Abstract
Over 30% of proteins are secreted across or integrated into membranes. Their newly synthesized forms contain either cleavable signal sequences or non-cleavable membrane anchor sequences, which direct them to the evolutionarily conserved Sec translocon (SecYEG in prokaryotes and Sec61, comprising α-, γ- and β-subunits, in eukaryotes). The translocon then functions as a protein-conducting channel1. These processes of protein localization occur either at or after translation. In bacteria, the SecA ATPase2,3 drives post-translational translocation. The only high-resolution structure of a translocon available so far is that for SecYEβ from the archaeon Methanococcus jannaschii4, which lacks SecA. Here we present the 3.2-Å-resolution crystal structure of the SecYE translocon from a SecA-containing organism, Thermus thermophilus. The structure, solved as a complex with an anti-SecY Fab fragment, revealed a ‘pre-open’ state of SecYE, in which several transmembrane helices are shifted, as compared to the previous SecYEβ structure4, to create a hydrophobic crack open to the cytoplasm. Fab and SecA bind to a common site at the tip of the cytoplasmic domain of SecY. Molecular dynamics and disulphide mapping analyses suggest that the pre-open state might represent a SecYE conformational transition that is inducible by SecA binding. Moreover, we identified a SecA–SecYE interface that comprises SecA residues originally buried inside the protein, indicating that both the channel and the motor components of the Sec machinery undergo cooperative conformational changes on formation of the functional complex.
SecY or Sec61α, the core component of the translocon, has ten transmembrane (TM1–TM10), six cytoplasmic (C1–C6) and five periplasmic/luminal (P1–P5) regions5 (see Supplementary Fig. 1). The amino-terminal (TM1–TM5) and carboxy-terminal (TM6–TM10) halves of SecY are assembled into a pseudo-symmetrical structure, having an hourglass-shaped channel in its interior that is gated from the periplasmic side by a ‘plug’ helix4. The translocon also has a lateral gate, through which a pre-protein’s signal peptide may enter the channel to initiate the translocation process, and the hydrophobic segment of a membrane protein may exit into the lipid phase to establish a transmembrane configuration. Although the reported M. jannaschii SecYEβ structure is in a closed, resting state, the channel that is formed within a single heterotrimer indeed mediates translocation6-8. Nevertheless, the fully functional translocon might require oligomerization of the SecY (Sec61) complex9, which provides the binding platform for the cytoplasmic partners (such as the ribosome10,11 and SecA12) or forms a larger channel13. In the dimer model of the Escherichia coli SecYEG–SecA system12, one copy of SecYEG provides a docking site for SecA, whereas another copy is used as a translocating pore. SecA2,3,5 interacts with SecYEG14, partially activating its ATPase15 (‘membrane ATPase’), which is then enhanced fully by a pre-protein (‘translocation ATPase’) during active, ongoing translocation. The C4 and C5 regions of SecY (see Supplementary Fig. 1), which are critical for the activation of SecA5,16,17, contain residues that contact the ATPase domain of SecA12,18. Although SecA is believed to function through large conformational changes19, its functional oligomeric state9,20 (see Supplementary Discussion), as well as the molecular details of the motor function, remain elusive. We chose to study the Sec machinery from Thermus thermophilus21, including the crystal structure determination of SecA20 and SecYE, the subject of this report.
Initial crystals of T. thermophilus SecYE showed a resolution limit of ~6Å. Resolution was improved when SecYE was in complex with the Fab fragment of a monoclonal antibody against T. thermophilus SecY. The crystal structure was determined by the multiple anomalous dispersion (MAD) method, in which SecYE had been labelled with selenomethionine (Supplementary Table 1). The refined model, at a 3.2Å resolution, includes most of the residues of Fab, SecY and SecE (Fig. 1). The electron density map displays most of the amino acid side chains of these components (Supplementary Fig. 2). The overall architecture of SecYE is similar to that of M. jannaschii SecYEβ4 (Supplementary Fig. 3), in that it has an inverted, pseudo-symmetrical arrangement of transmembrane helices, an hourglass-shaped conduit with the ‘pore ring’ constriction (Supplementary Fig. 2b), and a plug helix. The cytoplasmic C4–C5 loops, to which SecA and the ribosome bind4,10-12,18, protrude prominently from the membrane-embedded region. The Fab binds tightly to the highly conserved Ile 347–Phe 359 segment in the C5 loop of SecY (Supplementary Fig. 4a).
