Abstract
Spinocerebellar ataxia type 20 (SCA20) has been linked to chromosome 11q12, but the underlying genetic defect has yet to be identified. We applied single-nucleotide polymorphism genotyping to detect structural alterations in the genomic DNA of patients with SCA20. We found a 260 kb duplication within the previously linked SCA20 region, which was confirmed by quantitative polymerase chain reaction and fiber fluorescence in situ hybridization, the latter also showing its direct orientation. The duplication spans 10 known and 2 unknown genes, and is present in all affected individuals in the single reported SCA20 pedigree. While the mechanism whereby this duplication may be pathogenic remains to be established, we speculate that the critical gene within the duplicated segment may be DAGLA, the product of which is normally present at the base of Purkinje cell dendritic spines and contributes to the modulation of parallel fiber-Purkinje cell synapses.
INTRODUCTION
The most common cause of the autosomal dominant cerebellar ataxias is simple sequence repeat expansion (SCAs 1–3, 6–8, 10, 12, 17) (1–3). Missense mutations have also been identified in six SCAs (the Japanese 16q-linked SCA, SCAs 5, 11, 14, 15, 27) (4–9), and more recently genomic deletion at ITPR1 has been associated with SCA15 and SCA16 (9,10), which can now therefore be recognized as the same condition.
Single-nucleotide polymorphism (SNP) genotyping is a powerful method for detecting chromosomal duplications and deletions with high resolution and efficiency. This technique was employed to discover the mutational mechanism of SCA15 (9).
Spinocerebellar ataxia type 20 (SCA20) is a dominantly inherited cerebellar ataxia that is clinically distinct from the other SCAs. Notably, dysphonia is present together with dysarthria; and palatal tremor (‘palatal myoclonus’) is typical. In the eye movements, saccades are hypermetric, and there is no nystagmus. A unique neuroradiological finding is a progressive calcification of the dentate nucleus of the cerebellum, which likely precedes the onset of clinical manifestations. The age of onset of the disease ranges from 19 to 64 years (11,12).
Genetic linkage was found to the pericentromeric region of chromosome 11 (11). Since this region overlapped the SCA5 disease locus, locus homogeneity was considered, although the SCA5 and SCA20 clinical pictures differ considerably; but subsequent discovery of the SCA5 gene, β-III spectrin (SPTBN2) (6), allowed the demonstration that the two diseases are genetically distinct (13).
RESULTS
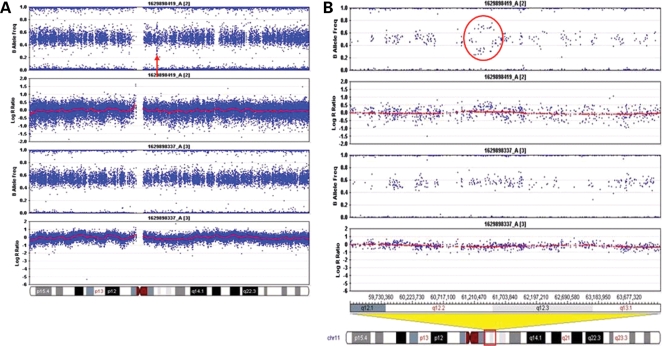
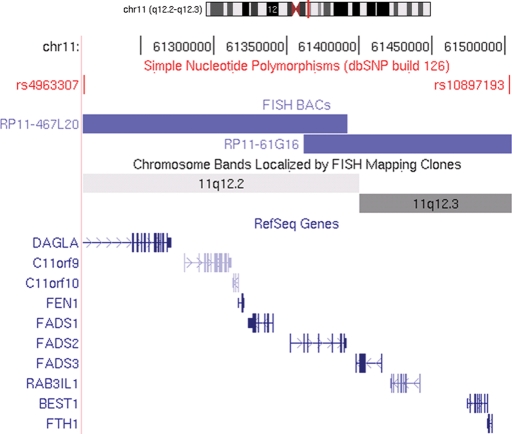
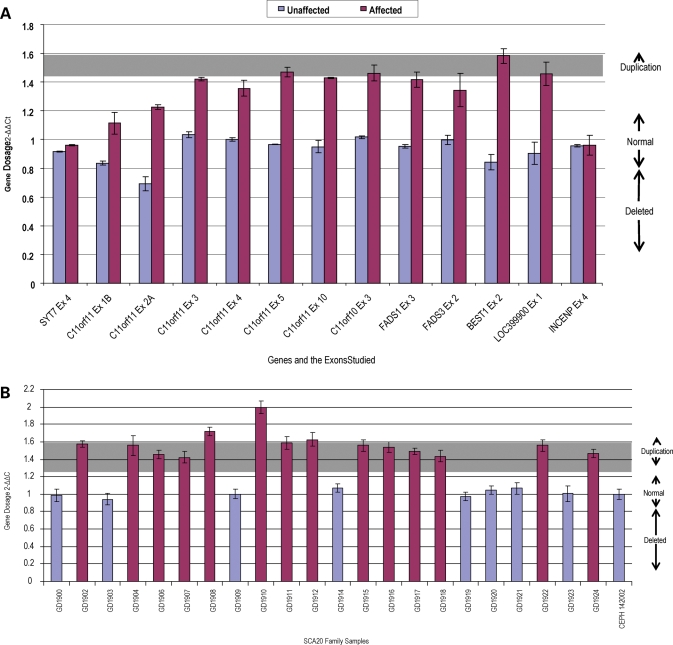
