Abstract
The homeotic Abdominal-B (Abd-B) gene expression depends on a modular cis-regulatory region divided into discrete functional domains (iab) that control the expression of the gene in a particular segment of the fly. These domains contain regulatory elements implicated in both initiation and maintenance of homeotic gene expression and elements that separate the different domains. In this paper we have performed an extensive analysis of the iab-6 regulatory region, which regulates Abd-B expression at abdominal segment A6 (PS11), and we have characterized two new polycomb response elements (PREs) within this domain. We report that PREs at Abd-B cis-regulatory domains present a particular chromatin structure which is nuclease accessible all along Drosophila development and both in active and repressed states. We also show that one of these regions contains a dCTCF and CP190 dependent activity in transgenic enhancer-blocking assays, suggesting that it corresponds to the Fab-6 boundary element of the Drosophila bithorax complex.
INTRODUCTION
The three homeotic genes of the Drosophila bithorax complex (BX-C), Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B), are responsible for specifying the identity of the posterior parasegments of the fly. The expression of these genes is regulated by a complex cis-regulatory region covering 300 kb of DNA divided into nine segment/parasegment-specific subregions, which control expression of a single homeotic gene in a particular parasegment. Different studies have led to the identification of specific cis-regulatory sequences responsible for establishment (initiator elements) and maintenance polycomb response element (PRE regulatory elements) of homeotic gene expression. Early in development, gap and pair-rule genes initiate parasegment-specific patterns of expression which are then maintained by two ubiquitously expressed groups of proteins, the polycomb group (PcG) and the trithorax group (TrxG), responsible for the silenced and activated states respectively. Both groups of proteins are thought to maintain the active or inactive state by modifying the chromatin structure of each cis-regulatory domain (1–3). A special class of cis-acting elements known as boundary elements or insulators are proposed to exist between each iab domain to allow their autonomy to properly specify segmental identity. Until now, only three of these elements, Miscadastral pigmentation (Mcp), Frontabdominal-7 (Fab-7) and Frontabdominal-8 (Fab-8), have been functionally identified (4–8), and the characterization of genomic deficiencies covering part of the iab-5 and iab-6 cis-regulatory domains has allowed to infer the existence of a putative Fab-6 boundary element (9). Although proteins responsible for the boundary function of these elements remain largely unknown, it has been reported that some insulator binding proteins, such as GAGA, dCTCF and CP190 are associated in vivo with some of these elements(10–14) and contribute to the enhancer-blocking activity (10,11,15).
Here, we report on the identification of two new PREs in the Abd-B iab-6 regulatory region and on the characterization of a dCTCF-CP190-dependent activity associated to one of them. Overall, our work constitutes an extensive analysis of the regulatory elements in the iab-6 domain of the BX-C and provides evidence that a boundary activity, always located near a PRE, exists between each of the cis-regulatory domains that control Abd-B expression.
MATERIALS AND METHODS
Transgenic constructs and Drosophila stocks
The enhancer of the white gene was obtained by PCR using the following primers: 5′AAGTCAACCCAGACCAACC3′ and 5′CACTTTCCCCTGCTTACCC3′ and, after digestion with EcoRV, the 1.6 kb fragment which includes the eye enhancer was cloned into a pCaSpeR vector containing the mini-white reporter gene. The following DNA fragments were cloned in between enhancer and promoter: from 3R:12724265 to 3R:12726608 for the B+P7 construct, from position 3R:12707732 to 3R:12708886 for the HS1 construct; from position 3R:12711412 to 3R:12713034 for HS2+3; from 3R:12711833 to 3R:12712760 for HS2, from 3R:12712760 to 3R:12713610 for HS3 and from 3R:12708102 to 3R:12708620 flanked by FRT sites for HS1*.
Fly strains and heat-shock treatments
Drosophila mutant alleles of PcG proteins, Psch27, Pc3 and ph-p410, were obtained from the Bloomington Stock Center. dCTCFy+6 and CP190H31–2 mutant flies (11,16) were obtained from V. G. Corces.
The enhancer-blocking assays in mutant backgrounds were performed using double balanced lines to obtain flies heterozygous for the transgen that carry the mutation in either heterozygous or homozygous states on a different chromosome for comparison to heterozygous transgenic flies.
Lines with HS1* excisions were obtained by crossing females expressing flp recombinase under the control of the heat-shock promoter (p[hsFLP],y1,w1118; Dr[Mio]/TM3,ry*,Sb1 from the Bloomington Stock Center) with transgenic males. Heat-shock of embryos ≤ 24 h old were carried out for 1 h/every day. In the following generation flies were selected for a change in eye colour and excisions were confirmed by PCR analysis. For each transgenic line the eye colour of heat-shocked and non-heat-shocked flies were compared.