Figure 1. Overall structure of T. thermophilus SecYE.
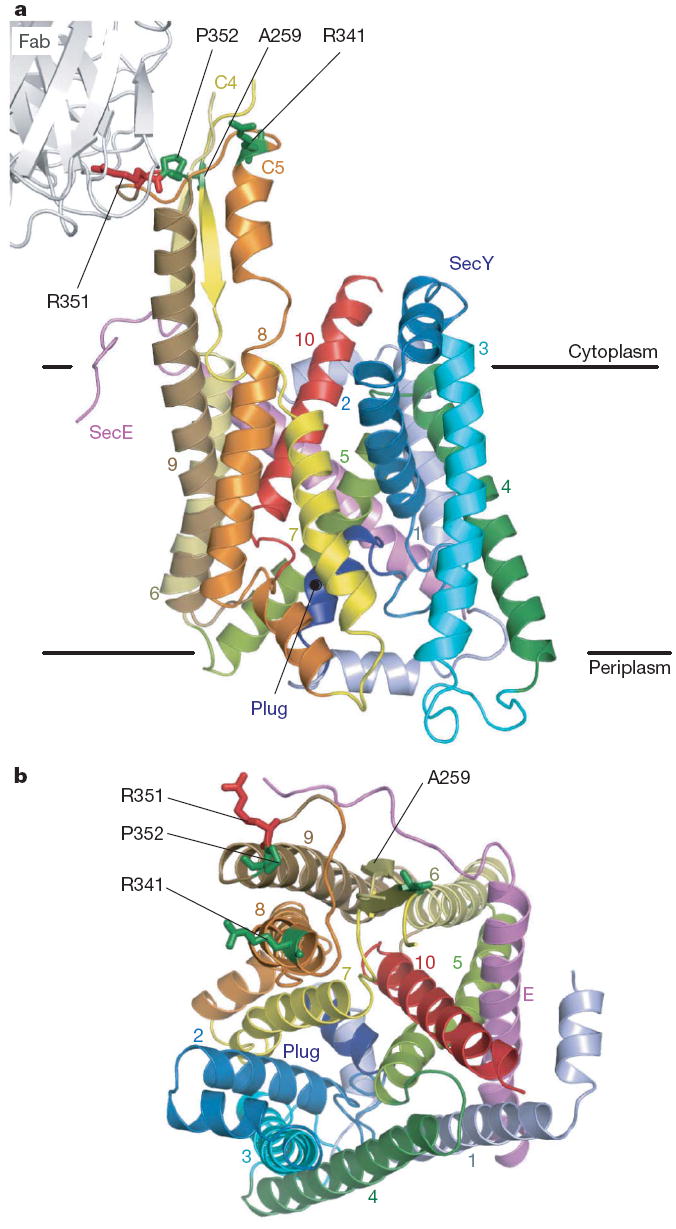
a, b, The SecYE complex viewed from the lateral gate side (a) and the cytoplasm (b). The SecY transmembranes are coloured light blue to red from the N to C termini, and SecE is coloured pink. Arg 351 (ref. 17) is coloured red and is shown in stick representation. The residues coloured green in stick representation were mutated to cysteine for intermolecular crosslinking experiments.
Despite the overall similarity, a closer comparison with M. jannaschii SecYEβ revealed that their C-terminal halves occupy different conformations (Supplementary Figs 3 and 5). The Cα root mean squared deviation was 1.36Å for TM1–TM5 and 2.85Å for TM6–TM10. TM6, TM8 and TM9 of T. thermophilus SecYE show displacements of 5Å, 4Å and 6Å, respectively, from those in M. jannaschii SecYEβ, expanding the TM2–TM8 distance in the lateral gate area of the former (arrows in Supplementary Fig. 5b). TM6 and TM8 are tilted by ~25° and ~10°, respectively, and TM9 shows a translational movement to the periphery of the channel (Supplementary Fig. 5c). Similar displacement of TM8 and TM9 was noted, when the SecYE structure was compared with the models of the E. coli SecY structure10,22 (Supplementary Fig. 6). The altered transmembrane arrangement creates a ~6 × 15Å hydrophobic crack formed by the cytoplasmic segments of TM2, TM7 and TM8 (Fig. 2a and Supplementary Fig. 7). SecYE with an exposed crack could represent an alternative conformation of the translocon that we term ‘pre-open’ as opposed to or to distinguish it form the closed state of M. jannaschii SecYEβ.