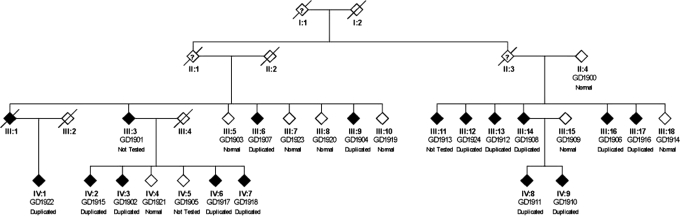
The SNP microarray genotyping identified a structural variation in two affected SCA20 individuals, in comparison to two unaffected family members. The profile obtained from the log R ratio and the B allele frequencies indicated a genomic duplication at chromosome 11q12.2–11q12.3, which is within the region to which SCA20 had previously been linked (11) (Fig. 1). The duplication is defined by the SNPs rs4963307 and rs10897193, which are 260 kb apart. We examined 62 contiguous SNPs across the duplicated region in two affected family members (GD1907 and GD1918); using the B allele frequency metric, we were able to assign the two affected family members a genotype at each of these SNPs of A/A/A (n = 16), A/A/B (n = 24), A/B/B (n = 20) and B/B/B (n = 48; we were unable to confidently call 16 genotypes). At 32 of these SNPs, one of the affected family members was called either A/A/B or A/B/B; in no instance did we observe an A/A/B genotype in one individual and an A/B/B genotype in the other. These data are consistent with the notion that the duplication originally occurred by intrachromosomal duplication. A search of repeats in NCBI Genome Viewer revealed a large number of repeats at the flanking ends of the duplicated region, but none at both ends that were identical. The duplicated region contains 10 known and 2 unknown genes (Fig. 2). Quantitative real-time polymerase chain reaction (PCR) was performed on eight different genes to evaluate the extent of the duplication and to assess the SCA20 family members. As expected, this analysis indicated that the duplication co-segregated with the disease haplotype in the SCA20 family (Figs 3 and 4). Based on examination of copy number metrics of the HumanHap550 BeadChip, none of the 1129 control samples showed a duplication in the same region.
Figure 1.
Infinium HumanHap550 SNP genotyping chips. (A) The top two panels show the results from one affected family member and the bottom two panels show the results from an unaffected family member. The horizontal band in each panel represents heterozygous signal from two-allele SNP markers distributed along chromosome 11. The arrow indicates a duplicated region that was shared by the two affected family members. (B) A higher magnification view showing the duplication more clearly (circled in red).
Figure 2.
A schematic diagram taken from the UCSC Genome Browser, Human March 2006 (v174) Freeze of chromosome 11 showing the ten genes within the SCA20 duplicated critical region and the two SNPs that delineate the duplication, rs4963307 and rs10897193. The schematic diagram also illustrates where the BACs lie in relation to the SNPs on chromosome 11.
Figure 3.
