Abstract
The RNA exosome processes and degrades RNAs in archaeal and eukaryotic cells. Exosomes from yeast and humans contain two active exoribonuclease components, Rrp6p and Dis3p/Rrp44p. Rrp6p is concentrated in the nucleus and the dependence of its function on the nine-subunit core exosome and Dis3p remains unclear. We found that cells lacking Rrp6p accumulate poly(A)+ rRNA degradation intermediates distinct from those found in cells depleted of Dis3p, or the core exosome component Rrp43p. Depletion of Dis3p in the absence of Rrp6p causes a synergistic increase in the levels of degradation substrates common to the core exosome and Rrp6p, but has no effect on Rrp6p-specific substrates. Rrp6p lacking a portion of its C-terminal domain no longer co-purifies with the core exosome, but continues to carry out RNA 3′-end processing of 5.8S rRNA and snoRNAs, as well as the degradation of certain truncated Rrp6-specific rRNA intermediates. However, disruption of Rrp6p–core exosome interaction results in the inability of the cell to efficiently degrade certain poly(A)+ rRNA processing products that require the combined activities of Dis3p and Rrp6p. These findings indicate that Rrp6p may carry out some of its critical functions without physical association with the core exosome.
INTRODUCTION
The process of transcription in eukaryotic cells produces a significant amount of RNA that is destroyed during maturation into functional transcripts (1). Most eukaryotic RNAs are synthesized as precursor-RNAs (pre-RNAs) that are cleaved, spliced and end-trimmed to produce mature, functional molecules. The RNA fragments removed by these internal cleavages are degraded in the nucleus and the cytoplasm by 5′–3′ and 3′–5′ exoribonucleases. The major 5′–3′ exoribonucleases include the nuclear Rat1p/Xrn2p and the cytoplasmic Xrn1p/Kem1p. Rat1p plays roles in rRNA 5′-end formation, mRNA transcription termination and the degradation of unprocessed pre-mRNAs and pre-rRNAs (2–7). Xrn1p functions in the major mRNA decay pathway and the nonsense-mediated mRNA decay pathway in yeast, as well as a pathway responsible for the degradation of some unspliced mRNAs and those targeted by siRNAs within the RISC complex (8–11).
The RNA processing exosome appears to provide the major 3′–5′ exoribonucleolytic activity in eukaryotes (12,13). The core exosome contains nine proteins with six of these arranged in a ring-like structure capped on the top by the remaining three subunits (14–16). The cap proteins contain putative RNA binding domains thought to interact with RNP substrates, thereby enhancing their insertion into the central core of the six-member ring. In archael exosomes, the central core contains the active sites for phosphorolytic cleavage of the RNA. Interestingly, recent evidence indicates that, despite considerable sequence similarity between the eukaryotic ring proteins and bacterial exoribonucleases, all of the human and yeast ring proteins have lost the active site residues required to carry out RNA cleavage (15,17). Nevertheless, two hydrolytic 3′–5′ exoribonucleases interact with the core exosome in eukaryotes and provide the essential RNA degradation activities of the complex. Dis3p/Rrp44p co-purifies with the core exosome from yeast and resides in the nucleus, nucleolus and the cytoplasm (18,19). Rrp6p also co-purifies with the core exosome and appears confined to the nucleus and nucleolus in yeast, but may have a cytoplasmic role in higher eukaryotes (20–22).
The core exosome plays an important role in the 3′–5′ mRNA degradation pathway in the cytoplasm. The complex is recruited to unstable adenosine-rich element (ARE) containing mRNAs by RNA binding proteins such as tristetraprolin and catalyzes their deadenylation and decay (23–27). Similarly, the Ski complex composed of Ski2p, Ski3p and Ski8p interacts via Ski7p with the core exosome in the cytoplasm of yeast leading to the degradation of LA viral RNAs and aberrant mRNAs containing nonsense codons or lacking a stop codon (10,22,28–31). In the nucleus, Rrp6p and the core exosome form the mature 3′-ends of 5.8S rRNA and many sn/sno-RNAs in a two-step process featuring initial 3′–5′ trimming by the core and Dis3p, followed by removal of the remaining nucleotides by Rrp6p (20,32–35). The nuclear exosome also acts in a RNA surveillance pathway that degrades incompletely processed RNAs. Thus, mRNAs that fail to complete some splicing or 3′-end processing steps accumulate in cells with defects in components of the nuclear exosome (21,36–38). Likewise, rRNA processing intermediates accumulate in exosome-deficient cells, as do noncoding RNAs and hypomodified pre-tRNAiMET. These noncoding RNA intermediates, which carry poly(A) tails synthesized by the poly(A)-polymerases Trf4p and Trf5p, which function in complexes termed TRAMP4 and TRAMP5, respectively, that also contain the RNA helicase Mtr4p and the zinc-knuckle proteins Air1p and Air2p (39–42). Recognition and polyadenylation of these RNAs by TRAMP is thought to facilitate interaction of the transcripts with the exosome, thereby enhancing the rate of RNA degradation.
Rrp6p also plays an important role in DRN (degradation of RNA in the nucleus), a surveillance pathway that requires the nuclear mRNA cap-binding complex (43). Loss of function of Rrp6p or Cbp1p, the large subunit of the cap-binding complex, stabilizes mRNAs trapped in the nucleus, as well as certain transcripts thought to exit more slowly from the nucleus than bulk mRNA (43,44). However, loss of Cbc1p activity does not lead to a general accumulation of polyadenylated rRNAs, suggesting that it does not function in concert with TRAMP to facilitate the degradation of this class of RNAs (45).