DNAseI assays
Nuclei extraction from Drosophila embryos and third instar larvae were performed as described before (12,17). About 30 µl of embryos and 20 larvae were used per sample and DNAseI digestions were performed with increasing amounts of enzyme (0–10 U for embryo and 0–50 U for larvae nuclei) for 3 min at 4°C. DNA samples were cleaved and labelled as indicated in the figure legends. Samples restricted with KpnI were hybridized with probe 1 (from position 3R:12717132 to 3R:12717707); samples restricted with HindIII with probe 2 (from position 3R:12713610 to 3R:12714662) and samples restricted with PstI (3R:12711182)/SacI (3R:12714662) were hybridized with probe 3 (from position 3R:12711233 to 3R:12711648).
ChIP assays
Dechorionated embryos 4–20 h old were crosslinked for 15 min in 50 mM Hepes pH 7.9, 1 mM EDTA, 0.8 mM EGTA and 100 mM NaCl with 2.5% formaldehyde and 3 v/v heptane. Embryos were resuspended in lysis buffer (1% SDS, 50 mM Tris pH 8, 10 mM EDTA, protease inhibitors) and sonicated in a Branson sonifier set at 35% output, 20 s for 4 times. Subsequent steps were done as previously described (12) except that immunoprecipitations were carried out in RIPA buffer (1% Triton-X100, 0.1% Na Deoxycholate, 0.1% SDS, 140 mM NaCl, 10 mM Tris HCl pH 8, 1 mM EDTA, 1 mM PMSF). Antibodies against Pc were purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA (sc-25762). PCR amplification of the resulting DNA was performed by standard procedures using the following primers: PRE7, 5′GTCGGCAATTCGGATTCCC3′ and 5′TTCGGTCGCTCACGTCGC3′; HS1, 5′TTTATCTTGCGATGGCTGC3′ and 5′GAAGATGACTAAATCCAATGC3′; HS2, 5′ATTCAAATTCCGTTAGGTGC3′ and 5′ATCTACTGGGTGTGCGAGG3′; control, 5′GTGCGATAGGATTGCTGC3′ and 5′GTTGGGGTACACGACAGC3′, corresponding to the dsap18 promoter. The amplified DNA was separated on 1% agarose gels and visualized by using ethidium bromide. Immunoprecipitated DNA was also analysed by Real-time qPCR (Thermocycler ABI 7000) using SYBR Green and standard settings (Applied Biosystems, Foster City, CA, USA). Immunoprecipitation reactions from two independently generated chromatin samples were performed and for each PCR fragment, the amount of DNA in the immunoprecipitated material was expressed as percentage of DNA present in the input material. Average values with the corresponding standard deviations are shown in the graph. Primers used are: PRE7, 5′GCAGCCATCATGGATGTGAA3′ and 5′GGAATACCGCACTGTCGTAGG3′; HS1, 5′GCGTGCGTTTATCTTGCGAT3′ and 5′GCAGCTAAACCCGATTTGCTT3′; HS2 5′GAATCGAAAACTCACGTAGCA3′ and 5′TGGTTGCTACTTTACTTGCGC3′; Control, 5′ACCTGAACCGTCTGATTGGC3′ and 5′GCAGAGAGGCGGTAATCGAG3′, corresponding to the α-tubulin gene.
RESULTS
Identification of DNAseI hypersensitive sites within the iab-6 domain of the BX-C
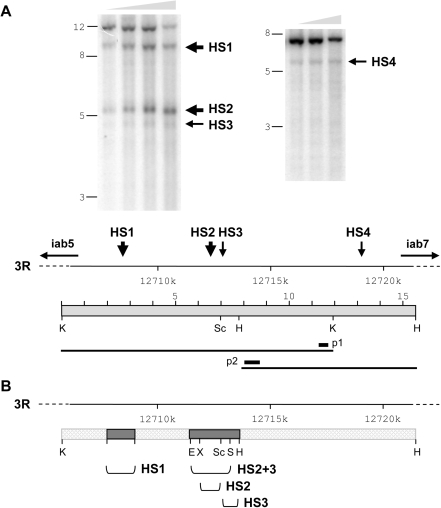
Regulatory regions that control the expression of eukaryotic genes are frequently hypersensitive to digestion with nucleases (18,19), as it has been shown for some regulatory elements implicated in homeotic gene expression (4,20,21). Since mapping DNAseI hypersensitive (HS) sites could be an accurate method for identifying the location of functional regulatory elements, we have analysed the accessibility to DNAseI over the whole regulatory iab-6 domain of the BX-C locus that controls Abd-B expression in the parasegment 11/abdominal segment 6 (PS11/A6). DNAseI digestion of Drosophila embryos revealed the existence of two major DNAseI HS sites (indicated by big arrows in Figure 1A) located around positions 3R:12709k (HS1) and 3R:12713k (HS2) along the BX-C sequence and two minor HS sites (indicated by small arrows in Figure 1A), HS3 located near the major HS2 site and HS4 located around position 3R:12719k. The scheme presented in Figure 1A shows the position, along the cis-regulatory iab-6 domain, of the different HS sites here characterized. Several types of regulatory elements implicated in homeotic gene regulation have been identified in the past that include initiator elements, maintenance elements (PREs), chromatin boundaries and promoter targeting sequences (1–3). So far, only one regulatory element has been characterized within the iab-6 cis-regulatory domain, which corresponds to an initiator element that shows a maintenance activity at later stages of embryogenesis (9). Since this element has been shown to be located between positions 3R:12716438 and 3R:12719280, we speculated that HS4 probably corresponds to the IAB6 initiator element previously characterized.
