Abstract
Bacteria are capable of “communicating” their local population densities via a process termed quorum sensing (QS). Gram-negative bacteria use N-acylated l-homoserine lactones (AHLs), in conjunction with their cognate LuxR-type receptors, as their primary signalling circuit for QS. In this critical review, we examine AHL signalling in Gram-negative bacteria with a primary focus on the design of non-natural AHLs, their structure-activity relationships, and their application in chemical biological approaches to study QS.
1. Introduction
The discovery of the cell by Robert Hooke in the late 1600s marked the beginning of a revolution in biology that continues to this day as scientists labour to fully understand the inner workings of living organisms.1,2 One of Hooke’s contemporaries, the Dutch scientist Anton van Leeuwenhook, observed the microbial world using specialized microscopes, and his discoveries represent the roots of microbiology.1,3 For nearly three centuries after van Leeuwenhook’s work, scientists believed that bacteria existed solely as single cell organisms and were incapable of any form of intercellular communication. This view of bacterial behaviour dramatically changed ~40 years ago.
In 1965, Tomasz and Beiser reported that an extracellular factor was produced by Streptococcus pneumoniae and released into the surrounding environment, and that this factor was required for the bacteria to enter into a competent state.4 The hypothesis that bacteria could signal to each other was further strengthened by the work of Hastings and co-workers in 1970. These researchers demonstrated that the Gram-negative marine bacterium Vibrio fischeri produces light at high (exponential growth phase), but not at low, cellular densities.5 This discovery indicated that V. fischeri could coordinate their light-producing capabilities based on population density. Subsequent studies by Eberhard et al. in 1981 revealed that this cell–cell signalling was facilitated by a low molecular weight organic compound, which was identified as an acylated homoserine lactone (AHL, Fig. 1).6 The combined efforts of these scientists established that bacteria are indeed capable of cell–cell signalling, and use signalling molecules to coordinate phenotypic responses in relation to their population densities.
Fig. 1.
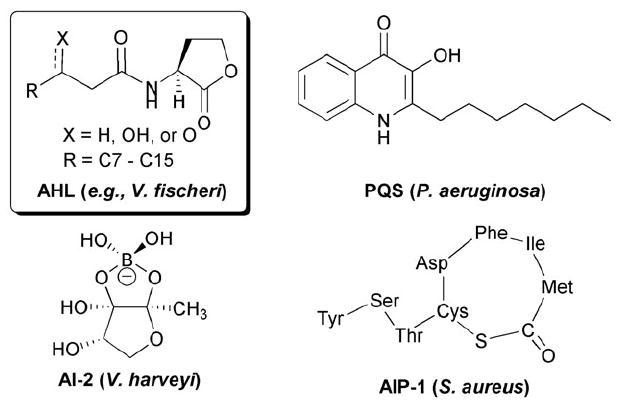
Selected QS signals listed with their corresponding bacterial species. AHL = N-acylated l-homoserine lactone; AI-2 = autoinducer 2; PQS = Pseudomonas quinolone signal; AIP-1 = autoinducing peptide 1.
The use of small molecule signals to coordinate gene expression with population density has now been termed quorum sensing (QS).7 Once bacteria reach a sufficiently high population density, they can undergo a lifestyle switch from that of a solitary cell to that of a multicellular group. As a group, bacteria alter gene expression levels and initiate processes that benefit the growing colony. These group behaviours are remarkable in their diversity, and can have significant impacts on their eukaryotic hosts. Examples include the production of virulence factors, swarming, biofilm formation, antibiotic production, bioluminescence, root nodulation, sporulation, and conjugation. Many of these QS-controlled outcomes have widespread and often devastating effects on human health, agriculture, and the environment.3,8-12 At high cell densities, bacteria also produce more QS signal, which in turn enhances the QS response and has led to these signals being named “autoinducers.”
Although originally thought to be limited to a small sub-set of bacterial species, it has become clear that QS is a fundamental process of the microbial world.3 Population dependent, cell–cell communication pathways can be found in both Gram-positive and Gram-negative bacteria, and in fungi.13 AHLs are the most common signal used by Gram-negative bacteria for cell–cell communication (Fig. 1). However, other small molecule signals have been associated with QS in Gram-negative bacteria, including the boronate ester autoinducer-2 (AI-2) used by V. harveyi and quinolone molecules (e.g., the Pseudomonas Quinolone Signal, PQS) used by the opportunistic pathogen Pseudomonas aeruginosa. Gram-positive bacteria, such as Staphylococcus aureus, use ~5–15-mer oligopeptides for QS (i.e., autoinducing peptides, or AIPs).14-16
To date, cell–cell signalling mediated by AHLs in Gram-negative bacteria represents one of the best-understood bacterial QS systems at the molecular level. Further, as QS plays a significant role in the establishment of infection by Gram-negative bacteria, AHL-based signalling has become the focal point of considerable scientific interest due to the impact it may have as a new anti-infective strategy (see section 2 below). This interest has spurred the development of a range of synthetic ligands that can intercept AHL-based QS and modulate QS-controlled behaviours. Such compounds represent valuable tools for the dissection of QS pathways. This critical review will provide an introduction to AHL-based QS and a systematic analysis of recent research on AHL mimics.
2. QS as a therapeutic strategy
The rapid growth of bacterial resistance underscores an urgent need for the development of new anti-bacterial strategies, and QS has attracted considerable recent attention as a new target.17-21 The interception of QS represents a potential strategy to slow bacterial virulence, such that the host could be treated with lower doses of antibiotics, or the infection could be cleared naturally by the host’s defenses. Such “anti-virulence” strategies may have an advantage over traditional antibacterial treatments, as the selective pressure is hypothesized to be reduced.22 Various research labs have shown the ability to inhibit the production of damaging virulence factors by P. aeruginosa, such as elastase B and pyocyanin, through either genetic mutations in QS genes or small molecule modulation of QS gene products.10,17-19,23 In addition, several animal infection models have shown that when mutations were made to QS specific genes in P. aeruginosa, less tissue damage occurred, and pneumonia rates and mortality decreased in comparison to wild-type P. aeruginosa infection.17-18,22 It is important to note, however, that although QS mutant strains displayed reduced virulence, none of the mutations led to avirulent strains. This suggests that, although QS plays an important role in pathogenesis, other factors are also key to infection.9,24 Nevertheless, due to the potential of QS as a new therapeutic target, this signalling phenomenon has been named “one of the most consequential molecular microbiology stories of the last decade.”25
3. QS circuits in Gram-negative bacteria: LuxI/LuxR
We start with an introduction to the basics of the QS signalling system in Gram-negative bacteria. These bacteria produce diffusible AHL ligands via an inducer synthase (or LuxI-type protein), and the ligand is sensed by its cognate cytoplasmic receptor (or LuxR-type protein) (Fig. 2). The AHL ligand is generated at low basal levels, and in general, high cell densities are required to achieve an intracellular ligand concentration sufficient for LuxR-type protein binding. Thereafter, the AHL–LuxR-type protein complex most often homodimerizes and binds adjacent to QS promoters (i.e., short palindromic sequences termed “lux boxes”), which activates the transcription of target genes required for bacterial group behaviour. (We note that examples of transcriptional repression upon AHL–receptor binding have also been reported, but this outcome appears to be far less common.)12 As AHL synthases and receptors were discovered in the bioluminescent bacterium V. fischeri (that uses QS to control expression of the lux operon), these protein families have been termed “Lux-type” synthases and receptors, respectively.
Fig. 2.
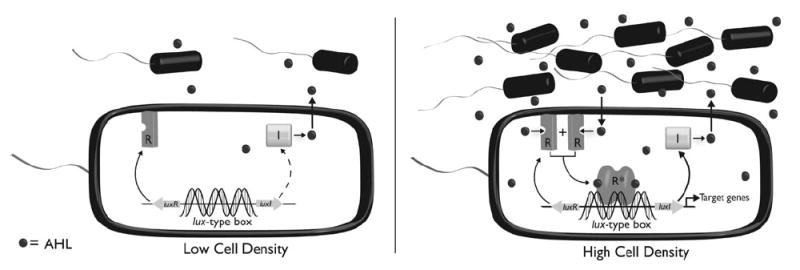
Simplified AHL signalling circuit in Gram-negative bacterial QS. R: LuxR-type receptor protein; I: LuxI-type autoinducer synthase protein; AHL: N-acylated l-homoserine lactone.
Several Gram-negative bacteria have been shown to use two or more AHL signals, along with other ligands and receptors, to “check and balance” QS. For example, P. aeruginosa uses N-(3-oxo-dodecanoyl)-l-homoserine lactone (OdDHL) and N-butanoyl-l-homoserine lactone (BHL), and two LuxR-type proteins, LasR and RhlR, respectively, for QS. This signalling circuit is complicated further by the fact that an orphan LuxR-type receptor, QscR, serves in part to repress the role of LasR,15 and that RhlR is also under the control of PQS (Fig. 1). Nevertheless, Fig. 3 shows the basic paradigm for Gram-negative QS: a signal is produced, binding of this signal to a receptor occurs at a threshold concentration, and this receptor complex then regulates (most commonly via activation) the transcription of genes that are involved in bacterial group processes.
Fig. 3.
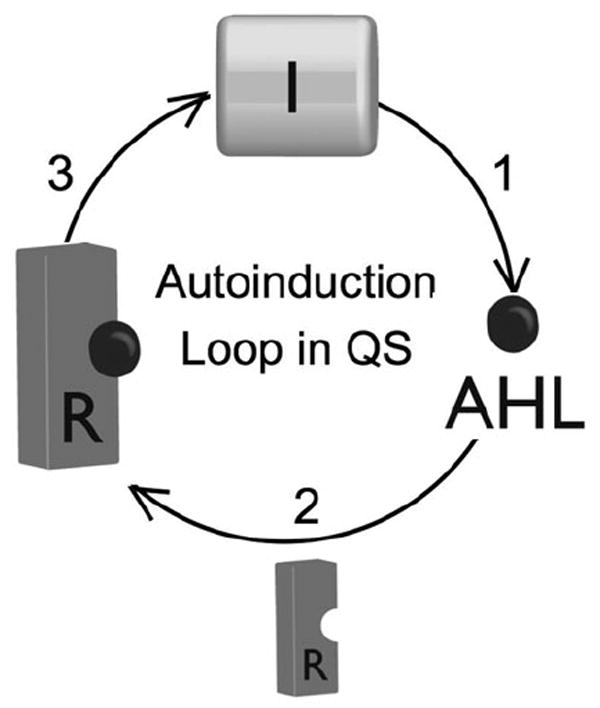
Autoinduction loop in QS: (1) AHLs are produced by LuxI-type synthase proteins (I); (2) above a critical concentration they bind to a LuxR-type receptor (R), (3) following dimerization, this receptor:ligand complex acts as a transcription factor to produce more I protein and control other essential functions of QS.
4. Three methods to intercept LuxI/LuxR-type QS
To modulate QS within the LuxI/LuxR-type pathway, there are three obvious targets: the synthase (I), the AHL ligand, and the receptor (R) (Fig. 3).17,23 Interception of any one of these three major components within the autoinduction circuit should lead to a bacterial “communication breakdown.” We discuss each approach in turn below.
4.1 Targeting the synthase (I)
Inhibition of the LuxI-type protein appears to be a straight-forward approach to thwart QS, as cell–cell signalling is clearly impossible without a signal. Surprisingly, however, there are few reported studies that specifically target the synthase protein.26 A relatively limited number of experiments with LuxI-type protein mutants indicate a dramatic drop in coordinated group behaviour. For example, P. aeruginosa LasI knockouts are almost completely deficient in virulence factor production. There are several reported X-ray crystal structures for LuxI-type proteins (including LasI) that could be used to guide the design of synthetic inhibitors; however, to our knowledge, such ligands are yet to be reported.12,27,28 As these crystal structures were only recently reported, we anticipate that this area of research will grow considerably in the near future.