Gene dosage analysis of the SCA20 duplicated region. Results are the mean of three replicates done in triplicate experiments and are expressed as 2−ΔΔCt ± SD. (A) Gene dosage results obtained for the entire SCA20 duplicated region comparing an unaffected family member to an affected family member. The genes outside the duplication are SYT7 and INCENP. (B) A representative gene dosage result for the entire SCA20 pedigree that was tested for one of the duplicated genes, DAGLA exon 5.
Figure 4.
The SCA20 family pedigree showing the individuals who carry the duplication that is segregating with the disease. Individuals in the family who do not carry the disease haplotype do not have the duplication (Normal). Three individuals were not tested as indicated because their DNA was unavailable.
To determine the orientation of the large genomic repeat, we used Genome-wide Analysis of Palindrome Formation (GAPF), a procedure that enriches for palindromes (i.e. inverted repeats) in genomic DNA (14). This approach is based on a relatively simple and efficient method to make ‘snap-back DNA’ from palindromic sequences by intra-molecular base-pairing, followed by elimination of non-palindromic single-strand DNA using S1 nuclease. This procedure has previously been used to show that DNA inverted repeats are non-randomly distributed and enriched in cancer cells (14). To analyze the SCA20 region in high-resolution in an affected individual, we modified the GAPF assay to use genomic tiling arrays, with probes spaced every 10 bp along the SCA20 region of the chromosome. We did not detect a GAPF-positive signal in the genomic area known to be duplicated, indicating that the duplication is unlikely to be an inverted repeat (data not shown), but presumably direct.
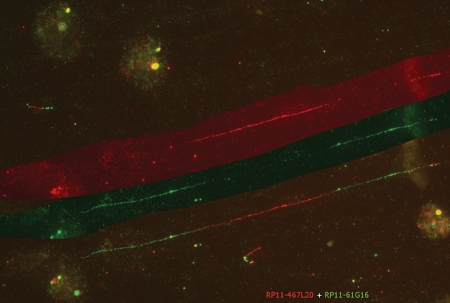
To confirm that the duplication is direct, we used fiber fluorescence in situ hybridization (FISH) with three BAC clones, RP11-467L20, RP11-61G16 and RP11-810P12, in different combinations. Most informative was the co-hybridization of RP11-467L20 in red and RP11-61G16 in green. Both clones map for the largest part within the duplication (Fig. 5); RP11-467L20 spans the proximal breakpoint, and RP11-61G16 spans the distal breakpoint of the duplication. The alternating red–green–red–green pattern of signal sequence is consistent with the duplication being of direct orientation (Fig. 5).
Figure 5.
Fiber FISH analysis on EBV-transformed lymphocytes of patient 00101063 from the family. The DNA fibers were hybridized with two BACs which span each end of the breakpoint, namely, RP11-467L20 visualized in red and RP11-61G16 visualized in green. In the lower part of the image the signals on one fiber are shown. The upper part of the figure shows only the RP11-467L20 (red) signal on this fiber; the middle part of the figure shows only the RP11-61G16 (green) signal on the fiber. The alternating green-overlap-red-green-overlap-red pattern is apparent. It is important to note that the red and green signals in the centre are not overlapping: this indicates a direct tandem duplication.
DISCUSSION
We have demonstrated by quantitative real-time PCR that all the affected individuals in the single known SCA20 family have a duplication on chromosome 11q12.2–11q12.3, within the previous linked SCA20 region (11). The orientation is direct: that is to say, the DNA of the duplicated segment runs in the same direction as the original segment. Since the duplication lay well within the linkage region, it was expected that all affected individuals with the disease haplotype would also carry the duplication; but it was useful to corroborate this. As no controls of similar ethnicity were found with this duplication, it is unlikely that this alteration is a common copy number variant (CNV). However, since many CNVs are rare, it is nonetheless possible that the observed duplication is a rare ‘private’, neutral (non-pathogenic) CNV. A pointer against this possibility is that benign CNVs do not generally affect multiple transcripts, but are more likely to involve only a single gene, or to be localized to intergenic regions of the genome. The balance of evidence thus strongly suggests that the observed duplication is truly pathogenic, and is the cause of SCA20 in this family. As SCA20 is the first SCA to implicate a CNV, it remains to be seen if CNVs have a role in other dominant or sporadic forms of ataxia.
