Abstract
Eukaryotic RNAs typically contain 5′ cap structures that have been primarily studied in yeast and metazoa. The only known RNA cap structure in unicellular protists is the unusual Cap4 on Trypanosoma brucei mRNAs. We have found that T. vaginalis mRNAs are protected by a 5′ cap structure, however, contrary to that typical for eukaryotes, T. vaginalis spliceosomal snRNAs lack a cap and may contain 5′ monophophates. The distinctive 2,2,7-trimethylguanosine (TMG) cap structure usually found on snRNAs and snoRNAs is produced by hypermethylation of an m7G cap catalyzed by the enzyme trimethylguanosine synthase (Tgs). Here, we biochemically characterize the single T. vaginalis Tgs (TvTgs) encoded in its genome and demonstrate that TvTgs exhibits substrate specificity and amino acid requirements typical of an RNA cap-specific, m7G-dependent N2 methyltransferase. However, recombinant TvTgs is capable of catalysing only a single round of N2 methylation forming a 2,7-dimethylguanosine cap (DMG) as observed previously for Giardia lamblia. In contrast, recombinant Entamoeba histolytica and Trypanosoma brucei Tgs are capable of catalysing the formation of a TMG cap. These data suggest the presence of RNAs with a distinctive 5′ DMG cap in Trichomonas and Giardia lineages that are absent in other protist lineages.
INTRODUCTION
Trichomonas vaginalis, an anaerobic protist that infects the urogenital tract of humans, is a divergent, deep-branching eukaryote (1,2). Transcriptional and post-transcriptional gene regulation in this and other unicellular protists is poorly understood. In T. vaginalis, transcription of protein coding genes is typically initiated only 5 to 20 nt upstream of the AUG translation initiator codon resulting in mRNAs with unusually short 5′ untranslated regions (UTRs) (3–6). Trichomonas vaginalis mRNAs possess 5′ cap structures and have 3′ poly(A) tails (7). Only few T. vaginalis pre-mRNAs are predicted to undergo splicing to remove a single intron (6,8). Recently, we have characterized T. vaginalis spliceosomal small nuclear RNAs (snRNAs) predicted to mediate pre-mRNA splicing. Although conserved in structure relative to other eukaryotic snRNAs, all five examined T. vaginalis snRNAs were found to lack a 5′ cap structure and appear to instead have 5′-monophosphate termini (7).
snRNAs, small nucleolar (sno) RNAs (9,10), telomerase RNA (11) and mRNAs that undergo trans-splicing in nematodes (12) typically contain a distinctive hypermethylated 5′-cap structure composed of a 2,2,7-trimethylguanosine (TMG) (13). TMG caps are formed by post-transcriptional dimethylation of monomethyl m7G caps (14). In contrast, the 5′ ends of mRNAs are not subject to hypermethylation and retain an m7G cap. The biological significance of hypermethylated cap structures remains unclear. In humans, m7G cap snRNAs are first exported to the cytoplasm, Sm proteins then bind, followed by a two-step cap methylation to produce the mature snRNA TMG cap (15). The return of these snRNAs to the nucleus, required for their participation in pre-mRNA splicing, is dependent on the presence of both a TMG cap structure and Sm protein assembly (16). Alternatively, some snoRNAs are not exported to cytoplasm (17), and their caps become trimethylated in the nucleolus, mediated by specific snoRNA structural motifs (18,19). The human RNA cap hypermethylase is consistently found in both nuclear Cajal bodies and in the cytoplasm of mammalian cells (20,21). Contrary to the situation observed in humans, the yeast RNA cap hypermethylase resides exclusively in the nucleolus, indicating that yeast snRNA and snoRNA hypermethylation occur in this compartment (22). In Trypanosoma cruzi, it has been recently demonstrated that a functional Tgs1p localizes throughout the nucleoplasm and in spots outside the nucleolus (23)
Trimethylguanosine synthase (Tgs) is the enzyme responsible for converting an m7G RNA cap to a TMG cap. Saccharomyces cerevisiae Tgs (ScTgs) is essential for hypermethylation of snRNAs and snoRNAs (22). Genetic depletion of ScTgs1p produces a cold-sensitive splicing defect that correlates with retention of the m7G-capped U1 snRNA in the nucleolus (22). Homology modelling and mutagenesis studies of ScTgs1 have identified residues required for the formation of the m7G-binding site and catalysis (24). Recombinant Tgs homologues from Schizosaccharomyces pombe (SpTgs) and the divergent protist Giardia lamblia Tgs2 (GlTgs) have likewise been biochemically characterized recently (25,26,27). These studies have shown that Tgs catalyses guanosine N2 methylation, requires S-adenosylmethionine (SAM)) as a methyl donor, has substrate specificity for m7G nucleotides, and does not require any RNA or protein partners for catalysis.
Here, we have biochemically characterized a T. vaginalis Tgs (TvTgs) that is encoded by a single gene in the genome of this parasite. Contrary to that observed for SpTgs, but similar to that found for GlTgs (25), this T. vaginalis RNA cap hypermethylase catalyses only a single round of methylation on a m7G cap to produce a 2,7-dimethylguanosine (DMG) instead of a TMG cap. In contrast, the predicted TMG cap was formed using recombinant Tgs from two other unicellular eukaryotes Entamoeba histolytica (EhTgs) and Trypanosoma brucei (TbTgs). Moreover, the end products of catalysis by TgS from the four different unicellular eukaryotes were the same whether an m7G cap or a m7G RNA transcript was used as substrate. The lack of 5′-end cap structures on T. vaginalis splicesomal snRNAs (7), typical substrates for Tgs, and the unusual methylation properties of both TvTgs and GlTgs suggest that an unidentified subset of RNAs with DMG cap structures are present in these divergent eukaryotes.
MATERIALS AND METHODS
Recombinant TvTgs expression and purification
The open reading frames encoding homologues of the T. vaginalis Tgs (TvTgs; GenBank™ accession number EAY18619), G. lamblia Tgs2 (GlTgs; GenBank™ accession number XP_001704513), E. histolytica Tgs (EhTgs; GenBank™ accession number XP_651698), T. brucei Tgs (TbTgs; GenBank™ accession number Tb11.02.5090) and S. pombe Tgs (SpTgs; GenBank™ accession number BAA13836) were cloned into the Escherichia coli expression vector pET200D (Invitrogen). Induction of expression and purification of the N-terminal His-tagged Tgs were carried out as described (25). Protein purity and concentration were evaluated by SDS–PAGE and densitometry using BSA standards and employing ImageJ software.