4.2 Targeting the AHL ligand
The second target is the AHL ligand itself. Degradation of the small molecule signal would be a complementary strategy to inhibiting its synthesis. It is worthwhile to note that several species, both prokaryotes and eukaryotes, are believed to degrade AHL signals in order to block the QS system of invading bacteria. There are several mechanisms by which this degradation takes place, including an increase of pH at infection sites in certain plant species (as basic conditions cause hydrolysis of the lactone), secretion of oxidized halogenated compounds that are capable of reacting with the 3-oxo-AHLs, use of AHLs as a carbon and nitrogen source, and breakdown of the AHL by lactonases and acylases.16,17,23 Although signal degradation does not represent a likely therapeutic approach in humans and animals, understanding AHL breakdown in the context of mixed microbial systems is essential to understanding the complex signalling networks that have evolved between bacteria and other organisms. Further, this approach could have significant value for agricultural applications, as transgenic plants producing AHL lactonase (e.g., tobacco) have been shown to exhibit heightened resistance to infection by several Gram-negative plant pathogens.17
4.3 Targeting the receptor (R)
The third and final target for the modulation of cell–cell communication in Gram-negative bacteria is the receptor, or LuxR-type protein. The largest body of work on QS control has centred on the receptor protein, and the majority of this review hereafter will focus on the interception of this critical ligand–receptor interaction. The LuxR-type protein family represents in some ways a traditional, medicinal chemistry target for receptor modulation, and a significant quantity of genetic, biochemical, and structural data have been amassed for this protein class.7,14 Similar to LuxI-type protein knockouts, LuxR-type protein knockouts show a marked decrease in QS-controlled outcomes, suggesting that inhibition of LuxR-type proteins represents a viable approach to control QS.17-19,21,23,24,29
In 2002, the first X-ray crystal structures of a LuxR-type protein, i.e., TraR from the plant pathogen Agrobacterium tumefaciens, were reported.30,31 Five years later the X-ray crystal structure of the N-terminal ligand-binding domain of LasR from P. aeruginosa was published.32 These structural data have the potential to significantly impact the design of synthetic ligands for the modulation of LuxR-type protein activity, and have already been put to use in recent studies (see section 8). Further, these data provide an entry point not only for the design of LuxR-type protein inhibitors, but presumably for activators as well. Activation of QS also could be desirable, namely it may have benefits in agricultural settings (e.g., activation of root nodulation by soil dwelling bacteria) or for environmental remediation.33
5. Advantages of small molecules to study QS
As introduced above, the design and synthesis of non-native ligands that can intercept QS has attracted considerable interest. It is valuable, however, to review why such molecular tools would be advantageous for the study of QS.34 First, small molecules allow for temporal control of a biological system, and this control is most often rapid, depending on the diffusibility of the compound. Understanding the importance of a gene during different growth phases is essential to understanding what role that gene plays in the life cycle of the cell. Genetic alterations, in contrast, only allow for steady-state observations. Second, small molecules allow for the study of the reversibility of a system. Such experiments are difficult to perform using genetic techniques, and are most frequently done via conditional gene alterations (i.e., light or temperature sensitive mutations). Third, small molecules can allow for quantitative control of phenotypes through the systematic alternation of compound concentration (or “dose”). We note, however, that there are also drawbacks to using small molecule probes to study bacterial QS. Probably the largest challenge will be finding compounds that exhibit high specificity for one LuxR-type protein due to the relatively high sequence homology of these proteins within their ligand-binding domains (~70–80%).7,14 With this issue aside, the diverse toolset of both natural AHL signals and synthetic AHL mimics is helping to break new ground in understanding the diverse nature of bacterial cell–cell communication.23
5.1 Review outline
In the following sections, we discuss the broad structure-activity trends that have been deduced for AHL activity from studies of AHL analogues over the past ~20 years. To start, we briefly review the natural AHL signals used by Gram-negative bacteria, their chemical synthesis, and the standard biological assays used to detect and evaluate AHLs. Thereafter, we provide an in depth review of the structure-activity relationships (SAR) for non-natural AHL agonists and antagonists of five different LuxR-type proteins (LuxR, LasR, RhlR, TraR, and CarR). This detailed analysis will reveal that the line between AHL-derived antagonists and agonists of LuxR-type proteins actually can be blurred. Recent studies have shown that certain antagonists have the ability to also activate LuxR-type proteins (depending on their concentration), and are more appropriately called partial agonists. This activity trend is valuable to keep in mind throughout the discussion of each case study in section 8, as this duality of activity was not probed in most studies. We close our SAR analysis with a summary of the general structural features of AHLs required for modulation of LuxR-type proteins, whether by inhibition or activation. Finally, we conclude this review by offering our perspective on the major challenges within the field, and on what the future may hold for the small molecule control of AHL-type QS.
6. Ligand-design starting point: natural AHLs
Over 50 species of Gram-negative bacteria have been shown to use AHLs in their QS signalling networks.Table 1 lists a subset of these systems. The biosynthesis of AHLs has been well-characterized and involves the catalytic reaction of an appropriately charged acyl carrier protein and S-adenosyl methionine facilitated by a LuxI-type protein.12,14 In general, natural AHLs have acyl groups that range in carbon number from 4 to 18, and have l-stereochemistry at their lactone ring. Many natural AHLs have carbonyl moieties (i.e., 3-oxo functionality) at the third carbon of the acyl chain; relatively fewer have hydroxyl groups at this position. In addition, some longer chain natural AHLs have cis alkene units at the seventh or ninth carbon in the acyl chain (Table 1, entries 8 and 9).15,23
Table 1.
Representative AHL systems, their corresponding bacteria, and effects on their host
| Entry | Bacterium | AHL | Nomenclaturea | Luxl/R | QS phenotype | Target/effects on host |
|---|---|---|---|---|---|---|
| 1 | Agrobacterium tumefaciens |
|
OOHL, 3-oxo-C8 | Tral/R | Plasmid conjugation | Crown gall tumour; large growths on plant host |
| 2 | Burkholderia cenocepacia |
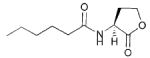
|
HHL, C6 | CepI/R
Ccil/R |
Biofilm, swarming motility, virulence | Opportunistic human pathogen; common infection in CF patients; may cause disease in plants |
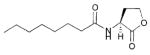
|
OHL, C8 | |||||
| 3 | Burkholderia pseudomallei |

|
OHL, C8 | PmlI1/PmlIR1 | Virulence, exoprotease | Melioidosis; affects lungs, heart, brain, liver, kidneys; commonly found in Southeast Asia |
|
|
3-Hydroxy-C8 | PmlI2/BpmR2 | ||||
| 4 | Chromobacterium violaceum |

|
HHL, C6 | CviI/R | Exoenzymes, cyanide, pigment | Rare human pathogen; can cause skin lesions and sepsis |
| 5 | Erwinia carotovora |
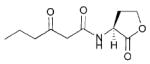
|
OHHL, 3-oxo-C6 | Expl/R
Carl/R |
Carbapenem, exoenzymes, virulence | Plant pathogen; common cause of decay in stored fruits and vegetables |
| 6 | Pseudomonas aeruginosa |
|
OdDHL, 3-oxo-C12 | Lasl/R QscRb | Exoenzymes, secretion, HCN, pigments (pyocyanin), biofilms | Opportunistic pathogen; major cause of hospital acquired infections (urinary tract, burn, external ear) |
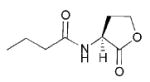
|
BHL, C4 | Rhll/R | ||||
| 7 | Pseudomonas putida |
|
OdDHL, 3-oxo-C12 | Ppul/R | Biofilms | Saprophytic soil bacterium; bioremediation of naphthalene contaminated soils |
|
|
ODHL, 3-oxo-C10 | |||||
| 8 | Rhizobium leguminosarum |
|
3-Hydroxy-7-cis-C14 | CinI/R
RhiI/R RaiI/R TraR BisR |
Root nodulation, symbiosis, plasmid transfer, growth inhibition | Symbiotic nitrogen fixing bacterium to legumes (e.g., peas) |

|
HHL, C6 | |||||
| 9 | Sinorhizobium meliloti |
|
DDHL, C12 | Sinl/R
ExpR TraR |
Nodulation and symbiosis | Symbiotic nitrogen fixing bacterium to legumes (e.g., alfalfa) |
|
|
3-oxo-9-cis-C16 | |||||
|
|
9-cis-C16:1 | |||||
| 10 | Vibrio fischeri |

|
OHHL, 3-oxo-C6 | Luxl/R | Symbiosis | Luminescent bacterium found in light organs of monocentrid fish and sepolid squid |

|
OHL, C8 | AinS/R |
AHL abbreviations used throughout the remainder of this review.
QscR is an orphan receptor that responds to OdDHL produced by LasI.
There are several insights that can be gained from studying Table 1. First, there is redundancy in the AHL signals that are used by different bacteria. For example, P. aeruginosa and P. putida (Table 1, entries 6 and 7) both use OdDHL as an AHL signal. This phenomenon is not just limited to bacteria of different species, but also different genera (e.g., Erwinia and Vibrio; Table 1). This signal overlap is interesting, because it may suggest that bacteria from different environments have a common evolutionary ancestor.15,35 Further, for bacteria that live in the same environment, this overlap may also provide a means for interspecies bacterial communication.15,36,37 Another important aspect evident in Table 1 is that, within a single species, one AHL can act as the signalling molecule for more than one receptor (e.g., OdDHL and the LasR and QscR receptors in P. aeruginosa). This duality could allow the organism to conserve valuable resources, or provide a means to carefully modulate the activity of receptors, for example, if the ligand binds to the two receptors with different affinities. Such differences in affinities may play a role in cross-species interactions as well.
Chemical synthesis of natural AHLs is relatively straight-forward and is shown in Fig. 4. The synthesis of “simple,” non-3-oxo natural AHLs is most commonly performed by coupling l-homoserine lactone (HSL) with an activated carboxylic acid. Synthesis of 3-oxo AHLs is more complex due to the reactivity of the 3-oxo moiety, and has been achieved either by using ketal-protected 3-oxo carboxylic acids and carbodiimide chemistry (Fig. 4A, route a), or by coupling l-HSL and acylated Meldrum’s acid (Fig. 4A, route b).23,38 Natural AHLs, as well as a large number of non-natural AHLs, have been synthesized using a solid-phase synthetic route starting from l-methionine-loaded amino-polystyrene (Met-PS) resin (Fig. 4B).39 Similar to traditional solution-phase routes, attachment of the acyl chain is achieved through the carbodiimide-mediated coupling of a carboxylic acid to an amine. This latter route is advantageous for the synthesis of sizable numbers of AHLs, due to the high purity of compounds that can be generated and its adaptability to parallel library construction.39
Fig. 4.