Further evidence that could be adduced were there to be known individuals, whose phenotype was recorded, and carrying a chromosomal duplication encompassing this region. However, we could find only one such duplication case in the cytogenetic literature (15,16), a 6-year-old child, who had a more severe neurological deficit, presumably due to the broader effects of his relatively large (11q11–11q13.3) duplication. A CT brain scan was reported as normal; but the inferred absence of dentate calcification cannot, at this young age, exclude SCA20.
The nature of the genes within the segment may support the proposition of pathogenicity of the duplication, and a gene normally expressed in cerebellum could be important to this disease mechanism. Of the 10 annotated genes within the 260 kb duplicated interval, diacylglycerol lipase α subunit (DAGLA) (17), a neural stem cell-derived dendrite regulator, is the most attractive in this respect. The remaining genes, listed following, have little support for candidacy: hypothetical protein LOC745, chromosome 11 open reading frame 9 (C11orf9) (18), hypothetical protein LOC746, chromosome 11 open reading frame 10 (C11orf10) (19), flap structure-specific endonuclease 1 (FEN1) (20), fatty acid desaturase 1 (FADS1), fatty acid desaturase 2 (FADS2), fatty acid desaturase 3 (FADS3) (21), RAB3A-interacting protein (rabin3)-like 1 (RAB3IL1) (22), bestrophin-1 (vitelliform macular dystrophy protein 2) (BEST1) (23), ferritin, heavy polypeptide 1 (FTH1) (24) and an otherwise undefined predicted gene (UCSC Genome Browser, Human Mar. 06 Assembly).
DAGLA is interesting as a SCA20 candidate because of its known function and its association with the ataxia interactome (25). Its protein product catalyzes the hydrolysis of diacylglycerol (DAG) to 2-arachidonoyl-glycerol (2-AG), an endocannabinoid with an important role in retrograde trans-synaptic suppression of synaptic transmitter release (26). From mouse studies, the gene is highly expressed in two particular neuronal classes of the brain, cerebellar Purkinje cells, where its protein product is present in the dendritic field, and pyramidal cells of the CA1 region of the hippocampus. The dendritic field of the Purkinje cell receives input from the parallel fibers of the granule cells, and this input is modulated at the synaptic junction; DAGLA has its highest expression at the base of the Purkinje cell dendritic spines near the anatomical site of this synapse. It is not clear why an incorrect amount (150%) of DAGLA production (if the duplication does indeed cause this) should lead to a gradual compromise of cerebellar function; but the observation in SCA15 provides a notable parallel, in which an incorrect amount (50%) of another factor, ITPR1, expressed at high level in the Purkinje cell, is associated with a quite similar very slowly progressive pure cerebellar degenerative phenotype (9).
The only known disease-associated gene within the region is BEST1, which is mutated in Best macular dystrophy (BMD) (MIM: 153700), also known as vitelliform macular dystrophy type 2. The pathogenesis is due to abnormal accumulation of lipofuscin within and beneath the retinal pigment epithelium cells. BEST1 forms calcium-sensitive chloride channels, and may conduct other physiologically significant anions such as bicarbonate (27). No obvious connection to the phenotype of SCA20 is evident.
The remaining genes do not have high cerebellar expression, and present no obvious features that would implicate them in the mechanism of SCA20. C11orf9, whose function is unknown, is not highly expressed in the cerebellum, although it is expressed in other regions of the brain, such as the brainstem and basal ganglia (18). FEN1 is an endonuclease that cleaves the 5′ end overhanging flap structure that is generated by displacement synthesis when DNA polymerase encounters the 5′ end of a downstream Okazaki fragment. FEN1 also possesses 5′ to 3′ exonuclease activity on nicked or gapped double-stranded DNA, and exhibits RNase H activity (20). Not much is known about C11orf10 (19), except that it is located immediately upstream of the FEN1 gene, but in the reverse orientation, with the 5′ ends overlapping. FTH1 stores iron in a soluble, non-toxic, readily available form and is important for iron homeostasis. FTH1 has ferroxidase activity. Iron is taken up in the ferrous form and deposited as ferric hydroxides after oxidation. Defects in ferritin proteins are associated with several neurodegenerative diseases. FADS1, FADS2 and FADS3 are fatty acid desaturases. Desaturase enzymes regulate desaturation of fatty acids through the introduction of double bonds between defined carbons of the fatty acyl chain. This cluster of fatty acid desaturases is thought to have arisen evolutionarily from gene duplication, based on the similar exon/intron of its constituent genes (21). RAB3IL1 (22) is a Ras-like GTPase that regulates synaptic vesicle exocytosis. RAB3IL1 is a physiologic guanine nucleotide exchange factor for Rab3A.
