Abstract
Double-stranded sections of mRNA are often inviting sites of interaction for a wide variety of proteins and small molecules. Interactions at these sites can serve to regulate, or disrupt, the homeostasis of the encoded protein products. Such ligand target sites exist as hairpin–loop structures in the mRNAs of several of the proteins involved in iron homeostasis, including ferritin heavy and light chains, and are known as iron responsive elements (IREs). These IREs serve as the main control mechanism for iron metabolism in the cell via their interaction with the iron regulatory proteins (IRPs). Disruption of the IRE/IRP interaction could greatly affect iron metabolism. Here, we report that anthracyclines, a class of clinically useful chemotherapeutic drugs that includes doxorubicin and daunorubicin, specifically interact with the IREs of ferritin heavy and light chains. We characterized this interaction through UV melting, fluorescence quenching and drug–RNA footprinting. Results from footprinting experiments with wild-type and mutant IREs indicate that anthracyclines preferentially bind within the UG wobble pairs flanking an asymmetrically bulged C-residue, a conserved base that is essential for IRE–IRP interaction. Additionally, drug–RNA affinities (apparent Kds) in the high nanomolar range were calculated from fluorescence quenching experiments, while UV melting studies revealed shifts in melting temperature (ΔTm) as large as 10°C. This anthracycline–IRE interaction may contribute to the aberration of intracellular iron homeostasis that results from anthracycline exposure.
The unique tertiary structures attainable by RNA, and the burgeoning knowledge of the ligand specificity of those tertiary structures, have revealed RNA to be an inviting target for a variety of proteins and small molecules (1–7). The interaction of these ligands with mRNA can alter the translation of the encoded protein. The iron responsive elements (IREs) of proteins involved in iron metabolism serve as the premier regulatory site of translation due to their unique tertiary structure. IREs are ∼30-nt long hairpin–loop structures in the untranslated regions (UTRs) of several iron metabolism proteins including both ferritin subunits (Figure 1), ferroportin and the erythroid isoform of aminolevulinate synthase, eALAS (8,9) The structure and sequence of IREs is highly conserved throughout all of the animal kingdom including early metazoan (10). The homeostasis of IRE containing proteins is regulated by the iron-dependent availability of the IRP proteins (8). Ligand interaction in the UTRs of mRNA can serve to stabilize or destabilize the mRNA, and thus increase or decrease translation. In general, binding of a protein or small molecule in the 5′-UTR inhibits translation by deterring the association of translation factors and preventing the assembly of the ribosome. Conversely, the binding of a ligand in the 3′-UTR upregulates translation by protecting against nuclease degradation (8). In the case of IRE–IRP interaction, at low intracellular iron concentrations, IRP1 is converted into its active binding state through the dissociation of its 4Fe-4S cluster. This active state then binds to the 5′ IRE of ferritin heavy and light chain mRNAs, serving to downregulate translation and reduce the iron-storage capacity of ferritin (8). Disruption of this IRE/IRP interaction could greatly affect intracellular iron handling.
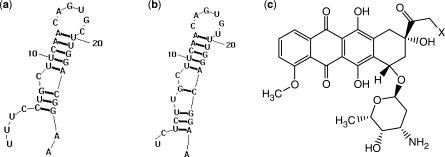
Figure 1.
(a) Human ferritin H-chain IRE 5 ' UTR, (b) human ferritin L-chain IRE 5 ' UTR, (c) anthracyclines investigated in this report, X = OH, doxorubicin; X = H, daunomycin.
Anthracyclines are powerful antineoplastic agents that are widely used for the treatment of leukemia, lymphoma, solid tumors and Hodgkin's disease (11). However, the clinical use of anthracyclines is limited by the occurrence of both chronic and acute cardiotoxicity that can often lead to cardiomyopathy, myocardial infarction and congestive heart failure (11–13). The mitigation of these often fatal side affects is of high interest and many attempts have been made to characterize the mechanism of anthracycline cardiotoxicity. However, there is much disagreement on this point in the literature. Theories include the suppression of transcription factors, production of reactive oxygen species, free radical-induced oxidative stress, decreased levels of intercardiac protective proteins and impaired intracellular iron handling and homeostasis (14–22). Elegant attempts have been made to tie all of the popular pathways listed above into a single complex model for anthracycline cardiotoxicity (23). However, the exact method by which anthracyclines lead to cardiomyopathy must still be elucidated in order to rationally design new therapeutic options for the successful treatment of cancer with anthracyclines.
The antitumor affects of anthracyclines are attributed to the ability of the molecules to inhibit topoisomerase by intercalating DNA, and the specifics of this interaction are very well-characterized (24–32). However, the ability of these drugs to intercalate double-stranded RNA has largely been neglected in the literature. It has previously been observed that anthracyclines cause an imbalance in the regulation of IRP and its binding to IREs, but it has yet to be investigated whether this perturbation is a direct result of anthracyclines interacting with the IREs (17,18). Toward unraveling the mechanism of anthracycline-induced disruption of IRE binding to IRP we investigated the possibility of a direct interaction between anthracyclines and IREs harbored at the 5′-UTR of ferritin heavy and light chain mRNAs. A direct interaction of a small molecule, such as an anthracycline, with the IRE could change the tertiary structure of the RNA drastically and alter the effectiveness of the IRE/IRP interaction. In this study, we characterized the isolated IREs of ferritin heavy and light subunit mRNAs in the absence and presence of doxorubicin (DOX) and daunorubicin (DAU), two widely used anthracyclines, via biophysical techniques including UV melting, fluorescence quenching and drug–RNA footprinting. Results from these experiments suggest that anthracyclines do indeed interact with the IRE hairpin–loop structures in a strong and specific manner. This interaction may contribute to the intracellular aberration of iron regulatory protein homeostasis upon anthracycline exposure.
