Abstract
Variation in cellular gene expression levels has been shown to be inherited. Expression is controlled at transcriptional and post-transcriptional levels. Internal ribosome entry sites (IRES) are used by viruses to bypass inhibition of cap-dependent translation, and by eukaryotic cells to control translation under conditions when protein synthesis is inhibited. We aimed at identifying genomic determinants of variability in IRES-mediated translation of viral [Encephalomyocarditis virus (EMCV)] and cellular IRES [X-linked inhibitor-of-apoptosis (XIAP) and c-myc]. Bicistronic lentiviral constructs expressing two fluorescent reporters were used to transduce laboratory and B lymphoblastoid cell lines [15 CEPH pedigrees (n = 205) and 50 unrelated individuals]. IRES efficiency varied according to cell type and among individuals. Control of IRES activity has a significant genetic component (h2 of 0.47 and 0.36 for EMCV and XIAP, respectively). Quantitative linkage analysis identified a suggestive locus (LOD 2.35) on chromosome 18q21.2, and genome-wide association analysis revealed of a cluster of SNPs on chromosome 3, intronic to the FHIT gene, marginally associated (P = 5.9E-7) with XIAP IRES function. This study illustrates the in vitro generation of intermediate phenotypes by using cell lines for the evaluation of genetic determinants of control of elements such as IRES.
INTRODUCTION
Initiation of protein synthesis in the eukaryotic cell is a complex process that leads to the assembly of the 80S ribosome at the start codon of the mRNA. This occurs through the interaction of the 7-methyl-guanilic acid residue at the 5′ terminus (5′-m7G) cap structure with the cap-binding complex, also known as eIF4F, which links mRNA and the 40S ribosomal subunit through eIF3. After recruitment to the mRNA 5′end, the 43S initiation complex (formed by 40S, eIF2.GTP.Met-tRNAi, and eIF3) scans the 5′-untranslated region (5′-UTR) for the start codon (1,2). Eukaryotic proteins can also be translated through an alternative mechanism, using internal ribosome entry sites (IRES), which requires the direct binding of the ribosome to a specific region of mRNA preceding the AUG sequence.
IRES were first described in the uncapped 5′-UTR of the genome of two picornaviruses: poliovirus and encephalomyocarditis virus (EMCV) (3,4). IRES have now been reported in viruses from different families, as well as in eukaryotic cells (5,6). They allow translation under conditions of inhibition of general protein synthesis, such as amino acid starvation (7), cell death (8–10), hypoxia (11,12), heat shock (13), and during the G2/M phase of the cell cycle (14–18).
IRES activity is under the control of general trans acting factors (eIFs) and IRES-specific trans acting factors (ITAFs) (19). ITAFs can modify ribosome recruitment and the structure of the IRES RNA. Known ITAFs of the X-linked inhibitor of apoptosis (XIAP) IRES are the La autoantigen (La), heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNP C1/C2), and heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) (20–22). ITAFs of EMCV IRES are the polypyrimidine tract-binding protein (PTB) and La (23,24).
Efficiency of IRES-mediated translation may vary according to cell type, host species, and the individual genetic background. We hypothesized that the study of gene expression from bicistronic constructs transduced into human B lymphoblastoid cell lines (LCLs) from three-generation families [the CEPH (Centre d'Etude du Polymorphisme Humain) resource] (25,26) would allow the identification of genes controlling the activity of viral and eukaryotic IRES.
In this study, we developed an experimental approach to assess genomic determinants of variability in EMCV, X-linked inhibitor-of-apoptosis (XIAP), and c-myc IRES activity in different cell lines and in CEPH LCLs. We identified that variation in the control of EMCV and XIAP IRES activity has a significant genetic component, and we completed (i) a genome-wide linkage analysis to detect quantitative trait loci (QTL), and (ii) a genome-wide association analysis that identified two suggestive loci involved in the control of the XIAP IRES activity.
MATERIALS AND METHODS
IRES sequences
IRES sequences were identified from the literature and from a dedicated IRES database (http://www.rangueil.inserm.fr/IRESdatabase/) (Supplementary materials). The EMCV IRES was subcloned from pWPI (provided by D. Trono, http://tronolab.epfl.ch/). The cloned EMCV IRES corresponds to that previously used by Pham et al. (27). XIAP and c-myc IRES were amplified from a cDNA library for the purpose of the study. The cloned XIAP and c-myc IRES correspond to those previously used by by Holcik et al. (28), and by the group of A. E. Willis (29,30), respectively.
Lentiviral vectors
All plasmids and lentiviral vectors used in this study are listed in Table 1.
Table 1.
