Abstract
Three Pakistani populations residing in northern Pakistan, the Burusho, Kalash and Pathan claim descent from Greek soldiers associated with Alexander’s invasion of southwest Asia. Earlier studies have excluded a substantial Greek genetic input into these populations, but left open the question of a smaller contribution. We have now typed 89 binary polymorphisms and 16 multiallelic, short-tandem-repeat (STR) loci mapping to the male-specific portion of the human Y chromosome in 952 males, including 77 Greeks in order to re-investigate this question. In pairwise comparisons between the Greeks and the three Pakistani populations using genetic distance measures sensitive to recent events, the lowest distances were observed between the Greeks and the Pathans. Clade E3b1 lineages, which were frequent in the Greeks but not in Pakistan, were nevertheless observed in two Pathan individuals, one of whom shared a 16 Y-STR haplotype with the Greeks. The worldwide distribution of a shortened (9 Y-STR) version of this haplotype, determined from database information, was concentrated in Macedonia and Greece, suggesting an origin there. Although based on only a few unrelated descendants this provides strong evidence for a European origin for a small proportion of the Pathan Y chromosomes.
Keywords: Population genetics, Pakistan, Greek, Y chromosome polymorphism
Introduction
Pakistan lies within a region that was invaded by Alexander the Great in 327-323 BC 1, although archeological evidence in northern Pakistan suggests that the Greek influence predates this invasion 2. Many ethnically and linguistically distinct populations inhabit this region, three of which (Burusho, Kalash and the Pathan) claim to be descendents of Greek soldiers who invaded the Indian sub-continent 3, 4, 5. A preliminary study using a limited number of Y-chromosomal markers found no evidence for admixture between the Greeks and Burusho or Pathan, and provided ambiguous evidence of genetic admixture between the Greek and Kalash populations 6. A subsequent analysis of autosomal loci gave no indication that the Kalash or other populations were genetically related to the Greeks 7 and in another study the Kalash population were shown as a genetic isolate 8 who might therefore have developed unusual genetic characteristics by drift 9.
The genetic material comprising the great bulk of the patrilinearally inherited human Y-chromosome is effectively haploid and does not undergo inter-chromosomal recombination, making it useful for evolutionary studies and forensic investigations 10. In particular, it can provide very high resolution haplotypes with known phylogenetic relationships, and these can reveal low levels of admixture that would not be detected by other methods. In the present study we have further investigated the origin and the genetic relationship of these three Pakistani populations with the extant Greek population by typing a large set of markers from the male-specific region of the Y chromosome in 77 Greeks and 875 Pakistani individuals, and applying analytical methods that should be more sensitive to low levels of admixture.
Material and Methods
DNA Samples
DNA samples of 952 unrelated males were analyzed in this study. The DNA was extracted directly from peripheral blood mononuclear cells in case of the Greek samples (n = 77) and from EBV-transformed lymphoblastoid cell lines for the Pakistani samples (n = 875). The Pakistani samples included Burusho, Kalash and Pathan individuals. Informed consent was obtained from all participants in this study.
Genotyping
All samples were typed for 91 binary markers 11 following a phylogenetic hierarchical approach to define evolutionary Y lineages or haplogroups 10. The markers and the method used for their genotyping are listed in Table 1. In addition, 16 multiallelic Y-STR markers were analyzed in these populations by three multiplex PCR reactions 12, 13 to identify Y haplotypes.
Table 1.