Figure 2. Comparison of the T. thermophilus SecYE and M. jannaschii SecYEβ structures.
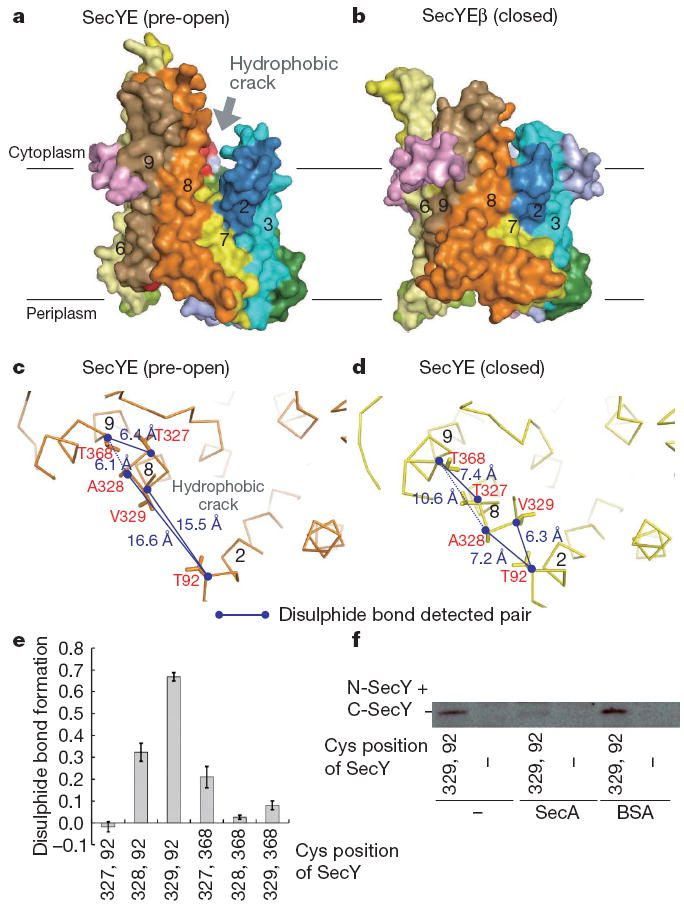
a, b, Molecular surfaces of SecYE (a) and SecYEβ (b), coloured as in Fig. 1a. Transmembrane regions are numbered. c, d, The cytoplasmic regions of TM2, TM8 and TM9 of SecY. Pre-open (c, crystal structure of Fab–SecYE) and closed forms (d, without Fab, molecular dynamics analysis at 72.93 ns) are shown. Numbers show distances between α carbons. e, Intramolecular disulphide bond formation in SecY as assessed by quantitative carboxymethylation. Averages of three analyses are shown with s.d. f, SecA-mediated inhibition of SecY (Thr92Cys–Val329Cys) intramolecular disulphide bond formation in the presence of AMP-PNP.
To address whether T. thermophilus SecY undergoes conformational transition between the pre-open and closed states, we performed a molecular dynamics simulation (Supplementary Discussion). During the 100-ns simulation, SecYE underwent large conformational changes, in which TM8 and TM9 moved from that of the open state towards the closed state (Supplementary Fig. 8 and Supplementary Movies 1 and 2). The low (6.0Å) resolution crystal structure of the antibody-free SecYE (Supplementary Table 1 and Supplementary Discussion) also showed a tendency towards the closed form, with the swinging and bending of TM8 (Supplementary Fig. 9). Taken together, the closed form seems to represent the energetically favoured, ground state of SecYE in the absence of any interacting components. In turn, it is conceivable that the pre-open form is a conformational sub-state induced by the antibody binding and possibly by the binding of a physiological ligand, SecA.
