Abstract
The IgM H chain gene organization of cartilaginous fishes consists of 15–200 miniloci, each with a few gene segments (VH-D1-D2-JH) and one C gene. This is a gene arrangement ancestral to the complex IgH locus that exists in all other vertebrate classes. To understand the molecular evolution of this system, we studied the nurse shark, which has relatively fewer loci, and characterized the IgH isotypes for organization, functionality, and the somatic diversification mechanisms that act upon them. Gene numbers differ slightly between individuals (~15), but five active IgM subclasses are always present. Each gene undergoes rearrangement that is strictly confined within the minilocus; in B cells there is no interaction between adjacent loci located ≥120 kb apart. Without combinatorial events, the shark IgM H chain repertoire is based on junctional diversity and, subsequently, somatic hypermutation. We suggest that the significant contribution by junctional diversification reflects the selected novelty introduced by RAG in the early vertebrate ancestor, whereas combinatorial diversity coevolved with the complex translocon organization. Moreover, unlike other cartilaginous fishes, there are no germline-joined VDJ at any nurse shark μ locus, and we suggest that such genes, when functional, are species-specific and may have specialized roles. With an entire complement of IgM genes available for the first time, phylogenetic analyses were performed to examine how the multiple Ig loci evolved. We found that all domains changed at comparable rates, but VH appears to be under strong positive selection for increased amino acid sequence diversity, and surprisingly, so does Cμ2.
All jawed vertebrates, from sharks to mammals, have circulating lymphocytes expressing a diverse Ab and TCR repertoire that is generated through somatic recombination (1, 2). The tetrapod IgH gene is usually a single locus consisting of multiple gene segments (VH, D, JH) that rearrange to form the joined VDJ variable region, followed by C region genes that are expressed at different stages of B cell development (3, 4). In contrast, the genes encoding IgM H chain genes in cartilaginous fishes are organized as multiple miniloci (clusters) (5, 6). Each cluster consists of few gene segments (VH-D1-D2-JH) and one C gene (see Fig. 1). Although most miniloci appeared individually capable of rearrangement, the intriguing discovery was that many other clusters are partially (VD-J, VD-DJ) or fully recombined (VDJ) in the germline (2, 6, 7). Because cartilaginous fishes represent the earliest vertebrates where RAG was established as V(D)J recombinase, such findings raised long-standing questions as to the relative significance of the role rearrangement plays in generating Ig diversity in sharks and skates and the manner in which elasmobranch Ig expression was regulated, for instance, whether H chain exclusion, if it existed, could be as dependent on rearrangement as it is in mammals. To approach these issues, it is necessary to define the individual IgH and establish their germline organization for comparison to the individual lymphocyte.
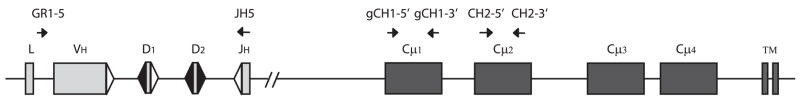
FIGURE 1.
IgM H chain locus in shark. Representation of functional H chain locus of Groups 1–5. Each consists of leader (L) and the gene segments VH, D1, D2, and JH in close proximity, the intersegmental regions each being ~400 bp. The triangles represent recombination signal sequences (RSS), with the 23 bp spacer (△) or 12 bp spacer (▲). The C region exons are located ~7 kb downstream in the G2 subfamily, and the exons are variously 0.5 to 3 kb apart. Universal PCR primer are shown above, as arrows; for instance, primer pair GR1–5/JH5 will amplify any nurse shark VH genes from G1–5.
Although the unique IgH organization in Chondrichthyes was reported many years ago (5), the high copy number in horn shark and other species, estimated to be 100–200, prevented a thorough analyses of the genes that constitute their expressed IgM Ab repertoire. Pulsed field gel electrophoresis analyses of Hydrolagus colliei were consistent with greater numbers of VH than Cμ4, but some isolated VH proved to be pseudogenes (8). Moreover, organizational information has been mainly from bacteriophage clones, which largely consisted of partial genes due to the size of the individual IgH (>16 kb from leader through the C region exons (9)). This is of some concern, as similar data obtained for bony fish IgL clusters proved to be misleadingly incomplete once a genomic overview was available. In the case of zebrafish IgL, multiple components could be found in L chain genes of all three isotypes, showing that the generally accepted simple IgL locus with 2–4 gene segments was neither the predominant nor sole possible organization in bony fishes (10).