List of 89 Y-SNPs typed in this study
| Y-SNP | Typing Method | Polymorphism | Y-SNP | Typing Method | Polymorphism |
|---|---|---|---|---|---|
| M91 | DHPLC | del T | M201 | DHPLC | G→T |
| M31 | DHPLC | G→C | M147 | DNA Sequencing | ins T |
| M6 | DHPLC | T→C | M177 | DNA Sequencing | C→T |
| PK1 | AFLP | C→A | M143 | DHPLC | G→T |
| M32 | DHPLC | T→C | M20 | AFLP | A→G |
| M60 | DHPLC | ins T | M11 | AFLP | A→G |
| M150 | DHPLC | C→T | M185 | DHPLC | C→T |
| M109 | DHPLC | C→T | M27 | ARMS-PCR | C→G |
| M152 | DHPLC | C→T | M76 | DHPLC | T→G |
| M218 | DHPLC | C→T | M349 | DHPLC | G→T |
| RPS4Y | AFLP | C→T | M357 | DHPLC | C→A |
| M8 | DNA Sequencing | G→T | PK3 | ARMS-PCR | T→C |
| M38 | DNA Sequencing | T→G | M214 | ARMS-PCR | A→G |
| M217 | DNA Sequencing | A→C | M175 | DNA Sequencing | del TTCTC |
| M48 | ARMS-PCR | A→G | M119 | DHPLC | A→C |
| PK2 | ARMS-PCR | T→C | M101 | DHPLC | C→T |
| YAP | PCR | Alu ins | M50 | DHPLC | T→C |
| SRY8299 | AFLP | G→A | M103 | DHPLC | C→T |
| sY81 | AFLP | A→G | M110 | DNA Sequencing | T→C |
| M35 | ARMS-PCR | G→C | P31 | DNA Sequencing | T→C |
| M78 | ARMS-PCR | C→T | M88 | DNA Sequencing | A→G |
| M148 | DHPLC | A→G | M111 | DNA Sequencing | del TT |
| M123 | ARMS-PCR | G→A | M122 | ARMS-PCR | T→C |
| M136 | DHPLC | C→T | L1Y | PCR | LINE1 ins |
| M89 | ARMS-PCR | C→T | M134 | DHPLC | del G |
| M9 | AFLP | C→G | M117 | DHPLC | del ATCT |
| 92R7 | AFLP | C→T | M133 | DHPLC | del T |
| M45 | DHPLC | G→A | SRY+465 | AFLP | C→T |
| M74 | DHPLC | G→A | PK4 | DHPLC | A→T |
| M207 | ARMS-PCR | A→G | LLY22g | AFLP | C→A |
| M124 | ARMS-PCR | C→T | M231 | DHPLC | G→A |
| M173 | ARMS-PCR | A→C | TAT | AFLP | T→C |
| M73 | DHPLC | del GT | M70 | ARMS-PCR | A→C |
| SRY-2627 | AFLP | C→T | M193 | DHPLC | ins CAAA |
| SRY-1532 | AFLP | A→G→A | 12f2 | PCR | del |
| M17 | ARMS-PCR | del G | M172 | ARMS-PCR | T→G |
| M56 | DHPLC | A→T | M12 | DHPLC | G→T |
| M157 | DHPLC | A→C | M67 | ARMS-PCR | A→T |
| M87 | DHPLC | T→C | M92 | ARMS-PCR | T→C |
| PK5 | AFLP | C→T | M267 | ARMS-PCR | T→G |
| M242 | ARMS-PCR | C→T | M62 | ARMS-PCR | T→C |
| M25 | DHPLC | G→C | M52 | ARMS-PCR | A→C |
| M36 | DHPLC | T→G | M69 | DHPLC | T→C |
| M97 | DHPLC | T→G | Apt | AFLP | G→A |
| M170 | ARMS-PCR | A→C | M82 | DHPLC | del AT |
Data Analysis
Population pairwise ΦST values were estimated by using the Arlequin package 14 based on STR variation within haplogroups weighted as described previously 6. Population pairwise ρ genetic distances were calculated according to Helgason et al. 15. Median-joining networks were constructed by Network 4.1.1.2 16 using the following five-fold range weighting scheme. The weights assigned were specific for the haplogroup and took into account the Y-STR variation across the haplogroup in the Pakistani and Greek populations: variance 0.00-0.14, weight 5; variance 0.15-0.29, weight 4; variance 0.30-0.44, weight 3; variance 0.45-0.59, weight 2; variance >0.59, weight 1. Network was also used to estimate the time to the most recent common ancestor (TMRCA).