Figure 1.
DNAseI hypersensitive sites in the iab-6 cis-regulatory domain. (A) DNA samples from DNAseI digests of 0–16 h embryo nuclei were restricted with either KpnI (left panel) or HindIII (right panel). Grey triangle on top indicates increasing amounts of DNAseI. Position of molecular markers in kb is indicated on the left. DNAseI HS sites are indicated by arrows. A schematic presentation of the iab-6 region is drawn below. Position of KpnI (K), ScaI (Sc) and HindIII (H) restriction sites, fragments used as hybridization probes (p1 and p2) and HS sites are indicated. (B) Position and length of fragments used in transgenic constructs are shown.
We have analysed the genomic regions containing the other hypersensitive sites here identified, HS1, HS2 and HS3, and we have found that they contain several consensus binding sequences for proteins known to be implicated in the maintenance phase of homeotic gene regulation (data not shown), such as the GCCAT sequence which is found at most pleiohomeotic binding sites and the GAGAG consensus sequence recognized by the GAGA factor (22,23). Thus, we asked whether these genomic regions correspond to regulatory elements implicated in maintenance of Abd-B expression.
Analysis of the PRE activity of iab-6 HS sites
To analyse the capacity of a DNA sequence to have PRE activity a P-element transgene whiteenhancer-mini-white has been used in the past (4,23,24). It has been shown that in the absence of a regulatory element the white enhancer drives a high level of white expression in the eye, giving a red eye colour. However, when a PRE is inserted between enhancer and promoter, it acts as a silencer and transgenic flies show repression of white and pairing-dependent silencing (silencing is enhanced in the presence of two copies of the transgene).
We used a similar whiteenhancer-mini-white construct to analyse the presence of a PRE in the different HS regions identified above. First, as a control, we inserted a 2.3 kb fragment with the previously characterized Fab-7 region containing both the boundary and PRE elements between the white enhancer and the mini-white reporter gene. When this construct was introduced into flies, 50% of the transgenic lines obtained showed silencing in heterozygous animals and 60% showed pairing-dependent silencing (Figure 2A). The percentage of silencing and pairing-sensitive silencing obtained in our assays was similar to that reported earlier for constructs carrying the Fab-7 or Fab-8 elements of the BX-C (4,24). Then, we introduced into flies constructs containing the different HS sites here identified in the iab-6 regulatory domain and tested for silencing and pairing-sensitive silencing.
Figure 2.
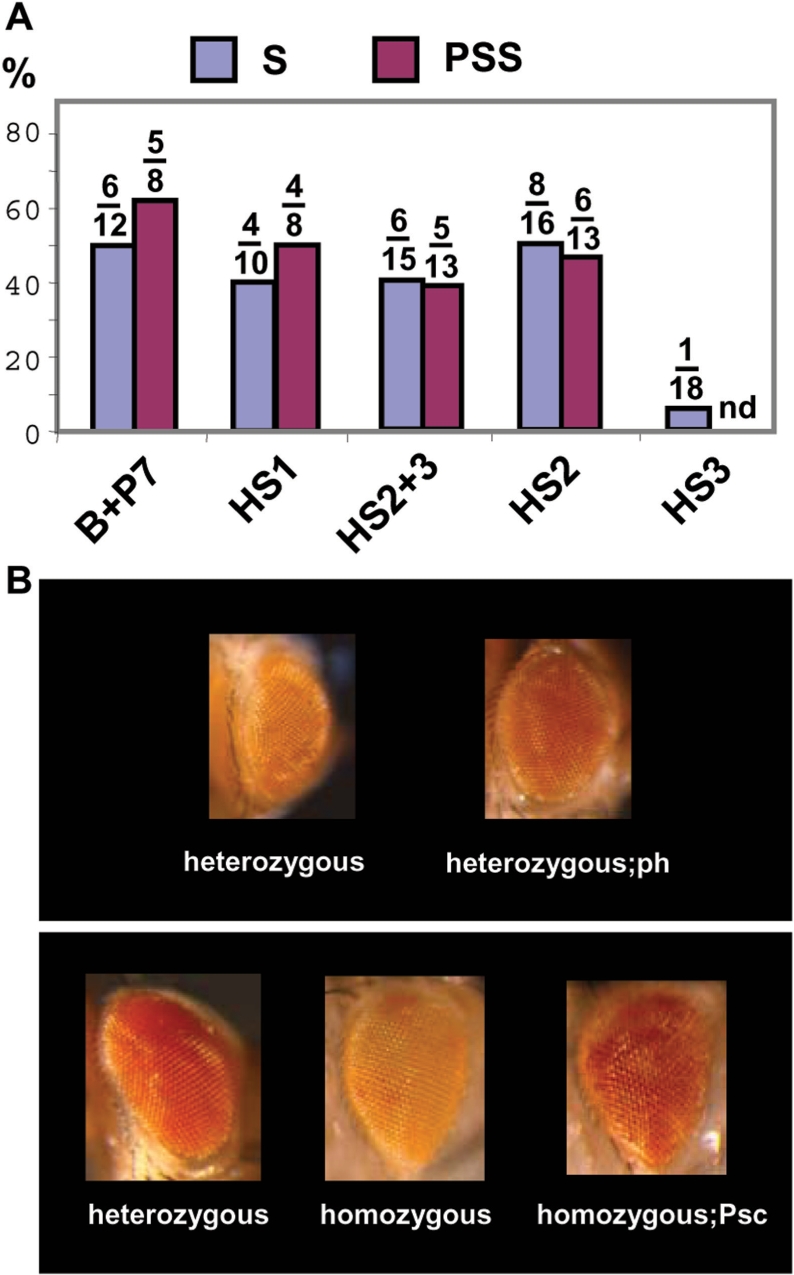
HS1 and HS2 fragments contain PRE activities. (A) The percentage of transgenic lines showing silencing (S) and pairing-sensitive silencing (PSS) for the different constructs is presented in the graphic. The number of independent transformant lines out of the total generated that show reduced white expression is presented on top of each bar. (B) Eye colour of representative flies of transgenic lines HS2+3 4.2 (top) and 3.1 (bottom) in mutant backgrounds as indicated below.