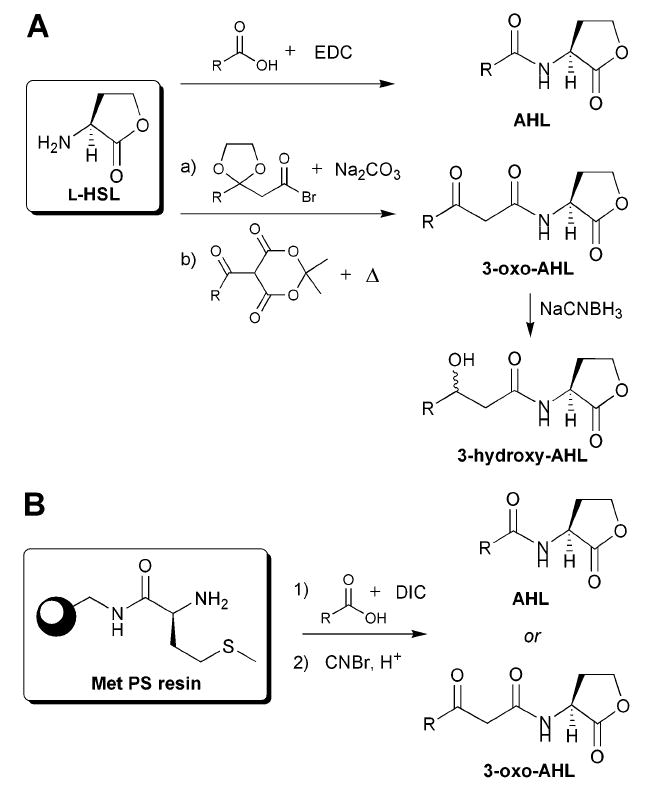
Common synthetic routes to natural AHLs. A: Solution-phase amide coupling conditions. HSL = homoserine lactone. EDC = 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride. B: Solid-phase synthetic route for the generation of AHLs from l-methionine-loaded amino-polystyrene (Met-PS) resin. DIC = N,N′-diisopropylcarbodiimide.
7. Methods to detect AHLs and evaluate their activities
The discovery and detection of natural AHLs can be achieved by several different methods. One common approach involves the use of biosensor strains containing a reporter gene under the control of a luxR-type promoter.40 Several examples of biosensor strains are listed in Table 2. When an AHL–LuxR-type protein complex binds to the promoter, this binding event, and thus ligand activity, can be read out in these biosensor strains as absorbance (β-galactosidase fusion), fluorescence (green fluorescent protein (GFP) fusion), or luminescence (luciferase fusion). These strains lack functional AHL synthases and therefore exogenous ligand must be added in order to activate the QS system. Overlay assays (e.g., on reverse-phase TLC plates) are commonly performed using these strains to detect AHLs in natural isolates. Although these overlay assays are generally robust detection methods, they also have drawbacks. For example, they only provide qualitative data, and are limited by the structures of AHLs that can be detected (i.e., long-chained AHLs do not elute well by reverse-phase TLC). Because of these issues, more quantitative methods to detect AHLs in natural isolates have recently been developed. Such techniques include GC-MS, nanoLC-ESI, and HPLC-MS.41
Table 2.
Representative AHL biosensor strainsa
| Entry | Source | QS system | Host and plasmid | Reporter system |
|---|---|---|---|---|
| 1 | V. fischeri | LuxI/R | E. coli pSB401 | luxCDABEb |
| 2 | V. fischeri | LuxI/R | E. coli pHV200I− | luxCDABE |
| 3 | P. aeruginosa | RhlI/R | P. aeruginosa PAO-JP2 prhlI-LVAgfp | GFP |
| 4 | P. aeruginosa | LasI/R | P. aeruginosa M71L7I− | β-Galactosidase |
| 5 | A. tumefaciens | TraI/R | A. tumefaciens NT1 pZLR4 | β-Galactosidase |
| 6 | A. tumefaciens | TraI/R | A. tumefaciens WCF47 pCF218+pCF372 | β-Galactosidase |
Although biosensor strains were initially developed to detect AHLs in natural isolates, they have also been utilized to evaluate the activities of non-native AHL analogues. These assays are most commonly performed in liquid culture, as opposed to overlay formats. Ligand activity is typically measured in three ways, described briefly below. We will make reference to these assay formats for the remainder of this review:
Agonism assays are performed in biosensor strains with exogenously added synthetic analogues at various concentrations. Agonists are able to activate the transcription of reporter genes and give a positive response.
Antagonism assays are performed in biosensor strains as competition experiments in the presence of both exogenously added natural AHL ligand and a synthetic compound. Antagonists are able to disrupt induction by the natural ligand and reduce the reporter gene read-out as compared to the natural ligand alone.
Competitive binding assays are likewise run as competition experiments, but the exogenously added natural ligand is typically radiolabelled. Competitive binders are able to displace the radiolabelled ligand and lower the observed radioactivity. Controls are typically run with unlabelled natural AHL as the competitor. (Note, these experiments do not rely on a transcriptional read-out, but are commonly performed in biosensor strains for comparison to agonism or antagonism assay data).
8. Designed AHL mimics
We now proceed to a systematic discussion of selected synthetic AHL mimics that have been designed, synthesized, and evaluated for antagonistic activity, agonistic activity, and/or competitive binding against LuxR-type proteins. For clarity, we have organized this section according to the Gram-negative QS systems in which each AHL mimic was studied. For each example, we outline: (1) the general AHL structural features tested, (2) the structures of the most active AHL mimics identified in the study, (3) the SARs that can be derived for these compounds from primary reporter gene assay data, and (4) the activity of these compounds in a secondary biological assay (if applicable). The majority of this work has centred on the canonical QS systems in V. fischeri (LuxR) and P. aeruginosa (LasR and RhlR). However, to provide a broader account, we also briefly discuss several pertinent examples of AHL mimics evaluated in other Gram-negative bacteria (A. tumefaciens and Erwinia carotovora).
8.1 LuxR from V. fischeri
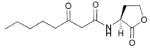
The first AHL analogue study was published in 1986 by Eberhard and co-workers.42 This initial work focused on a small collection of ~20 synthetic AHL derivatives. These compounds were evaluated for agonistic and antagonistic activity (against OHHL; Table 1, entry 10) in V. fischeri B-61, a naturally “dim” strain that produces more light with the addition of exogenous OHHL. The goal of this study was to determine whether the fatty acid side chain or the amino acid-derived lactone portion of AHLs was most important for activity. The AHLs varied in the length of their acyl chains, placement of the 3-oxo group in the chain, and the presence of alkene units in the chain. Certain derivatives contained replacements for the γ-lactone, including a γ-thiolactone and a caprolactam.
From this early study, the authors concluded that the γ-lactone ring was important for agonistic activity in LuxR, because the thiolactone and lactam derivatives either inhibited LuxR or showed no activity. In addition, they determined that the length of the fatty acid side chain was important for LuxR activation: the optimal chain length for agonistic activity was six carbons, and deviating from this length resulted in inhibitory activity, with the most antagonistic fatty acid side chain being nine carbons long (AHL 1, Fig. 5). The 3-oxo group was also determined to be essential for strong agonistic activity. Carbonyls placed at the 5-position decreased agonistic activity by 10-fold, and when located at the 2-position completely abolished agonistic activity. Otherwise, compounds that did not have a second acyl chain carbonyl displayed antagonistic activity against LuxR. Interestingly, one of the most active agonists discovered in this study was OOHL, the native AHL ligand for A. tumefaciens (Table 1, entry 1). We note that the bacterial reporter strain used in this study was not a LuxI synthase knockout, and therefore residual natural ligand was present in these assays and could complicate data analysis (see below).
Fig. 5.
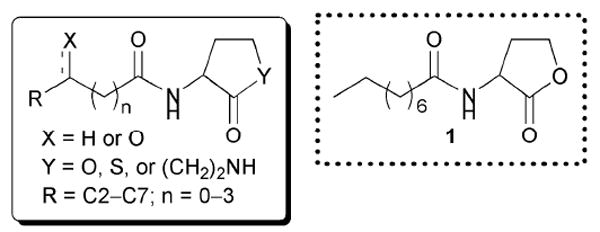
Structural features of AHLs tested against LuxR by Eberhard and co-workers summarized in shaded box; compounds were synthesized and tested as racemates. Their best antagonist is shown.
The next study exploring the activities of AHL mimics in the V. fischeri system did not appear until a decade later, when, in 1996, Greenberg and co-workers screened a set of AHLs very similar to those reported by Eberhard and co-workers.43 The difference between these two studies was that Greenberg and co-workers evaluated the activity of the AHL analogues in an E. coli reporter strain VJS533 pHV200I-, which harbours LuxR and the V. fischeri luminescence gene cluster with luxI inactivated (Table 2, entry 2). Use of this strain overcame the limitation of the previous study, as it lacked the OHHL synthase. The author’s main objectives were three-fold: to identify (1) competitive LuxR binders, (2) LuxR antagonists, and (3) LuxR agonists.
These studies revealed similar trends in ligand activity to those reported by Eberhard and co-workers, and the most active AHLs from this study are shown in Fig. 6 (AHLs 2, 3, and 4). Compounds with acyl chains longer than five carbons showed inhibitory activity against LuxR. Although the 3-oxo group was found to be non-essential for binding in ligand displacement assays, compounds that possessed this moiety were determined to be 100-fold better agonists in the luminescence assays. Except for a modest change to the γ-lactone ring, such as conversion to a γ-thiolactone, the lactone ring was found to be important for LuxR binding; for example, replacement with a caprolactam resulted in no LuxR binding. The researchers also demonstrated that introduction of alkene units into the acyl chain reduced agonistic activity, suggesting that a flexible fatty acid side chain is needed for binding to LuxR.
Fig. 6.
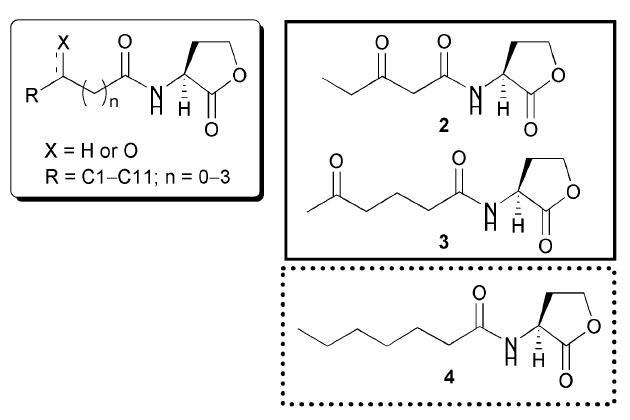
Structural features of AHLs tested against LuxR by Greenberg and co-workers summarized in shaded box; identified agonists and antagonist are outlined in solid and dashed lines, respectively.
It is important to note that, although Greenberg and co-workers saw AHL activity trends similar to those previously reported in V. fischeri by Eberhard, there were differences when using the E. coli LuxR reporter strain (e.g., decanoyl HL (DHL) was a far more potent inhibitor in the V. fischeri strain relative to the E. coli strain). These discrepancies, the reasons for which are not fully understood, underscore the need for caution when comparing small molecule screening data that were obtained from different QS reporter strains.
In 2002, Nielsen and co-workers reported a study of the activity of AHL analogues against LuxR that had various substitutions on the 3- and 4-positions of the γ-lactone ring (Fig. 7).44 This work was inspired by two factors: (1) such substitution had not been explored previously in AHL mimics, and (2) the discovery of halogenated furanone modulators of QS, which are similar in structure to AHLs and have substitution at the 3- and 4-position of the ring. These furanones were isolated from the macroalgae Delisea pulchra, and had been reported to inhibit several different QS systems and phenotypes.45 Nielsen and co-workers screened their AHL derivatives for agonistic or antagonistic activity in E. coli JM105 pUC18, which harbours the LuxR-based QS system of V. fischeri (with luxI knocked out) fused to a reporter gene for GFP.
Fig. 7.
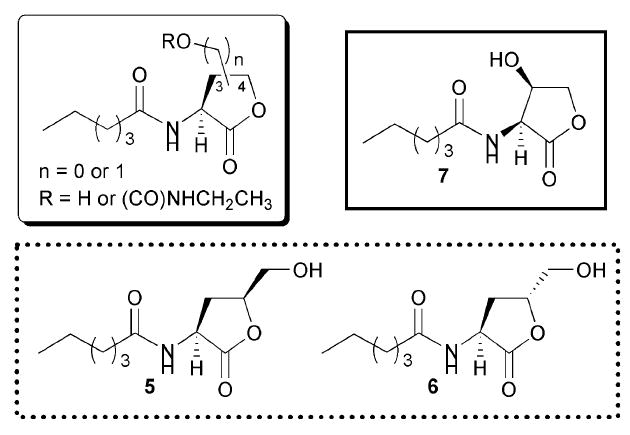
Structural features of AHLs tested against LuxR by Nielsen and co-workers summarized in shaded box; identified agonist and antagonists are outlined in solid and dashed lines, respectively.