Our model is the nurse shark, which we estimate to have 15 IgM clusters (11), and in this study we have characterized them in two unrelated individuals, in genomic and cDNA libraries from shark-33 and a bacterial artificial chromosome (BAC)3 library from the genetically unrelated shark-Y. We found there are 9–12 functional genes, and the gene number can differ slightly between individuals. Each minilocus consists of one VH, two Ds, one JH, and one set of Cμ exons, and must rearrange to be expressed. Each active cluster is capable of hypermutation. We found that the IgH clusters are at least 120 kb apart and formally show that the expressed B cell rearrangements are confined to the four gene segments within the minilocus. In the absence of combinatorial rearrangement events, we conclude that the principal source of heterogeneity in the shark primary repertoire is junctional diversification. There are no germline-joined Ig encoding IgM Abs in nurse shark, and we propose that such genes, when active, have evolved to be specialized in the different species.
Having characterized the complete set of germline μ clusters of nurse shark we went on to ask how the multiple Ig loci evolved in relation to each other. The variation in VH gene segments and, even more interestingly, the variation in C region sequence has not been extensively explored in this early, alternative Ig system. Divergent CH sequences and IgH arrangements observed in horn shark and Hydrolagus (8, 12) have long pointed to the plasticity of the chondrichthyan Ig system, but either the entire gene was not available for analysis or the functionality of the variant gene clusters were not definitively established. In the nurse shark we observed that pseudogenes with obvious structural defects can nonetheless rearrange and be transcribed; however, they are not represented at any significant level in adult lymphoid RNA. In this study we introduce the definition of an active nurse shark IgH: it rearranges, is transcribed, and is somatically mutated to reflect selection on and usage of a protein product. We then asked whether the genes encoding functional VH and Cμ sequences evolved at different rates and found, to our surprise, that not only VH but also Cμ2 show evidence for selection among the loci.
Materials and Methods
Animals
Nurse sharks (Ginglymostoma cirratum) were captured off the coast of the Florida Keys. Some animals had been used in other studies (shark-Y, shark-33, shark-JS). DNA was extracted from whole blood or from the erythrocyte pellet after centrifugation through Ficoll.
Libraries
Shark-33 erythrocyte genomic and epigonal organ cDNA libraries, constructed in λFix II and λZAP Express XR (Stratagene and Loftstrand Laboratories, respectively), were previously described (11), as was the shark-Y BAC library (13). The bacteriophage libraries were screened for μ genes with probes to VH and Cμ1–2, separately, whereas the high-density replica BAC library filters were hybridized with a probe to the highly conserved Cμ3-Cμ4 domains. The selected clones were purchased from Arizona Genomics Institute (http://www.genome.arizona.edu). The grid positions (plate addresses) of those clones containing the characterized IgH are available at (http://www.downstate.edu/pharmacology/faculty/hsu_pdf/ji2008.pdf, Table I). In this work, the BAC clone will referred to by the nurse shark library plate address.
Table I.
Germline VH genes: IgM and IgM-derived loci detected by vh probe
| Subfamily | Locus | Genomic V-D-D-J Clone Accession Number | cDNA | Remarks |
|---|---|---|---|---|
| IgM (rearranging): | ||||
| Group 1 | G1 | DQ857386 | EU106186 | |
| Group 2 | G2A | DQ192492, DQ192494 | AY583357, AY594650 | V1, V2 resp. may be allelesa |
| G2B | DQ192493 | EU106179 | ||
| ψG2C | DQ857389 | ND | Pseudogeneb | |
| Group 3 | G3 | DQ857384 | EU106181 | |
| Group 4 | G4A | Phage E2; DQ857390 | EU106182 | |
| G4C | Phage B3; DQ857388 | EU106184 | G4C and G4G share VH and Cμ1 but not Cμ2 | |
| G4G | Phage B2 | EU106185 | ||
| G4E | Phage S5; DQ857387 | EU106182 | ||
| Group 5 | G5 | DQ857385 | M92851, EU106180 | |
| Group 6 | ψG6 | ND | AY609274c | VH only; pseudogene |
| Group 7 | ψG7 | EU312153 | — | Pseudogened |
| Group 8 | ψG8 | EU312152 | EU106187 | Pseudogenee |
| IgM-related (germline-joined VDJ): | ||||
| IgM1gj | H1gj | ND | AF327520 | Prominent in neonates |
| IgM2gj | ψH2gj | EU312154 | — | Pseudogenef |
Complete H chains of the G2A members V1 and V2 differ by 1 bp in CDR2. These cDNA sequences carry no mutations.
G2C carries a one bp deletion in Cμ1 and a 13 bp deletion in Cμ2. Transcripts can be detected by RT-PCR.