When a 1.15 kb PCR fragment covering the distal HS1 site (see Figure 1B, HS1 construct) was inserted, 40% of the transgenic lines obtained showed silencing of white expression, heterozygous animals having orange or yellow eye colour (Figure 2A). Moreover, flies homozygous for the inserted transgene showed pairing-dependent repression of the white reporter gene in 50% of the lines (Figure 2A). Similar results were obtained with a 1.6 kb EcoRI-SacII restriction fragment covering both HS2 and HS3 hypersensitive sites (see Figure 1B, HS2+3 construct). In this case, 40% of the transgenic lines obtained had orange or yellow eyes and 40% of the lines showed enhanced silencing in homozygous animals when compared to their heterozygous siblings (Figure 2A), suggesting that these HS regions correspond to PREs.
To test whether silencing of the white reporter gene by the HS fragments depends on PcG genes, transgenic lines were placed in polyhomeotic, Polycomb and posterior sex combs mutant backgrounds. At least three different transgenic lines of both HS1 and HS2+3 constructs showed increased white expression in more than one mutant background. This is illustrated in Figure 2B for polyhomeotic (ph) and posterior sex combs (Psc) in representative lines showing repression of white and pairing-dependent repression respectively (transgenic lines HS2+3 4.2 and 3.1).
To further characterize the genomic region containing HS2 and HS3 sites, we took advantage of a ScaI restriction site located at position 3R:12712760 (see Sc in Figure 1). Since this site is flanked by both nuclease hypersensitive regions (data not shown), we inserted either a 0.93 kb XbaI-ScaI fragment containing the major HS2 site or a 0.85 kb ScaI-HindIII fragment that includes the minor HS3 site in the whiteenhancer-mini-white construct (see Figure 1B, HS2 and HS3, respectively). We examined 18 independent lines for construct HS3 but only one showed repression of white (Figure 2A) probably as a consequence of the genomic location of the transgene. These results indicate that the HS3 region is not important for PRE activity and further demonstrate that white expression in whiteenhancer-mini-white constructs is almost no dependent on the genomic location of the transgene. On the contrary, the eye phenotype of transgenic lines carrying the 0.93 kb HS2 fragment is similar to that of the lines containing the 1.6 kb HS2+3 fragment (compare HS2 and HS2+3 in Figure 2A). Furthermore, silencing in transgenic HS2 lines depend on Polycomb protein (data not shown). Thus, our results show that the HS2 fragment is necessary and sufficient for PRE function and that sequences covering the minor HS3 site are dispensable, showing no activity in these assays. Overall, our data indicate that these two regions HS1 and HS2 correspond to PREs.