Results
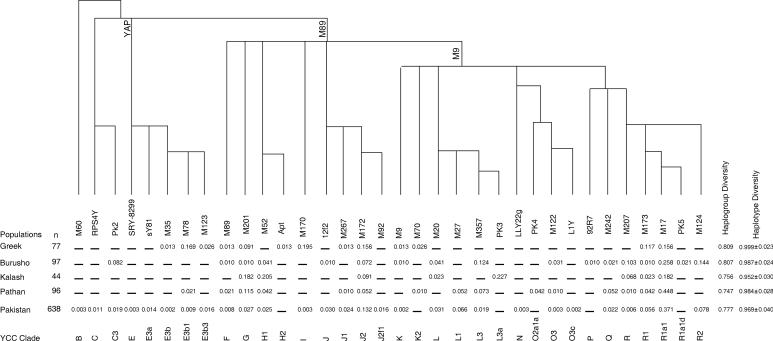
The Y-chromosomal lineages and their frequencies in the Greeks, Burusho, Kalash, Pathan and the rest of the Pakistani population are shown in Figure 1. The combination of biallelic markers identified 12 Y-chromosomal haplogroups or lineages in the Greeks, 17 in the Burusho and 15 in the Pathan populations. Only 8 Y lineages were found in the Kalash population. Principal component analysis of Y haplogroup frequencies incorporating published data from European17 and West Asian 18, 19 populations (data not shown) revealed that the Pakistani populations cluster together, separately from the Europeans, consistent with the previous conclusion that none of the Pakistani populations had a large male contribution from Greece, and demonstrating that this conclusion was not an artifact of the low phylogenetic resolution used before. The genetic distances between the populations were then calculated using measures that are more sensitive to recent events (Table 2). Pakistani-Greek population pairwise ΦST values based on the variation of STRs within haplogroups 6 ranged from 0.131 to 0.213, with the lowest value between the Pathan and the Greeks. Pairwise ρ genetic distances (the number of steps between a haplotype in one population and the closest haplotype in the second population, averaged over all comparisons) 15 ranged from 4.3 to 8.1, with the lowest value again between the Pathan and the Greeks. These results therefore suggest that there might have been a low degree of recent Pathan-Greek admixture. To investigate this possibility further, we have examined individual lineages.
Figure 1.
A. rooted maximum-parsimony tree of Y lineages found in the Greek, Burusho, Kalash, Pathan and Pakistan. The lineages were defined by binary markers whose designations and population frequencies are given below each branch. The YCC lineage names 10 are shown below the frequencies. Branch lengths are arbitrary.
Table 2.
Weighted population pairwise rho genetic distances (below diagonal) and ΦST values (above diagonal) based on STR variation within haplogroups
| Greek | Burusho | Kalash | Pathan | |
|---|---|---|---|---|
| Greek | 0.000 | 0.188 | 0.213 | 0.131 |
| Burusho | 5.659 | 0.000 | 0.214 | 0.196 |
| Kalash | 8.066 | 3.882 | 0.000 | 0.219 |
| Pathan | 4.277 | 2.451 | 3.254 | 0.000 |
Clade E lineages were more frequent in the Greeks (21%) as compared to Pakistan (4%). The majority of haplogroup E chromosomes belonged to clade E3b and all Greek and Pakistani samples were resolved into the branches E3b1 (M78) and E3b3 (M123). Among the three Pakistani populations claiming Greek descent, this clade was observed only in the Pathans. The Pathan samples belonged to clade E3b1 which constituted 17% of the Greek samples.
A median-joining network of clade E Y chromosomes was constructed in order to examine the genetic relationship between these Greek and Pathan samples. A duplication of 10 and 13 repeat units was observed in the clade-E-derived Y chromosomes for the trinucleotide repeat DYS425 and this locus was, therefore, excluded from the network. The most striking feature of this network was the sharing of haplotypes between the Pathan and Greek samples (Figure 2). One Pathan individual shared the same Y-STR haplotype with three Greek individuals, and the other Pathan sample was separated from this cluster by a single mutation at the DYS436 locus. This demonstrates a very close relationship between the Pathan and Greek E lineages, but how surprising is this?
Figure 2.