The cDNA contains only VH and JH germline sequence. No C sequence is spliced to JH, none was found in BAC library.
No cDNA found by RT-PCR (Malecek and Hsu, data not shown); stop in J gene segment.
Leader intron cannot be spliced out; confirmed in cDNA. JH may be deleted; also missing in cDNA.
Not found in BAC library with CH3–4 probe.
ND, Not done; —, not detected.
PCR and probes
The locations of the sequences targeted by some primers are shown in Fig. 1. Intradomain primers detecting all Cμ1 (gCH1–5′/gCH1–3′) and Cμ2 (CH2–5′/CH2–3′) exons were described previously (11). The combination GR5′ (20% GR1: 5′-GTTTCTCTACCTCAGCAAT-3′ and 80% GR2–5: 5′-GTTAGTCTMCCTCTGGAAT-3′) and JH5 (5′-TCACIGTCACCAT GGT-3′) detects all genomic VH from Groups 1–5. For Group 2 VH, a G2-specific primer in the leader and in FR3 (V18-1: 5′-ACCAGAATGAC GACGATG-3′; V18-2: 5′-TGTCTTCGATCTTCAGGC-3′) produced a 461 bp fragment. The pseudogene G2C was amplified by using an unique sequence 35 bp upstream of the leader (V4: 5′-AGAGCCTTCTCTCC TAAT-3′) in combination with JH5. Group 4-specific VH primers (F4L: 5′-ACCAGAATGACTACGTCG-3′; G4-VJINT: 5′-AGCGGTACTTAC GACAGC-3′) amplified a 718 bp fragment extending from leader to the VH-D1 intersegmental region. Primers S5-C2INT (5′-CCATTGATTTGACTGATG-3′) in the intron and CH2–3′ generated Group 4 Cμ2 PCR products for sequencing. The optimal annealing temperature was determined by a T-Gradient thermocycler (Biometra) and the extension time was usually 30 s for 300–500 bp products.
In this way, VH, Cμ1, and Cμ2 fragments can be generated from phage lysates or BAC DNA for restriction enzyme analyses or for direct sequencing (Genewiz). Cross-hybridizing probes to H chain were derived from Group 2 cDNA (cμ1, cμ2, and cμ3-cμ4 had been described elsewhere Ref. 11). The vh probe was obtained from G2A cDNA clones (for example, Q10, Ref. 11) using the V18-1/V18-2 primer combination to generate a 337 bp fragment. The 216 bp transmembrane probe (cmem) was amplified from the cDNA clone (accession number AY583356) using primers mem1: 5′-GATTCGATAGATCACACT-3′ and mem2: 5′-AAACAGGACTGATT GTAT-3′. Nurse shark RAG1 (rag1) probe was derived from the sequence in GenBank (accession number U13982).
For genomic Southern blotting, electrophoresed DNA samples were transferred to nylon filters (HyBond-N, Amersham Biosciences), and the membranes hybridized with radioactive probe under routine stringency conditions. This entailed incubating the blot with 105–106 cpm probe/ml hybridization buffer (0.2% polyvinylpyrolidone, 0.2% Ficoll, 0.2% BSA, 50 mM Tris (pH 7.5), 1 M NaCl, 0.1% sodium pyrophosphate, 1% SDS) at 68°C overnight and washing at 55°C with four changes of buffer (0.3 M NaCl, 30 mM sodium citrate (2× SSC), 0.1% SDS) and a final rinse (0.2× SSC) at room temperature. The blots were subjected to autoradiography, and signal intensities of bands were quantified using a Storm 860 phosphorimaging system with ImageQuant software (Molecular Dynamics).
Pulse field
BAC inserts were liberated by digesting with NotI and their sizes were determined on pulse field electrophoresis (CHEF-DRIII system, Bio-Rad). The 1% agarose gels were electrophoresed at 1–20 s linear ramp, 6 volts/cm, 120° angle, 14°C in 0.5× TBE buffer for 16–18 h. The gels were then stained with ethidium bromide to visualize the DNA fragments.
Phylogenetic analyses
We translated the VH gene segments and the four C exons for the nine functional loci and then aligned sequences across loci for each of the five domains by eye using Se-Al (v2.0a11, A. Rambaut, Institute for Evolutionary Biology, University of Edinburgh, Edinburgh, U.K., private distribution). The model of evolution for each domain was assigned using MrModelTest (14). Because the trees were small, we could perform branch-and-bound searches under Maximum Likelihood in PAUP* (version 4.0, Ref. 15), guaranteeing the optimal topology. Each data set was bootstrapped 1000 times to assess tree robustness.