Rrp6p plays a critical role in RNA 3′-end formation and degradation pathways in the nucleus, yet no experiments have addressed the important question of whether the activities of Rrp6p require its physical association with the core exosome. The current model for RNA processing and surveillance by the nuclear exosome implies that Rrp6p activity acts in concert with the core exosome complex, either through physical association or close coupling (13,46,47). Indeed, affinity purifications of core exosome subunits from yeast co-purify all of the other core polypeptides, as well as Dis3p/Rrp44p and Rrp6p, and affinity purification of Rrp6p co-purifies all of the core subunits and Dis3p/Rrp44p (48,49). Rrp6p also co-purifies with the human and Trypanosoma brucei core exosomes (25,50). Moreover, experiments in vitro that first characterized TRAMP activity did so in the context of its ability to enhance the activity of the nuclear exosome, or core exosome preparations likely containing Rrp6p (41,51). These considerations and the fact that Rrp6p and the core exosome appear to carry out distinct steps in RNA 3′ end formation prompted us to evaluate the relative contributions of Rrp6p and the core exosome to RNA degradation in vivo. We investigated the RNA processing and degradation phenotypes of strains depleted of Rrp6p and core exosome components and found evidence for Rrp6p-specific degradation substrates. Consistent with these findings, loss of the C-terminal domain of Rrp6p allows normal RNA 3′-end processing, but inhibits the degradation rRNA substrates that require Rrp6p and the core exosome for degradation.
MATERIALS AND METHODS
Northern blot analysis
Total RNA or poly(A)+ RNA was isolated from yeast strains grown to an A600 of 0.5–2.0 in YPD media as described (52). Depletion of the indicated cells was carried out by the addition of doxycyclin (10 μg/ml) for 5 h, before cells were collected. Northern blot analysis was carried out as previously described in Briggs et al. (33). The 5′ 32P oligonucleotide probes OSB138 and OSB267 were described in Phillips and Butler (34), OSB156 in Briggs et al. (33) and OSB157 in Fang et al. (54). Northern blots were analyzed by Phosphorimaging and quantitated using ImageQuant software (GE Healthcare, Piscataway, NJ).
Plasmid construction
Yeast centromere plasmids expressing each of the RRP6 mutants fused to green fluorescent protein (GFP) were constructed by gap repair of pGFP-RRP6H as previously described (34) or by direct cloning into the same vector. Plasmid pGFP-RRP6Δ210 (deletes amino acids 1–210) was constructed by inserting a PCR fragment amplified using OSB 620 and OSB431, into pGFP-RRP6H cut with XbaI and BglII. Plasmid pGFP-RRP6Δ34 (deletes amino acids 176–210) was constructed by inserting a PCR fragment amplified using OSB 709 and OSB41 to amplify hybridized templates produced by PCR with OSB707 and OSB709, and OSB708 and OSB41 into pGFP-RRP6H cut with XbaI and BglII. Plasmid pGFP-RRP6ΔExo (deletes amino acids 238–365) was constructed by inserting a PCR fragment generated using OSB41 and OSB42 to amplify hybridized templates produced by PCR with OSB 695 and OSB41, and OSB696 and OSB42 into pGFP-RRP6H cut with BglII. Plasmid pGFP-RRP6ΔC1+2 (delete amino acids 441–733) was constructed by inserting a PCR fragment generated using OSB706 and OSB726 to amplify hybridized templates produced by PCR with OSB697 and OSB706, and OSB698 and OSB726 into pGFP-RRP6ΔHRDC1 (34) with ClaI and XhoI. Plasmid pGFP-Rrp6pΔC2 (deletes amino acids 523–733) was constructed by inserting a PCR fragment (OSB733 and OSB734) into pGFP-RRP6ΔC1+2 cut with ClaI.
Physical association of proteins
Core exosomes was purified from 2 L of an RRP46-TAP rrp6-Δ strain constructed in a W303 derivative by the method of Rigaut et al. (53) (YSB232, MATa ade2-1 his3-11,5 leu2-3,112 trp1-1 ura3-1 pep4::HIS3 RRP46-TAP rrp6::LEU2) carrying derivatives of pGFP-RRP6H [CEN6 URA3 MET17-GFP-RRP6; (34)] grown to an OD600 of 2.0–3.0 in SCD–MET–URA at 30°C. Cells were collected by centrifugation at 7500g and suspended in 10 M K-HEPES pH 7.9, 10 M KCl, 1.5 M MgCl2, 0.5 M DTT, 2 mM benzamidine, 0.5 mM PMSF, 1 mM leupeptin, 2 mM pepstatin, 4 mM chymostatin and 2.6 mM aprotinin, and disrupted by two passes through a French pressure cell at 850 psi. The cell lysate was cleared by centrifugation at 34 000g for 22 min and then at 75 000g for 60 min. The supernatant was dialyzed against 20 mM K-HEPES pH 7.9, 50 mM KCl, 0.2 mM EDTA, 20% glycerol, 0.5 mM PMSF and 2 mM benzamidine for 3.5 h at 4°C. Five microliter of lysate was bound to 120 μl IgG beads (Amersham Biosciences, Piscataway, NJ) for 2 h at 4°C with gentle rocking. The IgG beads were pelleted by centrifugation and the supernatant was collected and labeled the ‘Free’ fraction. The beads were then washed with 50 ml of 10 mM Tris–Cl pH 8.0, 150 mM NaCl and 0.1% NP40 at 4°C. Core exosome complexes were released from the beads by incubation in 500 μl of 10 mM Tris–Cl pH 8.0, 150 mM NaCl and 0.1% NP40, 0.5 mM EDTA, 1 mM DTT with 70 U AcTEV enzyme (InVitrogen, Carlsbad, CA) for 3.5 h at 14°C with gentle rocking. The IgG beads were pelleted by centrifugation and 150 μl of the supernatant was precipitated with trichloroacetic acid, resuspended in SDS–PAGE sample buffer and called the ‘Bound’ fraction. Western blot analysis was performed as previously described (21) with monoclonal anti-GFP (1:1000; Roche), anti-Rrp6p (1:1000; Dr Phillip Mitchell), anti-Rrp4p (1:1000; Dr David Tollervey).