T. thermophilus SecA binds to the same (Pro-Gly-Ile-Arg-Pro-Gly) motif in C5 (see Fig. 3a) as Fab (Supplementary Fig. 4a), which inhibited the in vitro translocation activity of the translocon (Supplementary Fig. 4b). The arrangements of the transmembrane segments of SecYE were studied by intramolecular disulphide bond formation between cysteines placed at six different pairs of positions (Fig. 2c, d), which was assessed by reductive and non-reductive carboxymethylation. These data showed that SecY(Thr92Cys–Ala328Cys), SecY(Thr92Cys–Thr329Cys) and SecY(Thr327Cys–Val368Cys) form intramolecular disulphide bonds (Fig. 2e). The 92–328 and 92–329 linkages suggest that the isolated SecYE predominantly assumes the closed conformation, rather than the pre-open form (Fig. 2c, d). SecA decreased the 92–329 disulphide bond formation in the membrane-embedded SecYE (Fig. 2f and Supplementary Fig. 10). This would be consistent with a notion that SecA facilitates the conformational transition of SecYE from the closed to the pre-open states. The swinging of TM8 might initiate the opening of the channel gate through exposure of the hydrophobic crack (Fig. 2a, b and Supplementary Fig. 3). The crack contains some evolutionarily conserved hydrophobic residues (Ile85, Pro273, Phe276, Ala277, Phe322 and Tyr326) (Supplementary Figs 2a, 7 and 11a), which our mutation studies have shown to be functionally important (Supplementary Discussion and Supplementary Fig. 11b, c). Considering the observation that a signal peptide contacts TM2, TM7 and TM823, it is tempting to speculate that the crack participates in the reception of signal peptides from SecA.
Figure 3. Contacting residues between T. thermophilus SecA and SecYE.
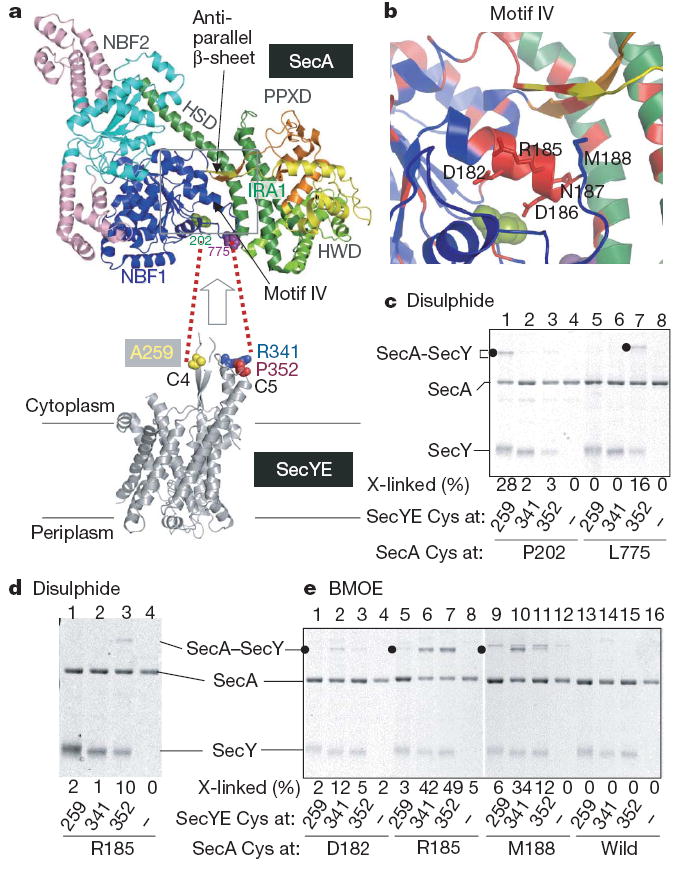
a, The SecA structure (Protein Data Bank 2IPC) is colour-coded for its domains30, except that light pink indicates SecA-specific regions. SecY contact residues identified in this study are space filled in green and purple. b, Close-up view of the α-helix in motif IV; evolutionarily conserved regions are shown in red, and the positions replaced by cysteines are indicated in stick representation. c, Disulphide crosslinking between SecA and SecY. The disulphide complex is indicated by solid circles. d, e, Disulphide and BMOE crosslinking of SecA and SecY.