EXPERIMENTAL PROCEDURES
Drugs and reagents
Doxorubicin hydrochloride and daunorubicin hydrochloride were procured from Oakwood Products Incorporated. All reagents were of molecular biology grade (Sigma, St. Louis, MO). Tubes and tips were sterile and autoclaved prior to use. Water was ultra pure, autoclaved for 1 h, and filtered through a 0.2 µm filter prior to use. All buffers used were also filtered prior to use.
mRNA sequence selection and preparation
The mRNA sequences of the human proteins of interest were retrieved from the Entrez Nucleotide Database (www.pubmed.org). The UTRs of these mRNA sequences were identified, and the folding of these sequences was modeled in the RNAstructure Version 4.3 program (rna.urmc.rochester.edu/rnastructure.html). The resulting secondary structures were surveyed to identify hairpin loop structures. The isolated sequence of the loop and stem region of these structures, on average 30 nt in length, was then folded via the RNAstructure program once again to ensure that the secondary structure was maintained in the absence of the rest of the sequence (Figure 1). The mRNA sequences of these structures were purchased from Dharmacon Research Inc., Lafayette, CO, deprotected according to the manufacturer's protocol, and their purity was verified via denaturing urea polyacrylamide gel electrophoresis. RNA concentrations were determined by UV-vis spectroscopy in 1 cm path length quartz cuvettes using the molar extinction coefficients provided by Dharmacon.
UV-vis melting and absorption experiments
UV melting experiments were performed in the absence of drug and in the presence of DOX or DAU at final drug concentrations of 5 μM and 20 μM. RNA and drug complexes were formed by the addition of an aliquot of stock drug solution in DMSO to a volume of RNA in BPE buffer (6 mM Na2HPO4, 2 mM Na2H2PO4, 1 mM EDTA, pH 7.1). The mixture was allowed to equilibrate for at least 20 min at 4°C to ensure complete association. The drug stock was prepared so that upon reaching the desired concentration of drug, the amount of DMSO in the sample was no >1% of the total volume. For absorption studies, a constant concentration of drug (7 µM) was titrated with increasing amounts of RNA (0.5–20 µM) and the absorbance was measured over the range of 400–600 nm on a Cary 50 Bio UV-vis spectrophotometer. Melting studies were performed over a temperature range of 25–90°C, using a melting rate of 1°C/min and data were collected every half degree. In order to favor hairpin formation and avoid self-association, constructs were heated at 99°C for 1 min, then slowly cooled (1°C/min) to 10°C prior to being melted from 25°C to 90°C (33). For melting experiments, absorbance was measured at 260 nm, the temperature was controlled with a Quantum Northwest TC 125 temperature control unit, drug concentration varied between 5 and 20 µM, and the concentration of RNA was held constant at 2.5 µM. The Tm was taken as the midpoint of the melting transition as determined by the maxima of first derivative plots. An example of the first derivative plots for ferritin H-chain IRE RNA in the absence and presence of both DOX and DAU can be found in the Supplementary Material (Supplementary Figure S1). All Tms were reproducible to within ± 1°C. Repeated recording of heating and cooling profiles indicates that no hysteresis occurs in the absence or presence of ligand.
Fluorescence quenching experiments
Quenching of the drug's natural fluorescence upon binding to RNA was monitored via titrations of a constant concentration of drug (7 µM) with RNA of increasing concentration (0.1–20 µM). Quenching experiments were performed in a 96-well black microplate following the protocol described by Hergenrother et al. (2). Fluorescence emission at 556 nm was recorded on a Molecular Devices SpectraMAX Gemini microplate reader after excitation at 480 nm and 485 nm for DOX and DAU respectively. All experiments were repeated at least four times. The data collected in the fluorescence quenching titrations was fit to a second-order decay curve utilizing TableCurve 2D v5.01, equation 8107 (2):
Where b is the apparent dissociation constant, x is the RNA concentration and a is the asymptotic limit.
Fluorescence quenching plots and curve fits are available in the Supplementary Material.
A stock solution of 4 M NaCl in BPE buffer was diluted with BPE to achieve the various salt concentrations listed in the paper (34).
RNase T1 footprinting
The protocol for drug–RNA footprinting was adapted from previously reported methods (35). RNA constructs were 5′-end-labeled with γ-32P ATP (PerkinElmer, Waltham, MA) utilizing T4 polynucleotide kinase (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Labeled RNA (1.25 µM), unlabeled RNA (11.5 µM) and drug (1–20 µM) were combined in a final volume of 9 µl of footprinting buffer (11 mM Tris–HCl, pH 7.0) and were allowed to equilibrate for 30 min at 25°C. The equivalent of 1 U of RNase T1 enzyme (USB) was added and allowed to proceed for 10 min at 25°C. The reaction was quenched by the addition of 5 µl of urea loading buffer (8 M urea, 20 mM EDTA and 2 mM Tris, pH 7.5). Samples were then heated at 95°C for 5 min, and 11 µl of each sample was run on a 12% denaturing acrylamide gel for 90 min at 290 V. The gel was then dried, exposed to an IP plate (Fuji Film) and was visualized on a Fuji Film FLA-3000 IP plate scanner. Quantification of band intensity was performed with the Fuji Film program Multi Gauge version 3.0 utilizing profile quantification in the direction of migration and a polygonal baseline for background compensation.
RESULTS
UV melting and absorbance experiments
The magnitude of ligand-induced change in nucleic acid Tm [ΔTm = Tm(RNA + ligand) −Tm(RNA)] is an indication of the affinity of the ligand for the nucleic acid target. Hence, we performed UV melting experiments to obtain the first indication of a direct association between the drugs and the IRE RNAs.
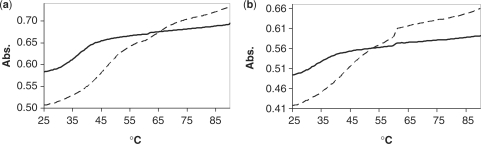
The UV melting curves of both ferritin H-chain and L-chain IREs (Figure 2) display bi-phasic melting profiles in the absence and presence of drug. The higher temperature transition (Tm = 61°C) is a minor transition and was found to remain constant under all conditions. The lower temperature transition revealed a concentration-dependent increase in Tm in the presence of DOX and DAU. The observed ΔTm for all construct and drug combinations are presented in Table 1. It was found that the addition of drug to the IRE RNAs caused an increase in Tm not only in a concentration-dependent manner but also in a drug-dependent manner, with DOX consistently causing a more substantial shift than DAU (Table 1, Supplementary Figure S1). It was also observed that the slight changes in sequence and structure of the constructs led to a graded degree of Tm shift in response to the presence of drug. In the presence of 20 μM DOX, both the ferritin heavy and light IREs produced a ΔTm of around 10°C (Table 1). Comparatively, the HIV-1 TAR RNA construct (6,36), which is also a hairpin-loop structure that has been shown to interact with small molecules, produced a maximum shift of 5°C under the same conditions (data not shown).