RESULTS
Identification of Rrp6p-specific RNA degradation intermediates
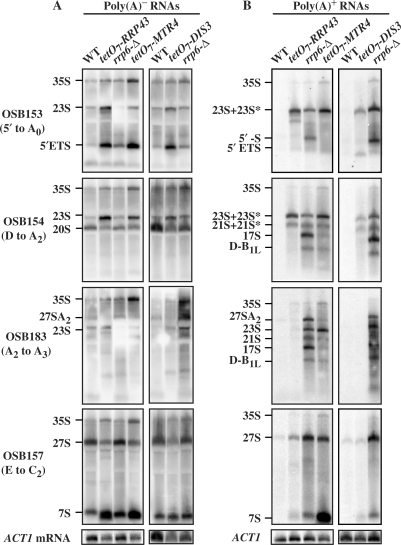
To determine the relative contribution of Rrp6p and the core exosome to RNA degradation, we analyzed the levels of poly(A)+ rRNAs in cells lacking Rrp6p, or after depletion of the core exosome component Rrp43p, the core exoribonuclease Dis3p/Rrp44p or the TRAMP component Mtr4p. Depletions of the essential Rrp43p, Dis3p or Mtr4p were accomplished using strains in which their genes are fused to the doxycycline repressible tetO7 promoter. Total RNA was isolated from these strains after 5 h of doxycycline treatment, the duration we previously determined to be the shortest necessary for maximal accumulation of RNA processing intermediates. The RNA samples were then separated into poly(A)- and poly(A)+ fractions using oligo(dT)-cellulose, separated by gel electrophoresis and rRNA processing intermediates detected by northern blot analysis. In this last step, we used radiolabeled oligonucleotides complementary to the external and internal transcribed spacers of the pre-RNA transcript containing the mature 18S, 5.8S and 25S rRNAs (Figure 1). Thus, only pre-rRNA processing intermediates are detected. Previous studies showed that this method detects those intermediates that become polyadenylated by TRAMP, but fail to be degraded in the absence of Rrp6p (39,45,54). To control for differences in gel loading and the efficiency of oligo(dT)-cellulose fractionation, we also analyzed the level of ACT1 mRNA in each sample (Figure 2). Note that ACT1 mRNA pools contain significant levels of poly(A)-deficient transcripts that do not bind to oligo(dT)-cellulose (Figures 2 and 3) (52,55). Analysis of the effects of these depletions on normal pre-rRNA processing intermediates, which do not bind oligo(dT)-cellulose, reveals that depletion of Rrp43p, Dis3p and Mtr4p results in the enhanced accumulation of 35S and 23S pre-rRNAs, consistent with an indirect inhibition of pre-rRNA processing, as previously shown (Figure 2A) (32). These depletions also cause the expected accumulation of 7S pre-rRNA and the accumulation of the 5′ETS, an rRNA processing byproduct degraded by the combined actions of the core exosome, Mtr4p and Rrp6p (Figure 2A). Other smaller changes in pre-rRNA levels seen in Figure 2 were not consistent from experiment to experiment, and are of questionable significance. We conclude that the depletions of Rrp43p, Dis3p and Mtr4p result in the expected defects in pre-rRNA metabolism.
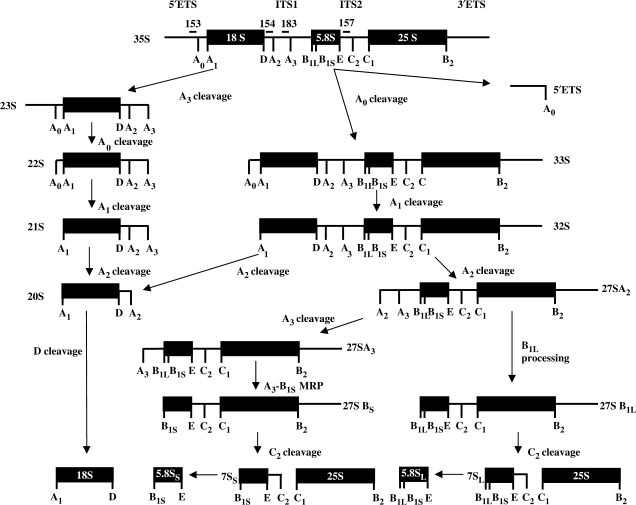
Figure 1.
Ribosomal RNA processing in Saccharomyces cerevisiae. The 35S rRNA primary transcript undergoes several internal cleavages to produce the processing intermediates shown. Additional cleavages produce the mature 18S and 25S rRNAs and 5′- and 3′-end trimming reactions participate in the formation of 5.8S rRNA. 5S rRNA is produced from an independent transcription unit and is not shown. Numbered bars above the 35S pre-rRNA indicate the approximate positions of oligonucleotide probes used in the experiments described.
Figure 2.
Cells deficient for core exosome, TRAMP and Rrp6p components accumulate specific polyadenylated pre-rRNAs. Total RNA was isolated from cells with the indicated genotypes grown in YPD at 30°C. The wild-type (WT) and tetO7 cells were treated for 5 h with 10 μg/ml doxycycline before harvest and RNA isolation. Total RNA was separated into (A) poly(A)- and (B) poly(A)+ fractions using two round of oligo(dT)-cellulose and analyzed by northern blotting with 5′-32P-labeled oligonucleotide probes as indicated to the left of each set of panels, except for ACT1 mRNA, which was detected with OSB400. The identity of the rRNA processing intermediates is indicated to the left of each panel.
Figure 3.
Synergistic effects on poly(A)+ rRNA levels caused by depleting Dis3p in the absence of Rrp6p. Poly(A)+ RNA was isolated, using two round of oligo(dT)-cellulose fractionation, from cells with the indicated genotypes grown in YPD at 30°C. The wild-type (WT) and tetO7 cells were treated for 5 h with 10 μg/ml doxycycline before harvest and RNA isolation. Only the poly(A)+ fractions are shown. RNA was analyzed by northern blotting with 5′-32P-labeled oligonucleotide probes; (A) OSB153, (B) OSB254 and (C) OSB183. ACT1 mRNA was detected with OSB184. The identity of the rRNA processing intermediates is indicated to the left of each panel. The graphs display the ratio of each poly(A)+ RNA to ACT1 mRNA and are the average of two independent experiments.
