Abstract
DNA double strand breaks (DSB) can be repaired either via a sequence independent joining of DNA ends or via homologous recombination. We established a detection system in Drosophila melanogaster to investigate the impact of sequence constraints on the usage of the homology based DSB repair via single strand annealing (SSA), which leads to recombination between direct repeats with concomitant loss of one repeat copy. First of all, we find the SSA frequency to be inversely proportional to the spacer length between the repeats, for spacers up to 2.4 kb in length. We further show that SSA between divergent repeats (homeologous SSA) is suppressed in cell cultures and in vivo in a sensitive manner, recognizing sequence divergences smaller than 0.5%. Finally, we demonstrate that the suppression of homeologous SSA depends on the Bloom helicase (Blm), encoded by the Drosophila gene mus309. Suppression of homeologous recombination is a novel function of Blm in ensuring genomic integrity, not described to date in mammalian systems. Unexpectedly, distinct from its function in Saccharomyces cerevisiae, the mismatch repair factor Msh2 encoded by spel1 does not suppress homeologous SSA in Drosophila.
INTRODUCTION
DNA double strand breaks (DSB) can be repaired in two fundamentally different ways, either via mechanisms involving homologous recombination, or without the use of extensive homology (1,2). Single strand annealing (SSA) is a mechanism based on homologous recombination. It can be used for repair if the DSB occurs between repeated sequences that have the same orientation (direct repeats). The repeats recombine with each other, whereby one repeat and the sequence between the repeats is lost.
In Escherichia coli (3,4), Drosophila melanogaster (5,6) and mammalian cells (7,8), the efficiency of homologous recombination was shown to depend on the length of uninterrupted sequence identity, as well as on the percentage of sequence identity within the region of homology. It was suggested that the strict homology requirement serves to avoid ectopic (nonallelic) recombination between related but nonidentical sequences (5). Based on this line of reasoning it was proposed that diverged introns may help to maintain the integrity of duplicated genes. According to this hypothesis, fast-evolving introns interrupt the homology between otherwise conserved paralogs, thereby inhibiting ectopic homologous recombination between them (9).
Recombination between similar, nonidentical sequences is termed homeologous recombination, to distinguish from homologous recombination between perfectly matching sequences. In several species the mismatch repair (MMR) pathway was shown to be involved in the suppression of homeologous recombination. In E. coli the MMR components MutL, MutS and MutH suppress interspecies recombination (10). In Saccharomyces cerevisiae the MutS homologs Msh2 and Msh3 prevent homeologous recombination (11,12), and Msh2 and Msh6 were shown to suppress homeologous SSA (13). In murine cells mutant for msh2, the frequency of homeologous, intrachromosomal recombination and the gene targeting frequency with non-isogenic constructs is enhanced (14,15).
Besides the MMR components, RecQ helicases also contribute to genomic stability. In E. coli, RecQ prevents illegitimate, ectopic recombination between short stretches of identical sequences (16), and in S. cerevisiae the sole RecQ homolog, Sgs1, suppresses homeologous recombination, notably spontaneously occurring, chromosomal translocations between related genes (13,17–19). In D. melanogaster the RecQ helicase system is more sophisticated, consisting of three paralogs (RecQ4, RecQ5 and Blm), similar to the situation in mammals with five RecQ members. Drosophila Blm was shown to partially rescue the sensitivity of sgs1 mutant strains to a DNA alkylating agent (20). Loss of Blm causes sterility in D. melanogaster, the sperm show high frequencies of chromosome nondisjunction and chromosome loss (21). Drosophila blm mutants are also impaired in DNA synthesis during homologous repair (HR) (22). In a system setup that allows the detection of different modes of DNA repair, homologous repair from the homologous chromosome (HR-h) was found to be decreased in blm mutants, while SSA frequency increased (23). Furthermore, blm mutants are more prone to crossing over, deletions of flanking sequences and template disruption during DSB repair (DSBR) (23,24). Mice and humans mutant for blm exhibit enhanced sister chromatid exchange (SCE) (25) and murine cell lines deficient for blm exhibit chromosomal instability (CIN) (26).
A recent model summarizes the complex role of Blm in HR (27). It postulates that Blm displays both pro- and anti-recombination activities. On the one hand it removes Rad51 from the single stranded 3′ overhang or disrupts the already formed D-loop, thereby inhibiting HR at an early stage (27). On the other hand Blm would also promote HR by unwinding the DNA double helix in front of the D-loop in order to allow for DNA synthesis (22); at a late stage it disrupts the D-loop during HR to facilitate DNA repair via synthesis dependent strand annealing (SDSA) (24), and it also dissolves double Holliday junctions (DHJ) during DSBR, thereby suppressing crossing over (28,29).
Here we have tested SSA frequencies in the fruitfly D. melanogaster, using a reporter system with two tandemly arranged genes and a rare-cutting restriction enzyme site in between. After intracellular cleavage, SSA (consisting of resection of DNA ends, annealing of remaining single strands and repair of gaps/protruding strands) results in the expression of a green fluorescent protein. We find that the shorter the spacer between the direct repeats and the more perfect their sequences are matching, the higher the SSA frequency. SSA is thus under tight control in D. melanogaster and highly sensitive to sequence divergence and distance between direct repeats. Besides the proposed functions of Bloom helicase in the repair of DSBs mentioned above, we found a novel role for Blm in the inhibition of homeologous recombination: it suppresses SSA between divergent sequences. This suggests an additional role for Blm in the maintenance of genomic integrity. In contrast to results in S. cerevisiae, spel1, the Drosophila homolog of the mismatch repair gene MSH2, did not show any involvement in the suppression of homeologous SSA.
MATERIALS AND METHODS
Plasmid sequences
All plasmids that contain tester constructs were generated using standard cloning procedures, and corresponding sequences are available on request. For S2 cell culture experiments, tester constructs were based on pDrBB2 from Michele Calos’ lab.