If indeed the duplication is the cause of SCA20, then its discovery has enabled us to place SCA20 among other neurological diseases caused by chromosomal duplication (Charcot–Marie–Tooth disease type 1A being the classic example). Whether an extra copy of the DAGLA gene is sufficient, of itself, to determine the SCA20 phenotype, or whether involvement of other genes within or near the duplicated region is necessary for the disease, awaits clarification. Identification of additional SCA20 families, or other adults with interstitial cytogenetic duplication involving proximal 11q, who have had neurological evaluation and brain imaging, would be very useful in this regard. In the absence of such supportive evidence from other families, animal models with overexpression of DAGLA and other candidate genes in the segment may be needed to confirm the mechanism of the duplication.
MATERIALS AND METHODS
Patient samples
Patient samples were collected as previously described (11).
SNP analysis
Structural alterations in genomic DNA were sought by SNP analysis. Two affected and two unaffected individuals from the original SCA20 pedigree were genotyped as per the manufacturer’s instructions, using the Infinium HumanHap550 SNP genotyping chips, which contain 555, 352 unique SNPs (Illumina Inc, San Diego, CA, USA). Data were collected on an Illumina BeadStation scanner, and genotypes generated from the genotyping module (v2.3.25, Illumina Inc). Signal intensities (log R ratio) and SNP B allele frequencies were assessed via the visualization tool in the BeadStudio package (Genome viewer), as outlined in Simon-Sanchez et al. (28). Allele frequencies (‘theta values’) were obtained for individual SNPs, and corrected for cluster position. These data then give the log R ratio, which is the log2 ratio of the observed normalized R value for each SNP, divided by the expected normalized R value for that SNP’s theta value.
Scores close to 1, 0.5 and 0, indicate B allele homozygosity, heterozygosity and A allele homozygosity, respectively, while deviations from these values, in contiguous SNPs, indicate a change in the copy number at that locus. An increase in copy number (>1) denotes a likely duplication, and a decrease (<1), a deletion.
Quantitative real-time PCR analysis
Quantitative real-time PCR for eight different genes was used to assess gene dosage and to determine the extent of the duplication in all available family members (Fig. 4). Twenty-two samples were studied with 13 probes across the region (Figs 3 and 4). Quantitative duplex PCR of genomic DNA was performed on the ABI Prism 7900 Sequence Detection System. β-globin was co-amplified as an internal control, using the following primers: 5′-TGGGCAACCCTAAGGTGAAG-3′ (β-Globin F, Exon 2) and 5′- GTGAGCCAGGCCATCACTAAA-3′ (β-Globin R, Exon 2), and a probe labeled with 6-FAM with the sequence: 5′-CTCATGGCAAGAAAGTGCTCGGTGC-3′. The probes for each region examined were labeled with VIC. Specific primers and probes (obtainable upon request) were designed using the Primer Express Program for TaqMan MGB probes (Applied Biosystems, Foster City, CA, USA). All primers were purchased from Integrated DNA Technologies (Integrated DNA Technologies, Coralville, IA, USA). Each PCR reaction was performed in a total reaction volume of 15 µl containing 25 ng genomic DNA, TaqMan Universal PCR Master Mix (Applied Biosystems), 900 nm primers and 250 nm probes. The PCR cycling conditions were standard, 95°C for 10 min, then 95°C for 15 s and 60°C for 1 min for 40 cycles. The plates for PCR each contained the genomic DNA samples, control DNA and a no-template water control in triplicate, and the experiments were performed in triplicate. The quantification of each amplicon was determined as the cycle at which the PCR amplification was in log phase in fluorescent signal (Ct), relative to β-globin as the internal control. The dosage of each region relative to β-globin was normalized to the mean of the unrelated controls using the 2−ΔΔCt method (29). Values of 0.8–1.2 were assumed to be normal, and values between 1.3 and 1.7 were considered indicative of heterozygous duplication.