To test if the five domains have the same evolutionary history of divergence leading to the nine sampled segments, the likelihood of each different gene tree given each of the five sets of DNA sequences were compared using the SH test (16, as implemented in PAUP*, version 4.0, Ref. 15). The SH test asks whether a candidate tree topology (e.g., the best tree estimated from the Cμ 1 gene-segment DNA data) is rejected by the focal DNA data set (e.g., by the VH gene segment data). If the reciprocal test also rejects the candidate tree, this is strong evidence that the two domains have not evolved along the same tree (i.e., together). We used the bootstrap consensus topologies, with branches with <50% support collapsed to form polytomies.
Finally, we used the codeml package from the PAML suite (version 4; Ref. 17) to ask whether the patterns of substitution differed among domains. For each segment, we estimated the global average dn and ds ratios (model = 0) using as our input tree the Maximum Likelihood topology from the PAUP* analyses. Genes (in our case, VH gene segments or C exons) with more elevated dn (the inferred number of non-synonymous substitutions per nonsynonymous site) relative to ds (the inferred number of synonymous substitutions per synonymous site) exhibit a stronger signature of positive selection (17; see also Ref. 18).
Results
Eight IgM groups
The genomic bacteriophage library was screened with probes to 1–2 (11) whereas the BAC library was screened both VH and Cμ with a Cμ3– 4 sequence. A comparison of the results allowed us to distinguish the functional genes that potentially encode a five-domain molecule. Table I lists all the μ-related genes.
We had demonstrated (11) that nurse shark H chain sequences could be distinguished by restriction endonuclease sites in Cμ2, and the exons thus defined were linked to VH gene segments previously classified into VH subfamilies (19). Although that study focused on Group 2 sequences, various other germline VH genes isolated from the shark-33 erythrocyte genomic library were identified and entered into the database, as listed in Table I. We will refer to the various IgM H chain genes by Group name. The VH sequences are members of one family (20), and the C regions conform to the definition of H chain subclasses. The active μ gene Groups thus encode IgM isotypes, because members of each Group are expressed in every animal we have so far studied.
All the μ clones contained gene segments (one VH, two D, one JH) that required somatic rearrangement to be functional (Fig. 2). These are classified as Groups 1–8 (G1-G8); G2 and G4 consisted of several members. Some genes have been designated pseudo-genes (prefaced with ψ) for reasons detailed in the footnotes to Table I. The characteristic restriction enzyme sites and patterns in the active genes (G1-G5) are summarized in Table II.
FIGURE 2.
Comparison of D and JH gene segments and their RSS. VH, D1, D2, and JH gene segments in G1, G2, G3, G4, G5, G7, and G8 all span an area of <2 kb, as shown in Fig. 1. The heptamer/nonamer sequences are bolded, being 3′ of the VH flank, 5′ of the JH gene segment, and straddling D1 and D2. Splice signal in JH in lower case. The “12–23 rule” permits several kinds of rearrangement possibilities, but the most common observed in cDNA is VH to D1 to D2 to JH. Many RSS do not conform to the canonical sequence but all the functional genes rearrange. Among pseudogenes (ψ), recombination has been looked for and found in G2C. G6 and G8 can be transcribed and are potentially open to RAG action, but the cDNA sequences are nonrearranged.
Table II.
Restriction endonuclease sites specific to genomic μ loci
| Southern Blotting
|
|||||
|---|---|---|---|---|---|
| VH Probe
|
Cμ1 Probe
|
||||
| Subfamily | Member | BamH I/Nco I | Ava II | Ava II | PCR Cμ2 Product Site Detected |
| G1 | 0.45, 0.7 kb | 1.8 kb | 10 kb | Cla I | |
| G2 | A(V1), B | 1.9 kb | 0.3, 0.37 kb | 0.9 kb | Hind III |
| A (V2) | 1.9 kb | 0.3, 5 kb | 0.9 kb | Hind III | |
| ψC | 1.9 kb | 0.3, 1.1 kb | 0.9 kb | Hind III | |
| G3 | 3 kb | 1 kb, 3.5 kb | 1.5 kb | EcoR I | |
| G4 | B, E, G | 1.9 kb | 1 kb (x2) | 1 kb | EcoR I, Hph I |
| A, C, D | 1.9 kb | 1 kb, 4 kb | 1 kb | EcoR I, Hph I | |
| Fa | 1.9 kb | 2 kb | 1 kb | EcoR I, Hph I | |
| G5 | 1.4, 7.8 kb | 3.9 kb | 0.7 kb | None of the above | |
The VH of G1 and G5 carry a BamHI site in FR2; those of G2, G3 and G4 Ava II sites. The VH of G4F is an exception in lacking an internal Ava II and so appears as a single band at 2 kb.