Plasmids used in this study: final (in bold) and intermediate constructs
| No. | Plasmids | Characteristics, component | Source |
|---|---|---|---|
| p1 | pSIN.cPPT.CMV.eGFP.WPRE | Lentiviral vector carrying CMV.eGFP transgene | Provided by D.Trono modified by S. Fleury |
| p2 | pCH9-mRFP | Plasmid carrying the mRFP sequence | Provided by M. Nassal |
| p3 | pBS | pBluescript SK+ | Stratagene, Basel, Switzerland |
| p4 | pWPI or pSIN.EF1.cPPT.IRESEMCV.eGFP.WPRE | Lentiviral vector carrying EF1.cPPT.IRESEMCV.eGFP transgene | Provided by D. Trono |
| p5 | pEF1_EMCV_ (pSIN.EF1.cPPT.mRFP.IRESEMCV.eGFP.WPRE) | mRFP from p2 in p4 | This study |
| p6 | pEF1_cmyc_ (pSIN.EF1.cPPT.mRFP.IREScmyc.eGFP.WPRE) | c-myc IRES (PCR on human cDNA) in p5 after deletion of EMCV IRES | This study |
| p7 | pEF1_XIAP_ (pSIN.EF1.cPPT.mRFP.IRESXIAP.eGFP.WPRE) | XIAP IRES (PCR on human cDNA) in p5 after deletion of EMCV IRES | This study |
| p8 | pEF1del_EMCV_ | p5 after ClaI and PacI deletion of EF1α promoter | This study |
| p9 | pEF1del_cmyc_ | p6 after ClaI and PacI deletion of EF1α promoter | This study |
| p10 | pEF1del_XIAP_ | p7 after ClaI and PacI deletion of EF1α promoter | This study |
pEF1_EMCV_ (pSIN.EF1.cPPT.mRFP.IRESEMCV.eGFP.WPRE): mRFP was amplified from pCH9-mRFP (kindly provided by M. Nassal) and added to PmeI sites. This fragment was inserted into the pSIN.EF1.cPPT.IRESEMCV.eGFP.WPRE (pWPI, provided by D. Trono) using the PmeI site present between the elongation factor alpha promoter (EF1-α) and the EMCV IRES sequence. Two additional restriction enzyme sites, Bsu36I and NdeI, were inserted before and after the IRES sequence, respectively. The EMCV IRES was substituted by c-myc and XIAP IRES by cloning between the Bsu36I and NdeI to generate pEF1_cmyc_, and pEF1_XIAP_.
Cell culture
Adherent cell lines 293T (human renal epithelial cell line), HeLa (human cervical cancer cells transformed by human papillomavirus 18), Huh-7 and Hep3B (human hepatocarcinoma cell lines) (31,32), SW1417 and HT-29 (human colorectal adenocarcinoma cell lines) were grown in Dulbecco's modified Eagle's medium (Gibco, Invitrogen, Carlsbad, CA, USA) supplemented with 100 U/ml of penicillin G, 100 μg/ml of streptomycin, and 10% fetal calf serum (FCS, Inotech, Labor AG, Basel Switzerland).
Raji (human B cells from Burkitt's lymphoma, ATCC CCL86) and Jurkat (human T cells from an acute T cell leukemia, ATCC TIB-152) cell lines were cultured in RPMI 1640/Glutamax-I medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FCS (Inotech). CEPH cell lines, obtained from the Coriell Institute for Medical Research (http://ccr.coriell.org/), were cultured in RPMI 1640/Glutamax-I supplemented with 15% FCS. CEPH pedigrees studied were 102, 884, 1328, 1331, 1332, 1333, 1334, 1340, 1341, 1345, 1346, 1347, 1362, 1408 and 13292, representing 205 cell lines. In addition, in genome association analysis, we investigated 50 unrelated HapMap LCLs.
Lentiviral production and transduction
Lentiviruses were produced in 293T cells by co-transfecting the Gag-Pol construct (pCMV_R8.92), the Rev expression plasmid (pRSVRev), the VSV G protein envelope construct (pMD.G), and the construct of interest expressing the 2 reporters genes (mRFP and eGFP) (Table 1). Transduction was performed by spinoculation of 2 × 105 non-adherent cells with 400 μl of lentivirus-containing supernatant or 1 × 105 adherent cells with 200 μl of supernatant, 3 h, 1500g, 22°C in 24-well plates. After 72 h, cells were harvested, washed and fixed with Cellfix (Becton Dickinson, Franklin Lakes, New Jersey, USA). For CEPH cells, transduction was performed by spinoculation of 5 × 104 cells with 100 μl of lentivirus-containing supernatant 3 h, 1500g, 22°C in 96-wells plate. After 72 h, cells were harvested, washed and fixed with Cellfix (Becton Dickinson). Expression of fluorescent proteins, mRFP and eGFP, was analyzed by flow cytometry (FACSCalibur, Cellquest software, Becton Dickinson). Experiments were performed twice in triplicate at one week interval.