Analysis of poly(A)+ RNA allows identification of transcripts targeted for degradation by TRAMP specific polyadenylation (39,41). The results from the depleted strains revealed varying levels of poly(A)+ pre-rRNA accumulation (Figure 2B). In these experiments, pre-rRNAs designated 23S and 23S* and 21S and 21S* migrate together, respectively. They are distinguished from one another by the fact that 23S* and 21S* hybridize to an oligonucleotide targeted between cleavage sites D and A2 (OSB154), but not to one targeted between cleavage sites A2 and A3 (OSB183), while 23S and 21S hybridize to both oligonucleotides (Figures 1 and 2B). The 23S* and 21S*, along with 17S (see below), pre-rRNAs were previously identified by Allmang et al. (32), who concluded that they accumulate specifically in exosome mutants. Depletion of Rrp43p and Dis3p showed similar patterns of poly(A)+ pre-rRNA accumulation, consistent with their function together in the core exosome complex. These depletions each cause the accumulation of poly(A)+ forms of 23S*, 21S* and 27S pre-rRNAs (Figure 2B). Depletion of Mtr4p also results in the accumulation of poly(A)+ 7S and 27S pre-rRNAs, consistent with its role as a co-factor in core exosome function (56,57). The rrp6-Δ strain differs from the Rrp43p and Dis3p depletions in that it accumulates polyadenylated forms of 27SA2, 23S, 21S, 17S, pre-rRNAs, an apparently novel poly(A)+ RNA designated D-B1L, and a previously identified species called 5′-S whose 5′-end probably corresponds to the 5′-end of the pre-rRNA transcript (58) (Figure 2B). Some of these Rrp6p-specific RNAs, notably 23S, 21S and D-B1L, also accumulate in the Mtr4p depletion suggesting that Mtr4p and Rrp6p may act together to degrade them.
Next, we tested the effects of depletion of Dis3p in the absence of Rrp6p to determine if synergistic effects on poly(A)+ rRNA processing occur. The intensity of the northern blot bands corresponding to the poly(A)+ 23S+23S*, 21S+21S* and 5′ ETS RNAs increased significantly after depletion of Dis3p in the tetO7-DIS3 rrp6-Δ strain compared to the rrp6-Δ and tetO7-DIS3 strains (Figure 3A and B). In contrast, the 27SA2, 23S, 21S, 17S, 5′-S and D-B1L RNAs showed no significant increase in the tetO7-DIS3 rrp6-Δ strain compared to the rrp6-Δ and tetO7-DIS3 strains (Figure 3C). These findings support the conclusion that Rrp6p plays a core exosome-independent role in the degradation of some of these poly(A)+ rRNAs.
Rrp6p-specific degradation of unadenylated rRNAs
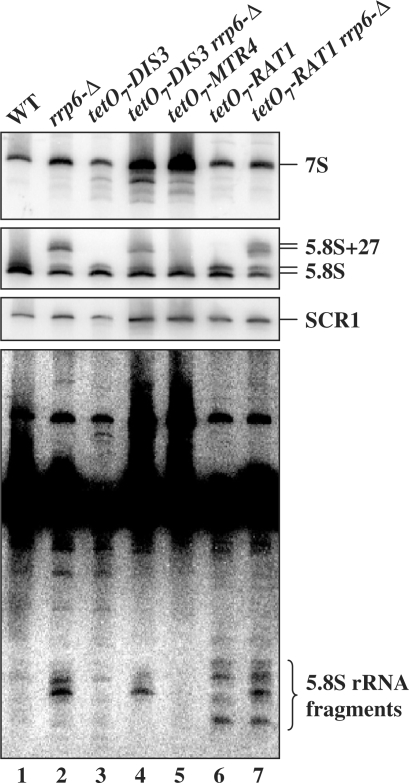
We asked if the specificity of Rrp6p RNA degradation is confined to poly(A)+ RNAs. We analyzed 5.8S rRNA degradation products on northern blots of total RNA from various strains (Figure 4A). We included RNA from an rrp6-Δ strain, and strains depleted of Dis3p, Mtr4p and the 5′–3′ exoribonuclease Rat1p. The blots were probed with OSB156, which hybridizes to the mature portion of 5.8S rRNA, and OSB157, which hybridizes to ITS2 just downstream of 5.8S rRNA (Figure 1). The absence of Rrp6p causes the accumulation of the 3′-extended 5.8S+27 pre-rRNA (Figure 4A, lanes 2, 4 and 7). Depletion of Dis3p and Mtr4p cause the accumulation of intermediates migrating faster than 7S pre-rRNA (Figure 4). Depletion of Rat1p is confirmed by the increase in the ratio of the long to short forms of 5.8S rRNA (Figure 4, lanes 6 and 7). Prolonged exposure of the blot probed with OSB156 revealed truncated 5.8S RNAs that accumulate in the rrp6-Δ and Rat1p depleted strains, but not in the Dis3p or Mtr4p depleted strains (Figure 4). These short RNAs do not bind to oligo(dT), suggesting that they do not carry poly(A) tails (data not shown). These findings suggest that these unadenylated 5.8S rRNAs fragments are degraded independently of the core exosome by the combined action of Rrp6p and Rat1p.
Figure 4.
Rrp6p ΔC2 degrades 5.8S rRNA fragments. Total RNA was isolated from cells with the indicated genotypes grown in YPD at 30°C. The wild-type (WT) and tetO7 cells were treated for 5 h with 10 μg/ml doxycycline before harvest and RNA isolation. The RNA was separated by electrophoresis on an 8% polyacrylamide 8 M urea gel, transferred to a membrane and northern blot analysis carried out using 5′-32P-labeled oligonucleotide probes OSB157 (top panel) and OSB156 (middle and bottom panels). SCR1 is an RNA polymerase III transcript used as a loading control. The bottom panel is a longer exposure of the middle panel.
Rrp6p requires its C-terminal domain for interaction with the core exosome
We constructed deletions of Rrp6p with the goal of identifying domains necessary for its interaction with the core exosome. The crystal structures of a fragment of yeast Rrp6p and the Escherichia coli Rrp6p homologue RNaseD, and amino acid sequence similarity analysis formed the basis for these deletions (33,59,60). Rrp6p has an N-terminal region found in eukaryotic, but not in bacterial homologs. This region is required for interaction of Rrp6p with its co-factor Lrp1p/Rrp47p (61). In addition, the Rrp6p and RNaseD structures indicated three independently folded domains. The first contains three motifs (Figure 5A, ExoI, II, II) that we and others previously showed to be necessary for the exoribonuclease activity of Rrp6p (34,59). The second contains conserved amino acids found in the HRDC domains of enzymes involved in the metabolism of nucleic acids (62). The third domain shows no significant sequence similarity to other RNaseD family members. Nevertheless, in RNaseD this domain folds into a structure very similar to the adjacent, conserved HRDC domain (60). Following the convention of Zuo et al., we refer to these two domains as HRDC1 and HRDC2. Finally, Rrp6p contains two sequences similar to nuclear localization signals (NLS) that we showed previously to play a modest role in localization of Rrp6p to the nucleus, but are unnecessary for other functions of Rrp6p (34). We constructed deletions of these domains, as well as several within the N-terminal region, and expressed them as N-terminal GFP fusions from low copy plasmids in yeast (Figure 5A). We showed previously that GFP-Rrp6p fusions expressed in this manner complement all of the known loss of function phenotypes caused by deletion of the RRP6 gene (34). We analyzed the function of the mutant Rrp6p proteins in a strain carrying a deletion of RRP6 and a C-terminal fusion of Rrp46p to the TAP affinity tag.