Genome-wide analysis of palindrome formation
In order to determine whether the SCA20 amplicon is in the form of a large inverted repeat, the GAPF procedure was performed as described previously (30), with modifications. Genomic DNA (1.5 µg) was treated with either KpnI or SbfI to enrich for DNA close to palindromic centers, or with no enzyme. The restriction enzymes were then heat-inactivated. To make snap-back DNA, genomic DNA was then boiled in 50 µl of water with 100 mm NaCl for 7 min and then placed in an ice-water bath to cool quickly. After snap-back treatment, DNA was treated with S1 nuclease (Invitrogen, Carlsbad, CA, USA) and amplified by ligation-mediated PCR. 7.5 µg DNA was labeled with biotin (GeneChip WT Double-stranded Target Labeling Kit, Affymetrix, Santa Clara, CA, USA) and hybridized to a Human Tiling 2.0R F array (Affymetrix). A comparison of GAPF profiles between affected and unaffected individuals was done with Tiling Analysis Software (Affymetrix, Version 1.1). The probe intensities were normalized using quantile normalization plus scaling, and bandwidth was set at 250 bp. Results were visualized with the Integrated Genome Browser (Affymetrix, Version 4.56).
Fiber FISH analysis
To confirm the GAPF interpretation of the duplication orientation, we applied fiber FISH, according to the methodology described previously (31,32), with some adaptations. In short, the cells of EBV-transformed cell line were suspended in water to a concentration of 1–5 × 105 cells/ml (5). Approximately 100 µl of cell suspension was pipetted onto a Repel-Silane (GE Healthcare)-coated slide, spread out over the entire surface of the slide, and quickly dried using a hair-dryer. Two 50 µl drops of 0.5% SDS, 50 mm EDTA, 0.2 M Tris–HCl, pH 7.0, were pipetted onto a 24 × 60-mm coverslip. The slide with the side containing the cells facing down was then lowered on top of the coverslip. The slide was turned upside-down and the coverslip was very carefully slid off. Again, the slide was dried using a hair-dryer, and then incubated for 5 min in methanol:acetic acid (3:1) to fix the DNA fibers. The dried slides were used directly in the denaturation and hybridization procedures. FISH was performed as described previously (33).
Analysis of control and population samples for duplication at SCA20
We analyzed the disease locus for copy number alteration in 1129 samples, previously typed by us using the Infinium HumanHap550 GeneChip product. These comprised 644 samples from neurologically normal subjects deposited at the National Institute of Neurological Disorders and Stroke Neurogenetics Repository (http://ccr.coriell.org/ninds/, unpublished data) and 485 samples from 29 world populations (34).
FUNDING
The research was funded in part by the Intramural Research Programs of the National Institute on Aging (project number 1 Z01 AG000949-02) and the National Institute of Neurological Disorders and Stroke, both of the National Institutes of Health, Department of Health and Human Services, USA. M.A.K. was supported by a NINDS Competitive Postdoctoral Fellowship. The National Institute on Aging project number associated with this work is Z01 AG000957-05. S.J.D. was supported by a Pediatric Oncology Research Training Program, National Institutes of Health 2T32CA009351-29.
ACKNOWLEDGEMENTS
The authors wish to thank the family members for their cooperation with this study. This study used samples from the NINDS Human Genetics Resource Center DNA and Cell Line Repository (http://ccr.coriell.org/ninds).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Ranum L.P., Day J.W. Dominantly inherited, non-coding microsatellite expansion disorders. Curr. Opin. Genet. Dev. 2002;12:266–271. doi: 10.1016/s0959-437x(02)00297-6. [DOI] [PubMed] [Google Scholar]
- 3.Schöls L., Bauer P., Schmidt T., Schulte T., Riess O. Autosomal dominant cerebellar ataxias: clinical features, genetics, and pathogenesis. Lancet Neurol. 2004;3:291–304. doi: 10.1016/S1474-4422(04)00737-9. [DOI] [PubMed] [Google Scholar]