Figure 2.
Overlay of the melting profiles of (a) ferritin H-chain mRNA and (b) ferritin L-chain mRNA in the absence (solid) and presence (dashed) of 20 µM DOX.
Table 1.
The average ΔTms observed for all drug and IRE combinations
| Ligand | H-chain | L-chain | G23I-IRE | U10C-IRE | U6C-IRE | C8A-IRE | U6C-U10C-IRE |
|---|---|---|---|---|---|---|---|
| 5 µM DOX | 2.5 | 2.7 | 4.7 | 3.7 | 2.6 | 3.9 | 1.0 |
| 20 µM DOX | 9.9 | 10.1 | 10.3 | 6.8 | 6.5 | 7.9 | 5.5 |
| 5 µM DAU | 0.5 | 3.5 | 3.8 | 1.7 | 1.5 | 2.9 | 0.5 |
| 20 µM DAU | 5.7 | 6.5 | 7.3 | 4.7 | 4.0 | 5.9 | 1.5 |
Average Tm values can be found in the Supplementary Material (Table S1).
Guided by the results from the RNase footprinting experiments on wild-type IREs (see details below), mutations were made to the native H-chain IRE that altered or removed certain structural features associated with drug binding and recognition (37,38). We constructed four point mutants designated U6C-IRE, C8A-IRE, U10C-IRE and G23I-IRE (I = inosine) with mutations at nucleotide positions 6, 8, 10 and 23, respectively; and one double mutant designated U6C-U10C-IRE, bearing double mutations that replaced the two UG pairs with CG pairs. These mutations were predicted by RNAstructure to preserve the IRE secondary structure. The difference in ΔTm between these mutants and the wild-type indicated the importance of both UG wobble base pairs found in the ferritin IRE. The observed ΔTm for the double mutant, U6C-U10C-IRE, which replaces both UG wobble base pairs, was 5.5°C and 1.5°C in the presence of 20 µM DOX and DAU, respectively. For the G23I-IRE mutant, the magnitudes of the observed ΔTm are slightly higher than that of the wild-type for both DOX and DAU at all concentration studied (see Supplementary Material for melting curves). Interestingly, U6C-IRE and U10C-IRE, mutants in which one of the UG pairs is replaced with a CG, have identical ligand-dependent ΔTm. However, their observed ΔTm is about 3°C less than that observed with the wild-type RNA in the presence of 20 μM DOX.
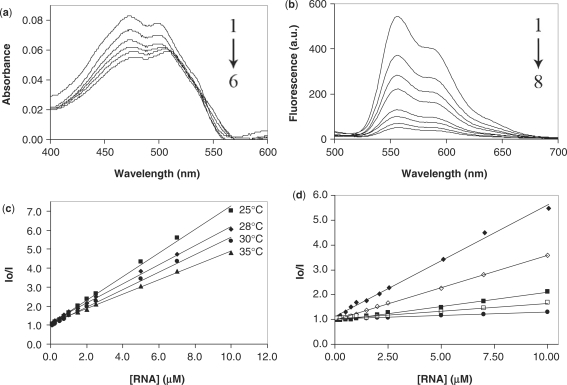
Figure 3a shows the changes that occur in the absorption spectrum of DOX in the visible region during the titration of a fixed concentration of DOX with increasing concentrations of the wild-type H-chain IRE. The resulting absorbance profile of DOX yielded two distinct absorbance maxima at ∼472 nm and 499 nm and a marked bathochromic shift was observed for both peaks resulting in a Δλ of 5 nm and 10 nm, respectively. Additionally, a hypochromism of 33.7% for the first maxima and 24.4% for the second was observed.
Figure 3.
(a) Absorbance profiles of 7 µM DOX in the presence of (from 1 to 6) 0, 0.5, 1.0, 2.5, 7 and 20 µM ferritin H-chain IRE RNA. (b) Fluorescence profiles of the titration of 7 µM DOX with (from 1 to 8) 0, 1, 1.5, 2.5, 5, 7, 10 and 15 µM ferritin H-chain IRE RNA. (c) Stern–Volmer plot of 7 µM DOX with ferritin H-chain IRE RNA at the indicated temperatures. (d) Stern–Volmer plot of 7 µM DOX with ferritin H-chain IRE RNA in BPE of the following Na+ concentrations: 16 mM (filled diamond), 46 mM (open diamond), 116 mM (filled square), 216 mM (open square) and 516 mM (filled circle). Each Stern–Volmer plot was obtained from an average of at least four independent experiments.
Fluorescence quenching
The natural fluorescence of DOX and DAU upon excitation was monitored as they were titrated with increasing amounts of RNA. The fluorescence spectra show a quenching of DOX fluorescence signal upon titration with IRE RNA (Figure 3b). The decay curves of the fluorescent signal were fitted, and apparent Kds were calculated. It is important to note that this apparent Kd is a total dissociation constant, and does not differentiate between the contribution of direct drug binding (e.g. intercalation) and the contribution of electrostatic interaction due to the charged nature of the anthracycline molecule. The apparent Kds and associated standard errors of RNA constructs are presented in Table 2.
Table 2.
Apparent Kds and standard errors of association of DOX with wild-type and mutant IREs
| H-chain | L-chain | G23I-IRE | U10C-IRE | U6C-U10C-IRE | |
|---|---|---|---|---|---|
| Apparent Kd (nM) | 904 | 837 | 903 | 991 | 1100 |
| Fit Std Error (nM) | ±30.9 | ±32.7 | ±22.0 | ±13.2 | ±38.5 |
Fluorescence quenching data at 25, 28, 30 and 35°C was used to perform a Stern–Volmer analysis (34,39,40) of ferritin H-chain IRE RNA quenching DOX according to the equation:
Where I0 and I are the fluorescence intensities in the absence and presence of RNA, respectively, [Q] is the concentration of quencher (RNA) and KSV is the dynamic quenching constant related to the bimolecular collision process. For the quenching experiment at 25°C, this analysis revealed a linear relationship between I0/I and [Q] (Figure 3c). The linearity of the Stern–Volmer plot indicates that the mode of quenching is either collisional or static. Collisional and static quenching can be distinguished by their different dependencies on temperature. An increase in temperature leads to an increase in the diffusion constant of the quencher and will result in an increase in collisional quenching. In contrast, an increase in temperature will lead to a decrease in the binding constant of quencher for fluorophore and will result in a decrease in quenching for a static quencher (40). To identify the quenching mechanism, we repeated the quenching experiments at 25, 28, 30 and 35°C. Stern–Volmer analysis of the resulting data revealed linear relationships between I0/I and [Q] and also showed that the KSV is inversely correlated with temperature (Figure 3c). Based on this result, it could be concluded that the probable quenching mechanism of the DOX-H-chain IRE–RNA complex is initiated by static quenching (39,40).