Tester constructs for in vivo experiments (including the attB site) were subsequently cloned into pCasper-4. I-SceI was expressed in cell cultures via pAc–SceI.
Calcium phosphate transfection of S2 cells
Transfection was performed in 12-well plates with Schneider's Drosophila Medium (GIBCO, Invitrogen Corporation, Carlsbad, CA, USA), containing 10% FBS (ICN Biomedicals, Irvine, CA, USA) and 1% PenStrep (GIBCO); 0.5 ml medium/well (1.5E6 cells/ml) was used; 1.2 μg tester construct plasmid and 1.2 μg I-SceI expressing plasmid were used per well. Plasmid DNA was mixed with 4× Ca mix and 2× Pi mix in this order, mixture was given to cells 4′ after 2× Pi mix addition (4× Ca mix: 0.5 M CaCl2, 0.1 M HEPES; 2× Pi mix: 0.05 M HEPES pH 7.05, 0.75 mM Na2HPO4, 0.75 mM NaH2PO4, 0.28 M NaCl). After 12 h 0.5 ml medium containing 500 μM CuSO4 was added to each well. Flow cytometry analysis was done 48 h after copper addition. All transfections were performed in triplicate.
Flow cytometry
Cells were picked and washed with TBS (25 mM Tris, 137 mM NaCl, 5 mM KCl, 0.7 mM CaCl2, 0.5 mM MgCl2, 0.6 mM Na2HPO4) and resuspended in 0.5 ml ice-cold BSS (0.14 M NaCl, 5.4 mM KCl, 0.3 mM Na2HPO4, 0.4 mM KH2PO4, 1 mM CaCl2, 0.8 mM MgSO4) containing 2% FBS. Analysis was performed at Cytomics FC 500 (Beckman Coulter, Inc., Fullerton, CA, USA) with excitation at 488 nm, green detection (500 nm < x < 525 nm) and red detection (600 nm < x < 615 nm). At least 20 000 cells were measured per sample. Green fluorescence, as a measure for SSA, was quantified in each sample by multiplying the average green fluorescence intensity from the EGFP positive cells (x̄EGFP) with the number of EGFP positive cells (nEGFP) and was then normalized to the corresponding product of the transfection control. Controls lacking an I-SceI expressing plasmid are not shown in here, but we included them in several pilot experiments. Background fluorescence in those experiments was not zero, but low (values were always lower than 5% of the sample values; data not shown).
FACS
EGFP positive cells were sorted for PCR analysis at the Zentrallabor für Zellsortierung ETH/UNI, Gloriastrasse 35, 8092 Zürich.
Plasmid DNA isolation
Plasmids were isolated from EGFP positive S2 cells applying GFXTM Micro Plasmid Prep Kit (Amersham, GE Healthcare, Waukesha, WI, USA).
Single fly DNA isolation
Flies were individually squashed in buffer (10 mM Tris–HCl pH 8.2, 1 mM EDTA, 25 mM NaCl, 0.2% Triton X 100) containing 200 μg/ml proteinase K and incubated for 30 min at 37°C followed by 15 min at 95°C.
PCR analysis of SSA products
For PCR amplification of SSA products MtnB5′-f1 (CCAGGCTTGCACACGACGTG) and EGFP-r (ACGTCGCCGTCCAGCTCGACCAG) were used. Sequencing of these PCR products was done with MtnB5′-f2 (GCAATTTTGCACTCGTTCG).
The contrast of the PCR product gel picture in Figure 3B was enhanced with Photoshop (+57).
Figure 3.
Verification of SSA in transiently transfected S2 cells. (A) Constructs for transfection are depicted: enhancer/promoter-containing mtnB copy (MtnBgen) and enhancer/promoter-less mtnb-EGFP fusion copy (MtnBgen-EGFP). For the positive control construct for intramolecular SSA (MtnBgen-gen), see Figure 1B. Constructs indicated in the bar diagram were always cotransfected with a plasmid expressing the endonuclease I-SceI. Green fluorescence as a measure for SSA was calculated as described in Materials and Methods section. Error bars represent standard deviations of three experiments. (B) Isolation of SSA products from S2 cells. EGFP positive cells were first sorted, and plasmid DNA was isolated. PCR to detect SSA products was performed. The indicated tester constructs are depicted inFigure 1B. PCR products of plasmid DNA from EGFP positive S2 cells after DSB induction (S2) and from E. coli without induction of DSB (Ec) are shown. Expected PCR bands for SSA products are 431 bp and 370 bp (with and without the intron). The three marker bands shown indicate the position of 300 bp, 400 bp and 500 bp.
Fly stocks
For C31 attP/attB integration we used y1 w1118;M{RFP.attP}zh86-Fb stocks with attP landing site on the third chromosome. For analysis of blm mutants w1118; p{v+ 70I-SceI}; mus309D2/TM2 y+ and mus309D3/TM6B were used. For analysis of the msh2 homolog spel1 the following stocks were used y1 w1118; Df(2L)b84h1 p{CaGal}/CyO and y1 w1118; Df(2L)k08712-rv21 p{v+ 70I-SceI}/CyO.
Generation of transgenic flies
attB containing pCasper plasmids were co-injected with C31 integrase capped mRNA into the fly stock mentioned above to generate stable fly lines as previously described (30).
Insertion of the same pCasper plasmids via P-element transformation was performed according to standard protocols.
SSA detection in male germline
To measure SSA frequencies in the male germline, experiments were generally performed as shown for the genomic mtnB direct repeat (MtnBgen-gen). Parental strains (P) y1 w1118; p{v+ 70I-SceI} and y1 w1118; M{MtnBgen-gen}zh86-Fb were crossed. Eggs were collected during 24 h before I-SceI endonuclease was induced by a heat shock at 37°C for 1 h. A second heat shock was applied 24 h later. Eclosing male flies (F1) were individually crossed with 3 y w virgins. Among the offspring (F2) the ratio of EGFP expressing flies to RFP positive flies was calculated to determine SSA frequency.