Figure 5.
Analysis of the ability of Rrp6p deletion derivatives to co-purify with the core exosome. (A) Polypeptide structure of Rrp6p and the deletion derivatives tested. The relevant regions of Rrp6p are listed at the top of the diagram. (B and C) Western blot analysis of bound proteins after IgG-Sepharose bead purification of core exosomes from cell lysates containing the indicated GFP-Rrp6p derivatives and TAP-tagged Rrp46p as described in Materials and methods section. The antibody used to probe the blots in (B and C) is indicated to the right of each panel. In each case, the ‘bound’ fraction represents a 150-fold enrichment of the ‘Input’ and ‘Free’ fractions.
Western blot analysis showed that all of the proteins were expressed at levels similar to wild type GFP-Rrp6p (Figure 5 and data not shown). The core exosome was purified from lysates of these cells by adsorption of Rrp46-TAP to IgG-Sepharose beads, followed by release of the core exosome complexes with the TEV protease. Western blot analysis of the released core exosomes using antibodies to the core component Rrp4p and to Rrp6p allowed us to determine if Rrp6p deletion derivatives bind the core exosome. One advantage of fusing Rrp6p-deletions to GFP is that it allowed us to use anti-GFP antibody to detect the N-terminal deletion Rrp6p-Δ210, which does not react with the anti-Rrp6p raised to an N-terminal fragment of Rrp6p (63). We included the Rrp6-3p mutant (D238A) mutant, which lacks exoribonuclease activity (34), to determine if loss of this activity interferes with interaction with the core exosome. The results show that all of the Rrp6p deletions bind the core exosome, except the ΔC1+2 and ΔC2 (Figure 5B and C). In some experiments, Rrp6p bands appear as doublets, apparently due to proteolysis. In the cases of the ΔC1+2 and ΔC2 mutants, Rrp6p was not detected in the bound fractions, despite significant enrichment of the core component Rrp4p (Figure 5C). We conclude that removal of the Rrp6p C-terminal domain causes a significant decrease in its ability to interact with the core exosome.
Previous studies showed that deletion of RRP6 results in a temperature sensitive growth phenotype (21). Interestingly, mutations in the exonuclease motifs that inactivate the RNA processing and degradation activities of Rrp6p still allow growth of the mutant strains at high temperature suggesting that Rrp6p possesses some uncharacterized activity necessary for growth at high temperature (34). We tested some of the deletion mutants for their ability to complement this growth defect and found that the Δ210 and ΔHRDC1 proteins allow significantly more growth at 37°C than the GFP negative control (Supplementary Figure S1). The ΔC1+2 mutation grows no better than the negative control suggesting the absence of some critical activity and/or that the protein may be misfolded and completely inactive. The ΔC2 mutant complements the temperature sensitivity quite well. This finding suggests that the temperature sensitive phenotype of an rrp6-Δ strain does not result from the lack of Rrp6–core exosome interaction.
RNA 3′-end formation does not require Rrp6p interaction with the core exosome
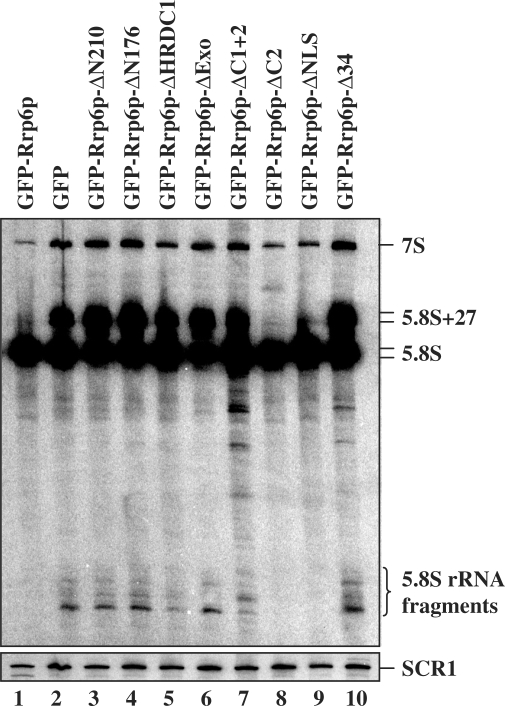
Next, we examined RNA processing (3′-end formation) associated with defects in Rrp6p. Previous studies showed that deletion of RRP6, or mutations inactivating its exoribonuclease activity, resulted in defects in the 3′-end processing of 5.8S rRNA and a variety of sn-/snoRNAs (34,35,57,64). We analyzed 5.8S rRNA and the snoRNAs snR72 and U24 in rrp6-Δ cells expressing each of the deletion derivatives of Rrp6p (Figure 6A, E and F). We included the previously characterized ΔNLS mutant as a positive control since it behaves normally for all tested RNA phenotypes, despite partial cytoplasmic localization (34). The results showed the expected 3′ extensions on 5.8S rRNA, snR72 and U24 in the strain expressing GFP only, and GFP-Rrp6p complements these defects (Figure 6B, E and F, compare lanes 1 and 2). All of the Rrp6p domain deletions except ΔC2 and ΔNLS show defects in 5.8S rRNA 3′-end processing similar to that observed in the absence of Rrp6p. These two deletion mutants also show normal 3′-end processing of snR72 and U24 RNAs. In contrast, all of the other deletion mutants, except ΔHRDC1, show defects in snR72 and U24 processing. The lack of an effect of the ΔHRDC1 mutation on snRNA 3′-end processing agrees with previous studies showing that a mutation in this region (rrp6-13; D457A) caused snRNA 3′-end formation defects for some, but not all snRNAs tested (34,59). Importantly, the ΔC2 mutation, which disrupts binding of Rrp6p to the core exosome, shows normal 3′-end formation for 5.8S rRNA and snRNAs. These findings indicate that deletion of the C-terminal domain does not inhibit the exoribonuclease activity of Rrp6p. We conclude that disruption of Rrp6p–core exosome interaction does not significantly inhibit the RNA 3′-end formation activity of Rrp6p.