- 4.Houlden H., Johnson J., Gardner-Thorpe C., Lashley T., Hernandez D., Worth P., Singleton A.B., Hilton D.A., Holton J., Revesz T., et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat. Genet. 2007;39:1434–1436. doi: 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]
- 5.Waters M.F., Minassian N.A., Stevanin G., Figueroa K.P., Bannister J.P., Nolte D., Mock A.F., Evidente V.G., Fee D.B., Muller U., et al. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat. Genet. 2006;38:447–451. doi: 10.1038/ng1758. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda Y., Dick K.A., Weatherspoon M.R., Gincel D., Armbrust K.R., Dalton J.C., Stevanin G., Durr A., Zuhlke C., Burk K., et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat. Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- 7.Chen D.H., Brkanac Z., Verlinde C.L., Tan X.-J., Bylenok L., Nochlin D., Matsushita M., Lipe H., Wolff J., Fernandez M., et al. Missense mutations in the regulatory domain of PKC gamma: a new mechanism for dominant nonepisodic cerebellar ataxia. Am. J. Hum. Genet. 2003;72:839–849. doi: 10.1086/373883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Swieten J.C., Brusse E., de Graaf B.M., Krieger E., van de Graaf R., de Koning I., Maat-Kievit A., Leegwater P., Dooijes D., Oostra B.A., Heutink P. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia. Am. J. Hum. Genet. 2003;72:191–199. doi: 10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van de Leemput J., Chandran J., Knight M.A., Holtzclaw L.A., Scholz S., Cookson M.R., Houlden H., Gwinn-Hardy K., Fung H.C., Lin X., et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:1076–1082. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwaki A., Kawano Y., Miura S., Shibata H., Matsuse D., Li W., Furuya H., Ohyagi Y., Taniwaki T., Kira J., Fukumaki Y. Heterozygous deletion of ITPR1, but not SUMF1, in spinocerebellar ataxia type 16. J. Med. Genet. 2008;45:32–35. doi: 10.1136/jmg.2007.053942. [DOI] [PubMed] [Google Scholar]
- 11.Knight M.A., Gardner R.J.M., Bahlo M., Matsuura T., Dixon J.A., Forrest S.M., Storey E. Dominantly inherited ataxia and dysphonia with dentate calcification: spinocerebellar ataxia type 20. Brain. 2004;127:1172–1181. doi: 10.1093/brain/awh139. [DOI] [PubMed] [Google Scholar]
- 12.Storey E., Knight M.A., Forrest S.M., Gardner R.J.M. Spinocerebellar ataxia type 20. Cerebellum. 2005;4:55–57. doi: 10.1080/14734220410019048. [DOI] [PubMed] [Google Scholar]
- 13.Lorenzo D.N., Forrest S.M., Ikeda Y., Dick K.A., Ranum L.P., Knight M.A. Spinocerebellar ataxia type 20 is genetically distinct from spinocerebellar ataxia type 5. Neurology. 2006;67:2084–2085. doi: 10.1212/01.wnl.0000247662.05197.59. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka H., Bergstrom D.A., Yao M.C., Tapscott S.J. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 15.Jehee F.S., Bertola D.R., Yelavarthi K.K., Krepischi-Santos A.C., Kim C., Vianna-Morgante A.M., Vermeesch J.R., Passos-Bueno M.R. An 11q11–q13.3 duplication, including FGF3 and FGF4 genes, in a patient with syndromic multiple craniosynostoses. Am. J. Med. Genet. 2007;143A:1912–1918. doi: 10.1002/ajmg.a.31863. [DOI] [PubMed] [Google Scholar]
- 16.Zarate Y.A., Kogan J.M., Schorry E.K., Smolarek T.A., Hopkin R.J. A new case of de novo 11q duplication in a patient with normal development and intelligence and review of the literature. Am. J. Med. Genet. 2007;143A:265–270. doi: 10.1002/ajmg.a.31519. [DOI] [PubMed] [Google Scholar]
- 17.Bisogno T., Howell F., Williams G., Minassi A., Cascio M.G., Ligresti A., Matias I., Schiano-Moriello A., Paul P., Williams E.J., et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 2003;163:463–468. doi: 10.1083/jcb.200305129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stöhr H., Marquardt A., White K., Weber B.H. cDNA cloning and genomic structure of a novel gene (C11orf9) localized to chromosome 11q12–>q13.1 which encodes a highly conserved, potential membrane-associated protein. Cytogenet. Cell Genet. 2000;88:211–216. doi: 10.1159/000015552. [DOI] [PubMed] [Google Scholar]