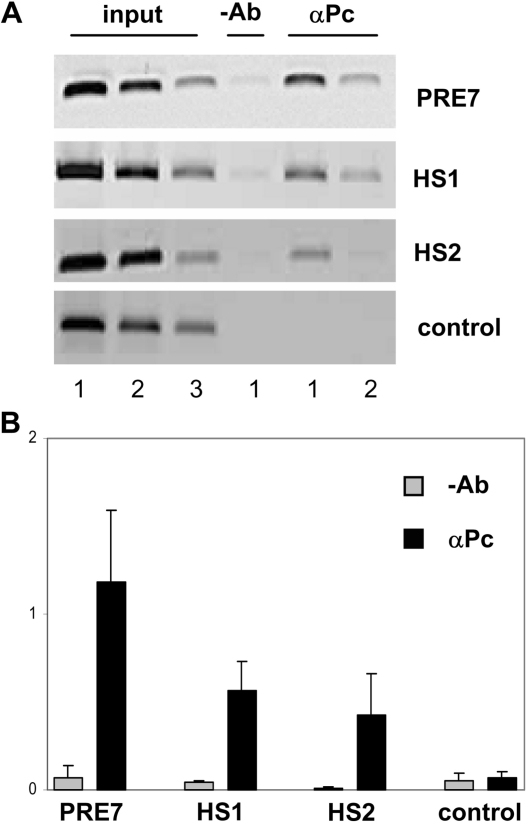
The distribution of PcG proteins over the entire BX-C complex has been analysed in Drosophila cultured cells and binding of several components of the PRC1 complex to a region between Mcp and Fab-7 has been reported (25,26). Sequences corresponding to the HS1 region were identified as PRC1 and Trx binding sites and a Trx site that could correspond to HS2 was shown to be present in cell lines that express Abd-B (26). To determine the in vivo binding of PcG proteins to these regions in Drosophila embryos, we performed ChIP experiments using a Polycomb-specific antibody. Immunoprecipitated DNA was analysed by either standard PCR procedures or real-time qPCR (Figure 3A and B, respectively). As shown in Figure 3, while a negative control sequence did not show any Polycomb association, Pc binding to both HS1 and HS2 regions was similar to that obtained for a previously characterized PRE of the BX-C (PRE7). In conclusion, we have identified two new PREs within the iab-6 cis-regulatory domain of the BX-C.
Figure 3.
Polycomb is associated with iab-6 PREs in Drosophila embryos. Cross-linked chromatin was immunoprecipitated with a Polycomb antibody (Pc) or no antibody (–Ab) and analysed by standard PCR (A) and real-time qPCR (B) using specific primers of the different PRE and control regions as indicated. (A) Lanes input correspond to PCR products obtained before immunoprecipitation from 10% of the material used for the immunoprecipitation. PCRs were performed with undiluted samples (lanes 1), 1/5 dilutions (lanes 2) and 1/25 dilutions (lanes 3). (B) Results are expressed as percentage of DNA input.
Analysis of the prevalence of DNAseI HS sites
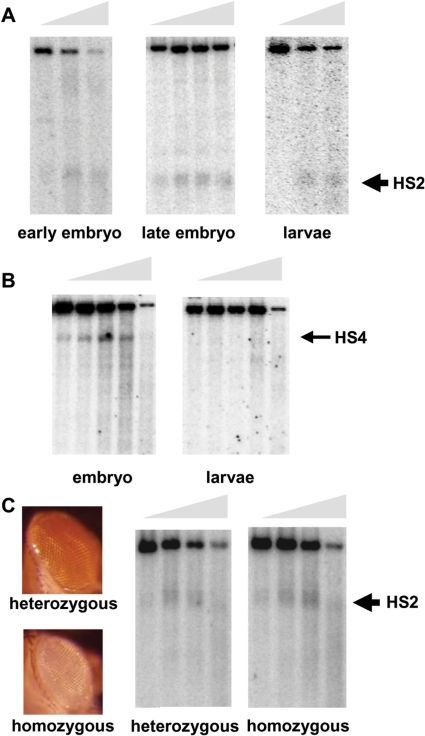
We have shown that Abd-B regulatory elements are nuclease accessible in Drosophila embryos. To investigate when these particular chromatin structures are established and whether they are present all along Drosophila development we performed DNAseI digestion of Drosophila 0–4 h embryos and third instar larvae. Our analyses showed that HS sites at either the iab-6 PRE element HS2 (Figure 4A) or the Fab-7 element (data not shown) are already present early in development and are maintained throughout development until late third instar larvae. On the contrary, the HS4 site, which corresponds to the IAB6 initiator element, is clearly seen in embryos whereas it is absent later in development in third instar larvae (Figure 4B).