The researchers discovered that substitution at the γ-lactone 4-position resulted in compounds that were only weak activators of LuxR. Indeed, lactones 5 and 6 exhibited inhibitory, as opposed to agonistic, activity at high concentrations (Fig. 7). In contrast, substitution at the 3-position was far less detrimental to agonistic activity, and these compounds showed activities similar to that of the native ligand, OHHL. Evaluation of these derivatives also showed that R stereochemistry at the 3-position was important for activity. Specifically, compound 7 showed agonistic activity against LuxR (Fig. 7), while inversion of stereochemistry at the 3-position resulted in a compound with 1000-fold reduced agonistic activity. Appending sterically bulky groups (e.g., carbamates) at the 3-position, however, yielded compounds that lost all activity against LuxR, regardless of stereochemistry at that position. Taken together, these assay data suggest that (assuming these compounds can target the LuxR-protein ligand-binding site) the binding area around the lactone ring is a narrow region within the LuxR ligand-binding domain that does not readily accommodate substitution at either the 3- or 4-positions of the lactone ring.
Since 2002, Doutheau and co-workers have reported a series of studies on AHL mimics and their activities against LuxR. This group has explored the effects of (1) incorporating greater structural diversity in the AHL acyl chain, and (2) replacing the amide group with alternate functional groups on ligand activity. In all three studies, the researchers tested their compounds in E. coli pSB401, which is a bioluminescent reporter strain containing LuxR and the lux box from V. fischeri and the luminescence gene cluster from Photorhabtus luminescens (Table 2, entry 1).46-48 All of their AHL derivatives were synthesized and tested in racemic form.
In the first study, Doutheau and co-workers explored adding steric bulk to the acyl chain of AHLs.46 They astutely noted that all of the previously reported studies on AHL mimics had primarily focused on probing acyl chain length, and thus they set out to make cycloalkyl, branched, and aryl substituted acyl chain AHL analogues. The majority of their analogues retained 3-oxo functionality.Fig. 8 shows two of the most active compounds found in this report (8 and 9). The authors demonstrated that branched alkyl or cycloalkyl AHLs with acyl chain lengths similar to OHHL were still able to activate the LuxR system; notably, the cyclopentyl AHL 8 was almost as potent as the natural ligand. When a phenyl group was added to the fourth carbon in the acyl chain, however, no agonistic activity was observed. Competitive antagonism assays revealed that many of these aryl derivatives were instead potent inhibitors of the LuxR QS system (e.g., halogenated derivatives of 9). In turn, the presence of very sterically bulky aryl or alkyl groups (e.g., napthyl, t-butyl, or adamantyl) on the fourth carbon yielded mimics with minimal activity against LuxR. From these data, the authors concluded that medium-sized, branched acyl derivatives were capable of LuxR agonism, while phenyl derivatives were capable of LuxR antagonism.
Fig. 8.
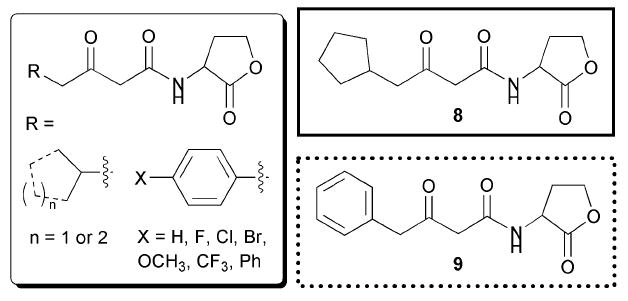
Structural features of AHLs tested against LuxR by Doutheau and co-workers summarized in shaded box; compounds were synthesized and tested as racemates. Their most active agonist and antagonist are outlined in solid and dashed lines, respectively.
The later reports of Doutheau and co-workers focused on replacing the AHL amide with different moieties. In their second report, the researchers made a series of sulfonyl AHLs (see Fig. 9), and determined that this substitution did not result in any compounds that showed agonistic activity against LuxR.47 However, they did discover several strong LuxR inhibitors (e.g., AHL 10). Furthermore, the researchers showed that sulfonyl AHLs with acyl chains close in length to OHHL (i.e., six atoms) and shorter were the strongest inhibitors. Through the examination of a model of LuxR (built by computational homology modelling of the TraR X-ray crystal structure), they determined that these sulfonyl AHLs could fit into the OHHL binding-domain of LuxR and that the sulfonyl group may be able to make new molecular contacts through hydrogen bonds. The authors went on to hypothesize that the sulfonyl inhibitors, once bound, could prevent LuxR from undergoing the structural rearrangements necessary for productive dimerization. We note, that while this hypothesis is certainly exciting, these computational results should be interpreted carefully, as they are based on a homology model of LuxR from TraR that inherently has associated error.
Fig. 9.
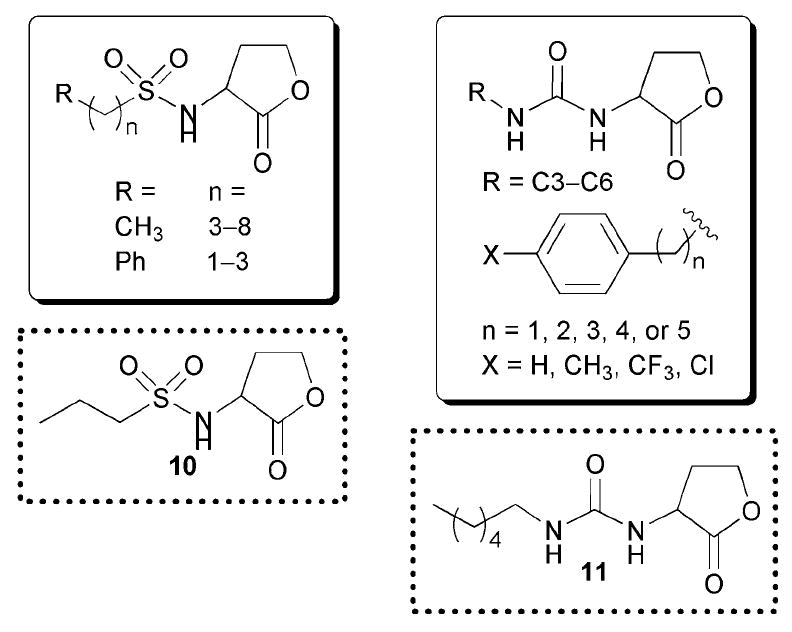
Structural features of AHLs tested against LuxR by Doutheau and co-workers summarized in shaded boxes. Compounds were synthesized and tested as racemates; antagonists are outline in dashed lines.
The third report by Doutheau’s group centred on replacing the AHL amide with a urea (Fig. 9).48 Again, the authors found that this substitution resulted in compounds that displayed no agonistic activity against LuxR, but instead were potent LuxR inhibitors (e.g., compound 11). In addition to reporter gene assays, they performed electrophoretic mobility shift assays on the inhibitors in the presence of a LuxR homologue (ExpR) and a short sequence of DNA containing the ExpR-binding site, and determined that these compounds were capable of inhibiting ExpR-DNA binding. Molecular modelling studies of these ligands with LuxR (using the same homology model described above) revealed that the urea derivatives could likewise fit into the OHHL-binding site and make hydrogen-bonding contacts with certain LuxR residues. Overall, there are two important outcomes of Doutheau and co-workers’ work: (1) adding some steric bulk to the AHL acyl chain is tolerated by LuxR and can lead to potent inhibitors, and (2) replacing the amide with alternate functional groups can confer antagonistic activity upon AHL analogues.
Recent studies in our laboratory have focused on the design and synthesis of combinatorial libraries of AHL derivatives, and the evaluation of these compounds for activity against LuxR-type proteins in a range of Gram-negative bacteria, including V. fischeri.39,49-51 We prepared a library of ~90 AHLs to probe the role of key features of AHL structure on ligand activity, including acyl chain length, lactone stereochemistry, and diversity of functionality on the acyl group. We sought to understand the distinctions in activity that each analogue exhibited in different species (i.e., in V. fischeri, A. tumefaciens, and P. aeruginosa), and thus determine a comprehensive set of SARs; such a comparative study had been lacking up until this work. Here, we will focus on our findings in V. fischeri. Later in this review we will comment on our conclusions toward understanding the activity of these AHL mimics across the three strains.
For our LuxR studies, we used a V. fischeri ES114 Δ-luxI derivative as a reporter strain, in which the native lux operon behaves as a bioluminescent reporter. We evaluated our library of AHL mimics in this strain, and uncovered several potent modulators of LuxR, ranging from antagonists to strong agonists.49-51 LuxR antagonists were identified that had IC50 values ranging from 7 nM to 5 μM (versus OHHL at 5 μM).Fig. 10 shows a sample of these active compounds (AHLs 12–15). We found that phenylacetanoyl HLs (PHLs) with electron-withdrawing substituents in the para position of the phenyl ring were the most potent LuxR inhibitors (e.g., PHL 13, Fig. 10). PHLs that did not possess electron-withdrawing groups, such as the tolyl derivative, had greatly reduced antagonistic activity.
Fig. 10.
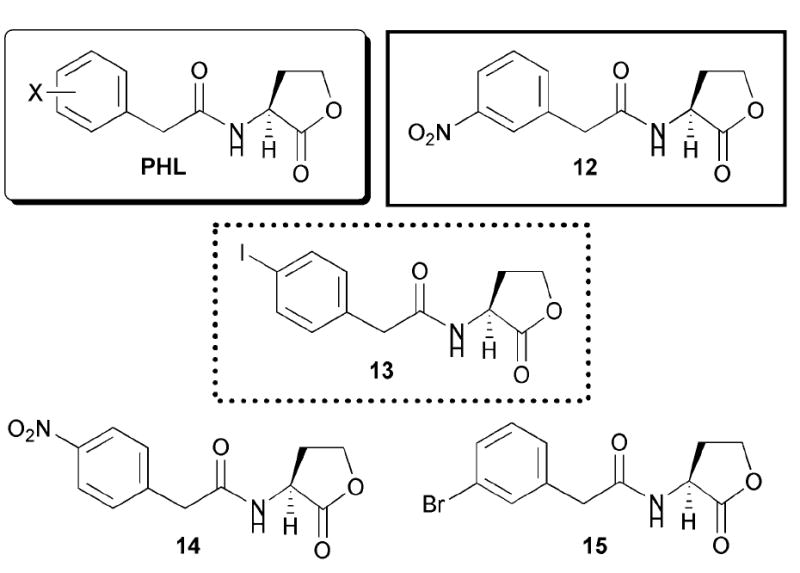
Structural features of AHLs tested against LuxR by Blackwell and co-workers summarized in shaded box. The most active agonist and antagonist are outlined in solid and dashed lines, respectively. Note, AHLs 14 and 15 behave as partial agonists.