Investigation of alternative splicing
Splicing was investigated by RT-PCR. Seventy-two hours post-spinoculation, DNA and RNA from 10 CEPH cell lines (C6, Pedigree:884; ID: GM13120; K1; Pedigree: 1346; ID: GM10857; L2, Pedigree:1340; ID: GM07019; HM4; Pedigree:1459; ID: GM12873; HM8, Pedigree:1454; ID: GM12813; HM10, Pedigree:1447; ID: GM12761; HM11; Pedigree:1447; ID: GM12762; HM12; Pedigree:1447; ID: GM12763; HM16, Pedigree:1444; ID: GM12751; HM33, Pedigree:1420; ID: GM12005), were isolated by using Qiagen, AG, Hombrechtikon, Switzerland, QIAamp DNA Mini Kit and Qiagen Rneasy Mini Kit according to the manufacturer instructions. After DNase I treatment, 800 ng purified cellular RNA was reverse-transcribed using Expand RT (Roche Diagnostics Ltd. Rotkreuz, Switzerland), dNTPs mix (Invitrogen) and RNase inhibitor (Roche). PCR was performed to amplify the region of interest by using the primers forward SG1667 (5′-ATCTTGGTTCATTCTCAAGCCTCAGAC-3′), and reverse SG1669 (5′-AGCTCGACCAGGATGGGCAC-3′). Detailed analysis of XIAP IRES used the IRES-specific primer MA.pr-216 (5′-ACACGACCGCTAAGAAACATTC-3′). For amplification, an initial denaturation step at 94°C for 15 min was followed by 35 cycles at 94°C for 30 s, 66°C for 30 s, and 72°C for 2 min, and a final extension step at 72°C for 10 min. PCR products were separated in a 1% agarose gel and visualized by ethidium bromide (EtBr) staining. Amplification products were gel-extracted and sequenced.
Heritability, linkage and association studies
To establish the heritability of the control of IRES activity, lymphoblastoid cell lines from CEPH 15 families were transduced with lentiviruses (EF1_EMCV_LVs and EF1_XIAP_LVs). IRES activity was calculated by the ratio of mRFP expressing cells to eGFP expressing cells, as determined by flow cytometry. SOLAR software allowed the calculation of heritability using the polygenic-screen command (http://www.sfbr.org/solar/) (33).
A genome-wide quantitative linkage analysis was performed on the CEPH pedigrees to identify chromosomal loci involved in the control of the IRES activity. Single nucleotide polymorphism (SNP) genotyping data, consisting on 2600 autosomal SNPs, available in public databases (http://snpdata.cshl.edu/population_studies/linkage_maps/) were used (34). This dataset consists of a scan of 22 autosomal chromosomes with a 3.9-cM resolution. Multipoint linkage with the SNP map was performed using MERLIN (35) with the –VC option, after elimination of Mendelian inconsistencies (PEDCHECK) (36) and unlikely genotypes (PEDWIPE). To calculate the empirical significance of the linkage results, we performed 500 simulations for each quantitative trait using the simulate command from Merlin with different seed numbers. We extracted the highest result from each simulation to build significance distributions. All simulations were performed using a cluster of 32 HP/Intel Itanium 2 based servers at the Vital-IT Center (http://www.vital-it.ch/).
Association analysis of quantitative phenotypes and corrections for multiple testing were performed using the PLINK software (http://pngu.mgh.harvard.edu/~purcell/plink/anal.shtml). Genotypes were downloaded from the HapMap project URL (http://www.hapmap.org/cgi-perl/gbrowse/gbrowse/hapmap), HapMap public release no. 19. Results of genome-wide association studies were visualized and annotated using WGAviewer (37).
RESULTS
IRES activity in different cell lines
The optimal arrangement of the bicistronic construct included the coding sequences for mRFP as first and for eGFP as second cistron, separated by the IRES of EMCV, c-myc or XIAP (data not shown).
The various lentiviruses delivering the constructs were used to transduce and generate seven stable cell lines of different origins. In addition to the common laboratory 293T and HeLa cell lines, we used two B lymphocyte cell lines [CEPH (HM8; Pedigree:1454; ID: GM12813) and Raji] in anticipation of the genetic study of loci involved in the control of IRES activity that uses CEPH LCLs. Jurkat cells were chosen as example of T-lymphocyte cells.