Figure 4.
iab-6 HS sites during development and in relation to their activity. (A) DNA samples from DNAseI digests of nuclei from 0 to 4 h (early), 10–24 h (late) embryos and third instar larvae were restricted with PstI/SacI and probed as indicated in Material and methods section. (B) DNA samples from DNAseI digests of 0–14 h embryo nuclei and third instar larvae were restricted with HindIII and analysed as in Figure 1. (C) DNAseI assays carried out in 0–24 h transgenic HS2 9.2 embryos probed with a PCR fragment from the mini-white promoter sequence. Eye colours of representative heterozygous and homozygous flies of transgenic line HS2 9.2 are presented on the left.
PREs are responsible for the activity of homeotic genes in some tissues and their repression in others. Thus, we wanted to establish whether DNAseI hypersensitivity correlates with activity of the PRE. For this purpose we analysed the pattern of DNAseI digestion of one of the transgenic lines described above carrying a P-element whiteenhancer-mini-white construct containing the HS2 fragment (transgenic line HS2 9.2) which showed silencing and pairing-sensitive silencing. As shown in Figure 4C, flies with one copy of the transgene had an orange eye colour while flies carrying two copies showed eye pigmentation close to that of white− animals, indicating that in HS2 9.2 homozygous animals white expression is totally repressed by the PRE. The assays performed showed the presence of a DNAseI HS site in the transgene at the genomic location corresponding to the ectopic HS2 fragment (Figure 4C), both in heterozygous and homozygous animals. Similar results were obtained in transgenic lines carrying a functional Fab-7 element (data not shown). These results indicate that HS sites are present at PREs that are responsible for partial or fully repressed reporter genes.
Characterization of a boundary activity in the HS1 genomic region
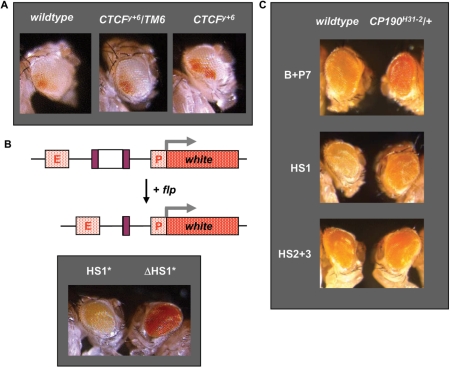
By genetic analysis with genomic deficiencies covering several kb of DNA, it has been suggested that there may be a boundary Fab-6 element, located between positions 3R:12708067 and 3R:12712604, responsible for the autonomy of iab-5 and iab-6 cis-regulatory domains (9). A well known enhancer-blocking factor in vertebrates, the CTCF protein, has been shown to be associated in vivo with several regions of the bithorax complex, one of them located around position 3R:12708500 (14). The HS1 site, here characterized as a PRE, is located at this position and we postulated that it might also contain a boundary activity. We have analysed the effect of dCTCF mutation on silencing of white reporter gene in heterozygous animals carrying the HS1 region. Any relief from enhancer-blocking should allow the communication between the whiteenhancer and the promoter of the white gene and increase eye colour. As shown in Figure 5A, a small but significant increase in eye colour was seen in a null dCTCF background, similar to data reported earlier for the Fab-8 boundary element of the BX-C (10,11,27).
Figure 5.
Boundary activity in the HS1 region. (A) Eye colour of representative heterozygous flies of transgenic line HS1 170.2 in wild-type and heterozygous and null dCTCF mutant background. (B) Schematic representation of the enhancer-blocking construct; E and P represent the enhancer and promoter of the white gene and HS1* and FRT sites are depicted by white and purple rectangles respectively. Eye phenotypes of representative heterozygous flies of transgenic line HS1* 34.2 before (HS1*) and after (▵HS1*) flp action are presented below. (C) Eye colour of representative heterozygous flies of transgenic lines B+P7 2.5, HS1 1.3 and HS2+3 3.1 in wild-type and heterozygous CP190 mutant background.
To delimit the sequence conferring blocking activity to the HS1 region we have used a FLP-FRT recombinational system in order to perform more accurate enhancer-blocking assays. A 0.5 kb fragment (HS1*) containing the dCTCF binding sites previously identified (14) was inserted between the whiteenhancer and the mini-white reporter gene. When this construct was introduced into flies, 40% of the transgenic lines obtained showed silencing of white expression. Since HS1* fragment was flanked by FRT sites, crosses with flies expressing flp recombinase would lead to recombination between the two FRTs and deletion of the fragment (Figure 5B). When the transgenes were subjected to flp recombination they showed increased white expression (Figure 5B). These results indicate that silencing of the reporter gene depends on the presence between enhancer and promoter of a fragment of only 0.5 kb that contains the two dCTCF binding sites. However, we have not been able to totally isolate the boundary element since some of these transgenic lines showed pairing-sensitive silencing, indicating that the HS1* fragment still contains some PRE activity (data not shown).