Interestingly, a simple change of substitution from the para position to the meta position on the PHL phenyl ring seemingly changed ligand activity from LuxR antagonist to LuxR agonist.49 The most remarkable finding in the PHL series occurred when the substituent in the meta position was a nitro group. The resulting compound (12, Fig. 10) was a more active LuxR agonist than the natural ligand, OHHL. In fact, the EC50 of this compound was 10-fold lower than that of OHHL (0.3 μM vs. 3 μM). We went on to demonstrate that PHL 12 could super-activate LuxR in the wild type V. fischeri strain (which contains the LuxI synthase) relative to the native ligand. This discovery is important, because PHL 12 is one of the first synthetic super-activators of QS to be reported, and to our knowledge the first in V. fischeri.52,53
Additional dose response analyses of several of the moderately strong PHL inhibitors of LuxR revealed an interesting activity trend.50 Specifically, at high concentrations, these PHLs were able to act as LuxR agonists, as opposed to antagonists. Further experimentation caused us to conclude that these antagonists are in fact partial agonists (e.g., 14 and 15, Fig. 10). This activity trend appears to extend beyond PHLs to other AHL derivatives, and to other LuxR-type proteins, and will be discussed later in this review (section 8.3).
In the course of these studies, we also found that AHLs with long aliphatic acyl chains bearing the 3-oxo moiety were good LuxR inhibitors (e.g., OdDHL, IC50 = 400 nM). These results were congruent with those of Greenberg and co-workers in LuxR.43 We speculate that these compounds could make hydrogen bond contacts similar to those of OHHL in the LuxR ligand-binding site, but the longer acyl chains could potentially interact with a hydrophobic site slightly removed from the ligand-binding site, and this interaction alters LuxR conformation and lowers its activity. Additional biochemical and structural experiments of course are required to support this hypothesis.
8.2 LasR and RhlR from P. aeruginosa
We next turn to an evaluation of AHL mimics in P. aeruginosa. There is tremendous interest in inhibiting AHL-mediated QS in P. aeruginosa due to the prevalence of this opportunistic bacterium in life threatening hospital-acquired infections and in chronic lung infections associated with cystic fibrosis.7-9,20-21 A number of recent studies have focused on the design of small libraries of AHL mimics capable of intercepting the LasR and RhlR QS systems, and this section reports the important findings stemming from this research.
In 1996, Iglewski and co-workers examined the activities of a range of AHL mimics in P. aeruginosa that varied in acyl chain length, substitution at the 3-position of the acyl chain, and lactone ring structure (Fig. 11).54 Several of these compounds were examined by the Eberhard and Greenberg groups against LuxR, as introduced above. E. coli MG4 pKDT17 was utilized as a reporter strain in the agonism assays; this strain lacks lasI while LasR activity is reported via the production of β-galactosidase. The authors found that AHLs with acyl chains similar in length to the natural ligand (i.e., 12 atoms; OdDHL) were more potent LasR agonists than those with shorter chains. In turn, the 3-oxo group was also found to be important, as removal of this group from AHLs resulted in compounds with reduced agonistic activity against LasR. When the OdDHL lactone ring was replaced with a γ-thiolactone, the AHL retained the same overall activity as the natural ligand (i.e., 16, Fig. 11). However, switching to a γ-lactam resulted in compounds lacking either agonistic or antagonistic activity against LasR.
Fig. 11.
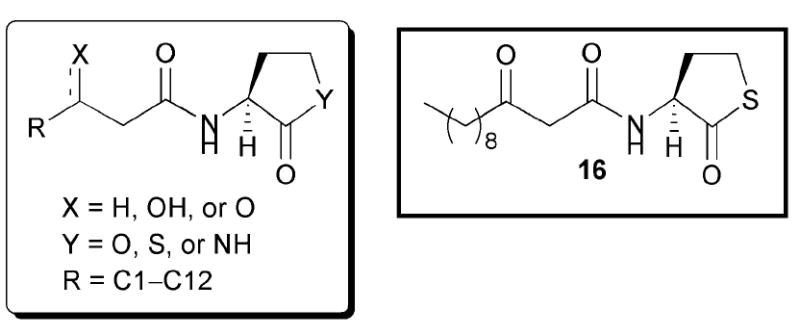
Structural features of AHLs tested against LasR by Iglewski and co-workers summarized in shaded box. AHL 16, the thiolactone analogue of OdDHL, acts as an agonist of LasR.
Iglewski and co-workers also examined their AHL analogues in a competitive binding assay with radiolabelled OdDHL using various E. coli MG4 strains.54 They found that their best synthetic LasR agonists were also their best competitive LasR binders. This result makes sense, as compounds that can activate the LasR system should be able to outcompete the natural ligand in a competitive binding assay. In addition, the researchers determined that AHLs with acyl chains greater than six atoms in length were likewise able to inhibit the binding of radiolabelled OdDHL (e.g., C9 AHL 1, Fig. 5). Overall, Iglewski’s team concluded that AHL acyl chain length was important for LasR binding, as compounds with chain lengths greater than six atoms displayed agonistic activity, and that the 3-oxo group heightens (but is not essential for) agonistic activity. Additionally, modest changes to the lactone ring, such as substitution of a thiolactone, did not abolish activity against LasR.
A 1999 study by Kline and co-workers explored the activities of constrained AHL analogues against LasR. The authors sought to probe rigid organic functional groups as mimics of the two possible tautomers of the β-ketoamide of OdDHL.55 They selected (1) salicylamides and β-nitrones to investigate the Z-enol tautomer, (2) furan and oxazoles to investigate the E-enol tautomer, and (3) gem-difluoronated analogues (e.g., 17) to test extended structures that would prevent tautomerization without introducing additional steric bulk. Selected compounds evaluated in this study are shown in Fig. 12. The authors examined the activity of these compounds using a P. aeruginosa β-galactosidase reporter strain deficient in both lasI and rhlI (PAO-JP2), and the E. coli reporter strain utilized by Iglewski and co-workers. Notably, none of their constrained enolic analogues showed appreciable agonistic activity in LasR. However, the gem-difluoro analogues were moderate agonists of LasR. For example, compound 17 showed the same level of LasR activation as the natural ligand OdDHL, but at 10-times higher the concentration (10 μM vs. 1 μM, respectively). Kline and co-workers concluded from these data that, in order to retain agonistic activity against LasR, 3-oxo AHLs must retain extended chain geometry and thus have some degree of flexibility.
Fig. 12.
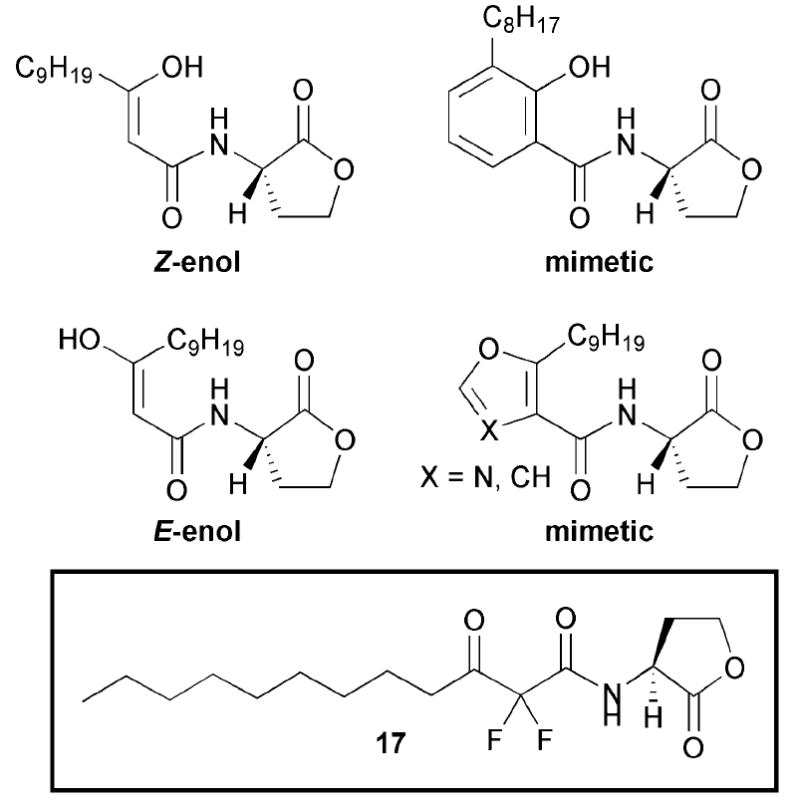
Structural features of AHLs tested against LasR by Kline and co-workers. AHL 17 is a non-enolizable agonist of LasR.
Starting in 2003, Suga and co-workers reported a series of studies concerning the effects of changing the AHL lactone to different types of ring structures on activity against LasR and RhlR (Fig. 13).56-58 The researchers prepared a library of 96 AHL mimics, in which the acyl chain remained unchanged relative to OdDHL or BHL, while the lactone ring was replaced with various carbocycles and heterocycles that had hydrogen-bonding capabilities at the same position as the carbonyl in the parent γ-lactone. They examined these compounds in a series of P. aeruginosa PAO-JP2 strains with GFP fusions (Table 2, entry 3).
Fig. 13.
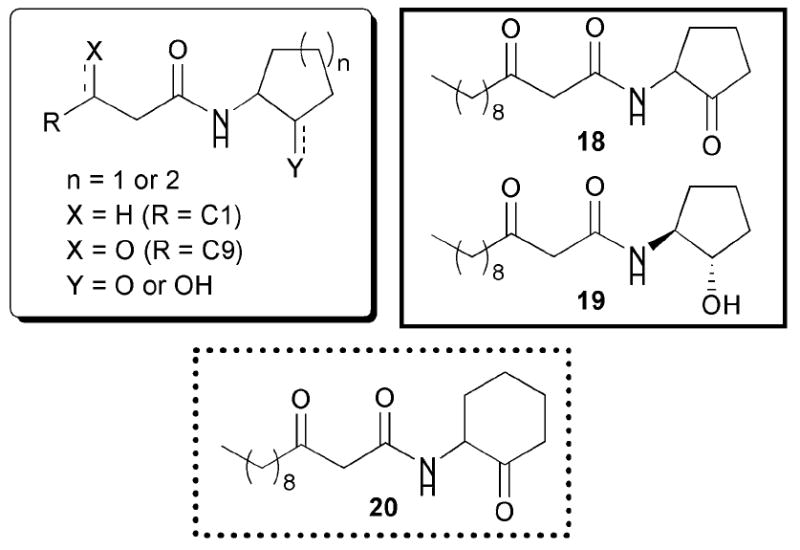
Structural features of AHLs tested against LasR and RhlR by Suga and co-workers summarized in shaded box; identified agonists and antagonist are outlined in solid and dashed lines, respectively.
The first study by Suga’s team reported several potent modulators of the LasR and RhlR proteins.56 A 3-oxo-C12 cyclopentanone derivative (18, Fig. 13) was found to activate the LasR system at levels close to that of OdDHL at 400 μM. Likewise, the same ring coupled to a C4 chain was shown to activate the RhlR system to the same level as BHL at 100 μM.55 The authors also demonstrated that a 3-oxo-C12 cyclohexanone derivative (20) was able to inhibit the LasR and RhlR systems by 35% and ~60% at 100 μM and 50 μM, respectively. Building on these results, they tested cyclohexanone 20 in secondary QS assays and found that this compound could strongly inhibit pyocyanin production, elastase B production, and biofilm growth in P. aeruginosa. Suga’s laboratory recently followed up on this work in a report that certain stereoisomers of their AHL analogues exhibited different agonistic and antagonistic activities against LasR and RhlR.58 Most notably, they found that the S,S derivative 19, a reduced variant of 18, displayed agonistic activity comparable to OdDHL at 50 μM and above in LasR (Fig. 13).
Based on these initial findings, Suga and co-workers later reported full details of the design, synthesis, and evaluation of their focused library of non-lactone AHL mimics (Fig. 14).57 The biological focus of this second study was primarily LasR, and the authors discovered three strong LasR agonists and eight LasR antagonists that lacked lactone functionality. The agonists were similar in part to previously reported structures (e.g., 16, which was included as an internal control; Fig. 11). Interestingly, they found a LasR agonist that contained an isoquinoline moiety (i.e., 21). Due to shared structural characteristics with the PQS signal in P. aeruginosa (Fig. 1), the authors speculated that 21 may not only be working through the LasR circuit, but also through the PQS circuit. While an interesting hypothesis, no data was provided to support this claim.