RESULTS
SSA frequency is reduced by sequence divergence and by increased spacer length between direct repeats in Drosophila S2 cells
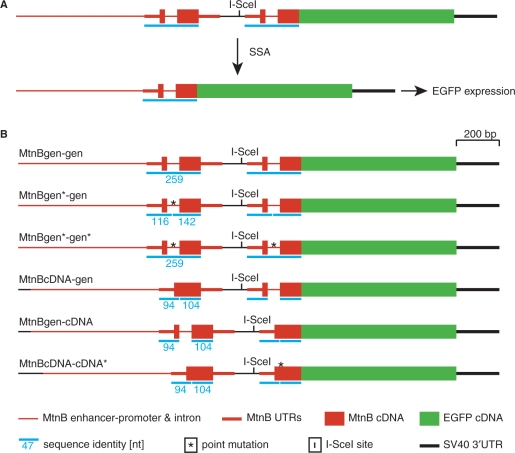
Recombination by SSA can take place if a DSB occurs between direct repeats. In order to determine parameters that influence the frequency of SSA, we developed an SSA detection system in which different parameters can be modified. The tester construct contains the 18-bp cleavage site for the rare cutting endonuclease I-SceI, which allows for the induction of a DSB, producing a staggered cut with a 4 bp 3′ overhang. A first copy of the metallothionein B gene (mtnB), including the regulatory enhancer/promoter DNA, is located upstream of this cleavage site. A second mtnB copy, without enhancer and promoter but fused to the enhanced green fluorescent protein (EGFP), is downstream of the cleavage site. Upon SSA of these two mtnB copies, expression of the MtnB-EGFP fusion protein is inducible via the mtnB enhancer/promoter (Figure 1A). We first analyzed SSA frequencies in a transient transfection assay in Drosophila S2 cells. Plasmids containing the tester constructs were cotransfected with a second plasmid expressing I-SceI under the actin promoter. Expression of MtnB-EGFP was induced by CuSO4 addition to the culture medium, and positive cells were detected by flow cytometry.
Figure 1.
Schematic representation of DNA constructs for SSA detection. (A) One of the two direct repeats, the one upstream of the I-SceI endonuclease cleavage site, is under the control of the mtnB enhancer/promoter. The coding region of the downstream repeat, lacking regulatory sequences for transcription, is fused in-frame to the EGFP cDNA. Induction of transcription does not lead to expression of EGFP protein, unless the DSB, generated by the endonuclease I-SceI, is repaired via SSA of the flanking metallothionein copies. (B) All constructs contain an I-SceI endonuclease recognition site (I-SceI), which is flanked by genomic (gen) or cDNA copies of mtnB. The length of sequence identity between tandem duplicates is indicated by blue bars and numbers, which represent nucleotide numbers. The longest stretch of sequence identity is 259 nt in the construct containing two genomic mtnB copies (MtnBgen-gen). This identity is disturbed in several constructs either by a silent point mutation (MtnBgen*-gen, MtnBcDNA-cDNA*) or the absence of the intron (MtnBcDNA-gen, MtnBgen-cDNA).
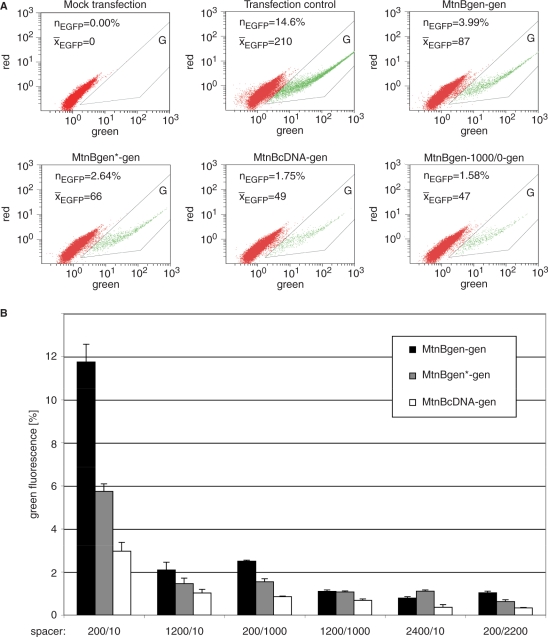
We measured SSA frequency in a construct with two identical genomic copies (259 bp) of mtnB (MtnBgen-gen) and compared it with two further constructs: one harboring a point mutation in the intron of the first mtnB copy (MtnBgen*-gen), and the other containing an intronless first mtnB copy (MtnBcDNA-gen) (Figure 1B). SSA in these constructs can form intermediate heteroduplex structures, including a non-matching DNA region, which consists of only 1 nt in MtnBgen*-gen, whereas in MtnBcDNA-gen the nonmatching region comprises 61 nt. We found that the single mismatch in the heteroduplex of MtnBgen*-gen reduces SSA frequency to 50% of that observed with MtnBgen-gen. SSA in MtnBcDNA-gen, with the nonmatching 61 bp intron, is even reduced to 25% (Figure 2). SSA in S2 cells therefore seems to be strictly regulated, sensing a 1-bp pair mismatch within a 259 bp DNA stretch. This points to the existence of a control mechanism to suppress homeologous SSA in D. melanogaster. Besides the impact of sequence divergence on SSA frequency, we also wished to examine the role of the spacer length between two direct repeats since in human cell culture experiments, efficiency of SSA decreases with increasing spacer length (31). The SSA constructs mentioned above have spacer sequences of 191 bp and 13 bp upstream and downstream of the I-SceI cleavage site, respectively. We added additional spacer sequences of 1 kb and 2.2 kb to either side of the I-SceI site and analyzed SSA frequencies. Identical as well as divergent repeats showed a decrease in SSA with increasing spacer length (Figure 2).
Figure 2.