Finally, we have analysed the requirement of another insulator protein, CP190, to the activity of these elements in transgenic assays. It has been previously reported that CP190 participates in the functionality of Fab-8 in enhancer-blocking assays (10,11). We have found that mutation of a single copy of CP190H31–2 gives rise to a significant relief in silencing induced by fragments containing the Fab-7 boundary element (Figure 5C, B+P7). In contrast, no effect was seen in HS2+3 transgenic flies which contain a PRE element (Figure 5C, HS2+3). These results suggest that CP190 should be a required component at the other boundary elements of the BX-C. It has been recently reported that CP190 is associated in vivo with several regions of the BX-C, one of them located in the HS1 region (10). Therefore, we have analysed the effect of CP190 mutation on HS1 enhancer blocking and we have found that transgenic flies carrying one copy of CP190H31–2 showed increased white expression (Figure 5C, HS1). Altogether these observations suggest that a boundary activity depending on dCTCF and CP190 proteins is likely to be present in the HS1 genomic region.
DISCUSSION
In this study we have used DNAseI HS sites to localize regulatory elements in the BX-C locus and we have characterized two new PREs in the Abd-B iab-6 regulatory region. Our results show that HS sites at PREs are present all along development while the HS site at the initiator element IAB6 is present in embryo but no at larval stages (9). Previous work had shown that several PREs from the BX-C are hypersensitive to nucleases in late embryos (4,20) and it has also been shown that the MCP regulatory element is hypersensitive to nucleases in early embryos (20) and the bxdPRE from the Ubx regulatory region is nuclease accessible in larvae (8). Altogether, these studies strongly suggest that these unusual chromatin structures can be associated to the functionality of the regulatory element being present at PREs not only in early embryos, when the activity state of the BX-C homeotic genes in each parasegment is established, but also at larval stages, well into the maintenance phase of BX-C regulation.
ChIP analyses performed over the entire Ubx region in Drosophila tissues that either express or not the gene have shown that PREs contain lower levels of core and linker histones, suggesting that either histone epitopes are masked by PcG or TrxG complexes or these regions are nucleosome free (28–30). We have found that Abd-B PREs are nuclease accessible in PRE copies carried by transposons independently of the silencing activity of the PRE, supporting the idea that these regulatory regions are depleted of nucleosomes in both ON and OFF transcriptional states. Therefore, although PcG silencing have been proposed to be associated to DNA compaction (31,32), our results indicate the contrary and favour a model, supported by recent data from other groups, that invokes interaction between PREs and core promoters (33–35), broad H3K27 trimethylation of the region, absence of H3K4 trimethylation from the promoter and transcription initiation blockage (36,37).
Based on data from several laboratories, a model has been proposed to summarize the current understanding of how BX-C functions. In this model, each distinct regulatory domain contains a modular arrangement of functional elements for the regulation of homeotic gene expression in a particular parasegment with boundary elements keeping each domain separate and autonomous (2). Although there are genetic evidences of the relevance of boundary elements in Abd-B gene regulation, the mechanism by which they function and the proteins implicated are still mostly unknown. dCTCF, which in vertebrates binds to and participate to the function of all insulators characterized to date (8), and CP190, which has been reported to be essential for gypsy insulator activity (16), have been shown to be associated in vivo with several Drosophila BX-C boundary elements (10) and to contribute to the enhancer-blocking activity of the Fab-8 element (10,11). Our observation that Fab-7 activity also depends on CP190 suggests that this protein must be a common player in all the boundary activities within the BX-C locus.
Although we have not been able to separate HS1 boundary and PRE elements, several observations point to a role of this region in maintaining the autonomy of iab-5 and iab-6 cis-regulatory regions; it has been reported earlier that dCTCF and CP190 insulator proteins bind to the HS1 region in Drosophila embryos (10,14) and our results indicate that both proteins are required for the function of this region in transgenic enhancer-blocking assays. These findings lead us to propose that this region may contain the Fab-6 boundary element of the BX-C. Boundary elements in both Fab-7 and Fab-8 regions are located in the vicinity of PREs, with each regulatory element marked by different DNAseI HS sites (4,6). In contrast, the boundary activity of the Mcp element is flanked on both sides by regulatory elements that cooperatively repress white expression in transgenic lines (5) and this regulatory region presents a single HS site (20). Our results suggest that the boundary activity of the HS1 fragment is likely to be similar to the Mcp element, since it corresponds to a single DNAseI hypersensitive site and boundary and PRE activities seem to be much more intermingled.