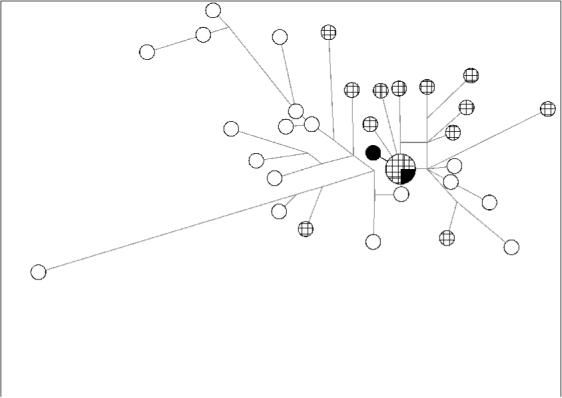
Median-Joining network of clade E lineages in Pakistan (open circles) and Greece (hatched circles). Circles represent haplotypes and have an area proportional to frequency. The Pathan individuals are shown in black.
Worldwide data for the 16-element haplotype are not available, but a subset of nine of the STRs are included in by the Y-STR Haplotype Reference Database (YHRD) 20 and were used to search this. The haplotype DYS19=13; 389I=13; 389II=30; 390=24; 391=10; 392=11; 393=13; 438=10; 439=12 was found in 53 individuals in a worldwide population sample of 7,897 haplotypes and was highly specific for the Balkans (Figure 3). The contour map shows a major concentration around Macedonia and Greece, with a low scattering in other European countries, Tunisia, West Africa and the Pathans. This gives a strong indication of a European, possibly Greek, origin of these Pathan Y chromosomes.
Figure 3.

Contour map showing the 9 Y-STR haplotype frequency distribution in Eurasia and northern Africa. This haplotype was shared between three Greeks and a Pathan individual belonging to clade E3b1.
Discussion
An extensive analysis of Y diversity within Greeks and three Pakistani populations ___ the Burusho, Kalash and Pathan ___ who claim descent from Greek soldiers allowed us to compare Y lineages within these populations and re-evaluate their suggested Greek origins. This study as a whole seems to exclude a large Greek contribution to any Pakistani population, confirming previous observations 7. However, it provides strong evidence in support of the Greek origins for a small proportion of Pathans, as demonstrated by the clade E network (Figure 2) and the low pairwise genetic distances between these two populations.
The Pathans were the only population among the three that claim Greek ancestry in which clade E was present. This branch is observed in Europe, Middle East, North and East Africa with a suggested origin in East Africa 21. Sub-clade E3b is common in Europe and probably originated in Africa 22. Compelling evidence in support of the genetic relationship between the Pathan and Greek E3b1 Y chromosomes was provided by the median-joining network (Figure 2). One Pathan shared a Y-STR haplotype, that included a duplication of 10 and 13 repeat units for the DYS425 locus, with three Greek individuals and the other was separated from this cluster by a single mutation, which enabled us to estimate the TMRCA (mean ± SD) using the Network software as between 2000 ± 400-5000 ± 1200 YBP depending upon the observed 23 or inferred mutation rates, 24 respectively. This coincides with the period of Alexander’s invasion during 327-323 B.C. This haplotype was not observed in any other E3b1 derived Pakistani Y chromosome but was highly specific for the Balkans ____ the highest frequency being in Macedonia.
It is worth emphasizing here that the chance of picking up rare events largely amplified by drift affecting a limited portion of the population can not be discounted. The genetic data alone do not tell us when the Balkan chromosomes arrived in Pakistan: it is necessary to turn to the historical record for this. There has been no known Greek admixture within the last few generations, but in addition to Alexander’s armies, the possibility of admixture between the Greek slaves who were brought to this region by Xerxes around one hundred and fifty years before Alexander’s arrival, and the local population, cannot be discounted. At that time Afghanistan and present day Pakistan were part of the Persian Empire 1. Nevertheless, Alexander’s army of 25,000-30,000 mercenary foot soldiers from Persia and West Asia and 5000-7000 Macedonian cavalry25 perhaps provides a more likely explanation because of their elite status and substantial political impact on the region.
Acknowledgments
We thank the donors for the samples used in this study. This work was supported by a Wellcome Trust Collaborative Research Initiative Grant to S. Qasim Mehdi. CT-S was supported by the Wellcome Trust. The authors would like to thank S Siddiqi and A Mansoor for technical assistance and Professors LL Cavalli-Sforza, SQM and anonymous reviewers for their helpful comments.