See erratum in: J. Immunol. 2008 Aug 15; 181(4): 2933-4
Screening of nurse shark BAC library
The nurse shark BAC library was screened with a Cμ3-Cμ4 probe, and 216 candidate clones were selected. Subsequent hybridizations with VH and with Cμ2 probes determined that 25 contained only C sequence and 156 carried both V and C. If the BAC library contains 11 genomes coverage of nurse shark DNA, then there are 14–16 IgM loci per genome, approximately what we previously estimated. We report on the detailed analyses of the first 96 clones, of which 10 contained C sequence and 68 clones carried both V and C. Of these 78 clones, 75 can be identified and represent μ genes from4.5 genomes (http://www.downstate.edu/pharmacology/faculty/hsu_pdf/ji2008.pdf, Table I).
G1-G5 clones were identified as shown in Table II. No G6 genes were found. G7 was present together with G8 in two clones (352J19, 269E20). By the combination of Southern blotting and Cμ2 restriction sites, 67/68 clones confirmed the linkage of V and Cμ2 within their Groups. The exception (075N10; Fig. 3) contains two partial IgH: Cμ1 to Cμ4 from Group 2A and far downstream, the VH gene segments from Group 5; they are at least 120 kb apart and located at either ends of the vector.
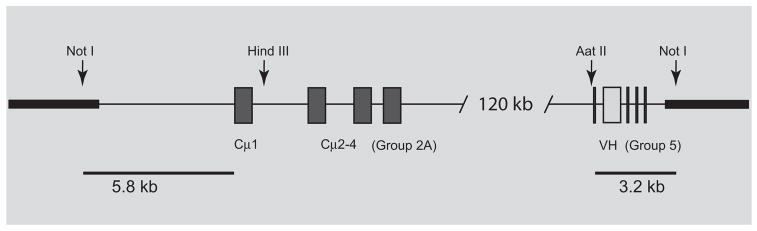
FIGURE 3.
BAC clone with two partial H chain loci. The 075N10 BAC clone was unique in containing VH gene segments from G5 and Cμ exons from G2A, as identified by Southern blotting (see Table II) and confirmed by sequenced PCR fragments of VH and Cμ2. The 075N10 contained an insert of ~140 kb DNA without internal NotI sites. The Ig genes were mapped in relation to the BAC vector sites NotI and NcoI, present on either side of the vector HindIII cloning site (accession no. U51113). A unique Aat II site 5′ of the G5 VH leader sequence is ~3.2 kb from the vector end. The G2A Cμ1 is present on an 8 kb NcoI fragment that proved to contain 1.3 kb vector after a double digest with NotI. The distance from the HindIII site in the vector to one that is 3′ of the Cμ1 exon is 6 kb. The four C exons are represented as filled boxes, and the VH gene as open box flanked by leader and D, JH gene segments.
With the exception of 075N10, ψG7, and ψG8, there was only one IgH per BAC clone, and whether there would have been more than one V or C exon per IgH, their restriction enzyme patterns would necessarily have to be identical for AvaII, BamH I, and NcoI to elude detection. There certainly are not more than one VH or CH from different groups per BAC clone because the additional bands would have been evident from the Southern blots. However, because G2 and G4 contain multiple members, we went on to characterize these BAC clones.
Group 2 genes
We had found three G2 loci, which we now call G2A, G2B, and a pseudogene G2C (formerly V1/V2, V3 and V4, respectively in Ref. 11). Among the 68 BAC clones with a complete set of H chain genes, 21 were scored as G2 by characteristics shown in Table II. These 21 (and 075N10) contained Cμ2 with a HindIII site; moreover, of the total 96 DNA samples subjected to amplification by G2-specific primers V18-1/V18-2, only the same 21 clones produced VH fragments of the expected 461 bp size. All others, including 075N10, were negative.
The 461 bp PCR fragments were digested with HinfI and classified according to the number of sites: twelve VH had two sites (G2A), nine had one site (G2B), and none had four sites (G2C). The clones with G2A VH carried a Cμ2 containing BstEII site, which distinguishes the C region of G2A from G2B (11). Some VH and C fragments were sequenced for confirmation. These results show that the BAC library is biased in G2 gene content in the absence of G2C, and in the 21 G2 genes, the linkage of VH gene segments and the CH in G2A were distinct from G2B.