SecA contains two nucleotide-binding folds (NBFs; also called NBD), a pre-protein crosslinking domain (PPXD) and C-terminal translocation domains (HWD and IRA1), which are all connected by a long α-helical scaffold domain (HSD) (Fig. 3a top panel and Supplementary Fig. 12). To identify the T. thermophilus SecA residues that contact the C4–C5 cytoplasmic domains of SecY (refs 12, 18), we introduced single cysteines into selected positions of SecA (Supplementary Fig. 12). We first mutated residues on the NBF1 surface, as NBF1 has been shown to be in the physical proximity of SecY (ref. 12). Single cysteines were also introduced into SecY positions 259, 341 and 352 (Figs 1 and 3a) for examination of the intermolecular disulphide bond formation (Fig. 3c, Supplementary Fig. 13 and Supplementary Discussion). Two combinations, SecA(Pro202Cys)–SecY(Ala259Cys) and SecA(Leu775Cys)–SecY(Pro352Cys), formed a disulphide bond on oxidation (Fig. 3a, c, lanes 1 and 7), in which SecYE must be proteoliposome-integrated (Supplementary Fig. 13a, c, lanes 9 and 15). Thus, residues 775 (Fig. 3a, purple) and 202 (Fig. 3a, green) of SecA are adjacent to the C5 and C4 residues of SecY, respectively (see Supplementary Discussion, Supplementary Fig. 13 and ref. 12 for the specificity of molecular interactions involving SecA).
Assuming that the molecular contacts between SecA at 202 and SecY at 259 and between SecA at 775 and SecY at 352 take place simultaneously, the two cytoplasmic protrusions of SecY must be accommodated between the NBF1 and C-terminal translocation domain of SecA (Fig. 3a). However, the SecA protomer structure lacks a large enough opening in this region (Fig. 3a, arrow), raising the possibility that the SecY-associated form of SecA has undergone a conformational change. We found that an evolutionarily conserved (Supplementary Fig. 14) region (180Gly-Phe-Asp-Tyr-Leu-Arg-Glu-Gln-Met188) corresponding to the C-terminal half of motif IV24 (Fig. 3b) interacts with SecY. Although the cysteines introduced into the motif IV positions of native SecA were not readily accessible to alkylation by a hydrophobic modifier (Supplementary Fig. 15 and Supplementary Discussion), they formed either a disulphide bond or a bis-maleimidoethane (BMOE; arm length of 8Å)-mediated cross-linkage with a specific cysteine at C5 of SecY (Fig. 3d, e and Supplementary Fig. 16). Thus, the interaction with the translocon leads to the exposure of residues 182, 185, 186 and 188 on the molecular surface (Supplementary Discussion). The motif IV region indeed undergoes a conformational change that is coordinated with the formation of a motor-translocon complex.
The ATPase of SecA is tightly downregulated in the resting state (intrinsic ATPase) through the interaction between the ATPase and the IRA1 domains25 (Fig. 3a). Our observation that binding to the translocon physically separates NBF1 and IRA1 excellently explains the translocon-mediated triggering of the membrane ATPase. The motif IV segment communicates with an anti-parallel β-sheet that is involved in the propagation of a pre-protein-binding signal to the ATPase domain26 (Fig. 3a, b). The surface exposure and the translocon interaction of motif IV might modulate the SecA function, not only by keeping IRA1 away, but also by enabling the β-sheet to activate the ATPase further, in response to pre-protein binding (translocation ATPase).
SecA interacts with SecY in at least two different modes18: the one involving the SecY C4–C5 domains is probably required for the ATPase activation, whereas the other, involving C6, accompanies the actual SecA-driven translocation. The SecA–SecY interface we identified in this study seems to correspond to the first interaction mode. According to the SecY dimer model by ref. 12, the non-translocating copy of SecY requires the essential Arg 357 residue17. The SecA–SecY interaction that we observed here directly involves Arg 351 (counterpart of E. coli SecY Arg 357) and might correspond to the one involving the non-translocating SecY copy (Fig. 4). In view of the occurrence of the second mode of SecA binding to the translocon, the translocating SecY copy should also interact with SecA.
Figure 4. Multiple modes of SecA–SecY interactions.