Methyltransferase activity assays and Thin-Layer Chromatography (TLC) analysis
With the use of nucleotide substrates, our standard methyltransferase reaction (10–20 μl) was performed at 37°C for 40 min in 50 mM Tris–HCl pH 7.5, 5 mM DTT, 1.25–5 μM [methyl-3H]AdoMet, up to 10 mM of nucleotide substrate, using variable amounts of recombinant enzyme. Except when otherwise indicated, m7GDP was the nucleotide substrate used. To measure substrate specificity, we utilized 5 μM [methyl-3H]AdoMet, 0.5 mM of the specified nucleotide and 3.5 μg recombinant TvTgs. For the inhibition assay using the end product of AdoMet-dependent methyltransferases S-adenosyl-l-homocysteine (AdoHcy), 2.5 μM [methyl-3H]AdoMet, 2 mM m7GDP and 0.35 μg of recombinant TvTgs was used with an incubation time of 20 min. To determine optimal pH, we used 1.25 μM [methyl-3H]AdoMet, 0.5 mM m7GDP and 7.0 μg recombinant TvTgs and adjusted the pH using 50 mM Tris–acetate (pH 7.0 and below) or 50 mM Tris–HCl (pH 7.5 and above). When evaluating enzyme concentration and time required to reach saturation, 5 μM [methyl-3H]AdoMet, 1 mM m7GDP substrate and either 0.875 μg, 1.75 μg or 3.5 μg recombinant TvTgs were utilized in a reaction incubated for 60 min, with 10-min time points taken for analysis. TvTgs affinities for the nucleotide substrates m7GDP, m7GTP and m7GpppA were measured by increasing concentration of each substrate. For the inhibition assay by GTP, reactions were done independently with one of the three methylated nucleotide substrates at 0.5 mM in the presence of increasing amounts of the inhibitor GTP. Aliquots of reactions, in triplicate, were then spotted on DEAE-cellulose filters (Whatman) and washed five times for 4 min each with 20-mM ammonium bicarbonate. Filters were air-dried and radioactivity was measured by liquid scintillation counting. Background was adjusted by subtracting the value obtained from a reaction without enzyme. As indicated aliquots of the reactions were also spotted on a PEI-cellulose TLC plates (Merck) and developed with 50 mM ammonium sulphate in one dimension (1D). TLC plates were treated with EN3HANCE (Perkin Elmer) and exposed to X-ray films (Biomax, Kodak). Relative migration of the methylated products were compared to products methylated by G. lamblia Tgs2 and S. pombe Tgs1 (25,14). Sodium periodate oxidation sensitivity was evaluated as previously described (25). The two sequential N2-methylation reaction protocol of Hausmann and Shuman (25) was used to compare TvTgs, S. pombe Tgs1 and G. lamblia Tgs2.
For evaluation of methyltransferase activity by Tgs using a m7G RNA substrate, T7 RNA polymerase promoter (sequence underlined below) was incorporated into a PCR product containing 352 bp of T. vaginalis β-tubulin (GenBank™ accession number XM_001582993) using TubF (GTC TCG GCA CAC TCC TTC TC) and TubR_T7 (TAA TAC GAC TCA CTA TAG GGA GAC GTG GGA ATG GAA CAA G) oligonucleotides. The gel-eluted PCR product was used as template for T7 RNA transcription (AmpliScribe T7-Flash Transcription™, Epicentre), purified by ProbeQuantTM G-50 Micro Column (GE Healthcare) and capped using Vaccinia Virus capping enzyme (Ambion) in the presence of [α-32P]GTP. The 32P-labelled m7G cap RNA was then purified from unincorporated [α-32P]GTP by three consecutive ProbeQuantTM G-50 Micro Column (GE Healthcare) leaving no detectable traces of unincorporated [α-32P]GTP. Our standard methyltransferase reaction (10–20 μl) was performed at 37°C for 40 min in 50 mM Tris–HCl pH 7.5, 5 mM DTT, with 2 fmol of 32P-labelled m7G cap RNA substrate and 0.5 mM AdoMet and 1 μg of recombinant Tgs. The RNA was subsequently purified by phenol/chloroform extraction and ethanol precipitation and the modified cap structure was released by digestion with Tobbaco acid pyrophosphatase (TAP) (Epicentre).
To measure substrate specificity using the m7G RNA substrate, increasing concentrations of AdoMet in a range of 0–3.2 mM, 0–0.78 mM and 0–5 mM were used for reactions analysing TvTgs, GlTgs and SpTgs, respectively. To calculate affinity of Tgs enzymes, reactions were analysed by 1D-TLC, as described above, and spots were quantified by liquid scintillation counting. For better analysis of the end products of reactions performed by GlTgs2, TvTgs, EhTgs and TbTgs, two-dimensional (2D) TLC was used. In addition, part of the reaction was further incubated with 1 μg of SpTgs at 37°C for 40 min, to drive the formation of a TMG. Plain cellulose TLC plates (Merck) were loaded with 1 μg of each one of the monophosphate ribonucleotide (AMP, CMP, GMP and UMP), and 400–1000 c.p.m. of the TAP-digested RNAs. Individual reactions were analysed on 10 × 10-cm TLC plates that were developed using either solvents A and B or A and C in a two-dimensional TLC system as described (28), and exposed to X-ray films (Biomax, Kodak). The composition of the solvents was: solvent A, isobutyric acid/concentrated ammonia/water [66/1/33 (v/v/v)]; solvent B, phosphate buffer/NH4 sulphate/n-propanol [100/60/2 (v/w/v)]; solvent C, isopropanol/concentrated HCl/water [68/18/14 (v/v/v)].
Sucrose gradient sedimentation
Recombinant TvTgs was analysed by zonal velocity sedimentation in a sucrose gradient (29) and the molecular mass of the active methyltransferase activity was determined by assaying fractions using a m7GDP substrate, as described above. Fifty micrograms of TvTgs was mixed with 50 mM Tris–HCl pH 7.5; 0.2 M NaCl; 1 mM EDTA, 2 mM DTT; 2% sucrose and 50 μg of the following protein standards: soy bean trypsin inhibitor (20 kDa), BSA (66 kDa) and catalase (248 kDa). The mix was loaded on the top of a 4–14% sucrose step gradient and centrifuged at 38 000 g for 16 h. Fractions (21 × 0.5 ml each) were collected from top to bottom of the gradient and analysed by SDS–PAGE and assessed for methyltransferase activity.
Structure modelling and mutagenesis
TvTgs sequence was compared with other Tgs sequences in the SWISSPROT database using the FASTA3 program (http://www.ebi.ac.uk/fasta33/). Secondary structure prediction was carried out by PROFSEC. The best templates for Tgs model construction were determined by threading methods, using Bio-Info Meta Server (http://bioinfo.pl/meta/) (30). In this site, the online services PDB-Blast (rscore 9e-46), FFAS03 (rscore -48.9), INUB (rscore 177.98) and 3DPSSM (rscore 3.7e-08) indicated that the E. coli HemK, a (N5)-glutamine methyltransferase (1T43) (31) was the best template for homology modelling. Using this template, ten models were constructed using MODELLER (32,33), and the PROSA II (34) was used to select the model with the most favourable packing and solvent exposure characteristics. After superposition of atomic coordinates, an energy minimization was done using Gromos96, a force field that predicts the dependence of a molecular conformation on the type of environment. This program calculates the relative binding constants by evaluating free energy differences between various molecular complexes using thermodynamic integration, perturbation and extrapolation. The software predicts energetic and structural changes caused by amino acid modifications using six subsequent rounds, minimizing backbone and side chains (3000 steps of steepest descent). Procheck (34) was used for additional analysis of stereochemical quality. Low PROSA II scores and high Procheck G-factors characterize high-quality models. Finally, Ramachandran plot and rmsd values were considered to validate the model. Based on the resulting structural model of TvTgs and the recent characterization of S. cerevisiae, S. pombe and G. lamblia Tgs (25,14,24,27), the following amino acid substitutions were made: S99I, S99R, D145E, D145R, S191R, P193A, W194A. Mutagenesis was introduced by PCR as described (36).