Fig. 14.
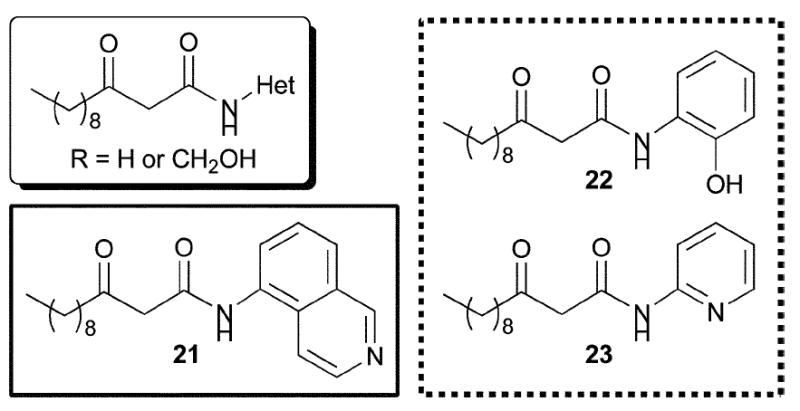
Structural features of AHLs tested against LasR by Suga and co-workers summarized in shaded box; identified agonist and antagonists are outlined in solid and dashed lines, respectively.
Five of the eight LasR antagonists reported in this second study were aniline derivatives (e.g., 22 and 23, Fig. 14). The most potent of these inhibitors, 22, also inhibited P. aeruginosa elastase B production. Unexpectedly, the authors found that this compound actually promoted, as opposed to inhibited, P. aeruginosa biofilm growth relative to an untreated sample in a static biofilm assay. The authors commented, however, that the morphology of the treated biofilm was changed. Suga’s team concluded that a five- or six-membered ring with a hydrogen bond acceptor displayed in a similar fashion as the carbonyl of the parent γ-lactone was necessary for LasR activation by AHL mimics. In turn, LasR inhibition could be achieved by compounds that contain an aromatic ring in place of the γ-lactone, as long as a hydrogen bond acceptor was present. Indeed, the largest difference between the LasR activators and inhibitors in this study appeared to be the introduction of aromaticity. The authors speculated, based on their findings and the fact that halogenated furanones are good inhibitors of P. aeruginosa QS (see above), that compounds with unsaturation (i.e., aromaticity) in the AHL lactone ring position alter the conformation of LasR upon binding. This conformational change then presumably reduces the ability of LasR to bind DNA, and inhibits transcription.14
Kato and co-workers recently reported several potent synthetic inhibitors of P. aeruginosa QS that also lack lactone functionalities.59 Similar to the studies by Suga outlined above, the authors changed the AHL lactone ring to a different structure. They simultaneously probed the effects of altering acyl chain length on the activity of their analogues against LasR. Their biological assays were performed in a range of different reporter strains, which are omitted here for brevity.59 First, the lactone was replaced with a cyclopentane ring, eliminating its hydrogen-bonding capability (Fig. 15). This switch yielded mimics with antagonistic activity against LasR, with the C10 cyclopentane AHL mimic (24) being the most potent inhibitor in the set (80% inhibition at 250 μM). Using 24 as their initial lead compound, the authors then substituted the ring with related carbocycles, such as cyclopropane, cyclobutane, cyclohexane, and cyclooctane. However, mimics with these other ring sizes displayed minimal inhibitory activity against LasR. The authors concluded that both acyl chain length and ring size were important for LasR inhibition for this class of analogues. They went on to demonstrate that 24 was capable of inhibiting pyocyanin, rhamnolipid, and elastase B production, and biofilm formation in P. aeruginosa.
Fig. 15.
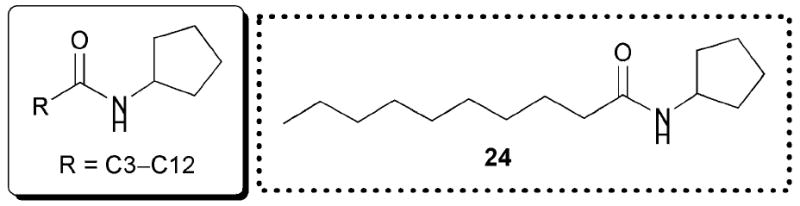
Structural features of AHLs tested against LasR by Kato and co-workers summarized in shaded box; their best antagonist is shown.
In 2005, our laboratory reported the synthesis of a small, focused library of AHLs with non-native acyl groups and the evaluation of these compounds as inhibitors of LasR in P. aeruginosa.39 We identified two compounds (PHL 25 and indole AHL 28, Fig. 16) that showed significant inhibition of the P. aeruginosa LasR circuit using a fluorescent PAO-JP2 reporter strain. Furthermore, these compounds were capable of strongly inhibiting static P. aeruginosa biofilm growth (~75%) in a dose-dependant manner. Based on these results, we designed a larger, focused combinatorial library of AHLs around these active leads and uncovered several inhibitors with heightened activity against LasR (26 and 27, Fig. 16).50 We note that these second generation compounds were discovered using an alternate E. coli LasR reporter strain (E. coli DH5α harbouring LasR and a lacZ fusion).
Fig. 16.
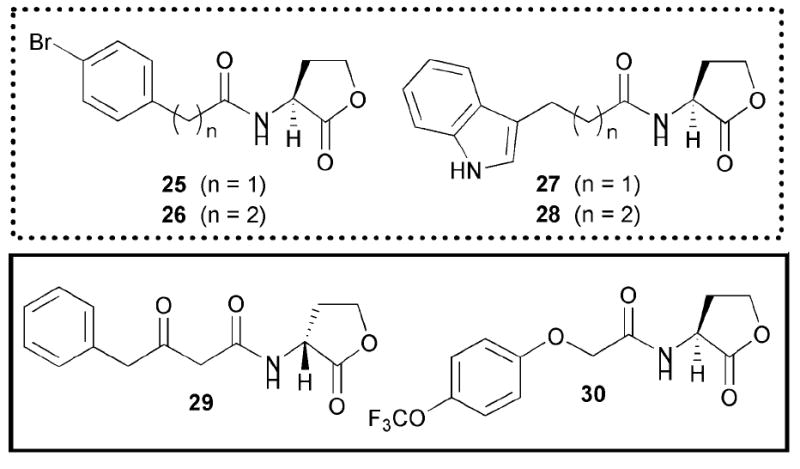
Active non-native AHLs identified by Blackwell and co-workers; significant agonists and antagonists are outlined in solid and dashed lines, respectively. Note, 26 behaves as a partial agonist.
In analogy to the earlier LasR studies described above, we found that AHLs with alkyl chains similar in length to OdDHL were good LasR inhibitors, with DHL being the most potent of the non-3-oxo, simple aliphatic AHLs tested.39,50,51,53 AHLs 26 and 27, the one-carbon longer or shorter variants of our initial leads 25 and 28, respectively, were the most potent LasR inhibitors identified in this study (Fig. 16). These compounds were found to moderately inhibit elastase B production in P. aeruginosa PAO1. Several PHLs were also identified as moderate to strong LasR antagonists. Similar to what we observed for LuxR in V. fischeri, simple structural changes to PHLs (i.e., moving a substituent from the para to the meta position on the PHL phenyl ring) yielded compounds with heightened inhibitory activity against LasR, with our most potent antagonist being meta-nitro PHL 12 (Fig. 10). Intriguingly, these same compounds were agonists, as opposed to antagonists, of LuxR.
Several other activity trends could be discerned from the LasR screening data for our expanded AHL library.50,51 First, LasR appears to tolerate AHLs with sterically bulky and structurally diverse acyl groups (e.g., AHL 30, Fig. 16). These data are supportive of LasR having a relatively large ligand-binding site, which is congruent with the reported LasR X-ray crystal structure.32 Second, the data revealed that not all d-AHL stereoisomers are inactive against LuxR-type proteins. A previous report by Ikeda and co-workers demonstrated that d-BHL fails to activate RhlR in P. aeruginosa, or inhibit RhlR in the presence of l-BHL.60 This work, as well as a recent study by Spring and co-workers,61 has prompted the hypothesis that only l-AHLs will display appreciable activity against LuxR-type proteins. In contrast, we found that d-AHL 29 (Fig. 16) showed similar agonistic activity to the natural ligand for LasR at the same concentration (10 μM).51 This is an important finding, as it illustrates that, even though the d-stereoisomer of the natural AHL ligand is inactive, not all d-stereoisomers are inactive, and suggests that AHL stereochemistry should be examined further in the design of next-generation AHL mimics.
8.3 TraR in A. tumefaciens
In 1996, Winans and co-workers reported a study on the effects of acyl chain length and composition on the activities of AHL mimics against TraR in A. tumefaciens (Fig. 17).62 Using a collection of AHL mimics similar to those reported by Greenberg in LuxR43 and Iglewski in LasR,54 the Winans team elucidated important trends for AHL activity in the TraR system. For their initial studies, they utilized an A. tumefaciens WCF47 Δ-traI derivative harbouring a lacZ fusion as their reporter strain. The researchers determined that TraR was best modulated by compounds that are similar in structure to its cognate AHL, OOHL (Table 1, entry 1). Agonistic activity was highest for AHLs with acyl chains of seven carbons or more and containing the 3-oxo moiety. In turn, antagonistic activity against TraR was highest for AHLs with acyl chains similar in length to the natural ligand yet lacked the 3-oxo group. The most active TraR antagonist in this study was OHL (Table 1, entry 2). However, antagonistic activity was displayed by other AHLs with substitutions at the 3-position of the acyl chain, such as a hydroxyl group. Based on this observation, the authors concluded that the 3-oxo group is not needed for binding to TraR, but rather provides an essential interaction for converting TraR to an active conformation.
Fig. 17.
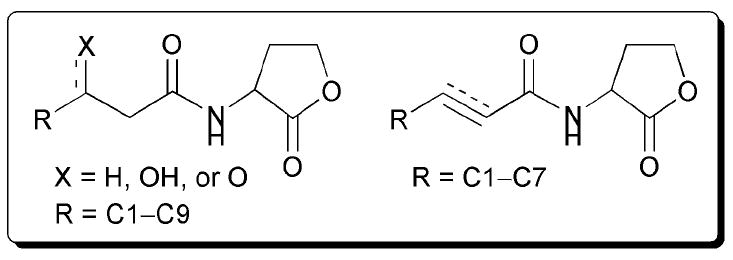
Structural features of AHLs tested against TraR by Winans and co-workers summarized in shaded box; compounds were synthesized and tested as racemates.
Winans and co-workers went on to examine the effect of TraR concentration on ligand activity, using an A. tumefaciens WCF47 reporter strain that overproduced TraR.62 Their studies revealed that at higher protein concentrations, a larger number of their synthetic AHLs were able to activate TraR relative to the number that could activate TraR produced at wild-type levels. The authors theorized that binding of an AHL molecule increases the affinity of TraR either for other TraR monomers or for DNA. If this model is true, then analogues of AHLs that are able to bind TraR (i.e., inhibitors) can also cause a small increase in affinity. At wild-type levels of TraR, this increase is insufficient to give activation. If TraR is overproduced, however, the amount of protein is such that these small increases are enough to drive activation. We have seen related activity trends in our lab while investigating the activities of TraR, LasR, and LuxR antagonists at high concentrations; we return to these phenomena below.50,51
In conjunction with our studies of LuxR and LasR outlined above, we have also reported detailed SARs for the modulation of TraR by AHLs. Except for structurally similar, natural AHLs, we found no synthetic AHL mimics that can activate TraR at native concentrations.39,50,51 However, we did identify a small set of compounds that are able to effectively inhibit TraR at nanomolar concentrations (vs. 100 nM OOHL), most notably PHLs. In analogy to LuxR, PHLs that contain electron-with-drawing groups in the para-position were the most active TraR antagonists. Similarly, we observed striking trends in inhibitory activity as these substituents were moved around the phenyl ring. For example, para-substituted PHLs were the most active TraR antagonists (e.g., PHL 25), with meta-substituted PHLs being the next most active, and finally ortho-substituted compounds showing little to no activity. This result is intriguing when we compare it to our data for LuxR and LasR, as substitutions in the meta-position on PHLs confer greater agonistic activity in LuxR and antagonistic activity against LasR. Perhaps the most consequential outcome of our TraR research, however, was the finding that certain TraR antagonists are actually partial agonists, such that at high concentrations (above 100 μM) they are able to act as agonists, but do not have the same efficacy as the natural ligand OOHL.