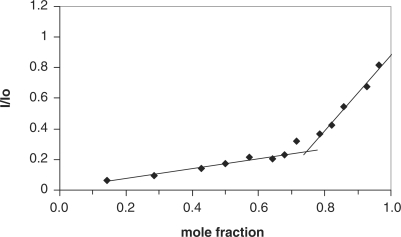
To determine the drug/RNA stoichiometry, we use the fluorescence quenching data to construct Job plots (Figure 4) (41,42). The x-axis of the Job plot, labeled as the mole fraction, is the ratio of the concentration of RNA to the total concentration of RNA and drug, or: Mole Fraction = [RNA]/([RNA] + [drug]). In this case, the sum of [RNA] and [drug] was held constant at 7 µM. The intersection of the two linear portions of the Job plot yield the mole ratio corresponding to the approximate binding stoichiometry between drug and RNA. This analysis revealed a mole fraction corresponding to ∼3.0 molecules of drug associating with each molecule of the H-chain IRE RNA. To further confirm the participation of the UG pairs in anthracycline binding, we also constructed Job plots using the fluorescence quenching data of the mutants IREs. We obtained a binding stoichiometry of ∼2.0 molecules of drug associating with each molecule of either U10C or the double mutant U6C, U10C (See Supplementary Figure S3).
Figure 4.
Job plot of the fluorescence decay of DOX (20 μM) being titrated with ferritin H-chain IRE RNA. The sum of [RNA] and [drug] was held constant at 7 µM in each experiment {Mole Fraction = [RNA]/([RNA] + [drug])}. Each point was obtained from an average of at least four independent experiments.
Binding energy
In an effort to partition the binding free energies of anthracyclines with the IRE RNAs into their nonelectrostatic and polyelectrolyte contributions, fluorescence quenching was monitored over a range of Na+ concentrations, and a Stern–Volmer plot for each was plotted (Figure 3d) (25). The quenching constant (Ksv) was determined for each curve and was then used to calculate the standard Gibbs free energy (ΔG°) using the equation:
Where R is the gas constant (1.987 cal/K/mol) and T is the temperature in Kelvin. The quenching constants estimated at 25, 28, 30 and 35°C are 6.2 × 105 M−1, 5.0 × 105 M−1, 4.6 × 105 M−1 and 3.8 × 105 M−1, respectively. A subsequent plot of log Ksv versus log [Na+] yielded the salt dependence of the binding constant defined by the slope (SK) of such plots. The SK value (−0.83 for DOX and ferritin H-chain IRE) was used in the following equation to determine the polyelectrolyte contribution (ΔGpe) at a given salt concentration (25,43):
To determine the nonelectrostatic contribution to binding energy, the thermodynamic free energy change (ΔGt) was calculated according to the equation (25):
The values obtained for the binding of DOX to H-chain IRE RNA (BPE buffer, 16 mM Na+, 25°C) are as follows: ΔG° = −7.7 kcal mol−1, ΔGpe = −2.0 kcal mol−1 and ΔGt =− 5.7 kcal mol−1. The error associated with these values is estimated to be ± 0.1 kcal mol−1 (25). A comparable value of ΔGpe has been reported for the binding of DOX to DNA under similar conditions (25).
Drug–RNA footprinting
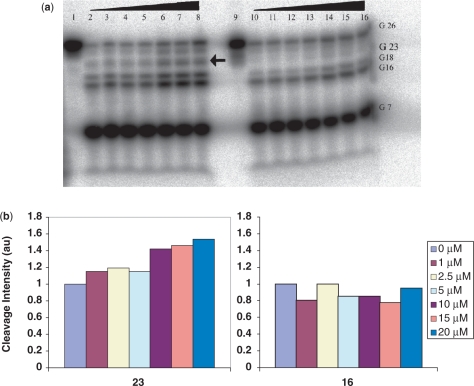
RNase footprinting was performed in order to probe the binding site(s) of DOX and DAU on the RNA constructs. Of the several RNases investigated, RNase T1 was found to provide the cleanest and most useful cleavage pattern. RNase T1 cleaves single-stranded RNA at the 3′-end of guanylic residues and, as expected, it caused strong cleavage of the RNA at G7, G16 and G18, all residues predicted by RNAstructure to be within the single-stranded region of the IRE secondary structure (Figure 5a). The most striking observation from this autoradiograph is that T1 cleavage at G22/G23 and G26/27, the nucleotide positions corresponding to the two UG sites, displayed a drug concentration-dependent increase in band intensity (Figure 5a). While G16 also showed some variation in band intensity, comparison of the quantitated G16 and G23 band intensities, after the footprinting of the H-chain IRE with DOX, revealed that G23 displayed a consistent and marked concentration-dependent increase in cleavage intensity compared to G16 (Figure 5b).
Figure 5.
(a) Autoradiograph of RNase T1 footprinting of γ-32P 5′-end-labeled ferritin H-chain IRE RNA (lanes 1–8) and H-chain U10C mutant (lanes 9–16). Lane: (1) H-chain RNA only; (2–8) RNA, RNase T1 and 0, 1, 2.5, 5, 10, 15, 20 μM DOX, (9) H-chain U10C mutant RNA only; (10–16) RNA, RNaseT1 and 0, 1, 2.5, 5, 10, 15, 20 μM DOX. The G 23 band, indicated by the arrow, is absent for the mutant. (b) Quantitations of the G16 and G23 bands intensities of ferritin H-chain IRE RNA after RNase T1 footprinting in the presence of DOX. DOX concentration for each band is indicated in the legend.