SSA recombination is impaired by mismatches and spacer DNA in transiently transfected S2 cells. S2 cells were transfected with two plasmids, one containing the SSA tester construct and the other encoding the endonuclease I-SceI under the actin promoter. SSA products resulted in EGFP cDNA driven by the copper inducible mtnB enhancer/promoter. 12 h after transfection EGFP expression was induced by addition of 250 μM CuSO4. (A) In order to reduce the background luminescence, green fluorescence was plotted against red. All cells with specific green fluorescence are shifted to the right. An area for EGFP positive cells was defined (G). For every sample the percentage of EGFP positive cells (nEGFP) and the mean fluorescence intensity of these cells (x̄EGFP) were determined. All indicated constructs are depicted in Figure 1B except MtnBgen-1000/0-gen, which contains an additional 1000 bp spacer upstream of the I-SceI recognition site. (B) On the x-axis the length of spacer sequences upstream/downstream of the I-SceI site are indicated. Green fluorescence as a measure for SSA was calculated as described in Materials and Methods section. Error bars represent standard deviations of three experiments.
To test whether the green fluorescence is truly caused by intramolecular SSA, rather than intermolecular recombination, we cotransfected Drosophila S2 cells with a first plasmid, containing mtnB (including promoter), and a second, separate plasmid, containing the promoterless mtnB-EGFP fusion construct. As usual, the I-SceI endonuclease gene was provided on a different plasmid. This resulted in only a few green cells, which confirms that most of the EGFP expression observed in the regular SSA tester constructs is caused by intra-plasmid recombination events (Figure 3A). To further analyze whether these intra-plasmid recombinations result in the predicted SSA products, we transfected S2 cells with MtnBgen-gen, MtnBgen-cDNA, MtnBcDNA-gen, or MtnBcDNA-cDNA plasmid constructs. We sorted EGFP positive cells, isolated plasmid DNA from these cells and performed PCR with primers spanning the direct repeats. Bands of the expected size for the SSA product were obtained with plasmid DNA retrieved from fluorescent S2 cells (Figure 3B).
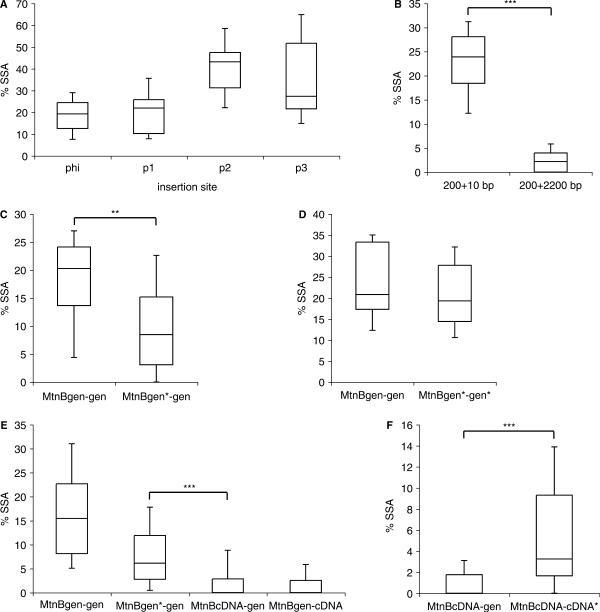
Sequence divergence and spacer sequence also decrease frequency of SSA in transgenic flies
To find out if the sensitive SSA control mechanism is also present in vivo, we analyzed SSA in the male germline of D. melanogaster. We expressed I-SceI during embryonic and larval stages in male flies harboring the SSA tester construct (as outlined in Materials and Methods section). Offspring of these males were then screened for green flies. We first investigated whether the integration of the same construct into different genomic loci influences SSA frequency. For this we inserted the MtnBgen-gen transgene into the genome either randomly with the aid of a P-element transposase, or at a predefined attP ‘landing site’ on the third chromosome by the phage C31 integrase (30,32). There was considerable variation in the percentage of green progeny of flies with insertions at different loci (Figure 4A). Therefore, to more accurately compare SSA frequencies of different tester transgenes, we chose to insert them at the same genomic locus via the C31 integrase system. We started out by testing if SSA frequency depends on the spacer length as in S2 cells and human cell lines. For this we compared MtnBgen-gen with and without the intervening 2.2 kb spacer DNA. Similar to the cell culture results, the spacer reduced the frequency by 91% (Figure 4B). We next investigated whether sequence divergence also reduces SSA frequency in vivo like in S2 cells. Indeed, SSA proved to be very sensitive to mismatches, since 1 nt difference in a 259 nt stretch reduced SSA frequency by 58% (Figure 4C). As a control to ensure that the point mutation itself does not influence the readout, we also tested MtnBgen*-gen*, where the same point mutation is present in both mtnB copies, thereby restoring a 259 nt repeat of identical sequence. The SSA frequency of MtnBgen*-gen* was as high as that of MtnBgen-gen (Figure 4D). We next analyzed the influence of a longer stretch of unpaired DNA, located within the duplication, on SSA. MtnBcDNA-gen showed a pronounced and highly significant reduction of SSA in comparison to MtnBgen*-gen. As expected the same reduction was also seen when we reversed the order of the genomic and the cDNA copy of mtnB (MtnBgen-cDNA) (Figure 4E). The reduction is probably due to the fact that in a heteroduplex generated by MtnBcDNA-gen the 61 bp intron of the genomic copy bulges out as a large non-matching loop, whereas in MtnBgen*-gen the heteroduplex is distorted only by a 1-bp mismatch. However, an alternative explanation is the reduced length of identical sequence flanking the site of interruption (Figure S1). We therefore analyzed SSA of MtnBcDNA-cDNA*, where we had introduced a silent point mutation in the mtnB cDNA at the site where the intron is normally located. Interestingly, the SSA frequency in MtnBcDNA-cDNA* was significantly higher than in the intron-containing construct MtnBcDNA-gen. Thus, not only the length of the identical sequence but also the size of the unmatched DNA sequence itself influences the efficiency of SSA. (Figure 4F). We also verified that repair occurred via SSA, rather than via unequal crossing over, by exploiting the fact that unlike SSA, DSB repair via homologous repair depends on Rad51 (33,34). In a rad51 mutant background, EGFP positive flies emerged with similar frequency as in controls, indicating that most, if not all repair occurred via SSA (data not shown).