RESULTS
TvTgs is an AdoMet- and m7G-dependent methyltransferase with strict substrate specificity
The T. vaginalis genome database (http://www.trichdb.org) was searched for putative Tgs genes and only a single gene (accession # XP_001579605) was found. This gene, which we call TvTgs, encodes a 32-kDa protein with 36% identity and 51% similarity to S. pombe Tgs (SpTgs), 34% identity and 46% similarity to S. cerevisiae Tgs (ScTgs) and 26% identity and 41% similarity to G. lamblia Tgs2 (GlTgs). For comparison, GlTgs shares 27% identity and 48% similarity to SpTgs, and 21% identity and 41% similarity to ScTgs. TvTgs contains the majority of the amino acids known to be necessary for AdoMet dependent methyltransferase activity of ScTgs and GlTgs (25,14,27) as well as those essential for ScTgs1 substrate binding (24) (Supplementary Figure 1). The TvTgs transcript was detected in T. vaginalis mRNA by reverse transcription and PCR (data not shown).
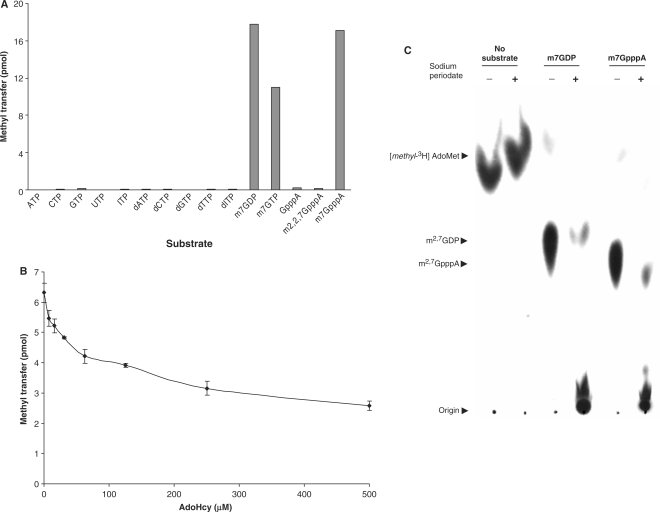
To evaluate substrate specificity, the protein-coding region of the TvTgs gene was cloned with a histidine-tag (His-tag) at its N-terminus and subsequently expressed and purified from E. coli using nickel chromatography. Tgs transfers a methyl-3H group from AdoMet to a nucleotide substrate producing an anionic methylated nucleotide that can be adsorbed to a DEAE filter and separated from the cationic AdoMet substrate. TvTgs was assayed for methyltransferase activity in the presence of [methyl-3H]AdoMet and a variety of potential nucleotide substrates (Figure 1A). We found that TvTgs can use m7GDP or m7GTP as a substrate but not non-methylated nucleotides, including GTP. Similarly, a guanine nucleotide cap analogue can only serve as a substrate if it has undergone prior N7 methylation, while nonmethylated or m2,2,7G cap analogues do not serve as substrates. Thus, TvTgs nucleotide substrate dependence matches that described for other Tgs (25,14). Additional analyses showed that TvTgs exhibits a bell-shaped pH dependence with optimal activity at pH 7.5 and highly reduced activity at pH 9.0 and pH 5.5 (data not shown). Methylation by TvTgs was also shown to increase with increasing enzyme concentration and time, and reactions reached saturation after 20–25 min of incubation with product formation reaching a stable plateau (data not shown). These findings mirror those previously reported for S. pombe and G. lamblia Tgs (25,14), consistent with the TvTgs gene encoding a functional Tgs.
Figure 1.
TvTgs is an RNA cap specific guanine N2 methyltransferase. (A) Nucleotide substrate specificity of TvTgs and (B) inhibition of TvTgs by AdoHcy. Reactions using [methyl-3H]AdoMet as the methyl donor were conducted as described in ‘Materials and methods’ section. Methyltransferase activity was subsequently quantified by measuring 3H incorporation into the reaction products using scintillation counting. (C) Sodium periodate sensitivity of the TvTgs methylated nucleotide product. Methylation reactions mediated by TvTgs using either m7GDP or m7GpppA as the nucleotide substrate (see ‘Materials and methods’ section) were further subjected to either 100 mM sodium periodate (+) or water (–). Products were spotted on DEAE-cellulose and subjected to TLC analyses. Arrows mark origin, substrate and products.
A hallmark of AdoMet-dependent methyltransferases is the formation of AdoHcy resulting from donation of the methyl group from AdoMet to its acceptor nucleotide. Thus, we tested whether AdoHcy is a product of this reaction by poisoning the enzyme with increasing amounts of AdoHcy. This was found to severely inhibit T. vaginalis methyltransferase with a half maximal (50%) inhibitory concentration value (or IC50) of ∼250 μM (Figure 1B), providing additional evidence that TvTgs is an AdoMet-dependent methyltransferase. To determine whether this enzyme is a specific guanine N2 and not a ribose O2′or O3′ methyltransferase, as predicted for a TgS (24), we evaluated the resistance of TvTgs methylated products to sodium periodate oxidation (Figure 1C). It has been shown that methylation at exocyclic N2 atom of m7G leaves the ribose O2′ or O3′ sensitive to oxidation, whereas ribose methylation renders these sites resistant to oxidation. When the ribose site is available for oxidation, the opened-ring 2′,3′-dialdehyde forms a covalent Schiff base adduct that binds PEI-cellulose at the origin and does not migrate during TLC analysis, allowing the oxidation state of the m7G ribose to be determined (25). As shown by TLC analyses, addition of 100 mM sodium periodate to reactions pre-incubated with m7GDP or m7GpppG, recombinant TvTgs and [methyl-3H]AdoMet resulted in retention of the labelled product at the origin (Figure 1C). This was also observed using m7GTP as the substrate (data not shown). The sensitivity of these TvTgs products to periodate oxidation indicates that ribose hydroxyls are not methylated, but instead that the N2 atom of m7G is the methylation site. These results are in agreement with the lack of the catalytic signature (the tetrad KDKE) present in 2′-O-ribose methyltransferases (37), as observed in our structure model (see below), further confirming that this enzyme is a homologue of other characterized eukaryotic Tgs.