We looked to see whether the G2C pseudogene could rearrange because its defects seemed restricted to the C exons (Table I, footnote b). Using a specific upstream primer (V4), we were able to find VDJ rearrangements from B cell genomic DNA (our unpublished results). G2C sequences can also be raised by RT-PCR but are primarily found in pup (data not shown). Thus, IgH pseudogenes can rearrange as well as be transcribed.
Group 4 genes
The 29 VH-positive G4 clones were classified according the criteria in Table II. After sequence analyses of several VH and Cμ2 PCR products from selected clones, all the G4 BAC clones were subjected to a battery of restriction enzymes and duly organized into six members, thereafter called G4A to G4F. They differ in both VH and Cμ2 sequence (Fig. 4); it is not known whether some are allelic variants.
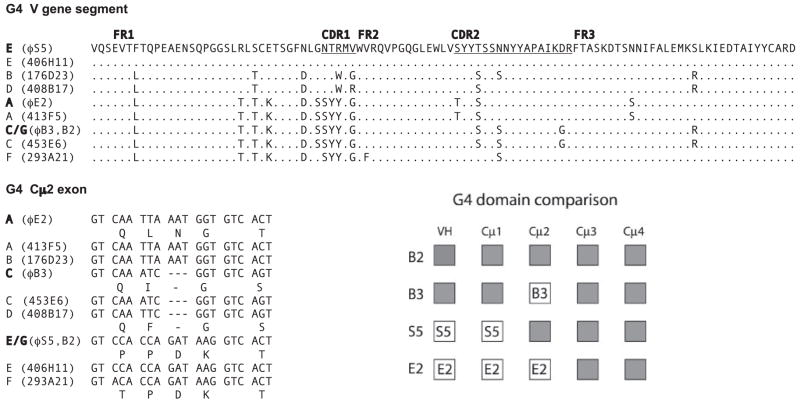
FIGURE 4.
Classification of G4 members by VH and Cμ2 sequences. Panel G4 V gene segment, The deduced amino acid sequences of VH sequences obtained from bacteriophage clones ϕE2, ϕB3/B2, and ϕS5 are compared among representatives of different BAC clones classified as G4A to G4F with their grid names. Dots indicate identity to reference VH sequence from ϕS5. Panel G4 Cμ2 exon, The 5′ end of the C2 exons of G4 genes are compared, and positions with different amino acids are indicated in the one-letter code. Dashes indicate gaps. Panel G4 domain comparison, The VH gene segments (VH, D1, D2, JH) are represented as a block, like the C exons. The domains of clones B3, E2, and S5 are compared with B2 to show the extent of shared sequence, indicated by shaded boxes. The domains of B2 and B3 are identical in all except Cμ2. E2 is identical to B2 in Cμ3 and Cμ4; S5 is identical to B2 in Cμ2, Cμ3, and Cμ4.
In comparisons with the shark-33 bacteriophage results, the shark-Y genes G4A, G4C, and G4E had their counterparts, ϕE2, ϕB3, and ϕS5, respectively. We have called G4G the locus that was prominent in the shark-33 genomic (ϕB2) and cDNA libraries (our unpublished results), but this was not present in the shark-Y BAC library. There are additional genes in the BAC library not found in shark-33 genomic or cDNA library: G4B, D, and F VH regions were not present among 58 G4 cDNA from shark-33. Thus, it appeared that members of Group 4 could be heterogeneous among nurse sharks.
As shown in Fig. 4 (panel G4 domain comparison), there is extensive sharing of sequence among the G4 sequences. The identity of entire domains suggests that exchange of C exons could have taken place by unequal crossing-over or gene conversion. This, however, is not universal; it has not occurred among G2 genes, where there are differences even in Cμ4 between G2A and G2B, or between the adjacent G2A and G5.
Although G3 and G4 are closely related, the VH are distinguishable by Southern blotting (Table II) and there are sequence differences throughout the H chain. There are a total of 35 clones that carry a Cμ2 with an EcoRI site, but only the six G3 lack the G4-specific Hph I site. G3 sequences are infrequent in adult cDNA libraries but recombined, hypermutated sequences can be raised by RT-PCR (data not shown).