We then used RNase T1 footprinting to follow the effects of the aforementioned mutations on cleavage at G23, the guanylic residue of one of the UG pairs (U10G23) that showed T1 cleavage enhancement in the presence of DOX. We chose to monitor G23 because it is the better resolved of the two G residues on the autoradiograph (Figure 5a). In a manner similar to the wild-type H-chain IRE, the footprinting of the G23I mutant with DOX displayed a drug concentration dependent increase in band intensity at G23 (data not shown). Conversely, the footprint of the U10C-IRE, the mutant which replaced the top UG pair, lacks the band corresponding to G23 when compared to that of the wild-type H-chain IRE (Figure 5a). Validation of the cleavage band designations can be found in the supplementary material (Supplementary Figure S4).
DISCUSSION
Anthracycline–DNA interactions are among the best characterized in the literature (24–32). Despite their chemical similarity to DNA and direct role in gene expression, little is known about the extent to which RNA interactions with anthracyclines contribute to anthracyclines’ biological activities. In this article, we demonstrate that DOX and DAU directly interact with the IREs hairpins (Figure 1a and b) located in the 5′-UTR of ferritin H- and L-chains mRNAs. DOX and DAU caused a concentration-dependent shift in the melting profiles of the H- and L-chains IREs (Figure 2 and Supplementary Figure S1). In most cases, we observed higher ΔTms for DOX compared to DAU for all IRE RNAs studied under the same conditions (Table 1). Since the magnitude of ΔTm is an indication of the affinity of a probe molecule for a nucleic acid target (44), the noted differences in the values of ΔTms suggest that DOX binds tighter to these RNAs compared to DAU. The lower ΔTms observed for DAU (Table 1, bottom half) is consistent with the literature report indicating that DAU binds to DNA between 1.5- to 4-fold less strongly than DOX (31). Furthermore, the only structural difference between the two drugs is the presence of a primary OH-group at C14 of DOX (Figure 1c); it is therefore possible that the OH-group at C14 is playing a role in the binding of anthracyclines to the RNAs, perhaps in a similar manner to its involvement in DNA binding and recognition (45).
The drug induced changes in the melting profiles of the mutant IREs gave further evidence for the base sequence preference of the interactions between the anthracyclines and the IRE RNAs. While the asymmetric bulge in the H-chain IRE is somewhat tolerant of a base change, the replacement of either or both of the UG wobble pairs flanking the asymmetric bulge with a CG significantly impaired DOX and DAU binding (Table 1). These results point to the requirement of the UG base pairs in facilitating the binding of DOX and DAU to the IRE RNAs. However, the UG base pair recognition by anthracyclines may be RNA sequence dependent. Preliminary evidence to that effect was seen in the melting profile of a C22U mutant TAR RNA. Relative to the wild-type TAR RNA, we observed almost no change in ΔTM of the C22U mutant in the presence of 20 μM DOX. This result may suggest that the microenvironment of the UG pairs within the RNA sequence may play important role in anthracycline recognition (see Supplementary Figure S2, for the melting profiles of wild-type and C22U mutant TAR RNA).
The ability of IRE RNAs to alter the spectral characteristics of anthracyclines gives further indication of an interaction of drug and RNA (Figure 3a and b). We observed a quenching of the natural fluorescence of DOX and DAU with increasing RNA concentration (Figure 3b). The quenching of DOX fluorescence by DNA has been ascribed to its intercalation into DNA (27,46). Thus, we attribute the extensive fluorescence quenching (>90% at a 2:1 ratio of DOX to H-chain IRE RNA) to the intercalation of one or more anthracycline molecules per RNA molecule (Figure 3b). A similar interpretation has recently been proposed to explain DOX fluorescence quenching by a SELEX generated DOX RNA aptamer (5). Additionally, the titration of a fixed concentration of DOX with increasing concentrations of the wild-type H-chain IRE results in a hyprochromic effect accompanied by a shift of the absorption maximum (λmax) to a longer wavelength (bathochromic shift). The observed maximal absorbance decrease is ∼33.7% (Figure 3a). These results are indicative of changes in the drug's environment and a perturbation of the DOX chromophore system upon binding to the IRE RNA, consistent with intercalative binding of DOX within the IRE RNA. Furthermore, the apparent Kd (Tables 2) extracted from the fluorescence quenching data give an indirect clue about the affinity of DOX for the IREs. Although we did not have sufficient information to differentiate between the contribution of direct drug binding and that of electrostatic interaction for all IRE constructs, the magnitudes of the apparent Kd's suggest strong association of the drug with the IREs.
We performed RNase footprinting experiments to gain more insight into the binding site(s) of DOX and DAU on the ferritin H-chain IRE and a series of mutant RNAs (47,48). Results from the drug–RNA footprinting experiments show that the T1 cleavage at two sites, G22/G23 and G26/27, is enhanced upon addition of DOX (Figure 5a). Chaires et al. (48) have attributed an analogous enhancement in nuclease cleavage of DNA, in the presence of DAU, to the perturbation of DNA structure in the immediate vicinity of a DAU intercalation site. If this explanation holds for RNA, our results suggest the existence of at least two main drug binding sites at or near the two UG wobble base pairs of the ferritin H-chain IRE (Figure 5). Interaction of anthracycline molecules at both of these proposed binding sites would not violate the nearest neighbor exclusion rule when applied to anthracycline–nucleic acid binding (27).