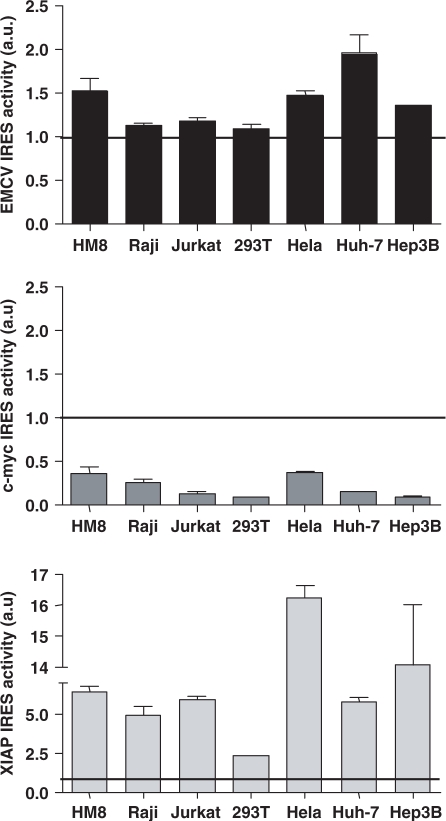
In all cell lines, XIAP constructs were found to harbor the most efficient IRES, followed by EMCV, and c-myc constructs (Figure 1). All three IRES exhibited cell type-dependent translational activity. IRES activity (eGFP/mRFP) ranged from 0.09 (293T) to 0.37 (HeLa) for c-myc IRES, 1.09 (293T) to 1.96 (Huh-7) for EMCV IRES, and 2.33 (293T) to 16.20 (HeLa) for XIAP IRES.
Figure 1.
IRES activity in cell lines of diverse origins. IRES activity was estimated as the ratio between eGFP expression (second cistron) and mRFP (first cistron) in the transduced population and normalized by values from mock transduced cells. Each panel is representative for one of two independent experiments performed in triplicate. Bars, SEM; a.u., arbitrary units.
Stringent conditions to assess IRES activity
To analyze the potential presence of a cryptic promoter in IRES constructs, we designed constructs without the EF1α promoter (Table 1). In this setting, expression of the second transgene implies that the DNA coding for the IRES possesses cryptic promoter activity. Such activity was excluded in the constructs under investigation (Supplementary materials). As control, we used a HCV IRES construct that presents cryptic promoter activity, as previously observed (38).
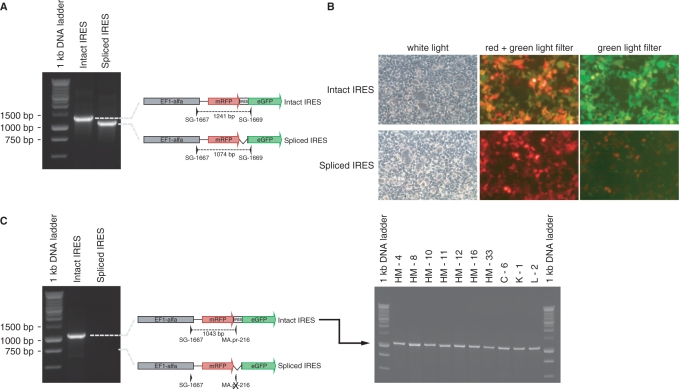
Alternative splicing would potentially bypass the first cistron while allowing the expression of the second cistron from the EF1α promoter (39). To check for this possibility, we analyzed cDNA by RT-PCR. Splicing was observed in XIAP IRES constructs. Two PCR products of 1241 bp corresponding to full length bicistronic RNA transcript and 1074 bp were sequenced (Figure 2A). The smaller transcript is a spliced form of the full length transcript missing the XIAP IRES sequence (Supplementary materials). Transfection of 293T cells with plasmids containing the full length or the spliced XIAP IRES sequence (Figure 2B) demonstrates that only the full length transcript is able to produce the green fluorescent protein. Although the transduced CEPH cell lines expressed both mRNAs forms, the full length transcript was present in all stable cell lines (Figure 2C).
Figure 2.
GFP expression requires an intact XIAP IRES. (A) PCR amplification using primers SG-1667 and SG-1669 on DNA constructs containing an intact XIAP IRES or a spliced XIAP IRES. (B) Fluorescence microscopy showing the expression of mRFP and eGFP in cells transfected with the construct containing an intact XIAP IRES or a spliced XIAP IRES. (C) PCR amplification using primers SG-1667 and MA.pr-216 (XIAP IRES specific) on DNA constructs (left panel) and donor cDNAs (right panel), showing the presence of an intact XIAP IRES in all cell lines.