Nucleotide substrate preference of TvTgs
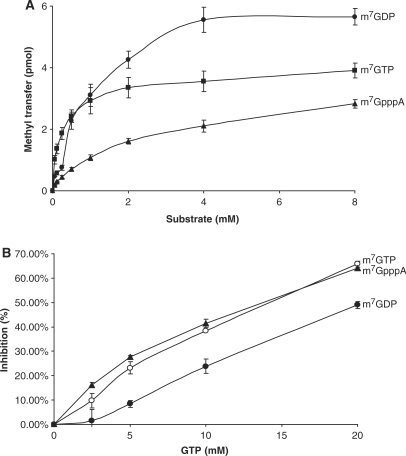
The substrate preference of TvTgs for transfer of [methyl-3H]AdoMet was assessed. The methyl transfer activity of TvTgs was found to display a hyperbolic dependence on an m7G substrate (Figure 2A). Km values were calculated using m7GDP, m7GTP and m7GpppG substrates and found to be 250 +/– 30.7 μM, 1.59 +/– 0.205 mM and 2.23 +/– 0.327 mM, respectively. These results indicate that TvTgs has a higher affinity for m7GDP. The observed specificity of TvTgs for these nucleotides substrates is similar to S. pombe Tgs but significantly lower than that reported for the G. lamblia enzyme (25,14). m7G-dependent methyltransferases bind to non-methylated nucleotides with reduced affinity without promoting N2 methylation (14). Thus we tested whether GTP inhibits TvTgs binding and methylation of the three m7G substrates and compared inhibition levels (Figure 2B). Under our assay conditions, we observed that the higher the Km value, the higher the inhibition by GTP. This result indicates a greater TvTgs affinity for m7GDP than m7GTP and/or m7GpppG. Contrary to G. lamblia Tgs2 (25), a γ-phosphate substrate (m7GTP) and a 5′-nucleoside substrate (m7GpppG) decreases TvTgs activity by 6- and 9-fold, respectively, as suggested by their relative Km values.
Figure 2.
Nucleotide preference of TvTgs. (A) The relative affinity of TvTgs for m7GDP, m7GTP and m7GpppA, indicated by the Km values, was measured as described in ‘Materials and methods’ section. (B) Inhibition of TvTgs activity by increasing amounts of GTP using equivalent concentrations of the three substrates (indicated on the right) was determined.
Structure-function analysis of TvTgs
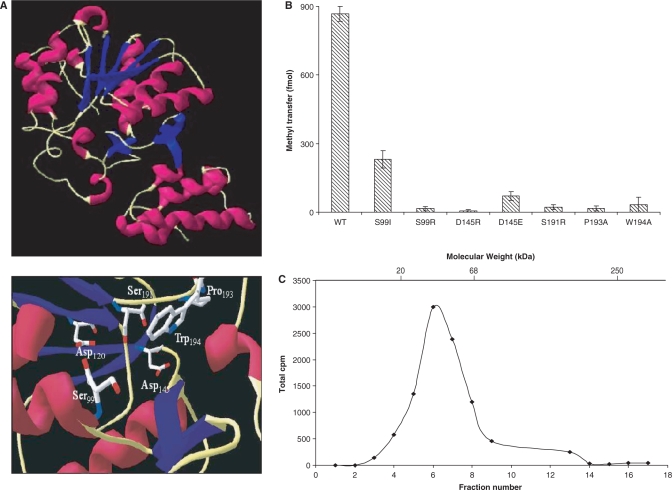
Comparing structural modelling and mutagenesis data from S. cerevisiae and G. lamblia Tgs (25,24,27) and E. coli HemK (1T43), a (N5)-glutamine methyltransferase (31), to that predicted for TvTgs, showed secondary structural similarity and conservation of critical amino acids (Figure 3A). Using E. coli HemK (1T43) as a model, a three-dimensional structure of TvTgs can be predicted including the identification of putative catalytic and donor sites (Figure 3A, bottom). The 280-amino-acid TvTgs is predicted to contain two structural domains: a four-helix N-terminal bundle (residues 1–64), and a C-terminal catalytic domain with a seven-stranded β-sheet (residues 65–280), characteristic of methyltransferases. Also as predicted a β-hairpin, which interacts with the C-terminal domain via hydrogen bonds and a salt bridge, connects the two domains (Figure 3A, top).
Figure 3.
Structure–function analysis of recombinant TvTgs. (A) A ribbon representation of the predicted TvTgs structure (top) and predicted catalytic and donor sites (bottom) indicated by homology modelling are shown. Conserved amino acids known to be indispensable for S. cerevisiae TvTgs activity (22,24) are presented (bottom). (B) Methyltransferase activity was measured using saturating amounts of m7GDP (10 mM) as substrate and equivalent amounts of wild-type (WT) and mutant TvTgs proteins as indicated. (C) Zonal velocity sedimentation analysis of TvTgs. Recombinant TvTgs and internal molecular weight standards (20-kDa soybean trypsin inhibitor, 66-kDa bovine serum albumin and 250-kDa catalase) were fractionated on a sucrose gradient. Aliquots of each fraction were then tested for methyltransferase activity, using [methyl-3H]AdoMet as the methyl donor. The peaks for separation of the protein standards, as determined using SDS–PAGE, are indicated at the top.
ScTgs belongs to a large family of Rossmann-fold AdoMet-dependent methyltransferases (24,38). Amino acids in motif I conserved among different Tgs, are involved in AdoMet binding, including glycine residues which are also present in E. coli HemK (31) (Supplementary Figure 1) and 2′-O-ribose methyltransferases (37). Catalytic amino acids within the nine Rossmann-fold AdoMet-dependent methyltransferase motifs can vary among different types of methyltransferase. For example, 2′-O-ribose methyltransferases have the catalytic tetrad KDKE distributed within motifs X, IV, VI and VIII (37). This tetrad is not observed in any described Tgs, including TvTgs (Supplementary Figure 1). Specific amino acids necessary for catalysis of ScTgs were recently determined (22,24). Many are in motif IV and are conserved among Tgs homologues. Based on our structure model, all of these except TvTgs S99 (ScTgs I83) are conserved in TvTgs (Supplementary Figure 1; Figure 3A, bottom). These include two aspartate residues involved in both water-mediated coordination of the methionine moiety of AdoMet and binding to the 2′- and 3′-OH groups of the ribose (ScTgs D103 and D126; TvTgs D128 and D145), and serine and proline catalytic residues (ScTgs S175 and P177; TvTgs S191 and P193) (Figure 3A, bottom). A tryptophan important for stabilization of the target base (ScTgs W75) was reported to be invariant in all eukaryotic homologues of Tgs except Plasmodium; however, it is not required for ScTgs activity (24). Like Plasmodium, TvTgs and GlTgs also contain a neutral non-polar amino acid substitution at this position (Supplementary Figure 1), consistent with ScTgs W75 not being necessary for catalysis, binding or correct folding of the protein (24).