Wobble pairs, especially GU or UG, are ubiquitous building blocks of higher order RNA structures. Compared to regular Watson–Crick base pairs, wobble pairs differ in both the array of functional groups displayed in the helical grooves and the orientation of the bases with respect to the phosphodiester backbone. The consequence of this is that a wobble pair causes weakening of the helix and local distortions which significantly increase the electronegative environment at or near the mismatch pair (49,50). The two UG pairs in the ferritin H-chain IRE could each provide an electronegative environment for nestling the electropositive DOX. In fact, the wobble pair has been shown to be a commonly conserved ligand binding site (49). To further probe this observation, we extensively studied the effects of mutation of the two UG pairs and bulge C on the binding affinity of DOX for the IRE RNA. Results from these experiments show that all mutations that preserved the U10-G23 wobble displayed essentially the same cleavage pattern as in the wild-type RNA. This suggests that these mutants are able to induce the structural changes necessary for DOX binding within the vicinity of U10-G23. More importantly, however, the G23I mutant sheds light on the specifics of the UG pair recognition by DOX. The G23I mutation results in the deletion of the guanosine N2 exocyclic amine, a very distinct functional group displayed in the minor groove of a wobble that often serves as a specific receptor recognition signal. The fact that the G23I-IRE mutant also displayed increase in Tm and enhancement of T1 cleavage at G23 in the presence of DOX suggest the G23 amine may not be essential for DOX recognition by the ferritin H-chain IRE (see Supplementary Material for the UV melting profiles for construct G23I, Supplementary Figure S5). An alternative mechanism of ligand recognition of a wobble pair is through its unique conformational features. This may well be exploited by DOX in its interaction with the H-chain IRE. This inference is supported by two key observations. First, the magnitudes of the observed ΔTMs for the G23I-IRE mutant are slightly higher than that of the wild-type in the presence of either DOX or DAU at all concentration studied (Table 1). These data suggest that the substitution of G23 for I may have resulted in slightly increased RNA ligand-binding affinity. Previous literature observation indicates that an IU base pair is slightly less stable than a GU pair (50). The subtle duplex instability as a result of the G23I mutation may enhance the structural changes essential for ligand binding, thereby resulting in increased interaction of ligand with the RNA. Second, the G23 band in the U10C, a mutant IRE in which the G23-U10 is substituted by a GC base pair, is protected from T1 cleavage (Figure 5). Based on the increased GC content, one will expect this mutation to increase the binding affinity of DOX for the 5-base paired segment of the IRE. The protection of G23 from T1 digestion may be due to diminished or lack of DOX binding, thereby preventing the necessary conformational changes that expose G23 to T1. Alternatively, it may also be that DOX binding persists but is not accompanied by a conformational change that is sensitive to T1 cleavage.
The Stern–Volmer analysis of the drug fluorescence quenching data supports a static mechanism for the quenching of the DOX fluorescence by the H-chain IRE RNA (Figure 3c and d). The static quenching could occur through the formation of a complex, stabilized mainly by hydrophobic interactions, between DOX (fluorophore) and RNA (quencher) (39,40). This complex could result from the intercalation of DOX into the IRE bases as suggested by the aforementioned evidence. In addition, the Stern–Volmer analysis allows an estimation of the equilibrium constant for the formation of the DOX-H-chain IRE RNA complex. Applying the polyelectrolyte theory (25,43), these equilibrium constants enable the dissection of the RNA binding free energies of anthracyclines into their nonelectrostatic and polyelectrolyte contributions. The calculated value of ΔGpe is very similar to that reported for the binding of DOX to DNA under similar conditions (25). This result suggests a similar polyelectrolyte contribution to the overall ΔG of binding of DOX to DNA and RNA. A Job plot of the fluorescence quenching of DOX with wildtype IRE RNA gives a drug to RNA binding ratio of ∼3.0:1 (Figure 4). A similar plot of the U10C IRE, a mutant in which one of the proposed binding sites has been replaced with a GC pair, resulted in a drug to RNA stoichiometry of 2.0:1 (Supplementary Figure S3). This result is consistent with our original inference on the participation of U10-G23 wobble pair in DOX binding to the IRE. However, the drug to RNA stoichiometry remained 2.0:1 when both UG pairs are replaced (Supplementary Figure S3). The lack of further change in the drug to RNA ratio upon the replacement of the proposed second DOX binding site may be due to the fact that the second mutation did not completely abolish DOX binding. Although footprinting only identified the two UG sites as possible binding sites, the nonelectrostatic binding energy, ΔGt, was found to account for only 74% of the total observed change in ΔG upon DOX binding to IRE RNA. Taken together, these data suggest that in addition to the intercalation of two anthracycline molecules at the UG sites, there is at least one additional site of interaction, most likely electrostatic in nature, on the IRE RNA.
The crystal structure of IRP1 bound to frog ferritin IRE has recently been solved (51). The structure reveals almost twice the number of specific interactions found in most protein–RNA associations. Despite the level of interaction, single mutations to just a few of the residues involved have been shown to greatly diminish the IRE/IRP interaction (37,52,53). This high degree of structural and sequence selectivity reveals the seemingly robust interaction to be in fact very sensitive to minute disturbances. The binding of anthracycline molecules to the IRE structure could cause changes to the tertiary structure of the IRE RNA that could diminish the affinity of IRP for the IRE. This may cause an aberration in the intracellular regulation of the translation of mRNAs harboring these IREs. Supporting this hypothesis is a recent observation from the Thorp group on up-regulation of the cellular expression of ferritin upon exposure to yohimbine, a small molecule that binds to the ferritin IRE (3).
The biochemical evidence presented in this study suggests that anthracyclines form strong and specific interactions with IRE RNAs. Furthermore, it sheds light on some of the structural requirements for this interaction. Although anthracyclines such as DOX and DAU have been reported to show a requirement for GC sequences in DNA binding (48,54), we have presented evidence that the binding of anthracyclines to ferritin IRE RNA demonstrates a strong preference for UG pairs. The prevalence of wobble base pairs in higher order RNA structures (49) suggests that this non-canonical pair, when structurally available, could serve as a ubiquitous recognition site for the binding of anthracyclines to other RNAs as well. Further studies are necessary to better understand the possible biological significance of the anthracycline-IRE interaction. Part of our ongoing effort is to probe the effects of DOX and DAU on IRE-IRP complex formation both in vitro and in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at the NAR Online.
FUNDING
Georgia Institute of Technology. Funding for open access charge: Blanchard Fellowship to AKO.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professors Scott Strobel and Nick Hud for helpful discussions. J. Canzoneri is a recipient of a GAANN predoctoral fellowship from the Georgia Tech Center for Drug Design, Development, and Delivery.