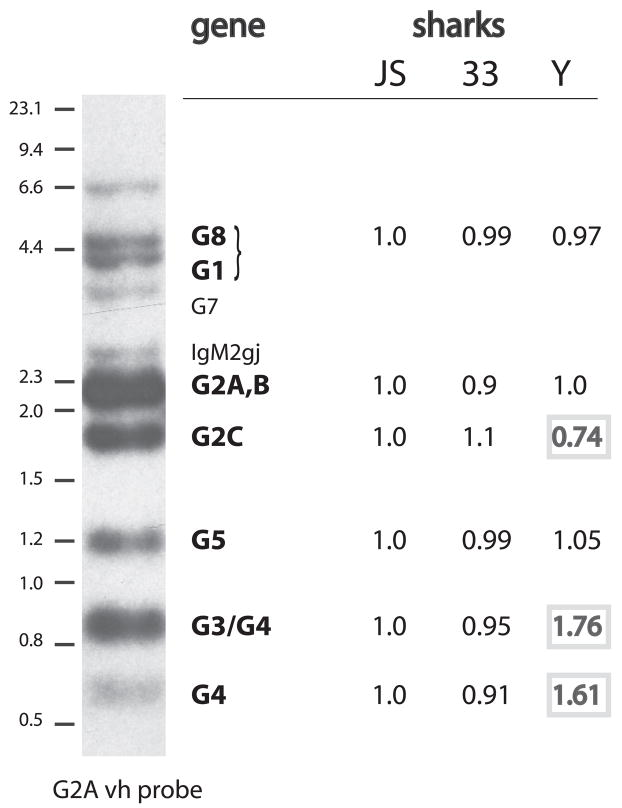
Differing numbers of H chain genes among sharks
Of the 10 vh-cross-hybridizing bands observed in HincII-digested genomic DNA, nine can be identified (Fig. 5, left) by correlation with individual phage or BAC clones. Genomic DNA from shark-JS, -33, and -Y were digested with HincII and with BamH I/NcoI, Southern blotting was performed, and the vh-hybridizing bands were compared among the individuals. Standardization of the three samples was done with a probe to nurse shark RAG-1, a single-copy gene, and all the values obtained through phosphorimaging were calculated, setting the shark-JS value as 1 (see Fig. 5, legend). The signals obtained for G1/G8, G2A/G2B, and G5 appear to be similar for all three individuals. However, there seems to be less G2C signal (the G2 pseudogene underrepresented in BAC library) and more G4 signal in shark-Y, probably reflecting the additional G4 genes absent in shark-33. The varying number of G4 IgH, together with the extensive sharing of C domain sequence, suggests that unequal crossing over occurs among G4 members, leading to exon shuffling and probably varying gene numbers in individual sharks.
FIGURE 5.
Comparison of VH gene content in three individuals by genomic Southern blotting. Erythrocyte DNA from shark-JS, -33, and -Y were digested with HincII and electrophoresed on a 1.2% agarose gel. The DNA was transferred to nylon filters and hybridized to a RAG1 probe to check for the comparative amounts of DNA present. The blot was scanned and analyzed by using a PhosphorImager; and the RAG1 signal at ca. 3.5 kb in each of the three lanes was quantitated in PhosphoImager units. The unit values of all three were divided by that of shark-JS, so that the shark-JS RAG-1 score is 1, while shark-33 was 1.17 and shark-Y, 1.1. These scores (1, 1.17, 1.1) were taken as reflecting the relative amounts of DNA transferred to the blot and used to adjust for comparative scoring of vh-hybridizing bands. The same blot was hybridized with the vh probe (Materials and Methods) and scanned. Six areas were quantitated in each lane (G8+G1, G2A/B, G2C, G5, G3+G4, and G4). A conventional x-ray autoradiographic example (shark-JS) of the spectrum of genomic bands is shown at the left, with size markers (λ HindIII and 100 bp marker, NEB) and all the bands identified except the one at 6.6 kb. Genes not observed include IgM1gj, at ~1 kb, which is not detected under the hybridization conditions. The value readings obtained from PhoshorImager analysis of the vh-hybridized blot were processed the following way. For instance, Shark-33 G5 score (0.99 at 1.2 kb) was derived from: [shark-33 G5 value ÷ 1.17] ÷ Shark-JS G5 value. Shark-Y G5 score (1.05 at 1.2 kb) was derived from Shark-Y G5 value ÷ 1.1] ÷ Shark-JS G5 value. Resulting scores differing by >20% are bold faced.
Germline-rearranged IgH
There are two genes encoding germline-rearranged, in-frame VDDJ in nurse shark that are derived from μ genes but, having diverged considerably in sequence and structure, they probably do not have IgM function. These are described as IgM-derived genes at the bottom of Table I. IgM1gj, previously characterized by Rumfelt and coworkers (21), lacks CH2 and exists as a monomer in the sera of neonates. No transcript carrying the transmembrane form has been isolated. The new one, IgM2gj, is a pseudogene. These two were generated by independent events in the germline because they differ in CDR3, and IgM2gj appears to be more closely related to G1–5 VH than to IgM1gj. IgM1gj and IgM2gj show, respectively, ~70 and 82% identity to other μ VH sequences at the nucleotide level and ~55 and 63% identity at the amino acid level from leader through FR4. One of four independent IgM2gj-type phage clones showed weak hybridization to Cμ1 probe and none with Cω sequence. Although the Cμ3-Cμ4 probe detected IgM1gj genes in the BAC library, it did not detect IgM2gj-bearing clones, suggesting that either it carried novel C sequence, or more likely, existed as an incomplete gene.