To test whether these conserved amino acids are required for TvTgs activity, residues were individually mutated, and mutant proteins were subsequently expressed and purified from E. coli. In each case, mutation resulted in either loss of or significantly diminished activity (Figure 3B). TvTsg mutants D145R, D145E, S191R, P193A and W194A were found to be inert in methyl transfer catalysis, consistent with these residues working cooperatively for substrate binding and catalysis (24). A conserved amino acid found in the m7G-binding pocket, shown to be required for yeast Tgs activity, ScTgs I83, (24) is absent in TvTgs. Instead a polar residue, TvTgs S99, has replaced the equivalent isoleucine or valine found in all other reported eukaryotic Tgs homologues. A polar amino acid substitution at this position is also seen in GlTgs (GlTgs Y44). Interestingly, mutation of TvTgs S99 and subsequent analysis of the mutant protein demonstrated it is critical for TvTgs activity, as S99R lost methyl transfer activity and S99I had highly reduced activity (Figure 3B).
Finally, we determined the molecular mass of the active, recombinant form of TvTgs, by zonal velocity sedimentation in a 2–14% sucrose gradient using native protein markers soybean trypsin inhibitor (20 kDa), bovine serum albumin (66 kDa) and catalase (250 kDa) as internal standards. The majority of His-tagged TvTgs sedimented in fractions 4–8 between the 20- and 66-kDa markers (data not shown). Fractions were tested for methyltransferase activity (Figure 3C) and the peak of activity was found in a fraction corresponding to a molecular mass of ∼35.1 kDa, in agreement with the predicted mass (∼36 kDa) of His-tagged TvTgs (Figure 3C). Thus, the active form of TvTgs appears to be monomeric, as previously described for other Tgs (25,14).
TvTgs and GlTgs are unique dimethylguanosine synthases
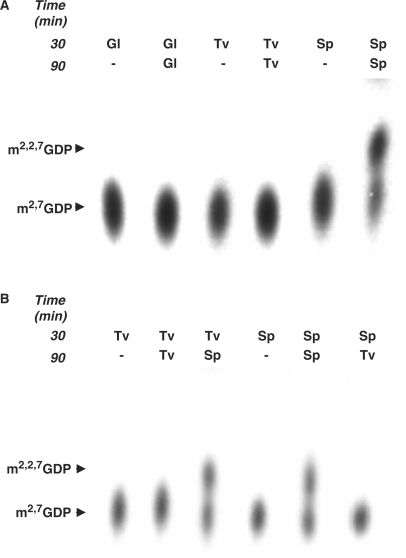
Synthesis of TMG in vitro occurs in two steps via a distributive mechanism. Using m7G as a substrate for methylation, first DMG is formed, which must then dissociate and compete with m7G for rebinding the enzyme to allow a second methylation forming TMG. To complete the reaction, it is necessary to add a large molar excess of AdoMet after the first methylation reaction reaches saturation (14). The recent observation that under these conditions G. lamblia Tgs2 can catalyse the addition of only one methyl group to m7G to form DMG (25) led us to evaluate whether TvTgs can produce both DMG and TMG. To this end, we expressed and purified His-tagged TvTgs, S. pombe Tgs and G. lamblia Tgs2 and compared the ability of the three enzymes to form DMG and TMG. Following a first round of methylation using 5 μM [methyl-3H]AdoMet and 0.5 μg of one of the three recombinant Tgs, the reaction was supplemented with 3.2 mM of unlabelled [methyl]AdoMet and 1 μg of the respective enzyme and the incubation was continued. Equal amounts of both reactions, using the three enzymes, were then subjected to PEI-cellulose TLC analyses. TvTgs was found capable of catalyzing only one round of methylation, similar to G. lamblia Tgs, and in contrast to S. pombe Tgs, which is capable of two consecutive rounds of methylation (Figure 4A). We then determined whether the DMG formed by TvTgs catalysis could act as a substrate for additional methylation, upon addition of S. pombe Tgs. After its formation by incubation with TvTgs, S. pombe Tgs was added during the second incubation in the presence of excess unlabeled [methyl]AdoMet. Under these conditions, TMG is formed (Figure 4B). Conversely, replacing S. pombe Tgs with TvTgs during the second round of incubation after formation of DMG by the former enzyme did not support TMG formation. Together, these data strongly indicate that TvTgs is not a trimethylguanosine synthase, as typically found in other eukaryotes, but is instead a dimethylguanosine synthase.
Figure 4.
Comparison of the distributive mechanism of RNA cap methyltransferase activity by S. pombe Tgs, G. lamblia Tgs2 and TvTgs. (A) Methyltransferase reactions (see ‘Materials and methods’ section) were first conducted for 30 min with 5 μM [methyl-3H]AdoMet, 0.1 mM m7GDP and 0.5 μg of the recombinant Tgs enzyme listed at the 30-min time. The reaction was then supplemented with 3.2 mM of unlabelled [methyl]AdoMet and 1 μg of the respective recombinant enzyme followed by incubation for an additional 60 min at 37°C, listed at the 90-min time. Equivalent amounts of the first and second reactions were then subjected to TLC analyses as described in Figure 1 legend and in ‘Materials and methods’ section. In this experiment, TvTgs (Tv), GlTgs (Gl) and SpTgs (Sp) were compared side by side. (B) Methyltransferase reactions, conducted as described in (A) above, contained either TvTgs (Tv) or SpTgs (Sp) during the first 30-min incubation, as indicated at the 30-min time. TvTgs reactions were then supplemented with either TvTgs or SpTgs, as indicated for the additional 60-min incubation step (90 min). SpTgs reactions were also supplemented with either SpTgs or TvTgs, as indicated for the additional 60-min incubation step (90 min). Reaction products were analysed using TLC as described above. Methylated products are indicated and marked by arrows.