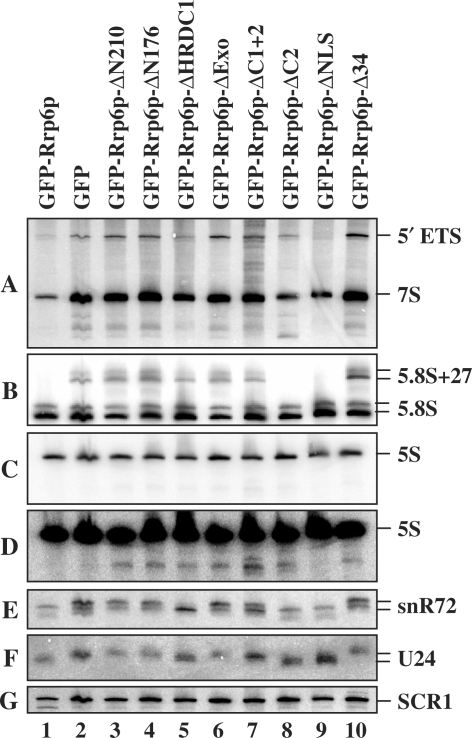
Figure 6.
Analysis of RNA processing and degradation phenotypes in strains expressing Rrp6p deletion derivatives. Total RNA was prepared from an rrp6-Δ strain expressing the indicated Rrp6p mutant, the RNA was separated by electrophoresis on an 8% polyacrylamide 8 M urea gel, transferred to a membrane and northern blot analysis carried out using various 5′-32P-labeled oligonucleotide probes. Each panel shows the result of probing the same blot with different labeled oligonucleotides; (A) OSB153 and OSB157, (B) OSB156, (C) OSB23, (D) longer exposure of (C), (E) OSB267, (F) OSB138 and (G) OSB151. The identity of the RNAs is listed to the right of each panel. SCR1 is an RNA polymerase III transcript used as a loading control.
Loss of the Rrp6p C-terminal domain inhibits the degradation of some rRNA processing products
Rrp6p also plays a role in the degradation of rRNA processing byproducts and intermediates (7,32,39). One by-product is the 5′-ETS released after cleavage of the rRNA precursor at site A0 (Figure 1). This RNA is normally degraded by the action of the core exosome and Rrp6p (Figures 2 and 3) (32). Analysis of the levels of 5′-ETS in the Rrp6p deletion mutants shows that all of the mutations, except ΔNLS, elevate 5′-ETS levels (Figure 6A). The fact that the ΔC2 mutation results in the accumulation of 5′-ETS RNA suggests that disruption of Rrp6p interaction with the core exosome hampers the ability of Rrp6p to efficiently degrade this by-product of rRNA processing.
Kadaba et al. (40) showed that defects in Rrp6p and the core exosome component Dis3p result in the accumulation of a 3′ truncated form of 5S rRNA. This 5S rRNA fragment appears to be an intermediate in the exosome degradation pathway since it accumulates in the absence of Trf4p, which activates RNAs for degradation by the core exosome and Rrp6p. We observe this 5S intermediate in all of the Rrp6p deletion mutants except the ΔNLS (Figure 6C and D; note that 5d is a longer exposure than 5c). In the case of the ΔC2 domain, we also observe a unique RNA fragment migrating faster than 7S pre-rRNA (Figure 6A; lane 8). Primer extension analysis of this fragment does not reveal new products compared to wild-type or other depletion mutants suggesting that its is shorter at its 3′- rather than its 5′-end (Supplementary Figure S2). The fact that the ΔC2 mutant fails to degrade these RNAs suggests that their disposal requires normal affinity of Rrp6p for the core exosome, or that recognition of these intermediates requires some other function of the C-terminal domain.
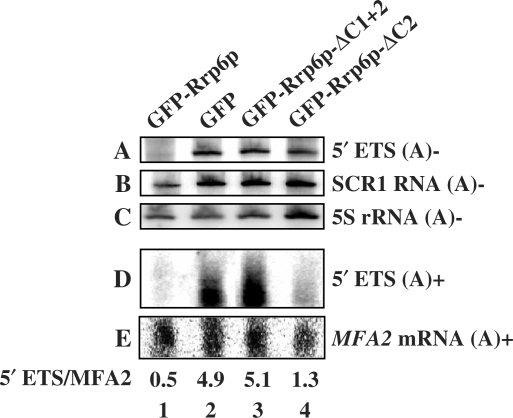
Previous studies showed that a fraction of 3′ truncated 5S and 5′-ETS RNAs accumulate in exosome mutants as polyadenylated species reflecting activation of their degradation by the TRAMP complex (39,40). Our analysis shows that poly(A)+ 5′-ETS RNA does not accumulate in the ΔC2 mutant to the same degree seen in the absence of Rrp6p, suggesting that efficient removal of the poly(A) tail does not require the interaction of Rrp6p with the core exosome (Figure 7). In contrast, the poly(A)- form of 5′-ETS accumulates in the ΔC2 mutant to the same extent as that seen in the absence of Rrp6p (Figure 7).
Figure 7.
Rrp6p ΔC2 removes the poly(A) tail from 5′ETS RNA. Northern blot analysis of RNA from an rrp6-Δ strain (YSB232) expressing the indicated Rrp6p deletion derivatives. The RNA was separated into poly(A)- (top three panels) and poly(A)+ (bottom two panels) fractions prior to northern blot analysis with 5′-32P OSB153 (A), OSB151 (B), OSB23 (C), OSB153 (D) and a random-primed hexamer 32P-labeled probe to MFA2 mRNA (E).
Loss of the Rrp6p C-terminal domain does not affect degradation of Rrp6p-specific RNAs
Next, we asked whether the degradation of 5.8S fragments requires the interaction of Rrp6p with the core exosome. The fragments appear in all of the Rrp6p deletion mutants except for the ΔC2 and ΔNLS (Figure 8). Their absence in the ΔC2 mutant supports the conclusion that Rrp6p degrades them, despite its reduced affinity for the core exosome.