Figure 4.
In vivo detection of SSA in the male germline. SSA frequency in flies was determined as described in Material and Methods section. Graphs show box plots for SSA frequencies with the middle line marking the median frequency of all tubes, boxes represent 25th and 75th percentiles and whiskers 10th and 90th percentiles. The two-tailed Mann Whitney U-Test was performed to assess statistical significance. **P < 0.01; ***P < 0.001. All constructs are depicted inFigure 1B. (A) SSA frequencies of MtnBgen-gen tester transgenes inserted at four different loci in the genome. The test lines were generated by targeted integration using the attP/attB system (phi) or random insertion using the traditional P-element system (p1, p2, p3). (B) SSA frequency of the MtnBgen-gen tester construct (with direct repeats spaced by 200 nt) compared to MtnBgen-gen containing an additional 2.2 kb spacer sequence between the repeats, downstream of the I-SceI recognition site. (C–F) SSA frequencies of the tester constructs indicated.
We also did an experiment to see if homologous repair from an ectopic donor sequence is suppressed by mismatches in the donor sequence. To this end, the transgene arrangement was chosen such that conversion to a functional GFP reporter required homologous recombination, rather than SSA (Figure S7). However, in spite of testing many offspring, recombination frequencies were too low for significant results. An identical donor sequence yielded two recombination events among 1650 flies, 1 nt mismatch gave four events in 2141 flies, while the deletion of the mtnB intron gave one event in 3142 flies. Successful recombination events were verified by PCR (data not shown).
Drosophila Bloom helicase (Blm) suppresses homeologous SSA in vivo
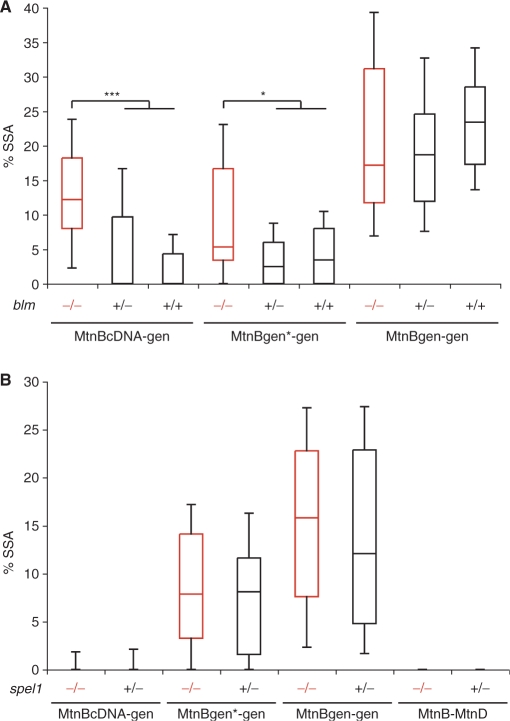
In budding yeast the RecQ helicase Sgs1 and the mismatch repair factors Msh2 and Msh6 were shown to be involved in the suppression of homeologous SSA (13). As mentioned, Drosophila mus309, encoding Blm, is an ortholog of yeast sgs1, while spel1 is an ortholog of msh2. We were interested to know if in D. melanogaster these orthologs also have a similar role in SSA. For this purpose we crossed heterozygous mutants and assayed DSB repair via SSA in the offspring's germline. Using this approach, offspring consisted of homozygous mutants as well as heterozygous and wildtype controls.
For the analysis of Blm, we used compound heterozygotes with two mutant alleles, mus309D2 and mus309D3 (Figure S2), since both alleles behave as genetic nulls (21). PCR of the SSA product with subsequent sequencing was performed in all EGFP positive flies to subtract NHEJ events leading to rare random in-frame ligated mtnB-EGFP fusion products. The values below are based on corrected numbers. No significant change in SSA frequency between blm mutant flies and the controls could be seen with 100% sequence identity (MtnBgen-gen). A small increase of SSA frequency was seen in mutant flies, when we analyzed repair of a construct with a 1-bp mismatch (MtnBgen*-gen). With the more divergent construct MtnBcDNA-gen, a strong increase of SSA frequency was detected in mutants (Figure 5A).
Figure 5.
Homeologous SSA in male germline of blm (mus309) mutants and spel1 mutants. Box plots for SSA frequencies are depicted with the middle line marking the median of all tubes, boxes represent 25th and 75th percentiles and whiskers 10th and 90th percentiles. (A) Analysis of blm mutants. The rare cutting endonuclease I-SceI was expressed under control of the heat shock promoter in blm compound heterozygotes (mus309D3/mus309D2), in heterozygotes (mus309D3/+) and wt controls; they are labeled in the graph with −/−, +/− and +/+, respectively. All constructs (MtnBgen-gen, MtnBgen*-gen and MtnBcDNA-gen) are depicted inFigure 1B. SSA PCR products of all EGFP positive flies were sequenced to exclude random in-frame NHEJ events, graphs represent corrected data. The two-tailed Mann Whitney U-Test was performed to test for statistical significance between flies deficient (red) and proficient (black) for Blm. *P < 0.05; ***P < 0.001. In this experiment, tester constructs were inserted with the help of P-elements at different genomic loci on the second chromosome. SSA frequencies can therefore only be compared within the same tester construct. (B) Analysis of SSA in flies mutant for the msh2 homolog spel1. SSA was analyzed in spel1 compound heterozygotes (−/−) and heterozygous controls (+/−), which were both derived from a cross of spel1 heterozygous parents. The tester constructs MtnBgen-gen, MtnBgen*-gen and MtnBcDNA-gen (Figure 1B) are integrated at the attP landing site on the third chromosome, and MtnB-MtnD (Figure 6A) was P-element inserted on the X-chromosome (line px-3).