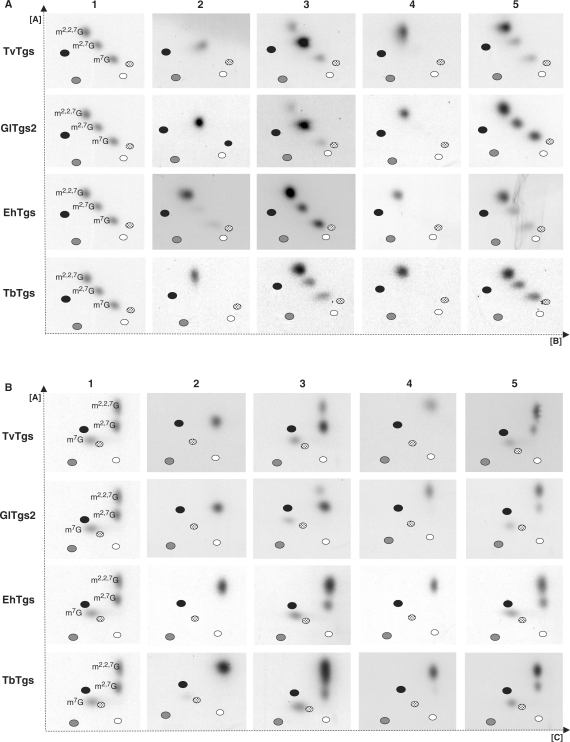
The inability of TvTgs or GlTgs to convert m7G to TMG could be attributed to the lack of an appropriate substrate. Therefore, instead of using nucleotide substrates, we prepared transcripts that have a [32P]m7G cap structure using the Vaccinia Virus capping enzyme (VVC). The m7G cap RNA substrate was then incubated with the Tgs, and products were analysed by TLC following RNA purification and tobacco acid pyrophosphatase (TAP) treatment to release the guanosine cap. As predicted, we observed that RNA that is not treated with VVC (to generate m7G) does not serve as a substrate for TvTgs, GlTgs or SpTgs (data not shown). In contrast the m7G-capped RNA substrate was utilized by all three Tgs with higher affinity than mononucleotides, as suggested with the Km values of 54.8 +/– 4.3 μM, 9.4 +/– 0.8 nM and 34.5 +/– 15.4 nM calculated for TvTgs, GlTgs and SpTgs, respectively (data not shown). To further examine the cap structure formed when incubating an m7G-capped RNA substrate with TvTgs, we subjected the end products of these assays to 2D-TLC analyses, using two different solvents in the second dimension (Figure 5A and B). In addition to testing TvTgs, we included Tgs from other unicellular parasites, G. lamblia (GlTgs), E. histolytica (EhTgs) and T. brucei (TbTgs) to compare TgS specificity among unicellular eukaryotes (Figure 5). The use of the higher avidity, 32P-labelled mRNA substrate in these assays improved detection of Tgs activity and allowed us to determine whether di- and trimethylguanosines end products were formed by comparing their migration with mono, di and trimethylguanosine standards (Figure 5A and B, columns 1, 3 and 5). A 1000-fold molar excess of the methyl-donor substrate AdoMet relative to the 32P-labelled m7G RNA acceptor substrate was used to drive the methyltransferase reaction to completion. Reaction products were then either directly analysed using 2D-TLC following TAP treatment to remove the cap structure (Figure 5A and B, columns 2 and 3) or were also further incubated with SpTgs to drive formation of a TMG prior to TAP treatment and 2D-TLC analyses (Figure 5A and B, columns 4 and 5). Our results clearly demonstrate that TvTgs and GlTgs are capable of catalysing the formation of a DMG, but not a TMG cap structure. Furthermore, these DMG caps can be converted to TMG upon subsequent incubation with SpTgs, confirming its identity as a bona fide DMG. Interestingly, we found that the ability to form only a DMG cap is limited to G. lamblia and T. vaginalis as both E. histolytica and T. brucei Tgs can convert a m7G capped RNA to a TMG capped RNA as observed for S. pombe and thought to be standard for eukaryotic Tgs. The ability of TvTgs to promote dimethylation of m7G cap RNAs and our previous observation that T. vaginalis spliceosomal snRNA are not endogenous substrates for TvTgs (7) suggests that T. vaginalis may contain a novel subset of DMG capped RNAs.
Figure 5.
2D-TLC analysis of cap structures formed using Tgs from four unicellular parasites. The first dimension was run using solvent A (28; see ‘Materials and methods’ section) and the second dimension used either solvent B or solvent C as shown in (A) or (B), respectively. 32P-labelled m7G cap RNA was the substrate for methyltransferase activity with recombinant Tgs from T. vaginalis (TvTgs), G. lamblia (GlTgs2), E. histolytica (EhTgs) and T. brucei (TbTgs) as indicated on the left. The guanosine capped end-products of these reactions were then immediately released by TAP treatment (columns 2 and 3) or were further incubated with S. pombe TgS (SpTgS) prior to release by TAP treatment (columns 4 and 5). Mono, di and trimethylguanosine standards were included as markers to reveal the relative migration of these cap structures (column 1). Black, grey, white and dotted ovals also indicate migration of the four monophosphate nucleotides—AMP, GMP, UMP and CMP, respectively. Samples were analysed in the absence (columns 2 and 4) or the presence of mono, di and trimethylguanosine standards (column 3 and 5) for clarity. In the latter case, the cap structure formed resulted in an increased intensity of the corresponding cap standard.
DISCUSSION
We have identified and characterized a homologue of Tgs, an RNA cap-specific m7G-dependent N2 methyltransferase, in the divergent eukaryotic parasite T. vaginalis. This RNA methyltransferase, called TvTgs, can utilize a variety of m7G substrates, with different affinities, to form a m2,7G cap and displays a strict dependence on prior methylation of guanine N7, a unique property of Tgs. Substrate affinity, inhibition by AdoHcy and sensitivity to sodium periodate oxidation indicate that TvTgs is a N2 guanine methyltransferase. As previously shown for GlTgs and SpTgs, recombinant TvTgs can methylate m7G nucleotides to form a DMG cap in the absence of any RNA or protein co-factor. Both TvTgs and GlTgs also convert the m7G cap on an RNA substrate to a DMG cap, having a significantly higher affinity for this substrate relative to nucleotide substrates.
The predicted structure and key amino acids in the active site that are required for activity of yeast and G. lamblia Tgs (25,24,27) are conserved in TvTgs. Mutagenesis of selected residues results in a substantial decrease or loss of enzyme activity, similar to that observed for S. cerevisae Tgs (ScTgs). A significant difference observed in the predicted TvTgs m7G-binding pocket is the presence of a serine (TvTgs S99) replacing an isoleucine (ScTgs I83). Except for GlTgs which also contain a polar residue at this position (GlTgs Y44), the corresponding residue in all previously characterized Tgs is either an isoleucine or valine (24). All three of these residues are short-chain amino acids; however, serine (TvTgs S99) and tyrosine (GlTgs Y44) are polar whereas isoleucine (ScTgs I83) and valine (SpTgs V59) are non-polar. In T. vaginalis, replacement of this serine residue with an isoleucine in TvTgs resulted in greatly reduced activity. In S. cerevisiae, replacement of ScTgs I83 to a polar arginine did not rescue cold-strain phenotype and did not lead to formation of m2,2,7G cap sn(o)RNAs in the yeast mutant (26).
Recombinant SpTgs is an RNA methyltransferase capable of transferring in vitro two methyl groups to N2 of a guanosine previously methylated at the N7 position (25,24,27). Despite similarity in predicted structure and conserved catalytic residues, recombinant TvTgs is limited to a single round of N2 methylation forming a 2,7-dimethylguanosine as end product. 2D-TLC analyses using high avidity and 32P-labelled m7G cap RNA substrate indicate the complete absence of a trimethylated guanosine as an end product for either TvTgs or GlTgs. It is notable that T. vaginalis and G. lamblia Tgs are similar in this regard and divergent from all other characterized eukaryotic Tgs, including E. histolytica and T. brucei Tgs analysed here, which catalyse the formation of an m2,2,7G cap (TMG).