REFERENCES
- 1.Tok J, Cho J, Rando R. Aminoglycoside antibiotics are able to specifically bind the 5′- untranslated region of thymidylate synthase messenger RNA. Biochemistry. 1999;38:199–206. doi: 10.1021/bi9819428. [DOI] [PubMed] [Google Scholar]
- 2.Thomas J, Liu X, Hergenrother P. Size-specific ligands for RNA hairpin loops. J. Am. Chem. Soc. 2005;127:12434–12435. doi: 10.1021/ja051685b. [DOI] [PubMed] [Google Scholar]
- 3.Tibodeau J, Fox P, Ropp P, Theil E, Thorp. H. The up-regulation of ferritin expression using a small-molecule ligand to the native mRNA. Proc. Natl Acad. Sci. 2006;103:253–257. doi: 10.1073/pnas.0509744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xavier K, Eder P, Giordano T. RNA as a drug target: methods for biophysical characterization and screening. Trends Biotechnol. 2000;18:349–356. doi: 10.1016/s0167-7799(00)01464-5. [DOI] [PubMed] [Google Scholar]
- 5.Bagalkot V, Farokhzad O, Langer R, Jon S. An aptamer–doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 6.Mei H, Cui M, Heldsinger A, Lemrow S, Loo J, Sannes-Lowery K, Sharmeen L, Czarnik A. Inhibitors of protein-RNA complexation that target the RNA: specific recognition of human immunodeficiency virus type 1 TAR RNA by small organic molecules. Biochemistry. 1998;37:14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 7.Leibold E, Munro H. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5′ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc. Natl Acad. Sci. USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hentze M, Muckenthaler M, Andrews N. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–297. doi: 10.1016/s0092-8674(04)00343-5. [DOI] [PubMed] [Google Scholar]
- 9.Oliveira C, Goossen B, Zanchin N, McCarthy J, Hentze M, Stripecke R. Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5316–5322. doi: 10.1093/nar/21.23.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–966. doi: 10.1261/rna.464807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singal P, Iliskovic N. Doxorubicin-induced cardiomyopathy. N. Engl. J. Med. 1998;339:900–905. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 12.Thorburn A, Frankel A. Apoptosis and anthracycline cardiotoxicity. Mol. Cancer Ther. 2006;5:197–199. doi: 10.1158/1535-7163.MCT-06-CO1. [DOI] [PubMed] [Google Scholar]
- 13.Jones R, Swanton C, Ewer M. Anthracycline cardiotoxocity. Expert Opin. Drug Saf. 2006;5:791–809. doi: 10.1517/14740338.5.6.791. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Ma A, Kitta K, Fitch S, Ikeda T, Ihara Y, Simon A, Evans T, Suzuki Y. Anthracycline-induced suppression of GATA-4 transcription factor: implication in the regulation of cardiac myocyte apoptosis. Mol. Pharmacol. 2003;63:368–377. doi: 10.1124/mol.63.2.368. [DOI] [PubMed] [Google Scholar]
- 15.Fogli S, Nieri P, Breschi M. The role of nitric oxide in anthracycline toxicity and prospects for pharmacologic prevention of cardiac damage. FASEB J. 2004;18:664–675. doi: 10.1096/fj.03-0724rev. [DOI] [PubMed] [Google Scholar]
- 16.Jeyaseelan R, Poizat C, Baker R, Abdishoo S, Isterabadi L, Lyons G, Kedes L. A novel cardiac-restricted target for doxorubicin. CARP, a nuclear modulator of gene expression in cardiac progenitor cells and cardiomyocytes. J. Biol. Chem. 1997;272:22800–22808. doi: 10.1074/jbc.272.36.22800. [DOI] [PubMed] [Google Scholar]
- 17.Kwok J, Richardson D. Unexpected anthracycline-mediated alterations in iron-regulatory protein-RNA-binding activity: the iron and copper complexes of anthracyclines decrease RNA-binding activity. Mol. Pharmacol. 2002;62:888–900. doi: 10.1124/mol.62.4.888. [DOI] [PubMed] [Google Scholar]
- 18.Kwok J, Richardson D. Anthracyclines induce accumulation of iron in ferritin in myocardial and neoplastic cells: inhibition of the ferritin iron mobilization pathway. Mol. Pharmacol. 2003;63:849–861. doi: 10.1124/mol.63.4.849. [DOI] [PubMed] [Google Scholar]
- 19.Hentze M, Kuhn L. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl Acad. Sci. USA. 1996;93:8175–8182. doi: 10.1073/pnas.93.16.8175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gewirtz D. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 21.Saad S, Najjar T, Al-Rikabi A. The preventive role of deferoxamine against acute doxorubicin-induced cardiac, renal and hepatic toxicity in rats. Pharmacol. Res. 2001;43:211–218. doi: 10.1006/phrs.2000.0769. [DOI] [PubMed] [Google Scholar]
- 22.Rabbani A, Iskandar M, Ausio J. Daunomycin-induced unfolding and aggregation of chromatin. J. Biol. Chem. 1999;274:18401–18406. doi: 10.1074/jbc.274.26.18401. [DOI] [PubMed] [Google Scholar]
- 23.Minotti G, Cairo G, Monti E. Role of iron in anthracycline cardiotoxicity: new tunes for an old song? FASEB J. 1999;13:199–212. [PubMed] [Google Scholar]
- 24.Zeman S, Phillips D, Crothers D. Characterization of covalent adriamycin-DNA adducts. Proc. Natl Acad. Sci. 1998;95:11561–11565. doi: 10.1073/pnas.95.20.11561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaires J, Satyanarayana S, Suh D, Fokt I, Przewloka T, Priebe W. Parsing the free energy of anthracycline antibiotic binding to DNA. Biochemistry. 1996;35:2047–2053. doi: 10.1021/bi952812r. [DOI] [PubMed] [Google Scholar]
- 26.Rabbani A, Finn R, Thambirajah A, Ausio J. Binding of antitumor antibiotic daunomycin to histones in chromatin and in solution. Biochemistry. 2004;43:16497–16504. doi: 10.1021/bi048524p. [DOI] [PubMed] [Google Scholar]
- 27.Chaires J, Dattagupta N, Crothers D. Studies on interaction of anthracycline antibiotics and deoxyribonucleic acid: equilibrium binding studies on the interaction of daunomycin with deoxyribonucleic acid. Biochemistry. 1982;21:3933–3940. doi: 10.1021/bi00260a005. [DOI] [PubMed] [Google Scholar]