FUNDING
The Ministerio de Educación y Ciencia (MEC) (BMC2002-00905, BMC2006-01627 and CSD2006-00049); the Comissió Interdepartamental de Recerca i Innovació Tecnològica (CIRIT) (2001SGR00344). Funding for open access charge: MEC, BMC2006-01627.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Josep Casacuberta, Jordi Casanova and Elena Casacuberta for helpful comments on the manuscript. We also thank Ester Fuentes for technical assistance. We are thankful to Victor G. Corces and Bloomington Stock Center for providing fly stocks. S.P.-L. and S.C. acknowledge receipt of doctoral fellowships from the MEC.
REFERENCES
- 1.Akbari OS, Bousum A, Bae E, Drewell RA. Unraveling cis-regulatory mechanisms at the abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev. Biol. 2006;293:294–304. doi: 10.1016/j.ydbio.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- 3.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the polycomb and trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 4.Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- 5.Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 7.Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 9.Mihaly J, Barges S, Sipos L, Maeda R, Cleard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development. 2006;133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- 10.Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RAH, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canudas S, Perez S, Fanti L, Pimpinelli S, Singh N, Hanes SD, Azorin F, Espinas ML. dSAP18 and dHDAC1 contribute to the functional regulation of the Drosophila Fab-7 element. Nucleic Acids Res. 2005;33:4857–4864. doi: 10.1093/nar/gki776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16:3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweinsberg S, Hagstrom K, Gohl D, Schedl P, Kumar RP, Mishra R, Karch F. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics. 2004;168:1371–1384. doi: 10.1534/genetics.104.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pai C-Y, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 17.Cartwright IL, Cryderman DE, Gilmour DS, Pile LA, Wallrath LL, Weber JA, Elgin SCR. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 1999;304:462–496. doi: 10.1016/s0076-6879(99)04028-8. [DOI] [PubMed] [Google Scholar]
- 18.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 19.Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RDG, Chenoweth JG, Tesar PJ, Furey TS, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Sched P. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dellino GI, Tatout C, Pirrotta V. Extensive conservation of sequences and chromatin structures in the bxd polycomb response element among Drosophilid species. Int. J. Dev. Biol. 2002;46:133–141. [PubMed] [Google Scholar]
- 22.Busturia A, Lloyd A, Bejarano F, Zavortink M, Xin H, Sakonju S. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128:2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- 23.Mishra RK, Mihaly J, Barges S, Spierer A, Karch F, Hagstrom K, Schweinsberg SE, Schedl P. The iab-7 Polycomb Response Element maps to a nucleosome-free region of chromatin and requires both GAGA and pleiohomeotic for silencing activity. Mol. Cell. Biol. 2001;21:1311–1318. doi: 10.1128/MCB.21.4.1311-1318.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146:1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz YB, Kahn TG, Nix DA, Li X-Y, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 26.Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc. Natl Acad. Sci. USA. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moon H, Filippova G, Loukinov D, Pugacheva E, Chen Q, Smith S, Munhall A, Grewe B, Bartkuhn M, Arnold R, et al. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO rep. 2005;6:165–170. doi: 10.1038/sj.embor.7400334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahn TG, Schwartz YB, Dellino GI, Pirrotta V. Polycomb complexes and the propagation of the methylation mark at the Drosophila Ubx gene. J. Biol. Chem. 2006;281:29064–29075. doi: 10.1074/jbc.M605430200. [DOI] [PubMed] [Google Scholar]
- 29.Mohd-Sarip A, van der Knaap JA, Wyman C, Kanaar R, Schedl P, Verrijzer CP. Architecture of a Polycomb nucleoprotein complex. Mol. Cell. 2006;24:91–100. doi: 10.1016/j.molcel.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ONand OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald DP, Bender W. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol. 2001;21:6585–6597. doi: 10.1128/MCB.21.19.6585-6597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Francis NJ, Kingston RE, Woodcock CL. Chromatin Compaction by a Polycomb Group Protein Complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 33.Cleard F, Moshkin Y, Karch F, Maeda RK. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 2006;38:931–935. doi: 10.1038/ng1833. [DOI] [PubMed] [Google Scholar]
- 34.Comet I, Savitskaya E, Schuettengruber B, Negre N, Lavrov S, Parshikov A, Juge F, Gracheva E, Georgiev P, Cavalli G. PRE-Mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell. 2006;11:117–124. doi: 10.1016/j.devcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 36.Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SCR, Pirrotta V. Polycomb silencing blocks transcription initiation. Mol. Cell. 2004;13:887–893. doi: 10.1016/s1097-2765(04)00128-5. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr. Opin. Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]