IRES activity in CEPH cell lines
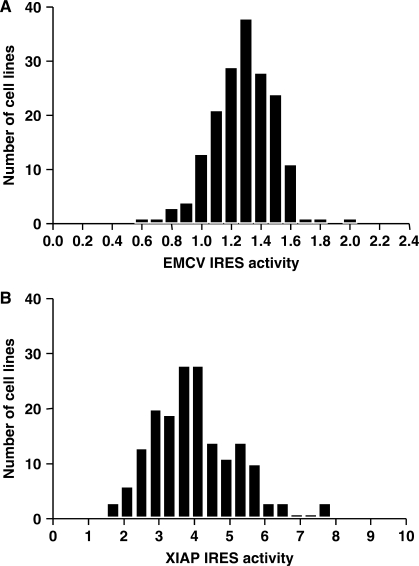
EMCV and XIAP IRES constructs were used for the analysis of CEPH cell lines. We excluded c-myc IRES constructs because of limited activity in B cells. Upon transduction, 178 of the 205 cell lines remained viable. We observed inter-individual differences of 3.08-fold and 4.74-fold for EMCV and XIAP IRES activity, respectively (Figure 3). Some individuals consistently presented extreme (low and high) IRES activity for both constructs investigated. However, we did not observe a correlation between EMCV and XIAP IRES activity among individuals (r2: 0.035; P value: 0.0127). EMCV and XIAP IRES activities in 50 unrelated HapMap cell lines ranged from 0.657 to 1.638 and from 2.200 to 9.271 respectively.
Figure 3.
EMCV and XIAP IRES activity. Distribution of IRES activity values for EMCV (A), and XIAP (B) in transduced CEPH cell lines.
Heritability of IRES activity
To determine the fraction of the total phenotypic variation in the CEPH population that was genetically determined, i.e. that is attributable to variation among individual genotypes (40), we assessed the heritability (h2) of the various studied phenotypes in the 15 CEPH families. The heritability estimates were h2 of 0.47 and 0.36 for EMCV and XIAP IRES, respectively.
Genome-wide linkage analysis
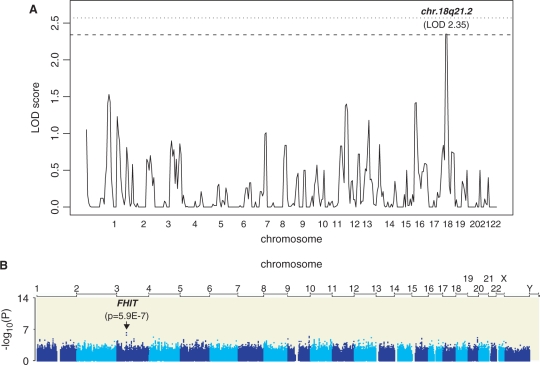
We performed a genome-wide linkage analysis on IRES expression values from 15 CEPH pedigrees. No significant QTL could be identified for EMCV IRES activity. XIAP IRES activity was associated with a suggestive LOD score in chromosome 18 [2.35, p = 5E-04 (marker rs1144098)]. We determined empirical genome-wide significance levels by multiple permutation analysis. This allowed us to determine that the chromosome 18 peak is significant at the 90% significance threshold, thus not reaching nominal genome-wide significance (Figure 4).
Figure 4.
Genome scan and association analyses. (A) Genome-wide linkage analysis results in 15 CEPH families. Horizontal lines show significance thresholds as determined by multiple permutations. Dashed line shows 90% and dotted line 95% significance threshold, respectively. (B) Genome-wide association analysis using the 2.2 million HapMap SNPs.
Genome association analyses
In order to further dissect the linkage analysis result, we assayed LCLs from 50 unrelated CEPH individuals that had been genotyped at a high density within the framework of the HapMap project (41). The association analysis was performed in a 1, 5, 15 and 30 Mb region centered on the initial linkage assignment using 218, 800, 3283 and 5954 tag SNPs, respectively. No SNP could be statistically associated with the XIAP IRES activity after correction for multiple testing. Thus, we used the initial SNP (rs1144098) identified by the linkage analysis for further analysis of the region.
Genotype data for rs1144098 were not available for the Caucasian HapMap population. However, rs1144098 was tagged by SNP rs948590 in the Yoruba HapMap population. The latter SNP is in complete linkage disequilibrium (r2 = 1.0) with rs1144097, rs1144099, rs9965768 and is in high linkage disequilibrium with rs12607660 (r2 = 0.843), rs1221882 (r2 = 0.872) and rs1221862 (r2 = 0.902) in the Caucasian HapMap population. All associated SNPs are localized in a ∼23kb genomic region in intron 1 of DCC (deleted in colorectal cancer) on Chromosome 18q21.3. This gene extends for over 1 Mb.