References
- 1.Wolpert S. A new history of India. Oxford University Press; New York: 2000. [Google Scholar]
- 2.Jettmar K. Beyond the Gorges of the Indus: Archaeology before Excavation. Oxford University Press; Karachi, Pakistan: 2001. [Google Scholar]
- 3.Biddulph J. Tribes of the Hindoo Koosh. Indus Publications; Karachi, Pakistan: 1977. [Google Scholar]
- 4.Lines M. The Kalasha People of North-western Pakistan. Emjay Books International; Peshawar, Pakistan: 1999. [Google Scholar]
- 5.Caroe O. The Pathans. Oxford University Press; Karachi, Pakistan: 1958. [Google Scholar]
- 6.Qamar R, Ayub Q, Mohyuddin A, et al. Y-chromosomal DNA variation in Pakistan. Am J Hum Genet. 2002;70:1007–1024. doi: 10.1086/339929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mansoor A, Mazhar K, Khaliq S, et al. Investigation of the Greek ancestry of populations from northern Pakistan. Hum Genet. 2004;114:484–490. doi: 10.1007/s00439-004-1094-x. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg NA, Mahajan S, Ramachandran S, Zhao C, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 2005;1:e70. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jobling MA, Tyler-Smith C. The human Y chromosome:an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 11.Underhill PA, Shen P, Lin AA, et al. Y chromosome binary haplotypes and origins of modern human populations. Ann Hum Genet. 2001;26:358–361. doi: 10.1046/j.1469-1809.2001.6510043.x. [DOI] [PubMed] [Google Scholar]
- 12.Thomas MG, Bradman N, Flinn HM. High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet. 1999;105:577–581. doi: 10.1007/s004399900181. [DOI] [PubMed] [Google Scholar]
- 13.Ayub Q, Mohyuddin A, Qamar R, et al. Identification and characterisation of novel human Y-chromosomal microsatellites from sequence database information. Nucleic Acids Res. 2000;28:e8. doi: 10.1093/nar/28.2.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider S, Kueffer J-M, Roessli D, Excoffier L. Arlequin ver 1.1: a software for population genetic data analysis. Genetics and Biometry Laboratory, University of Geneva; Switzerland: 1997. [Google Scholar]
- 15.Helgason A, Sigurdardottir S, Nicholson J, et al. Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet. 2000;67:697–717. doi: 10.1086/303046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 17.Francalacci P, Morelli L, Underhill PA, et al. Peopling of three Mediterranean islands (Corsica, Sardinia, and Sicily) inferred by Y-chromosome biallelic variability. Am J Phys Anthropol. 2003;121:270–279. doi: 10.1002/ajpa.10265. [DOI] [PubMed] [Google Scholar]
- 18.Flores C, Maca-Meyer N, Larruga JM, et al. Isolates in a corridor of migrations:a high-resolution analysis of Y-chromosome variation in Jordan. J Hum Genet. 2005;50:435–441. doi: 10.1007/s10038-005-0274-4. [DOI] [PubMed] [Google Scholar]
- 19.Cinnioglu C, King R, Kivisild T, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- 20.Roewer L, Krawczak M, Willuweit S, et al. Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci Int. 2001;118:106–113. doi: 10.1016/s0379-0738(00)00478-3. [DOI] [PubMed] [Google Scholar]
- 21.Semino O, Magri C, Benuzzi G, et al. Origin, diffusion, and differentiation of Y-chromosome haplogroup E and J:Inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet. 2004;74:1023–1034. doi: 10.1086/386295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruciani F, La Fratta R, Santolamazza PG, et al. Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet. 2004;74:1014–1022. doi: 10.1086/386294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kayser M, Roewer L, Hedman M, et al. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet. 2000;66:1580–1588. doi: 10.1086/302905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhivotovsky LA, Underhill PA, Cinnioglu C, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engels DW. Alexander the Great and the Logistics of the Macedonian Army. University of California Press; Berkeley, CA: 1981. [Google Scholar]