Our data do not preclude the presence of additional in vivo factors that might drive TvTgs and GlTgs to a second round of methylation or additional Tgs genes missing from the T. vaginalis genome database (6) that are capable of two rounds of methylation. Indeed, there are two Tgs genes in G. lamblia, only one of which (Tgs2) has been characterized as a dimethylguanosine synthase (25) while the other (Tgs1) remains uncharacterized. Giardia has been reported to have snRNAs with trimethylguanosine (TMG) caps (39), consistent with the uncharacterized G. lamblia Tgs1 being the trimethylguanosine synthase responsible for the formation of TMG cap structures.
We have recently demonstrated that T. vaginalis snRNAs are uncapped and contain free phosphates at their 5′ ends (7). Other small RNAs in T. vaginalis, such as intronic snoRNAs, also appear to lack TMG caps (our unpublished data). The lack of a classical Tgs in T. vaginalis is consistent with the lack of snRNAs with TMG caps. This, nevertheless, begs the question of what RNA population(s) is the target of TvTgs in T. vaginalis. What is the consequence of these RNAs containing dimethylguanosine 5′ caps? What is the subcellular localization of these RNAs?
Sindbis and Semliki Forest eukaryotic viruses are reported to contain a significant fraction of 2,7-dimethylguanosine caps (40,41). In addition, m2,7G cap reporter mRNAs are translated with a higher efficiency than m7G cap transcripts in vitro, whereas m2,2,7G cap transcripts are translated with very low efficiency (42–44). Recently, two eukaryotic translation initiation factor 4Es (eIF4E) were found in G. lamblia and shown to have preferential affinity for either m7G or m2,2,7G caps. Knockdown of the eIF4E that specifically binds a m7G cap blocked translation, whereas Giardia m2,2,7G cap mRNAs transfected into the parasite were not translated, consistent with G. lamblia mRNAs containing a 5′ m7G cap (45). In light of our data and those demonstrating the presence of a dimethylguanosine synthase in G. lamblia (25) it would be interesting to determine whether either of the G. lamblia eIF4E proteins bind to m2,7G capped RNA and whether similar eIF4E proteins that bind m2,7G capped RNAs are present in T. vaginalis. Although both G. lamblia and T. vaginalis mRNAs were shown to be capped (7,26), their cap structure is not known.
The ability of the TvTgs studied here and the previously described G. lamblia Tgs2 (25) to catalyse only a single round of methylation is reminiscent of a subset of tRNA-specific N2 methyltransferases that execute only one cycle of methylation producing 2-methylguanosine and the rRNA-specific guanine-N2 methyltransferase, RsmC, which is also limited to a single round of methylation (47–51). tRNA-specific N2 methyltransferases are found in archaea and eukaryotes (51); whereas RsmC has only been reported in eubacteria (52,53). For example, the tRNA methyltransferase Trm-G10 from the archeae Pyrococcus abyssi exhibits a characteristic Rossmann fold and a SAM-dependent methyltransferase domain, both of which are found in archea and eukaryota and are absent in eubacteria (50). The Tgs gene family appears to be present exclusively in eukaryotes; however, an uncharacterized family of methyltransferases from archaea and Gram-positive bacteria exhibits significant similarity to ScTgs (24). The relationship of Tgs in divergent eukaryotes is not well resolved in evolutionary trees (24) making it difficult to trace their relatedness and evolutionary origin. Nonetheless, given the common features shared by TvTgs, G. lamblia Tgs2 and several eubacterial and archaeal methyltransferases, it is tempting to speculate that TvTgs and G. lamblia Tgs represent the basal state of eukaryotic Tgs (one capable of only a single round of methylation; a dimethylguanosine synthase) and that the acquisition of catalysing a second round of methylation, as observed in yeast, is a derived state. Alternatively, TvTgs and G. lamblia Tgs may have once been capable of generating an m2,2,7G cap independent of accessory factors, as observed for yeast Tgs, but have evolved to require additional cytosolic factors in the current state. An understanding of why purified Tgs from these basal unicellular eukaryotes acting as dimethylguanosine synthases and not trimethylguanosine synthasese awaits further analyses.
FUNDING
The National Institutes of Health (AI30537); a postdoctoral fellowship from CNPq-Brazil (PDE 200065/2004-1). Funding for open access charges: National Institutes of Health (AI30537).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Maria Delgadillo-Correa for technical assistance, Dr Bidyottam Mittra for advice, Drs Natalia de Miguel, Cheryl Okumura and Nancy Sturm for critical comments on the manuscript and our colleagues in the lab for helpful discussions.
REFERENCES
- 1.Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 1992;267:9805–9815. [PubMed] [Google Scholar]
- 2.Simoes-Barbosa A, Meloni D, Wohlschlegel JA, Konarska MM, Johnson PJ. Spliceosomal snRNAs in the unicellular eukaryote Trichomonas vaginalis are structurally conserved but lack a 5′ cap structure. RNA. 2008;14:1617–1631. doi: 10.1261/rna.1045408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hausmann S, Shuman S. Giardia lamblia RNA cap guanine-N2 methyltransferase (Tgs2) J. Biol. Chem. 2005;280:32101–32106. doi: 10.1074/jbc.M506438200. [DOI] [PubMed] [Google Scholar]
- 4.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 5.Simpson AG, Roger AJ, Silberman JD, Leipe DD, Edgcomb VP, Jermiin LS, Patterson DJ, Sogin ML. Evolutionary history of “early-diverging” eukaryotes: the excavate taxon Carpediemonas is a close relative of Giardia. Mol. Biol. Evol. 2002;19:1782–1791. doi: 10.1093/oxfordjournals.molbev.a004000. [DOI] [PubMed] [Google Scholar]
- 6.Liston DR, Carrero JC, Johnson PJ. Upstream regulatory sequences required for expression of the Trichomonas vaginalis alpha-succinyl CoA synthetase gene. Mol. Biochem. Parasitol. 1999;104:323–329. doi: 10.1016/s0166-6851(99)00137-1. [DOI] [PubMed] [Google Scholar]
- 7.Liston DR, Lau AO, Ortiz D, Smale ST, Johnson PJ. Initiator recognition in a primitive eukaryote: IBP39, an initiator-binding protein from Trichomonas vaginalis. Mol. Cell. Biol. 2001;21:7872–7882. doi: 10.1128/MCB.21.22.7872-7882.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher MA, Lau AO, Johnson PJ. Structural basis of core promoter recognition in a primitive eukaryote. Cell. 2003;115:413–424. doi: 10.1016/s0092-8674(03)00887-0. [DOI] [PubMed] [Google Scholar]
- 9.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanacova S, Yan W, Carlton JM, Johnson PJ. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc. Natl Acad. Sci. USA. 2005;102:4430–4435. doi: 10.1073/pnas.0407500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell ES, Fournier MJ. The small nucleolar RNAs. Annu. Rev. Biochem. 1995;64:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 13.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 14.Liou RF, Blumenthal T. trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 1990;10:1764–1768. doi: 10.1128/mcb.10.4.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busch H, Reddy R, Rothblum L, Choi YC. SnRNAs, SnRNPs, and RNA processing. Annu. Rev. Biochem. 1982;51:617–654. doi: 10.1146/annurev.bi.51.070182.003153. [DOI] [PubMed] [Google Scholar]
