hufbauer M, Koerner I, Martinez-Sobrido L, Solorzano A, Garcia-Sastre A, Haller O, Kochs G. Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. Proc. Natl Acad. Sci. USA. 2007;104:6806–6811. doi: 10.1073/pnas.0701849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puthavathana P, Auewarakul P, Charoenying PC, Sangsiriwut K, Pooruk P, Boonnak K, Khanyok R, Thawachsupa P, Kijphati R, Sawanpanyalert P. Molecular characterization of the complete genome of human influenza H5N1 virus isolates from Thailand. J. Gen. Virol. 2005;86:423–433. doi: 10.1099/vir.0.80368-0. [DOI] [PubMed] [Google Scholar]
- 16.Nayak DP. Defective interfering influenza viruses. Annu. Rev. Microbiol. 1980;34:619–644. doi: 10.1146/annurev.mi.34.100180.003155. [DOI] [PubMed] [Google Scholar]
- 17.Von Magnus P. Propagation of the PR8 strain of influenza A virus in chick embryos. II. The formation of incomplete virus following inoculation of large doses of seed virus. Acta Pathol. Microbiol. Scand. 1951;28:278–293. doi: 10.1111/j.1699-0463.1951.tb03693.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann E, Mahmood K, Yang CF, Webster RG, Greenberg HB, Kemble G. Rescue of influenza B virus from eight plasmids. Proc. Natl Acad. Sci. USA. 2002;99:11411–11416. doi: 10.1073/pnas.172393399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dauber B, Heins G, Wolff T. The influenza B virus nonstructural NS1 protein is essential for efficient viral growth and antagonizes beta interferon induction. J. Virol. 2004;78:1865–1872. doi: 10.1128/JVI.78.4.1865-1872.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]