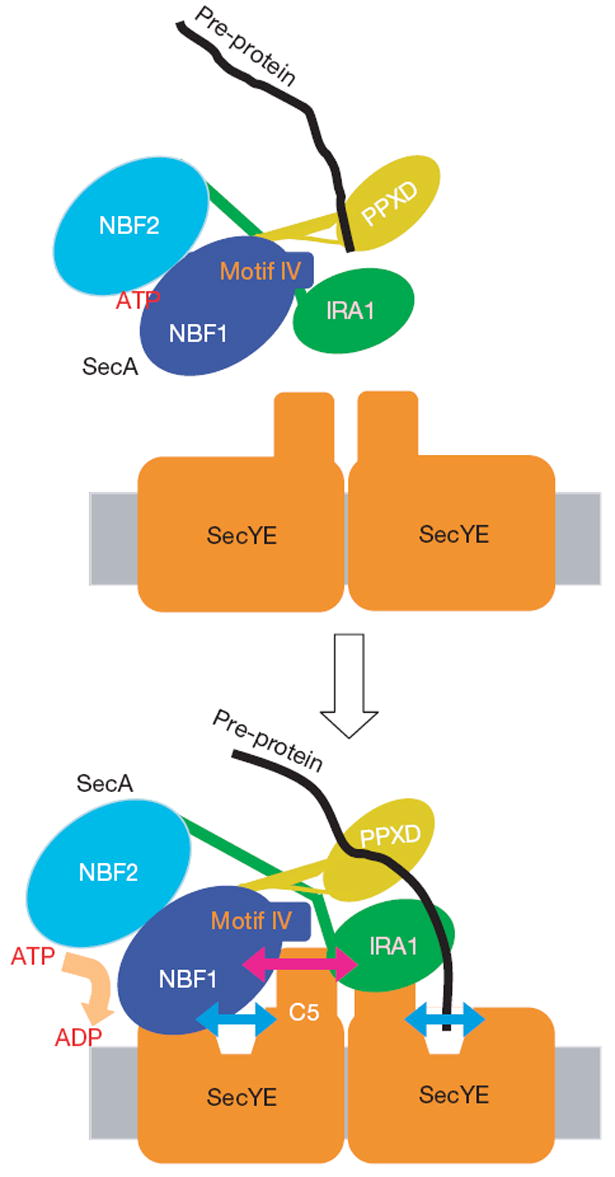
According to the dimer model12, one copy of SecY serves as a SecA-docking site, and the other functions as a translocation pore. The SecA–SecY interaction observed here should represent the one between the non-translocating copy of SecY and SecA, and is crucial for the SecA ATPase activation. Both the SecA and SecY components undergo conformational changes on their interaction, as shown by bidirectional arrows. The orientation of SecY protomers in the dimeric assembly is shown arbitrarily.
The binding of the Fab fragment or SecA to the cytoplasmic region of SecY seems to induce the pre-open state of the translocon. The opening may occur in both copies of SecY, and have a role in the initial reception of the signal-peptide–SecA complex, which could be followed by further propagation of conformational changes within the translocon (Fig. 4). The pre-opening could also be important for the activation of the SecA ATPase (Fig. 4). Our proposal that the translocon and the translocation-driving motor undergo cooperative conformational changes should be substantiated by the structural determination of the SecA–translocon complex with a bound pre-protein substrate.
METHODS SUMMARY
The T. thermophilus SecYE complex was overproduced in E. coli, solubilized from the membrane with n-dodecyl-β-d-maltoside and purified by three successive chromatography steps. It was mixed with the Fab fragment of a monoclonal antibody against SecY, and the Fab–SecYE complex was isolated by gel filtration chromatography. Crystals of the complex were grown by vapour diffusion. The structure of the SecYE complex was determined by the MAD method, using the selenomethionine-labelled SecYE crystals, with refinement to Rwork/Rfree of 24.4%/28.0% at 3.2Å resolution (Supplementary Table 1). Molecular graphics were generated with PyMol27. An all-atom molecular dynamics simulation of the pre-open form of SecYE with explicit solvent and phospholipids was performed using the NAMD2 program28, as described in Supplementary Information.
To quantify the intramolecular disulphide bond formation in the purified double-cysteine mutants of SecYE, the mutants were subjected to reductive or non-reductive carboxymethylation in the presence of 6 M guanidine hydrochloride, as described previously29. For intra- or intermolecular disulphide cross-linking, the samples were treated with an oxidant, Cu(phen)3, and were analysed by non-reducing SDS–PAGE. To assess the effect of T. thermophilus SecA on the conformation of SecYE, inverted membrane vesicles, prepared from E. coli cells overproducing the SecYE double cysteine mutants, were pre-incubated with SecA or BSA, as specified, before oxidation. For intermolecular crosslinking between SecYE and SecA, a single cysteine mutant of SecA was mixed with proteoliposomes containing SecYE with a single cysteine at a specific SecY position. For the BMOE-mediated crosslinking of SecY and SecA, single cysteine samples were incubated with BMOE.