Figure 8.
Total RNA was isolated from an rrp6-Δ strain (YSB232), grown at 30°C in SCD–URA–MET media, which carried plasmids that express each of the indicated Rrp6p mutants. The RNA was separated by electrophoresis on an 8% polyacrylamide 8 M urea gel, transferred to a membrane and northern blot analysis carried out using 5′-32P-labeled oligonucleotide probes OSB157 (top panel) and OSB151 (bottom panel). SCR1 is an RNA polymerase III transcript used as a loading control.
Finally, we investigated whether disruption of Rrp6p–core exosome interaction interferes with the degradation of poly(A)+ rRNAs that accumulate in the absence of Rrp6p (Figure 2B). Northern blot analysis of oligo(dT)-selected rRNAs shows that these transcripts accumulate in strains lacking the N-terminus of Rrp6p, its HRDC1 or C1+2 domains (Figure 9) In contrast, they do not accumulate in the absence of C-2. These findings support the conclusion drawn from the analysis of the effects of depleting Dis3p in absence of Rrp6p (Figure 2B), that the degradation of these poly(A)+ RNAs by Rrp6p occurs without significant influence by the core exosome.
Figure 9.
Rrp6p ΔC2 degrades poly(A)+ rRNAs. Northern blot analysis of oligo-(dT)-selected RNAs from an rrp6-Δ strain expressing the indicated Rrp6p deletion derivatives. The RNA was separated in to poly(A)+ and poly(A)- fractions and the poly(A)+ analyzed by northern blotting with the indicated 5′-32P-labeled oligonucleotide probes.
DISCUSSION
The experimental results reported here shed light on the question of whether the activity of the exoribonuclease Rrp6p requires physical association with the core exosome. Our findings indicate that Rrp6p functions in a core exosome-independent manner for some activities in vivo. Previous studies that analyzed polypeptides associated with affinity purified core exosomes showed that Rrp6p co-purifies with these complexes (48,49,65,66). Likewise, affinity purification of yeast Rrp6p in some of these studies co-purified the core exosome components (25,50). However, these studies did not address the relative stoichiometries of the proteins or the possibility that some of these proteins might exist in sub-complexes or free pools of individual subunits. More recent analyses in human cells, plants and flies suggested that functionally important sub-complexes of core exosome subunits may exist (67–69). Initial evidence for functional independence of Rrp6p from the core exosome comes from the fact that the core exosome functions prior to Rrp6p in the 3′-end processing of 5.8S rRNA and many snRNAs and snoRNAs (20,33,35,57,70). Indeed, our results show that deletion of the C-terminal domain of Rrp6p significantly impairs it ability to interact with the core exosome, but has no effect on these 3′-end formation activities. Moreover, Dis3p, the active component of the core exosome, and Rrp6p exhibit significant differences in their exoribonuclease activities. Dis3p appears to degrade RNA processively, while Rrp6p uses a more distributive mode (15,17,21). Rrp6p also showed better activity than Dis3p in the removal of poly(A) tails from model substrates in vivo (15). Interestingly, the interaction of Dis3p with the rest of the core proteins significantly inhibited its activity relative to free Dis3p, while Rrp6p showed similar activity in the presence or absence of the core exosome.
The findings reported here suggest that Rrp6p may function in vivo without direct interaction with the core exosome. Deletion of the C-terminal domain of Rrp6p disrupts its ability to co-purify with the core exosome. Nevertheless, the mutant enzyme retains its exoribonuclease activity as shown by the fact that it carries out Rrp6p-specific 3′-end processing of 5.8S rRNA and snoRNAs. In contrast, the ΔC2 mutant fails to efficiently degrade some rRNA intermediates that appear to require the action of Rrp6p and the core exosome. For example, the 5′-ETS by-product of rRNA processing accumulates in rrp6- mutants and in strains depleted of core exosome components, and combination of these defects causes a synergistic increase in 5′-ETS levels (Figures 2, 3 and 5) (20,34). The ΔC2 mutant fails to degrade this RNA suggesting that efficient degradation requires physical interaction between Rrp6p and the core exosome, or that the C-terminal domain plays a specific role in the recognition of this RNA. The 5′-ETS RNAs also accumulate as poly(A)+ forms in Rrp6p- and core exosome-deficient cells implying that the TRAMP complex polyadenylates them prior to degradation by Rrp6p and the core exosome [Figures 2 and 7, (39)]. However, poly(A)+ forms of 5′-ETS do not accumulate to a significant extent in the ΔC2 mutant, suggesting that Rrp6p removes their poly(A) tails in an core exosome-independent manner (Figure 7, lane 4). We observed similar results for the 3′ truncated form of 5S rRNA, shown previously to require Rrp6p, TRAMP and the core exosome for its degradation (data not shown) (40). Thus, in the cases where Rrp6p and the core exosome co-operate to degrade an RNA, it appears that Rrp6p may remove the poly(A) tails in an exosome-independent manner and then participate with the core exosome in the degradation of the body of the transcript.
Our experiments also identified rRNA processing intermediates that appear to be degraded by Rrp6p in a core exosome-independent manner. These poly(A)+ RNAs (27SA2, 23S, 21S, 17S, 5′-S and D-B1L) accumulate in the absence of Rrp6p and in certain Rrp6p deletion mutants, but not upon depletion of core exosome components (Figures 2, 3 and 8). Moreover, while the depletion of Dis3p in an rrp6-Δ strain results in a synergistic enhancement of the accumulation of some poly(A)+ rRNAs, these Rrp6p-specific transcripts are unaffected (Figure 3). Thus, unlike the 5′-ETS and 3′ truncated 5S rRNAs, these intermediates seem to require only Rrp6p for their degradation. Indeed, they do not accumulate in the ΔC2 mutant suggesting that their degradation does not require the participation of the core exosome. However, the detection of these rare intermediates requires that they bind to oligo(dT) and therefore carry poly(A) tails. Thus, we cannot exclude the possibility that their presence in any of our experiments might reflect the inability of Rrp6p mutants to remove the tails rather than their inability to degrade the body of the RNA. Regardless of the mechanism, the fact that these RNAs do not accumulate in the ΔC2 mutant indicates that disruption of Rrp6p-core exosome interaction does not affect their degradation.