We then performed a genome-wide association study on the 50 unrelated HapMap individuals using all 2.2 million SNPs that have been generated for these samples (42). Results of this analysis revealed that the top association (p = 5.9E-7) localized to chromosome 3 (Figure 4) within intron 7 of the FHIT (‘fragile histidine triad’) gene. The second and third ranked SNPs also mapped to the same linkage disequilibrium block, thus FHIT is the best candidate gene resulting from this screen. Given the large number of SNPs tested, the association does not survive multiple testing correction either by Bonferroni or by multiple permutation tests. However, given the known functions of FHIT as a signaling and tumor suppressor gene, it remains an interesting candidate for mediating IRES function.
DISCUSSION
The main results of this study include the observation that (i) IRES efficiency depends on the cellular background; (ii) differences in EMCV and XIAP IRES efficiency are observed for lymphoblastoid B cell lines from multiple individuals and family pedigrees; (iii) variability of IRES activity has a genetic, heritable component; (iv) a quantitative linkage analysis scan identified a suggestive chromosomal locus that mapped to intron 1 of the DCC gene on chromosome 8 and (v) genome-wide association studies revealed that a SNP intronic to the FHIT gene on chromosome 3 showed the highest association. Both of these loci are candidates for involvement in IRES function.
All three IRES exhibited cell type-dependent translational activity. Differences in IRES-mediated translation in cells of different origins may reflect different titration of general trans acting factors and ITAFs. XIAP IRES had the highest level of activity in the experimental system.
A more precise assessment of activity attributed to IRES-mediated translation included analyses for the presence of cryptic promoter activity and RNA splicing, that would render spurious estimates of IRES activity. Cryptic promoter activity, previously described in IRES DNA constructs using the Hepatitis virus C IRES, was not observed in the constructs under study. Alternative splicing would potentially bypass the first cistron while allowing the expression of the second cistron from the EF1α promoter. Specifically, previous reports have described skipping of the first cistron in XIAP IRES constructs (43,44). We also identified an alternative transcript generated by a proportion of the XIAP IRES vectors in the present study. Splicing used the same 3′ splice site as in Van Eden et al. (44) and Baranick et al. (39); however, it led to loss of expression of the second cistron. Thus, the eGFP expression observed in the different cell lines reflects the XIAP IRES activity.
Variation in gene expression (transcription) has been reported to occur within and among populations (45). Such variation does not only depend on inter-species differences but also on differences between individuals from the same species (46–48). The determinants of gene expression levels can be studied through genetic approaches (49) by using cells from pedigrees (linkage analysis) such as those from CEPH (50) or by using cell lines from unrelated individuals (association studies). We decided to apply both approaches to the analysis of variation of translation levels.
In order to dissect the genetic determinants of IRES activity, we used B LCLs immortalized by Epstein-Barr virus (EBV) from the CEPH collection to perform genome-wide quantitative linkage analysis. The DNA from each LCL has been genotyped at high resolution and is publicly available allowing linkage and association studies to be performed. CEPH LCLs have proven to be useful in the identification of genomic markers influencing sodium-lithium counter transport (51), variation in gene expression (52–57), transcriptional response to ionizing radiation (58), sensibility to chemotherapy cytotoxicity (59), and we have used it successfully to map a susceptibility locus to HIV-1 infection (60).
Evaluation of EMCV and XIAP IRES activity in CEPH cell lines identified inter-individual differences of 3.08-fold and 4.74-fold, respectively. EMCV and XIAP IRES-related phenotypes were heritable. Thus, variation in IRES activity is determined by the individual genetic background. Some individual cell lines presented extreme (low and high) phenotypes of IRES activity for both IRES elements (EMCV or XIAP), suggestive of dependence on the same cofactors modulating IRES activity. Given the evidence of significant heritability of the trait in the in vitro system, we progressed to a genome scan. However, no significant QTL was identified for EMCV IRES activity despite high h2 values. We can hypothesize that EMCV phenotypes are regulated by multiple loci contributing modestly to the trait.