We believe that the complementary observations of our lab (activation of TraR (and other LuxR-type proteins) by AHLs at high ligand concentration)50 and Winans and co-workers (activation by AHLs at high TraR concentration),62 can be explained through a simple equilibrium as depicted in Fig. 18. Compounds similar in structure to the natural AHL have some innate affinity for the LuxR-type protein ligand-binding site (Keq). As the concentration of either “reactant” is increased (i.e., protein or ligand), the product (or protein–ligand complex) concentration must likewise increase. This receptor–ligand complex is then a reactant for the dimerization event, and subsequent processes leading to transcription. Therefore, no matter if the protein concentration or ligand concentration is increased, the outcome is the same—increased protein–ligand binding and, therefore, increased activation. We note that transcriptional activation by LuxR-type proteins is a complicated process involving several other players and binding events; however, this model provides a simplified vantage point from which to initiate further study of the mechanism of action of AHL mimics.
Fig. 18.
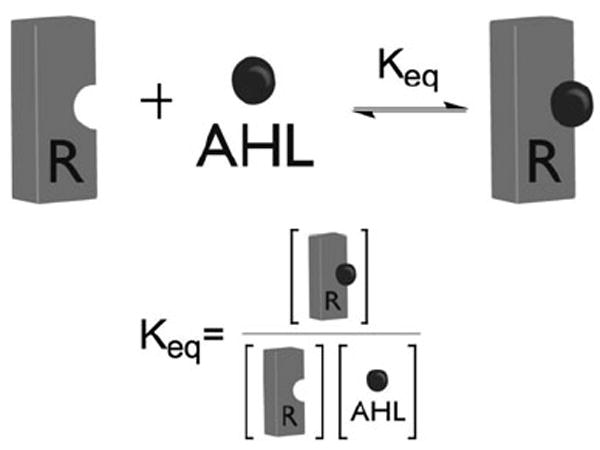
Simplified schematic of the AHL–LuxR-type protein binding equilibrium.
8.4 CarR in E. carotovora
In 1993, Bycroft and co-workers reported the examination of a set of AHL analogues for their ability to regulate antibiotic (carbapenem) production in the plant pathogen E. carotovora. This process is under the control of OHHL and its cognate LuxR-type protein, CarR.63 The authors studied the effects of altering the AHL acyl chain length, switching the lactone to a γ-thiolactone or γ-lactam, and inverting the stereochemistry of the lactone ring from l to d (Fig. 19) in an AHL-negative mutant of E. carotovora. They found that replacement of the oxygen in the lactone ring with sulfur or nitrogen resulted in compounds that were far less active against CarR than OHHL, reinforcing what other researchers have found for LuxR and LasR, as described above. The non-natural stereoisomer of the native lactone, (i.e., d-OHHL) was found to be 10-times less active than l-OHHL at inducing antibiotic production. In addition, the authors determined that the 3-oxo group was essential for agonistic activity and that changing the acyl chain length (by as little as one carbon) resulted in at least a 50% reduction in potency relative to the natural ligand. Together, these data suggested that CarR is highly selective for OHHL.
Fig. 19.
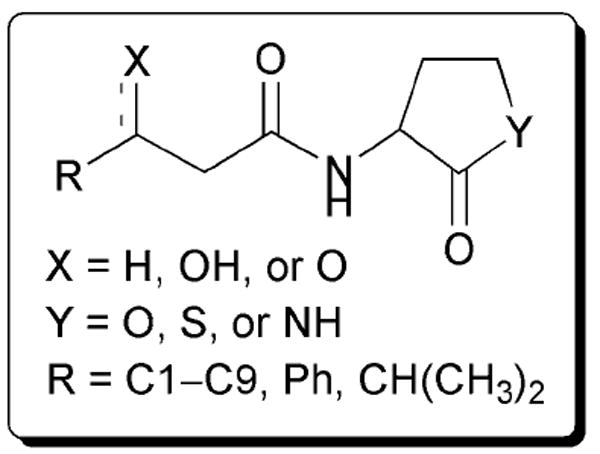
Structural features of AHLs tested by Bycroft and co-workers in E. carotovora summarized in shaded box.
Recently, Spring and co-workers published another study on the activity of AHL mimics in E. carotovora (Fig. 20).64 Specifically, the authors sought to determine the activity of non-hydrolysable AHL mimics (i.e. ketones), as the hydrolyzed, ring-open form of AHLs has been shown to exhibit little, if any, activity.61 Such non-hydrolysable AHLs would have significant value as chemical probes of QS due to their longer lifetimes, and thus prolonged activities. Similar to Bycroft and co-workers, the researchers determined that even small changes to the head group of OHHL were detrimental to its ability to agonize the production of carbapenem, again suggesting that CarR is highly sensitive to AHL structure. Spring and co-workers went on to examine AHL mimics that were difluorinated at the 2-position of the acyl chain, and determined that these derivatives were able to elicit some antibiotic production. Interestingly, they found that AHLs with non-native head groups, while unable to stimulate antibiotic production, retained some ability to stimulate exoprotease production in E. carotovora (an output that is at least partly under the control of CarR). This finding suggested that there may be a different LuxR-type homologue involved in regulation of exoenzyme production in E. carotovora.65
Fig. 20.
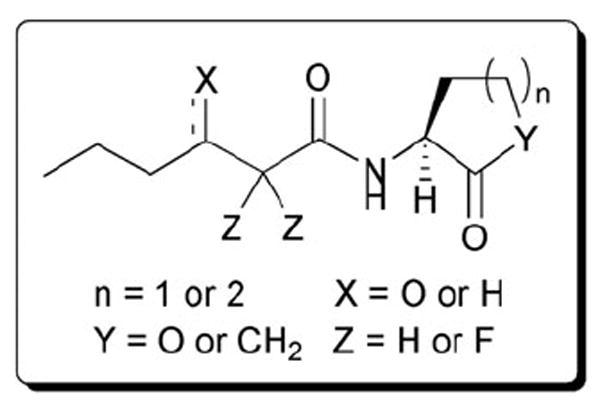
Structural features of AHLs tested by Spring and co-workers in E. carotovora summarized in shaded box.
9. Conclusions, challenges, and the future
This review has provided an overview of AHL analogue activity in a range of bacterial species, strains, and assays. From these analyses, some general conclusions can be made about the AHL structural features that are necessary for activity against LuxR-type proteins. Again, we note that when discussing active agonists and antagonists, it is valuable to consider activity against LuxR-type proteins as a continuum from activation to inhibition, as many AHLs have been identified that can both slightly activate and inhibit a QS circuit (depending on their concentration). Therefore, for the purposes of this final analysis, we will group activators and inhibitors together as necessary, and simply discuss “activity.”
Five broad activity trends that we have delineated from these past studies of AHL mimics are listed below:
In general, the length of the acyl chain is critical for the activity of AHL mimics. Compounds that have acyl chain lengths that are close to that of the natural AHL have heightened activity.
For species where the natural AHL possesses a modification at the 3-carbon of the acyl chain (e.g., a carbonyl), this group is important for activity, but not essential. Elimination of this group most often results in an AHL mimic that has inhibitory activity rather than agonistic activity.
Stereochemistry of the lactone ring is important. In general the natural l-stereoisomer of the lactone ring is needed for activity. There are few exceptions, as we have discovered an AHL LasR activator with d stereochemistry. Because of this exception, and the relatively small number of d-AHLs that have been studied to date, it is important not to assume that the l-stereoisomer is the most active compound in studies where racemic mixtures of lactone are evaluated.
Direct modifications to the lactone ring are tolerated, but not in all systems. In general, this modification results in compounds that are less active. However, lactone ring replacements that retain hydrogen bond acceptors, mimicking the lactone (e.g., a thiolactone), are most often active.
Lastly, the incorporation of aromatic functionality into AHLs, either as lactone replacements or acyl chain substituents (e.g., PHLs), most commonly yields analogues with inhibitory activity.
There are a number of challenges to face as we continue to study Gram-negative QS with AHL analogues. First, one of the largest hurdles we confront is the need for the standardization of assays. Many of the reports that we have examined in this review, although focused on the same LuxR-type protein, were performed using different reporter strains. This is problematic, as our group and others have shown that not all compounds display similar activities in different strains. This discrepancy could potentially be overcome if the field moves beyond cell-based assays towards in vitro assays with purified LuxR-type proteins. We note that this approach has been hampered to date by the difficult purification and manipulation of these proteins in the absence of a natural ligand.7 Second, although several non-native AHLs have been identified that can strongly modulate Gram-negative QS, there is relatively little known about their exact mechanisms of action. One significant question is how an inhibitor is able to block the natural AHL from binding and how this ultimately results in QS inhibition. Such mechanistic studies are essential for both the development of new chemical probes of QS and the ongoing study of QS intervention as a potential anti-infective strategy.
To close, bacterial QS is a new and rapidly expanding field, and we have only recently joined the “conversation.” Continued efforts toward the discovery of new QS modulators will only serve to expand this dialogue. We contend that the development of structures more diverse than the simple AHL mimics discussed herein will be an important advance for the field. Ligands with enhanced chemical stability relative to AHLs will also be of value. Several groups have recently reported approaches to the discovery of such non-AHL scaffolds, including the isolation and characterisation of natural products that modulate QS (e.g., from marine algae and garlic) and high-throughput screens of commercial small molecule libraries.53,66-72 Overall, the chemical nature of QS makes this research area intrinsically accessible and attractive to synthetic chemists, chemical biologists, and biochemists, to just name a few. Whether initiating, translating, or intercepting bacterial communication, the chemical community is certainly poised to play an important role in the QS field for years to come.
Acknowledgments
We apologize to the authors of studies on AHL analogues that we could not include in this review due to length restrictions. Financial support for quorum sensing research in our laboratory is supported by the NIH (AI063326-01), NSF (CHE-0449959), Greater Milwaukee Foundation, Burroughs Wellcome Foundation, Camille & Henry Dreyfus Foundation, Research Corporation, Johnson & Johnson, and DuPont. H.E.B. is an Alfred P. Sloan Foundation Fellow. Sarah Fowler is acknowledged for her critical review of this manuscript. Background image for graphical abstract provided and copyrighted by Dennis Kunkel Microscopy, Inc.
Biographies
Grant D. Geske was born in LaCrosse, WI and attended St. Norbert College for his undergraduate studies in chemistry (BS, 1998). He pursued theoretical chemistry at Utah State University with Professor Alexander Boldyrev (MS, 2001). He then returned to Wisconsin in 2003 to work with Professor Helen Blackwell, where he led a project on the design of synthetic quorum sensing modulators (PhD, 2008)
Jennifer C. O’Neill is a native of Phoenix, AZ and pursued her undergraduate studies in biochemistry at Brigham Young University (BS, 2003). She came to UW-Madison in 2003 and is currently a fifth year graduate student in the lab of Professor Helen Blackwell. Her PhD thesis, which she will defend in summer 2008, has focused in part on delineating the role of diketopiperazines in quorum sensing
Helen E. Blackwell attended Oberlin College for her undergraduate studies (BA, 1994), and pursued her graduate studies in chemistry at Caltech with Professor Robert Grubbs (PhD, 1999). From 1999–2002, she was as postdoctoral fellow with Professor Stuart Schreiber at Harvard University. Thereafter, she joined the faculty of UW-Madison in 2002, where she is currently an Associate Professor of Chemistry
Footnotes
Part of a thematic issue examining the interface of chemistry with biology.