- 16.Hausmann S, Shuman S. Specificity and mechanism of RNA cap guanine-N2 methyltransferase (Tgs1) J. Biol. Chem. 2005;280:4021–4024. doi: 10.1074/jbc.C400554200. [DOI] [PubMed] [Google Scholar]
- 17.Plessel G, Fischer U, Luhrmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol. Cell. Biol. 1994;14:4160–4172. doi: 10.1128/mcb.14.6.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J. Cell. Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narayanan A, Eifert J, Marfatia KA, Macara IG, Corbett AH, Terns RM, Terns MP. Nuclear RanGTP is not required for targeting small nucleolar RNAs to the nucleolus. J. Cell. Sci. 2003;116:177–186. doi: 10.1242/jcs.00176. [DOI] [PubMed] [Google Scholar]
- 20.Terns MP, Grimm C, Lund E, Dahlberg JE. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Speckmann WA, Terns RM, Terns MP. The box C/D motif directs snoRNA 5′-cap hypermethylation. Nucleic Acids Res. 2000;28:4467–4473. doi: 10.1093/nar/28.22.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouaikel J, Narayanan U, Verheggen C, Matera AG, Bertrand E, Tazi J, Bordonne R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 2003;4:616–622. doi: 10.1038/sj.embor.embor863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 2002;9:891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 25.Ruan JP, Ullu E, Tschudi C. Characterization of the Trypanosoma brucei cap hypermethylase Tgs1. Mol. Biochem. Parasitol. 2007;155:66–69. doi: 10.1016/j.molbiopara.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mouaikel J, Bujnicki JM, Tazi J, Bordonne R. Sequence-structure-function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 2003;31:4899–4909. doi: 10.1093/nar/gkg656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hausmann S, Altura MA, Witmer M, Singer SM, Elmendorf HG, Shuman S. Yeast-like mRNA capping apparatus in Giardia lamblia. J. Biol. Chem. 2005;280:12077–12086. doi: 10.1074/jbc.M412063200. [DOI] [PubMed] [Google Scholar]
- 28.Hausmann S, Ramirez A, Schneider S, Schwer B, Shuman S. Biochemical and genetic analysis of RNA cap guanine-N2 methyltransferases from Giardia lamblia and Schizosaccharomyces pombe. Nucleic Acids Res. 2007;35:1411–1420. doi: 10.1093/nar/gkl1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 30.Martin RG, Ames BN. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J. Biol. Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- 31.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, Shipman L, Zhang M, Anton BP, Roberts RJ, Cheng X. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J. Mol. Biol. 2004;340:695–706. doi: 10.1016/j.jmb.2004.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sali A. Comparative protein modeling by satisfaction of spatial restraints. Mol. Med. Today. 1995;1:270–277. doi: 10.1016/s1357-4310(95)91170-7. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez R, Sali A. Evaluation of comparative protein structure modeling by MODELLER-3. Proteins. 1997;68(Suppl 1):50–58. doi: 10.1002/(sici)1097-0134(1997)1+<50::aid-prot8>3.3.co;2-w. [DOI] [PubMed] [Google Scholar]
- 35.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 37.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 38.Mittra B, Zamudio JR, Bujnicki JM, Stepinski J, Darzynkiewicz E, Campbell DA, Sturm NR. The TbMTr1 spliced leader RNA cap 1 2′-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity. J. Biol. Chem. 2008;283:3161–3172. doi: 10.1074/jbc.M707367200. [DOI] [PubMed] [Google Scholar]
- 39.Cheng X, Blumenthal RM. Singapore: World Scientific Inc.; 1999. S-Adenosylmethionine-dependent methyltransferases: structures and functions. [Google Scholar]
- 40.Niu XH, Hartshorne T, He XY, Agabian N. Characterization of putative small nuclear RNAs from Giardia lamblia. Mol. Biochem. Parasitol. 1994;66:49–57. doi: 10.1016/0166-6851(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 41.HsuChen CC, Dubin DT. Di-and trimethylated congeners of 7-methylguanine in Sindbis virus mRNA. Nature. 1976;264:190–191. doi: 10.1038/264190a0. [DOI] [PubMed] [Google Scholar]
- 42.van Duijn LP, Kasperaitis M, Ameling C, Voorma HO. Additional methylation at the N(2)-position of the cap of 26S Semliki Forest virus late mRNA and initiation of translation. Virus Res. 1986;5:61–66. doi: 10.1016/0168-1702(86)90065-1. [DOI] [PubMed] [Google Scholar]
- 43.Darzynkiewicz E, Stepinski J, Ekiel I, Jin Y, Haber D, Sijuwade T, Tahara SM. Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 1988;16:8953–8962. doi: 10.1093/nar/16.18.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grudzien E, Stepinski J, Jankowska-Anyszka M, Stolarski R, Darzynkiewicz E, Rhoads RE. Novel cap analogs for in vitro synthesis of mRNAs with high translational efficiency. RNA. 2004;10:1479–1487. doi: 10.1261/rna.7380904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cai A, Jankowska-Anyszka M, Centers A, Chlebicka L, Stepinski J, Stolarski R, Darzynkiewicz E, Rhoads RE. Quantitative assessment of mRNA cap analogues as inhibitors of in vitro translation. Biochemistry. 1999;38:8538–8547. doi: 10.1021/bi9830213. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Wang CC. Identification in the ancient protist Giardia lamblia of two eukaryotic translation initiation factor 4E homologues with distinctive functions. Eukaryot. Cell. 2005;4:948–959. doi: 10.1128/EC.4.5.948-959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J. Biol. Chem. 1986;261:9703–9709. [PubMed] [Google Scholar]
- 49.Constantinesco F, Motorin Y, Grosjean H. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J. Mol. Biol. 1999;291:375–392. doi: 10.1006/jmbi.1999.2976. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Straby KB. The human tRNA(m(2)(2)G(26))dimethyltransferase: functional expression and characterization of a cloned hTRM1 gene. Nucleic Acids Res. 2000;28:3445–3451. doi: 10.1093/nar/28.18.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004;279:37142–37152. doi: 10.1074/jbc.M403845200. [DOI] [PubMed] [Google Scholar]
- 52.Tscherne JS, Nurse K, Popienick P, Ofengand J. Purification, cloning, and characterization of the 16 S RNA m2G1207 methyltransferase from Escherichia coli. J. Biol. Chem. 1999;274:924–929. doi: 10.1074/jbc.274.2.924. [DOI] [PubMed] [Google Scholar]
- 53.Bujnicki JM, Rychlewski L. RNA: (guanine-N2) methyltransferases RsmC/RsmD and their homologs revisited–bioinformatic analysis and prediction of the active site based on the uncharacterized Mj0882 protein structure. BMC Bioinformatics. 2002;3:10. doi: 10.1186/1471-2105-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.