We identified a suggestive QTL associated with XIAP IRES activity, and identified a marker SNP (rs1144098) located in intron 1 of DCC. Given the large size of the DCC gene, these results could suggest a role of this locus in the control of XIAP IRES activity, although the resolution of quantitative linkage studies is usually in the order of 5–10 Mb, and other genes in the vicinity cannot be excluded. DCC, a human receptor of netrin-1 (61), contributes to pro-apoptotic signaling when not bound to its ligand whereas an anti-apoptotic signal is induced in presence of netrin-1 (62). Similarly, XIAP is an important regulator of apoptosis (63). Although XIAP is not in the same pathway as DCC, these observations could suggest a mechanistic interplay between DCC and XIAP.
We also performed a genome-wide association study on unrelated individuals from the HapMap collection. From this we identified SNPs in intron 1 of the FHIT gene as showing the highest association. FHIT has been shown to participate in the Src and Wnt signaling pathways (64,65), and is thus a plausible candidate to be involved in the modulation of IRES expression. Studies on a larger set of cell lines from unrelated individuals will be needed to validate this observation.
The need to progress from mapping data to functional data is however complex, as well identified in recent genome studies that search the genetic basis of complex traits. In contrast from prior studies of monogenic disorders, the step from linkage to precise cloning cannot be readily done. In the present study, the linkage intervals are still very large, the associated or linked markers may fall within a gene, but this does not necessarily imply that the causal variant is within short distance. Given current mapping precision, test of the involvement of the candidate loci in controlling or modulating IRES activity is still premature.
In conclusion, we demonstrate for the first time the heritable control of IRES activity, and propose two suggestive loci associated with the control of XIAP IRES activity. The study also illustrates potential uses and limitations of in vitro cellular phenotypes by using CEPH pedigrees—even for the evaluation of genetic elements of small size such as a 198 bp IRES sequence.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation (31-110012 to A.T.). NCCR Frontiers in Genetics (to S.E.A.). Funding for open access charge: Swiss National Science Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stanley Lemon, Didier Trono, Michael Nassal for materials, and Sylvia Rothenberger for suggestions. Vital-IT (http://www.vital-it.ch/) is gratefully acknowledged for support in data analysis.
REFERENCES
- 1.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 2.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 3.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 5.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 6.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez J, Bode B, Koromilas A, Diehl JA, Krukovets I, Snider MD, Hatzoglou M. Translation mediated by the internal ribosome entry site of the cat-1 mRNA is regulated by glucose availability in a PERK kinase-dependent manner. J. Biol. Chem. 2002;277:11780–11787. doi: 10.1074/jbc.M110778200. [DOI] [PubMed] [Google Scholar]
- 8.Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell Biol. 2000;20:496–506. doi: 10.1128/mcb.20.2.496-506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holcik M, Yeh C, Korneluk RG, Chow T. Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene. 2000;19:4174–4177. doi: 10.1038/sj.onc.1203765. [DOI] [PubMed] [Google Scholar]
- 10.Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, Willis AE. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coldwell MJ, deSchoolmeester ML, Fraser GA, Pickering BM, Packham G, Willis AE. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene. 2001;20:4095–4100. doi: 10.1038/sj.onc.1204547. [DOI] [PubMed] [Google Scholar]
- 14.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis S, Bruynooghe Y, Denecker G, Van HS, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell. 2000;5:597–605. doi: 10.1016/s1097-2765(00)80239-7. [DOI] [PubMed] [Google Scholar]
- 16.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell GA, Lemon SM. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 17.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol. Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 18.Qin X, Sarnow P. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J. Biol. Chem. 2004;279:13721–13728. doi: 10.1074/jbc.M312854200. [DOI] [PubMed] [Google Scholar]
- 19.Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 20.Holcik M, Korneluk RG. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: role of La autoantigen in XIAP translation. Mol. Cell Biol. 2000;20:4648–4657. doi: 10.1128/mcb.20.13.4648-4657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holcik M, Gordon BW, Korneluk RG. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell Biol. 2003;23:280–288. doi: 10.1128/MCB.23.1.280-288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis SM, Veyrier A, Hosszu UN, Bonnal S, Vagner S, Holcik M. Subcellular relocalization of a trans-acting factor regulates XIAP IRES-dependent translation. Mol. Biol. Cell. 2007;18:1302–1311. doi: 10.1091/mbc.E06-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YK, Jang SK. Continuous heat shock enhances translational initiation directed by internal ribosomal entry site. Biochem. Biophys. Res. Commun. 2002;297:224–231. doi: 10.1016/s0006-291x(02)02154-x. [DOI] [PubMed] [Google Scholar]