- 19.Adachi N., Karanjawala Z.E., Matsuzaki Y., Koyama H., Lieber M.R. Two overlapping divergent transcription units in the human genome: the FEN1/C11orf10 locus. OMICS: J. Integr. Biol. 2002;6:273–279. doi: 10.1089/15362310260256927. [DOI] [PubMed] [Google Scholar]
- 20.Hiraoka L.R., Harrington J.J., Gerhard D.S., Lieber M.R., Hsieh C.L. Sequence of human FEN-1, a structure-specific endonuclease, and chromosomal localization of the gene (FEN1) in mouse and human. Genomics. 1995;25:220–225. doi: 10.1016/0888-7543(95)80129-a. [DOI] [PubMed] [Google Scholar]
- 21.Marquardt A., Stöhr H., White K., Weber B.H.F. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 22.Luo H.R., Saiardi A., Nagata E., Ye K., Yu H., Jung T.S., Luo X., Jain S., Sawa A., Snyder S.H. GRAB: a physiologic guanine nucleotide exchange factor for Rab3A, which interacts with inositol hexakisphosphate kinase. Neuron. 2001;31:439–451. doi: 10.1016/s0896-6273(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 23.Stöhr H., Marquardt A., Rivera A., Cooper P.R., Nowak N.J., Shows T.B., Gerhard D.S., Weber B.H. A gene map of the Best’s vitelliform macular dystrophy region in chromosome 11q12–q13.1. Genome Res. 1998;8:48–56. doi: 10.1101/gr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentze M.W., Keim S., Papadopoulos P., O’Brien S., Modi W., Drysdale J., Leonard W.J., Harford J.B., Klausner R.D. Cloning, characterization, expression, and chromosomal localization of a human ferritin heavy-chain gene. Proc. Natl Acad. Sci. USA. 1986;83:7226–7230. doi: 10.1073/pnas.83.19.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim J., Hao T., Shaw C., Patel A.J., Szabó G., Rual J.F., Fisk C.J., Li N., Smolyar A., Hill D.E., et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida T., Fukaya M., Uchigashima M., Miura E., Kamiya H., Kano M., Watanabe M. Localization of diacylglycerol lipase-alpha around postsynaptic spine suggests close proximity between production site of an endocannabinoid, 2-arachidonoyl-glycerol, and presynaptic cannabinoid CB1 receptor. J. Neurosci. 2006;26:4740–4751. doi: 10.1523/JNEUROSCI.0054-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun H., Tsunenari T., Yau K.W., Nathans J. The vitelliform macular dystrophy protein defines a new family of chloride channels. Proc. Natl Acad. Sci. USA. 2002;99:4008–4013. doi: 10.1073/pnas.052692999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon-Sanchez J., Scholz S., Fung H.C., Matarin M., Hernandez D., Gibbs J.R., Britton A., de Vrieze F.W., Peckham E., Gwinn-Hardy K., et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum. Mol. Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔ C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka H., Cao Y., Bergstrom D.A., Kooperberg C., Tapscott S.J., Yao M.C. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol. Cell. Biol. 2007;27:1993–2002. doi: 10.1128/MCB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Datson N.A., Semina E., van Staalduinen A.A., Dauwerse H.G., Meershoek E.J., Heus J.J., Frants R.R., den Dunnen J.T., Murray J.C., van Ommen G.J. Closing in on the Rieger syndrome gene on 4q25: mapping translocation breakpoints within a 50-kb region. Am. J. Hum. Genet. 1996;59:1297–1305. [PMC free article] [PubMed] [Google Scholar]
- 32.Giles R.H., Petrij F., Dauwerse H.G., den Hollander A.I., Lushnikova T., van Ommen G.J., Goodman R.H., Deaven L.L., Doggett N.A., Peters D.J., Breuning M.H. Construction of a 1.2-Mb contig surrounding, and molecular analysis of, the human CREB-binding protein (CBP/CREBBP) gene on chromosome 16p13.3. Genomics. 1997;42:96–114. doi: 10.1006/geno.1997.4699. [DOI] [PubMed] [Google Scholar]
- 33.Dauwerse J.G., Jumelet E.A., Wessels J.W., Saris J.J., Hagemeijer A., Beverstock G.C., van Ommen G.J., Breuning M.H. Extensive cross-homology between the long and the short arm of chromosome 16 may explain leukemic inversions and translocations. Blood. 1992;79:1299–1304. [PubMed] [Google Scholar]
- 34.Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]