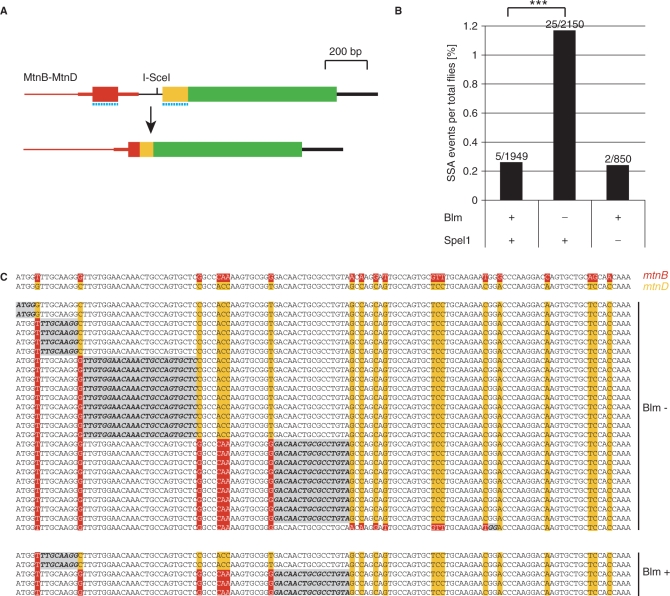
We then wondered if Bloom helicase is also responsible for the suppression of recombination between even more divergent sequences like mtnB and mtnD cDNA, derived from the most closely related paralogs of the four metallothionein genes in D. melanogaster. These cDNAs are 129 nt long and exhibit only 81% sequence identity. Analysis of the MtnB-MtnD construct (Figure 6A and Figure S3) revealed that also in this case, suppression of homeologous SSA is relaxed in Blm deficient flies, resulting in 4.5-fold elevated SSA frequencies (Figure 6B). Sequencing analysis of the repair products revealed an interesting fact. Neither mutant, nor control flies showed any evidence for discontinuous mismatch repair of the heteroduplex DNA (540 potential mismatches in total), since all the MtnB-MtnD SSA repair products contained pure mtnB type sequence upstream and mtnD type sequence downstream of the recombination site; in other words, discontinuous repair patterns were never seen (Figure 6C). In control PCRs with DNA from flies with uncleaved DNA, we always detected only the longer product corresponding to the uncleaved DNA, thus PCR artefacts can be excluded.
Figure 6.
Homeologous SSA between two related genes is enhanced in blm mutant male germline. (A) Schematic representation of an SSA event between two metallothionein paralogs, mtnB (red) and mtnD (orange). Dotted blue line marks region of 80% sequence identity. (B) The frequency of SSA events between mtnB and mtnD in blm and spel1 mutants. The MtnB-MtnD construct tested here was inserted via P-elements on the X-chromosome (px-6). I-SceI was expressed under control of the heat shock promoter in blm mutant flies (mus309D3/mus309D2), as well as in controls (mus309D3/+ and +/+). In an additional experiment it was also expressed in a Spel1 deficient background. From the Blm experiment, SSA products of all EGFP positive offspring were sequenced. Identical SSA product sequences from the same fly tube were considered to originate from a single SSA event. Bars represent frequencies of distinguishable SSA events per total offspring flies analyzed, as indicated by numbers. The χ2-test was performed to test for statistical significance between blm mutant flies and controls. ***P < 0.001. (C) List of detected SSA products. The top two lines represent cDNA sequences of mtnB and mtnD. Divergent nucleotides are highlighted in red (mtnB) and orange (mtnD). All detected SSA events from blm mutant flies (−) and controls (+) are shown. DNA segments where recombination occurred are highlighted in grey.
To investigate the influence of the msh2 ortholog spel1 on homeologous SSA, we analyzed compound heterozygous flies, harboring two different deletions, which overlap at the spel1 locus (35). Both deletions are homozygous lethal. Analysis of the four constructs, MtnBgen-gen, MtnBgen*-gen, MtnBcDNA-gen (Figure S4) and MtnB-MtnD (Figure S5), did not reveal a change in SSA frequency between spel1 deficient flies and heterozygous controls (Figure 5B). Because we already induce DSB formation in embryos, we performed a control experiment with spel1 deficient parents to exclude any contribution of spel1 gene product to the embryo by heterozygous parents (Figure S6). If spel1 suppresses SSA between divergent sequences, the effect should be most evident in the divergent MtnB-MtnD construct. However, analysis of this construct in flies with spel1 deficient parents showed that SSA frequency (2 events/850 flies) was as low as in wildtype flies (5 events/1950 flies) (Figure 6B).
DISCUSSION
Here we describe some critical sequence constraints for recombination via single strand annealing (SSA) in D. melanogaster. SSA frequency was highest with identical tandem repeats and a minimal length of spacer DNA between them. A single nucleotide difference between 259 nt long direct repeats is recognized and reduces SSA frequency by 50%. We identified blm, coding for the Bloom helicase of the RecQ helicase family, as a gene responsible for the suppression of SSA between divergent repeats (termed homeologous SSA), thereby preventing potentially deleterious recombination events such as chromosomal translocations between paralogous genes. In S. cerevisiae, the sole RecQ helicase Sgs1 is involved in inhibiting SSA between divergent sequences (13). In contrast to results in yeast, the gene product of the D. melanogaster Msh2 ortholog spel1 did not show any suppression of SSA between divergent sequences.
Most obviously, SSA can reduce any fortuitously duplicated DNA sequence back to one copy. Recent studies in both humans (36,37) and drosophilids (38) have revealed an unexpected genomic variability within one and the same species, notably duplications of DNA segments, both coding and noncoding. In most cases these are thought to be stochastic events without any benefit for the carrier, but such duplications undoubtedly also allow for the generation of gene families whose members have overlapping but distinct, important functions. Thus an organism faces the task of removing useless, if not detrimental DNA duplications yet preserving gene family members, which are particularly abundant in higher eukaryotes including insects and mammals. A typical example is the Drosophila metallothionein family with the closely related, chromosomally linked members mtnB and mtnD that must have arisen by duplication. Fortunately, the tendency of introns to diverge faster than coding sequences mounts an additional barrier against such undesired gene losses (9); in agreement with this notion, the introns are most divergent between mtnB and D. Compared to identical repeats, we indeed find SSA between mtnB and mtnD transgenes to be strongly reduced, and also show that the homolog of human Bloom helicase (but not the MSH2 homolog spellchecker), is an important discriminatory component. The guardian function of Bloom helicase in preventing ectopic SSA recombination appears particularly important in higher organisms, including Drosophila, which harbor many gene families.