- 25.Cann HM, de TC, Cazes L, Legrand MF, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon—Thomsen A, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 26.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 27.Pham HM, Arganaraz ER, Groschel B, Trono D, Lama J. Lentiviral vectors interfering with virus-induced CD4 down-modulation potently block human immunodeficiency virus type 1 replication in primary lymphocytes. J. Virol. 2004;78:13072–13081. doi: 10.1128/JVI.78.23.13072-13081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holcik M, Lefebvre C, Yeh C, Chow T, Korneluk RG. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1999;1:190–192. doi: 10.1038/11109. [DOI] [PubMed] [Google Scholar]
- 29.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 30.Le Quesne JP, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 31.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 32.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 33.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matise TC, Sachidanandam R, Clark AG, Kruglyak L, Wijsman E, Kakol J, Buyske S, Chui B, Cohen P, de TC, et al. A 3.9-centimorgan-resolution human single-nucleotide polymorphism linkage map and screening set. Am. J. Hum. Genet. 2003;73:271–284. doi: 10.1086/377137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, Telenti A, Goldstein DB. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dumas E, Staedel C, Colombat M, Reigadas S, Chabas S, stier-Gin T, Cahour A, Litvak S, Ventura M. A promoter activity is present in the DNA sequence corresponding to the hepatitis C virus 5′ UTR. Nucleic Acids Res. 2003;31:1275–1281. doi: 10.1093/nar/gkg199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, Logg CR. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc. Natl Acad. Sci. USA. 2008;105:4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era–concepts and misconceptions. Nat. Rev. Genet. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 41.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holcik M, Graber T, Lewis SM, Lefebvre CA, Lacasse E, Baird S. Spurious splicing within the XIAP 5′ UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA. 2005;11:1605–1609. doi: 10.1261/rna.2158605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Eden ME, Byrd MP, Sherrill KW, Lloyd RE. Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA. 2004;10:720–730. doi: 10.1261/rna.5225204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat. Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 46.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 47.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 48.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 49.Deutsch S, Lyle R, Dermitzakis ET, Attar H, Subrahmanyan L, Gehrig C, Parand L, Gagnebin M, Rougemont J, Jongeneel CV, et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum. Mol. Genet. 2005;14:3741–3749. doi: 10.1093/hmg/ddi404. [DOI] [PubMed] [Google Scholar]
- 50.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d'etude du polymorphisme humain (CEPH): collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 51.Schork NJ, Gardner JP, Zhang L, Fallin D, Thiel B, Jakubowski H, Aviv A. Genomic association/linkage of sodium lithium countertransport in CEPH pedigrees. Hypertension. 2002;40:619–628. doi: 10.1161/01.hyp.0000037131.41957.a8. [DOI] [PubMed] [Google Scholar]
- 52.Cheung VG, Conlin LK, Weber TM, Arcaro M, Jen KY, Morley M, Spielman RS. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat. Genet. 2003;33:422–425. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 53.Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monks SA, Leonardson A, Zhu H, Cundiff P, Pietrusiak P, Edwards S, Phillips JW, Sachs A, Schadt EE. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 2004;75:1094–1105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, Lyle R, Hunt S, Kahl B, Antonarakis SE, Tavare S, et al. Genome-wide associations of gene expression variation in humans. PLoS. Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, Redon R, Bird CP, de GA, Lee C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jen KY, Cheung VG. Transcriptional response of lymphoblastoid cells to ionizing radiation. Genome Res. 2003;13:2092–2100. doi: 10.1101/gr.1240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watters JW, Kraja A, Meucci MA, Province MA, McLeod HL. Genome-wide discovery of loci influencing chemotherapy cytotoxicity. Proc. Natl Acad. Sci. USA. 2004;101:11809–11814. doi: 10.1073/pnas.0404580101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loeuillet C, Deutsch S, Ciuffi A, Robyr D, Taffe P, Munoz M, Beckmann JS, Antonarakis SE, Telenti A. In vitro whole-genome analysis identifies a susceptibility locus for HIV-1. PLoS Biol. 2008;6:e32. doi: 10.1371/journal.pbio.0060032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leonardo ED, Hinck L, Masu M, Keino-Masu K, Ackerman SL, Tessier-Lavigne M. Vertebrate homologues of C. elegans UNC-5 are candidate netrin receptors. Nature. 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- 62.Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat. Rev. Cancer. 2004;4:978–987. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- 63.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 64.Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc. Natl Acad. Sci. USA. 2007;104:20344–20349. doi: 10.1073/pnas.0703664105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pekarsky Y, Garrison PN, Palamarchuk A, Zanesi N, Aqeilan RI, Huebner K, Barnes LD, Croce CM. Fhit is a physiological target of the protein kinase Src. Proc. Natl Acad. Sci. USA. 2004;101:3775–3779. doi: 10.1073/pnas.0400481101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.