- 28.Chaires J. Biophysical chemistry of the daunomycin-DNA interaction. Biophys. Chem. 1990;35:191–202. doi: 10.1016/0301-4622(90)80008-u. [DOI] [PubMed] [Google Scholar]
- 29.Fritzsche H, Berg H. Analysis of equilibrium, kinetic and structural data of anthracycline-DNA interaction. Gazz. Chim. Ital. 1987;117:331–352. [Google Scholar]
- 30.Priebe W. Mechanism of action-governed design of anthracycline antibiotics: a “turn-off/turn-on” approach. Curr. Pharmaceut. Design. 1995;1:51–68. [Google Scholar]
- 31.Graves DE, Krugh TR. Adriamycin and daunorubicin bind in a cooperative manner to deoxyribonucleic acid. Biochemistry. 1983;22:3941–3947. doi: 10.1021/bi00285a033. [DOI] [PubMed] [Google Scholar]
- 32.Pullman B. Sequence specificity in the binding of anti-tumour anthracyclines to DNA: a success of theory. Anticancer Drug Des. 1991;6:95–105. [PubMed] [Google Scholar]
- 33.Proctor D, Kierzek E, Kierzek R, Bevilacqua P. Restricting the conformational heterogeneity of RNA by specific incorporation of 8-bromoguanosine. J. Am. Chem. Soc. 2003;125:2390–2391. doi: 10.1021/ja029176m. [DOI] [PubMed] [Google Scholar]
- 34.Tang K, Pan N, Zhang Y, Zou G. Studies of adriamycin binding to histone H1 by resonant mirror biosensor and fluorescence spectroscopy. Anal. Lett. 2005;38:2151–2164. [Google Scholar]
- 35.McPike M, Goodisman J, Dabrowiak J. Drug-RNA footprinting. Methods Enzymol. 2001;340:431–449. doi: 10.1016/s0076-6879(01)40435-6. [DOI] [PubMed] [Google Scholar]
- 36.Dassonneville L, Hamy F, Colson P, Houssier C, Bailly C. Binding of Hoechst 33258 to the TAR RNA of HIV-1. Recognition of a pyrimidine bulge-dependent structure. Nucleic Acids Res. 1997;25:4487–4492. doi: 10.1093/nar/25.22.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaffrey S, Haile D, Klausner R, Harford J. The interaction between the iron-responsive element binding protein and its cognate RNA is highly dependent upon both RNA sequence and structure. Nucleic Acids Res. 1993;21:4627–4631. doi: 10.1093/nar/21.19.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ke Y, Wu J, Leibold E, Walden W, Thiel E. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding. Fine-tuning of mRNA regulation? J. Biol. Chem. 1998;273:23637–23640. doi: 10.1074/jbc.273.37.23637. [DOI] [PubMed] [Google Scholar]
- 39.Wang C, Wu Q, Li C, Wang Z, Ma J, Zang X, Qin N. Interaction of tetrandrine with human serum albumin: a fluorescence quenching study. Anal. Sci. 2007;23:429–433. doi: 10.2116/analsci.23.429. [DOI] [PubMed] [Google Scholar]
- 40.Lakowics JR. Principles of Fluorescence Spectroscopy,. 2nd. Plenum Publishers, New York: Kluwer Academic; 1983. pp. 257–265. [Google Scholar]
- 41.Job P. Formation and stability of inorganic complexes in solution. Ann. Chim. 1928;9:113–203. [Google Scholar]
- 42.Horowitz E, Hud NV. Ethidium and proflavine binding to a 2′,5′-linked RNA duplex. J. Am. Chem. Soc. 2006;128:15380–15381. doi: 10.1021/ja065339l. [DOI] [PubMed] [Google Scholar]
- 43.Record MT, Ha J, Fisher M. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 44.Wilson WD, Tanious FA, Fernandez-Saiz M, Rigl CT. In Drug-DNA Interaction Protocols. Totowa, NJ: Humana Press Inc.; 1997. Evaluation of drug–nucleic acid interactions by thermal melting curves; pp. 219–240. [DOI] [PubMed] [Google Scholar]
- 45.Wang AH, Ughetto G, Quigly G, Rich A. Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-ANG. resolution. Biochemistry. 1987;26:1152–1163. doi: 10.1021/bi00378a025. [DOI] [PubMed] [Google Scholar]
- 46.Haj H, Salerno M, Priebe W, Kozlowski H, Garnier-Suillerot A. New findings in the study on the intercalation of bisdaunorubicin and its monomeric analogues with naked and nucleus DNA. Chem. Biol. Interact. 2003;145:349–358. doi: 10.1016/s0009-2797(03)00061-9. [DOI] [PubMed] [Google Scholar]
- 47.Li K, Davis TM, Bailly C, Kumar A, Boykin DW, Wilson WD. A heterocyclic inhibitor of the Rev-RRE complex binds to RRE as a dimer. Biochemistry. 2001;40:1150–1158. doi: 10.1021/bi002338b. [DOI] [PubMed] [Google Scholar]
- 48.Chaires J, Herrera J, Waring M. Preferential binding of daunomycin to 5′ATCG and 5′ATGC sequences revealed by footprinting titration experiments. Biochemistry. 1990;29:6145–6153. doi: 10.1021/bi00478a006. [DOI] [PubMed] [Google Scholar]
- 49.Varani G, McClain W. The GU wobble base pair. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strobel S, Cech T, Usman N, Beigelman L. The 2,6-diaminopurine riboside.5-methylisocytidine wobble base pair: an isoenergetic substitution for the study of G.U pairs in RNA. Biochemistry. 1994;33:13824–13835. doi: 10.1021/bi00250a037. [DOI] [PubMed] [Google Scholar]
- 51.Walden W, Selezneva A, Dupuy J, Volbeda A, Fontecilla-Camps J, Theil E, Volz K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- 52.Erlitzki R, Long J, Theil E. Multiple, conserved iron-responsive elements in the 3′-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J. Biol. Chem. 2002;277:42579–42587. doi: 10.1074/jbc.M207918200. [DOI] [PubMed] [Google Scholar]
- 53.Ke Y, Theil E. An mRNA loop/bulge in the ferritin iron-responsive element forms in vivo and was detected by radical probing with Cu-1,10-phenantholine and iron regulatory protein footprinting. J. Biol.Chem. 2002;277:2373–2376. doi: 10.1074/jbc.C100614200. [DOI] [PubMed] [Google Scholar]
- 54.Qu X, Wan C, Becker HC, Zhong D, Zewail AH. The anticancer drug-DNA complex: femtosecond primary dynamics for anthracycline antibiotics function. Proc. Natl Acad. Sci. USA. 2001;98:14212–14217. doi: 10.1073/pnas.241509698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.