In addition to these general considerations, a specific substrate for naturally occurring SSA could be the hybrid element insertion (HEI) process in Drosophila; the corresponding P-elements located on sister chromatids recombine in concert with a nearby locus on the homologous chromosome, resulting in a deletion and a duplication product. The generated duplication is prone to undergo SSA because the P-element is still located between the duplicates (39). In this process, SSA would preserve the original genome sequence.
An alternative way of looking at SSA is in the context of the synthesis dependent strand annealing (SDSA) mechanism, which was demonstrated to be a major pathway for DSB repair in D. melanogaster (34). In the SDSA model, a DSB is followed by 5′ resection of the DNA ends, as it is the case in SSA. The resulting 3′ overhangs may invade homologous templates to copy the DNA sequence spanning the DSB. The invasive strand is subsequently unwound and anneals with the other overhang to restore the original sequence. Thus, SSA resembles SDSA without DNA synthesis. In addition, the factors involved in SSA are a subset of those in SDSA. Therefore, one might also consider SSA as a side reaction of SDSA, which could explain the tight sequence constraints for SSA. If a DSB is going to be repaired via SDSA in a region that contains divergent DNA repeats, the 3′ overhangs may anneal without DNA synthesis, form heteroduplexes and undergo SSA. In this context, rejection of heteroduplexes promotes faithful DNA repair via SDSA instead of SSA.
Another possible benefit of strict sequence constraints during heteroduplex rejection may become apparent when two or more DSBs occur simultaneously in similar sequences. Recombination between these sequences would lead to chromosomal translocations, which would most likely be deleterious (40).
So far no evidence existed for the involvement of Blm in the control of sequence constraints during homologous recombination, despite an established role of Blm in safeguarding the genome during DSB repair. A model published recently summarizes the complex involvement of Blm in homologous repair (HR) (27). It includes the removal of Rad51 from the single stranded 3′ overhang, dissociation of the D-loop, unwinding of the DNA double helix in front of the D-loop and dissolution of double Holliday junctions (DHJ) (27). This essential role in HR makes it difficult to investigate a possible additional function of Blm in the suppression of HR between divergent sequences. Different from the situation in HR, such an analysis is possible in SSA, where Blm is not essential.
We show here with several substrate constructs that Blm suppresses SSA between divergent sequences but not between identical ones. As discussed before, this might be important during SDSA in regions containing repeat sequences. In addition one might speculate that Blm also suppresses strand invasion at ectopic templates during HR, which would be in line with the yeast data, where Sgs1 inhibits chromosomal translocations between similar but divergent genes (17). However, analyzing homologous repair with an ectopic donor sequence, we could not detect significant differences in repair frequencies between constructs that contained an identical donor sequence and others that contained donor sequences harboring mismatches since repair events occurred very rarely.
In Drosophila blm mutants, a general shift from HR towards SSA was reported, but only if HR used the homologous chromosome as a template (HR-h) and not if the sister chromatid was used (HR-s) (23,33). In our experiments the SSA tester constructs were always in a heterozygous state, hence HR-h was not possible. As observed by others under comparable conditions, we did not see a general shift towards SSA in blm mutants if the repeats were identical. We find however that the SSA frequency was only elevated in blm mutants when heteroduplexes could be generated between divergent repeats. Data from S. cerevisiae suggest that heteroduplexes are unwound rather than degraded (13). We therefore propose that the Blm helicase unwinds heteroduplexes during DSB repair in D. melanogaster.
Although we saw a pronounced, robust effect in blm mutants, it is likely that we still underestimate the impact of Blm on the suppression of homeologous recombination. Firstly, S. cerevisiae has only one RecQ helicase, Sgs1, which suppresses recombination between divergent sequences, whereas D. melanogaster has three RecQ helicases: RecQ4, RecQ5 and Blm. Redundancy between these helicases in the suppression of homeologous recombination appears possible. Secondly, the parents of the investigated flies were heterozygous for blm since Blm deficient females are sterile. Thus a parental contribution of Blm mRNA or protein cannot be excluded. Indeed we have observed that maternal contribution of DSB repair factors can improve DNA repair efficiency in embryos (unpublished results).
The mismatch repair (MMR) system was found to suppress recombination between divergent sequences in various species, such as E. coli (10), budding yeast (11–13) and the mouse (14,15). The E. coli MMR factor MutS as well as the MutS homologs Msh2 in yeast and mice suppress homeologous recombination. Thus it was quite unexpected that spel1 mutant flies, encoding the Drosophila Msh2 homolog, did not show increased SSA frequency between divergent sequences. In these experiments residual spel1 activity can be excluded because the spel1 gene is deleted. Furthermore, a control experiment where both parents were homozygous mutants yielded the same result. This suggests that in contrast to the situation in other model organisms, MMR is not involved in the suppression of homeologous recombination in D. melanogaster. However, we cannot exclude that another component of the MMR machinery in D. melanogaster, Msh6, contributes to the suppression of homeologous recombination.