Supplementary Material
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgments
We thank K. Inaba, Y. Akiyama and M. Hattori for useful suggestions about sample preparation and crystallization; T. Sakamoto and T. Saika for their assistance in the purification of T. thermophilus SecYE; K. Mochizuki, M. Sano, K. Yoshikaie, T. Adachi and Y. Echizen for technical support; and the beamline staff members at BL41XU of SPring-8 (Sayo, Japan) and NW12 of PF (Tsukuba, Japan) for technical help during data collection. We also thank I. Artsimovitch for critically reading the manuscript. This work was supported by a SORST program grant from JST (Japan Science and Technology) to O.N., by a CREST grant from JST to K.I. and N.D., by a BIRD grant from JST to H.M. and Y.S., by Global COE Program (Center of Education and Research for Advanced Genome-Based Medicine) and a grant for the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) to O.N., by NIH grants to D.G.V., by grants from MEXT to H.M., S.F., R.I., K.I. and O.N., and by Mitsubishi Foundation grants to O.N.
Footnotes
Author Contributions T.T. carried out the structural determination and the biochemical experiments of T. thermophilus SecYE. H.M. carried out biochemical analyses of SecA–SecY interactions. A.P. and D.G.V. assisted with the crystallization and data collection of SecYE as well as with manuscript preparation. S.F., R.I. and O.N. assisted with the structural determination. T.M. and Y.S. performed the molecular dynamics simulation. N.D. performed disulphide-bond quantification and mass spectrometry. All authors discussed the results and commented on the manuscript. O.N. and K.I. supervised the work and wrote/ edited the manuscript.
Author Information The coordinates and structure factors have been deposited in the Protein Data Bank, under the accession codes 2ZJS for Fab–SecYE and 2ZQP for SecYE. Reprints and permissions information is available at www.nature.com/ reprints.
References
- 1.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 2.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 3.Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004;1694:67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 4.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 5.Mori H, Ito K. The Sec protein-translocation pathway. Trends Microbiol. 2001;9:494–500. doi: 10.1016/s0966-842x(01)02174-6. [DOI] [PubMed] [Google Scholar]
- 6.Cannon KS, Or E, Clemons WM, Jr, Shibata Y, Rapoport TA. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, et al. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Rusch SL, Kendall DA. Oligomeric states of the SecA and SecYEG core components of the bacterial Sec translocon. Biochim Biophys Acta. 2007;1768:5–12. doi: 10.1016/j.bbamem.2006.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra K, et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature. 2005;438:318–324. doi: 10.1038/nature04133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menetret JF, et al. Ribosome binding of a single copy of the SecY complex: implications for protein translocation. Mol Cell. 2007;28:1083–1092. doi: 10.1016/j.molcel.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 13.Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douville K, Price A, Eichler J, Economou A, Wickner W. SecYEG and SecA are the stoichiometric components of preprotein translocase. J Biol Chem. 1995;270:20106–20111. doi: 10.1074/jbc.270.34.20106. [DOI] [PubMed] [Google Scholar]
- 15.Lill R, et al. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori H, Ito K. Biochemical characterization of a mutationally altered protein translocase: proton motive force stimulation of the initiation phase of translocation. J Bacteriol. 2003;185:405–412. doi: 10.1128/JB.185.2.405-412.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori H, Ito K. An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc Natl Acad Sci USA. 2001;98:5128–5133. doi: 10.1073/pnas.081617398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA. 2006;103:16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 20.Vassylyev DG, et al. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol. 2006;364:248–258. doi: 10.1016/j.jmb.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 21.Mori H, et al. Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J Biol Chem. 2003;278:14257–14264. doi: 10.1074/jbc.M300230200. [DOI] [PubMed] [Google Scholar]
- 22.Breyton C, Haase W, Rapoport TA, Kuhlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 23.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 24.Sianidis G, et al. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20:961–970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karamanou S, et al. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- 26.Karamanou S, et al. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26:2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delano WL. The PyMOL molecular graphics system v.0. 97. 2002 < http://pymol.sourceforge.net/>.
- 28.Kalé L, et al. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:283–312. [Google Scholar]
- 29.Shindo N, et al. Separation of 18 6-aminoquinolyl-carbamyl-amino acids by ion-pair chromatography. Anal Biochem. 1997;249:79–82. doi: 10.1006/abio.1997.2155. [DOI] [PubMed] [Google Scholar]
- 30.Hunt JF, et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.