References
- 1.Gest H. Notes Rec R Soc. 2004;58:187. doi: 10.1098/rsnr.2004.0055. [DOI] [PubMed] [Google Scholar]
- 2.Gest H. Perspect Biol Med. 2005;48:266. doi: 10.1353/pbm.2005.0053. [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL, Losick R. Cell. 2006;125:237. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Tomasz A, Beiser SM. J Bacteriol. 1965;90:1226. doi: 10.1128/jb.90.5.1226-1232.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nealson KH, Platt T, Hastings JW. J Bacteriol. 1970;104:313. doi: 10.1128/jb.104.1.313-322.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Biochemistry. 1981;20:2444. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 7.Fuqua C, Greenberg EP. Nat Rev Mol Cell Biol. 2002;3:685. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 8.Bassler BL. Cell. 2002;109:421. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- 9.Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen TB, Givskov M. Microbiology. 2006;152:895. doi: 10.1099/mic.0.28601-0. [DOI] [PubMed] [Google Scholar]
- 11.Camilli A, Bassler BL. Science. 2006;311:1113. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pappas KM, Weingart CL, Winans SC. Mol Microbiol. 2004;53:755. doi: 10.1111/j.1365-2958.2004.04212.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen H, Fink GR. Genes Dev. 2006;20:1150. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP. FEMS Microbiol Rev. 2001;25:365. doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 15.Williams P, Winzer K, Chan WC, Cámara M. Philos Trans R Soc LondonSer B. 2007;362:1119. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González JE, Keshavan ND. Microbiol Mol Biol Rev. 2006;70:859. doi: 10.1128/MMBR.00002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasmussen TB, Givskov M. Int J Med Microbiol. 2006;296:149. doi: 10.1016/j.ijmm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Bjarnsholt T, Givskov M. Philos Trans R Soc London, Ser B. 2007;362:1213. doi: 10.1098/rstb.2007.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raffa RB, Iannuzzo JR, Levine DR, Saeid KK, Schwartz RC, Sucic NT, Terleckyj OD, Young JM. J Pharmacol Exp Ther. 2005;312:417. doi: 10.1124/jpet.104.075150. [DOI] [PubMed] [Google Scholar]
- 20.Smith RS, Iglewski BH. Curr Opin Microbiol. 2003;6:56. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 21.Suga H, Smith KM. Curr Opin Chem Biol. 2003;7:586. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Clatworthy AE, Pierson E, Hung DT. Nat Chem Biol. 2007;3:541. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 23.Chhabra SR, Philipp B, Eberl L, Givskov M, Williams P, Camara M. Top Curr Chem. 2005;240:279. [Google Scholar]
- 24.Persson T, Givskov M, Nielsen J. Curr Med Chem. 2005;12:3103. doi: 10.2174/092986705774933425. [DOI] [PubMed] [Google Scholar]
- 25.Winzer K, Hardie KR, Williams P. Curr Opin Microbiol. 2002;5:216. doi: 10.1016/s1369-5274(02)00304-1. [DOI] [PubMed] [Google Scholar]
- 26.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Proc Natl Acad Sci U S A. 1999;96:4360. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould TA, William T, Schweizer HP, Churchill ME. Acta Crystallogr Sect D: Biol Crystallogr. 2004;60:518. doi: 10.1107/S0907444903028300. [DOI] [PubMed] [Google Scholar]
- 28.Gould TA, Schweizer HP, Churchill ME. Mol Microbiol. 2004;53:1135. doi: 10.1111/j.1365-2958.2004.04211.x. [DOI] [PubMed] [Google Scholar]
- 29.Welch M, Mikkelsen H, Swatton JE, Smith D, Thomas GL, Glansdorp FG, Spring DR. Mol BioSyst. 2005;1:196. doi: 10.1039/b505796p. [DOI] [PubMed] [Google Scholar]
- 30.Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneaux JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Nature. 2002;417:971. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 31.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, de Francesco F, Neddermann P, di Marco S. EMBO J. 2002;21:4393. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bottomley MJ, Muraglia E, Bazzo R, Carfí A. J Biol Chem. 2007;282:13592. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez JE, Marketon MM. Microbiol Mol Biol Rev. 2003;67:574. doi: 10.1128/MMBR.67.4.574-592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spring DR. Chem Soc Rev. 2005;34:472. doi: 10.1039/b312875j. [DOI] [PubMed] [Google Scholar]
- 35.Sandoz KM, Mitzimberg SM, Schuster M. Proc Natl Acad Sci U S A. 2007;104:15876. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams P. Microbiology. 2007;153:3923. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- 37.Manefield M, Whiteley AS. Philos Trans R Soc London, Ser B. 2007;362:1235. doi: 10.1098/rstb.2007.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas GL, Böhner CM, Williams HE, Walsh CM, Ladlow M, Welch M, Bryant CE, Spring DR. Mol Biosyst. 2006;2:132. doi: 10.1039/b517248a. [DOI] [PubMed] [Google Scholar]
- 39.Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J Am Chem Soc. 2005;127:12762. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 40.Steindler L, Venturi V. FEMS Microbiol Lett. 2007;266:1. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 41.Fekete A, Frommberger M, Rothballer M, Li X, Englmann M, Fekete J, Hartmann A, Eberl L, Schmitt-Kopplin P. Anal Bioanal Chem. 2007;387:455. doi: 10.1007/s00216-006-0970-8. [DOI] [PubMed] [Google Scholar]
- 42.Eberhard A, Widrig CA, McBath P, Schineller JB. Arch Microbiol. 1986;146:35. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 43.Schaefer AL, Hanzelka BL, Eberhard A, Greenberg EP. J Bacteriol. 1996;178:2897. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olsen JA, Severinsen R, Rasmussen TB, Hentzer M, Givskov M, Nielsen J. Bioorg Med Chem Lett. 2002;12:325. doi: 10.1016/s0960-894x(01)00756-9. [DOI] [PubMed] [Google Scholar]
- 45.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. EMBO J. 2003;22:3803. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reverchon S, Chantegrel B, Deshayes C, Doutheau A, Cotte-Pattat N. Bioorg Med Chem Lett. 2002;12:1153. doi: 10.1016/s0960-894x(02)00124-5. [DOI] [PubMed] [Google Scholar]
- 47.Castang S, Chantegrel B, Deshayes C, Dolmazon R, Gouet P, Haser R, Reverchon S, Nasser W, Hugouvieux-Cotte-Pattat N, Doutheau A. Bioorg Med Chem Lett. 2004;14:5145. doi: 10.1016/j.bmcl.2004.07.088. [DOI] [PubMed] [Google Scholar]
- 48.Frezza M, Castang S, Estephane J, Soulère L, Deshayes C, Chantegrel B, Nasser W, Queneau Y, Reverchon S, Doutheau A. Bioorg Med Chem. 2006;14:4781. doi: 10.1016/j.bmc.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 49.Geske GD, O’Neill JC, Blackwell HE. ACS Chem Biol. 2007;2:315. doi: 10.1021/cb700036x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geske GD, O’Neill JC, Miller DM, Mattmann ME, Blackwell HE. J Am Chem Soc. 2007;129:13613. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geske GD, O’Neill JC, Miller DM, Mattmann ME, Lin Q, Wezeman RJ, Blackwell HE. ChemBioChem. 2008;9:389. doi: 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Janssens JCA, Metzger K, Daniels R, Ptacek D, Verhoeven T, Habel LW, Vanderleyden J, De Vos DE, De Keersmaecker SCJ. Appl Environ Microbiol. 2007;73:535. doi: 10.1128/AEM.01451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Müh U, Hare BJ, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, Greenberg EP. Proc Natl Acad Sci U S A. 2006;103:16948. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Passador L, Tucker KD, Guertin KR, Journet MP, Kende AS, Iglewski BH. J Bacteriol. 1996;178:5995. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kline T, Bowman J, Iglewski BH, de Kievit T, Kakai Y, Passador L. Bioorg Med Chem Lett. 1999;9:3447. doi: 10.1016/s0960-894x(99)00626-5. [DOI] [PubMed] [Google Scholar]
- 56.Smith KM, Bu Y, Suga H. Chem Biol. 2003;10:81. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 57.Smith KM, Bu Y, Suga H. Chem Biol. 2003;10:563. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 58.Jog GJ, Igarashi J, Suga H. Chem Biol. 2006;13:123. doi: 10.1016/j.chembiol.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Appl Environ Microbiol. 2007;73:3183. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ikeda T, Kajiyama K, Kita T, Takiguchi N, Kuroda A, Kato J, Ohtake H. Chem Lett. 2001;30:314. [Google Scholar]
- 61.Glansdorp FG, Thomas GL, Lee JK, Dutton JM, Salmond GP, Welch M, Spring DR. Org Biomol Chem. 2004;2:3329. doi: 10.1039/B412802H. [DOI] [PubMed] [Google Scholar]
- 62.Zhu J, Beaber JW, Moré MI, Fuqua C, Eberhard A, Winans SC. J Bacteriol. 1998;180:5398. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salmond G, Stewart GS, Williams P, Bycroft BW. J Antibiot. 1993;46:441. doi: 10.7164/antibiotics.46.441. [DOI] [PubMed] [Google Scholar]
- 64.Welch M, Dutton JM, Glansdorp FG, Thomas GL, Smith DS, Coulthurst SJ, Barnard AM, Salmond GP, Spring DR. Bioorg Med Chem Lett. 2005;15:4235. doi: 10.1016/j.bmcl.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 65.Barnard AML, Salmond GPC. Anal Bioanal Chem. 2007;387:415. doi: 10.1007/s00216-006-0701-1. [DOI] [PubMed] [Google Scholar]
- 66.Hjelmgaard T, Persson T, Rasmussen TB, Givskov M, Nielsen J. Bioorg Med Chem. 2003;11:3261. doi: 10.1016/s0968-0896(03)00295-5. [DOI] [PubMed] [Google Scholar]
- 67.Wu H, Song Z, Hentzer M, Andersen JB, Molin S, Givskov M, Høiby N. J Antimicrob Chemother. 2004;53:1054. doi: 10.1093/jac/dkh223. [DOI] [PubMed] [Google Scholar]
- 68.Persson T, Hansen TH, Rasmussen TB, Skindersø ME, Givskov M, Nielsen J. Org Biomol Chem. 2005;3:253. doi: 10.1039/b415761c. [DOI] [PubMed] [Google Scholar]
- 69.Choo JH, Rukayadi Y, Hwang JK. Lett Appl Microbiol. 2006;42:637. doi: 10.1111/j.1472-765X.2006.01928.x. [DOI] [PubMed] [Google Scholar]
- 70.Müh U, Schuster M, Heim R, Singh A, Olson ER, Greenberg EP. Antimicrob Agents Chemother. 2006;50:3674. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taha MO, Al-Bakri AG, Zalloum WA. Bioorg Med Chem Lett. 2006;16:5902. doi: 10.1016/j.bmcl.2006.08.069. [DOI] [PubMed] [Google Scholar]
- 72.Vattem DA, Mihalik K, Crixell SH, McLean RJC. Fitoterapia. 2007;78:302. doi: 10.1016/j.fitote.2007.03.009. [DOI] [PubMed] [Google Scholar]