Recapitulating the results presented in this work, one can state that in D. melanogaster Blm revealed a novel function in suppressing DSB repair between divergent sequences. In contrast to the situation in mammals and yeast where Msh2 also suppresses homeologous recombination, in D. melanogaster, the Msh2 ortholog, Spel1, was not suppressing recombination between divergent sequences. Based on these data we propose that Bloom helicase, by increasing recombination fidelity, helps to inhibit chromosomal rearrangements, and thus might specifically prevent the loss of related genes that originated by duplication. Our studies have shed light on the way different species handle the difficult task of maintaining genome integrity while repairing damage to their DNA.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
‘Controlled Gene Integration’ project of the European Union (LSHG-CT-2003-503303 to W.S.); the Schweizerischer Nationalfonds; the Kanton Zürich. Funding for open access charge: Kanton Zürich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Amy C. Groth and Michele P. Calos for the plasmid pDrBB2, Johannes Bischof and Konrad Basler for attP flies, Carlos Flores and William Engels for fly strains mutant for mus309 and deficient for spellchecker1. We further thank Eva Niederer for fluorescence activated cell sorting and George Hausmann and Albert Pastink for valuable discussions.
REFERENCES
- 1.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 2.Pastink A, Eeken JC, Lohman PH. Genomic integrity and the repair of double-strand DNA breaks. Mutat. Res. 2001;480:37–50. doi: 10.1016/s0027-5107(01)00167-1. [DOI] [PubMed] [Google Scholar]
- 3.Shen P, Huang HV. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonda DK, Radding CM. By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell. 1983;34:647–654. doi: 10.1016/0092-8674(83)90397-5. [DOI] [PubMed] [Google Scholar]
- 5.Nassif N, Engels W. DNA homology requirements for mitotic gap repair in Drosophila. Proc. Natl Acad. Sci. USA. 1993;90:1262–1266. doi: 10.1073/pnas.90.4.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coveny AM, Dray T, Gloor GB. The effect of heterologous insertions on gene conversion in mitotically dividing cells in Drosophila melanogaster. Genetics. 2002;161:249–258. doi: 10.1093/genetics/161.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldman AS, Liskay RM. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc. Natl Acad. Sci. USA. 1987;84:5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldman AS, Liskay RM. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol. Cell. Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuo K, Clay O, Kunzler P, Georgiev O, Urbanek P, Schaffner W. Short introns interrupting the Oct-2 POU domain may prevent recombination between POU family genes without interfering with potential POU domain ‘shuffling' in evolution. Biol. Chem. Hoppe-Seyler. 1994;375:675–683. doi: 10.1515/bchm3.1994.375.10.675. [DOI] [PubMed] [Google Scholar]
- 10.Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 11.Datta A, Adjiri A, New L, Crouse GF, Jinks Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailis AM, Rothstein R. A defect in mismatch repair in Saccharomyces cerevisiae stimulates ectopic recombination between homeologous genes by an excision repair dependent process. Genetics. 1990;126:535–547. doi: 10.1093/genetics/126.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl Acad. Sci. USA. 2004;101:9315–9320. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott B, Jasin M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol. Cell. Biol. 2001;21:2671–2682. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 16.Hanada K, Ukita T, Kohno Y, Saito K, Kato J, Ikeda H. RecQ DNA helicase is a suppressor of illegitimate recombinationin Escherichia coli. Proc. Natl Acad. Sci. USA. 1997;94:3860–3865. doi: 10.1073/pnas.94.8.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt KH, Wu J, Kolodner RD. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol. Cell. Biol. 2006;26:5406–5420. doi: 10.1128/MCB.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 19.Spell RM, Jinks-Robertson S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168:1855–1865. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusano K, Berres ME, Engels WR. Evolution of the RECQ family of helicases: a drosophila homolog, Dmblm, is similar to the human bloom syndrome gene. Genetics. 1999;151:1027–1039. doi: 10.1093/genetics/151.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kusano K, Johnson-Schlitz DM, Engels WR. Sterility of Drosophila with mutations in the Bloom syndrome gene—complementation by Ku70. Science. 2001;291:2600–2602. doi: 10.1126/science.291.5513.2600. [DOI] [PubMed] [Google Scholar]
- 22.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 23.Johnson-Schlitz D, Engels WR. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc. Natl Acad. Sci. USA. 2006;103:16840–16845. doi: 10.1073/pnas.0607904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McVey M, Larocque JR, Adams MD, Sekelsky JJ. Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl Acad. Sci. USA. 2004;101:15694–15699. doi: 10.1073/pnas.0406157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl Acad. Sci. USA. 1974;71:4508–4512. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chester N, Babbe H, Pinkas J, Manning C, Leder P. Mutation of the murine Bloom's syndrome gene produces global genome destabilization. Mol. Cell. Biol. 2006;26:6713–6726. doi: 10.1128/MCB.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl Acad. Sci. USA. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raynard S, Bussen W, Sung P. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J. Biol. Chem. 2006;281:13861–13864. doi: 10.1074/jbc.C600051200. [DOI] [PubMed] [Google Scholar]
- 30.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl Acad. Sci. USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schildkraut E, Miller CA, Nickoloff JA. Gene conversion and deletion frequencies during double-strand break repair in human cells are controlled by the distance between direct repeats. Nucleic Acids Res. 2005;33:1574–1580. doi: 10.1093/nar/gki295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groth AC, Fish M, Nusse R, Calos MP. Construction of Transgenic Drosophila by Using the Site-Specific Integrase From Phage {phi}C31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wei DS, Rong YS. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics. 2007;177:63–77. doi: 10.1534/genetics.107.077693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ. Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics. 2004;167:699–705. doi: 10.1534/genetics.103.025411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flores C, Engels W. Microsatellite instability in Drosophila spellchecker1 (MutS homolog) mutants. Proc. Natl Acad. Sci. USA. 1999;96:2964–2969. doi: 10.1073/pnas.96.6.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science. 2008;320:1629–1631. doi: 10.1126/science.1158078. [DOI] [PubMed] [Google Scholar]
- 39.Preston CR, Sved JA, Engels WR. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics. 1996;144:1623–1638. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Preston CR, Engels W, Flores C. Efficient repair of DNA breaks in Drosophila: evidence for single-strand annealing and competition with other repair pathways. Genetics. 2002;161:711–720. doi: 10.1093/genetics/161.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.