Analysis of unadenylated degradation intermediates of 5.8S rRNA also showed that Rrp6p degrades these RNAs in a core exosome-independent manner. The hydrolysis of these RNAs requires Rrp6p, but not the core exosome or the TRAMP complex (Figure 7A). The fact that these intermediates do not appear in the ΔC2 mutant indicates that their degradation may not require the interaction of Rrp6p with the core exosome. Notably, these RNAs do not appear in poly(A)+ fractions of RNAs fractionated by oligo(dT) indicating that they do not carry significant poly(A) tails (data not shown). Thus, unlike the RNAs discussed above, they may never have entered, or they may have already exited, the TRAMP-dependent RNA degradation pathway.
The conclusion that the C-terminal portion of Rrp6p plays a critical role in its interaction with the core exosome agrees with experiments on the structure of the human exosome. Two-hybrid system studies of the interaction of human Rrp6p (PMScl-100) with other core exosome components indicated that the C-terminal 276 amino acids of hRrp6p interact with hRrp43 (Oip2) (71). This fragment includes a region corresponding to the E. coli HRDC2 domain and shows significant similarity between the yeast and human proteins (33,60). Thus, the human interaction data support our conclusion that the C-terminal domain plays an important role in the interaction of Rrp6p with the core exosome.
In summary, the findings reported here provide evidence that Rrp6p may act independently of the core exosome during the degradation of some rRNA processing products. Structure-function analysis of Rrp6p revealed that it requires its C-terminal domain for interaction with the core exosome. When this interaction is disrupted, Rrp6p continues to carry out critical nuclear RNA 3′-end processing and degradation steps, yet fails to degrade RNAs that appear to require the combined action of Rrp6p and the core exosome. These results suggest a functional sub-specialization of core exosome and Rrp6p functions in the nucleus.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: NSF grant MCB-0817324 to JSB.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank David Tollervey and Phil Mitchell for providing antibodies, and Amir Taslimi for help generating Rrp6p deletions. We are grateful to the members of our laboratory for stimulating discussions.
REFERENCES
- 1.Moore MJ. Nuclear RNA turnover. Cell. 2002;108:431–434. doi: 10.1016/s0092-8674(02)00645-1. [DOI] [PubMed] [Google Scholar]
- 2.Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 3.Geerlings TH, Vos JC, Raue HA. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′–>3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 6.Lee CY, Lee A, Chanfreau G. The roles of endonucleolytic cleavage and exonucleolytic digestion in the 5′-end processing of S. cerevisiae box C/D snoRNAs. RNA. 2003;9:1362–1370. doi: 10.1261/rna.5126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang F, Phillips S, Butler JS. Rat1p and Rai1p function with the nuclear exosome in the processing and degradation of rRNA precursors. RNA. 2005;11:1571–1578. doi: 10.1261/rna.2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilleren PJ, Parker R. Cytoplasmic degradation of splice-defective pre-mRNAs and intermediates. Mol. Cell. 2003;12:1453–1465. doi: 10.1016/s1097-2765(03)00488-x. [DOI] [PubMed] [Google Scholar]
- 9.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′–>5′ degradation. Mol. Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 11.Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butler JS. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. doi: 10.1016/s0962-8924(01)02225-5. [DOI] [PubMed] [Google Scholar]
- 13.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 14.Buttner K, Wenig K, Hopfner KP. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 16.Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- 17.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 18.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–>5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 20.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ –> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burkard KT, Butler JS. A nuclear 3′–5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 23.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 26.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J. Cell Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- 28.Anderson JSJ, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex [In Process Citation] EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown JT, Johnson AW. A cis-acting element known to block 3′ mRNA degradation enhances expression of polyA-minus mRNA in wild-type yeast cells and phenocopies a ski mutant. RNA. 2001;7:1566–1577. [PMC free article] [PubMed] [Google Scholar]
- 30.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 31.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 32.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 34.Phillips S, Butler JS. Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA. 2003;9:1098–1107. doi: 10.1261/rna.5560903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- 37.Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- 38.Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell. 2002;9:1285–1296. doi: 10.1016/s1097-2765(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 39.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 42.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 43.Das B, Butler JS, F. S. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell Biol. 2003;16:5502–5512. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuai L, Das B, Sherman F. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:13962–13967. doi: 10.1073/pnas.0506518102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schilders G, van Dijk E, Raijmakers R, Pruijn GJ. Cell and molecular biology of the exosome: how to make or break an RNA. Int. Rev. Cytol. 2006;251:159–208. doi: 10.1016/S0074-7696(06)51005-8. [DOI] [PubMed] [Google Scholar]
- 47.Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 49.Krogan NJ, Peng WT, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 50.Estevez AM, Kempf T, Clayton C. The exosome of Trypanosoma brucei. EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patel D, Butler JS. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell Biol. 1992;12:3297–3304. doi: 10.1128/mcb.12.7.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 54.Fang F, Hoskins J, Butler JS. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell Biol. 2004;24:10766–10776. doi: 10.1128/MCB.24.24.10766-10776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrick D, Parker R, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell. 2007;26:217–229. doi: 10.1016/j.molcel.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc. Natl Acad. Sci. USA. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005;13:973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 61.Stead JA, Costello JL, Livingstone MJ, Mitchell P. The PMC2NT domain of the catalytic exosome subunit Rrp6p provides the interface for binding with its cofactor Rrp47p, a nucleic acid-binding protein. Nucleic Acids Res. 2007;35:5556–5567. doi: 10.1093/nar/gkm614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem. Sci. 1997;22:417–418. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 63.Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell Biol. 2003;23:6982–6992. doi: 10.1128/MCB.23.19.6982-6992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Briggs MW, Butler JS. RNA polymerase III defects suppress a conditional-lethal poly(A) polymerase mutation in Saccharomyces cerevisiae. Genetics. 1996;143:1149–1161. doi: 10.1093/genetics/143.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 66.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 67.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 68.Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol. Biol. Cell. 2006;17:1399–1409. doi: 10.1091/mbc.E05-08-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lehner B, Sanderson CM. A protein interaction framework for human mRNA degradation. Genome Res. 2004;14:1315–1323. doi: 10.1101/gr.2122004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.