Shark-33 IgM rearrangements
The epigonal organ cDNA library of shark-33 was described in a previous publication (11). The phages were screened with VH and CH probes, and Ig sequence from selected lysates were subjected to amplification using T3 primer and a universal oligo targeting CH2. The PCR products of ~1.2 kb were sequenced and classified. Of 136 sequences, there were 20 G1, 43 G2, 2 G3, 58 G4, 12 G5, and 1 non-spliced, non-rearranged G8. Most of the 135 rearranged sequences carried the typical single and tandem substitutions found in other nurse shark Ig and Ig-like serum proteins (22, 23), demonstrating that all nine functional genes in shark-33 (G1, G2A, G2B, G3, G4A, G4C, G4E, G4G, G5) could be targets for somatic hypermutation.
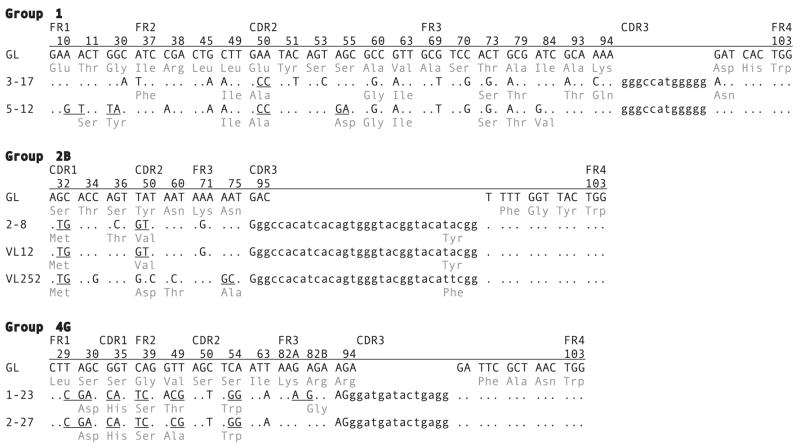
Nonfunctional sequences isolated during this screening were either not rearranged at all or partially rearranged (data not shown). In brief, 135 cDNA were in-frame and most were mutated. The substitution patterns showed evidence for mutant selection. The 15 G2A cDNA we previously analyzed showed overall a lower R:S ratio in the FR compared with the CDR. We found some sequences that share CDR3, the indication that they are from progeny of a single B cell clone. The sequences in these sets were mutated, and while many mutations were shared, others reflect changes that occurred after cell division (Fig. 6). The divergence ranged from 1 (2– 8/VL12) to 14 differences (3–17/5–12), introduced over varying generations of clonal expansion. Overall, non-conservative changes are primarily found in the CDR, suggesting that selection acted upon mutating receptors.
FIGURE 6.
Comparisons of intraclonally derived cDNA sequences. cDNA sequences were selected from shark-33 epigonal organ library after hybridization with probes to VH and CH. Group 1 cDNA sequences 3–17 and 5–12, which have the same CDR3, are compared with the reference G1 VH germline gene and JH flank. The codon, the codon positions, and the names of regions (FR1, CDR1, FR2, CDR2, FR3, CDR3, and FR4) are shown and compared with the substituted positions in the mutants. The portion of CDR3 consisting of N/P region and D segment sequences is indicated in lower case. Dots indicate identity with the germline, point mutations are shown, and tandem (contiguous) changes are indicated by underlining. For example, codons 10 and 11, GAA ACT, are changed to GAG TCT in cDNA 5–12. This change caused a replacement substitution in codon 11, which is shown in the three-letter amino acid code. In addition to CDR3, 3–17 shares eight point mutations and one tandem doublet with 5–12. Group 2B cDNA sequences 2–8, VL12, and VL252 share CDR3 and one doublet mutation. Whereas 2–8 differs from VL12 by one point mutation, both of them had diverged much earlier from VL252. Group 4G cDNA sequences 1–23 and 2–17 share CDR3, two point mutations and five tandem mutation sets.
The various JH of the Groups are distinguishable (Fig. 2), and it was determined that all G1, G2A, G2B, G3, and G5-bearing VDJ recombined within a minilocus. Although G2A and G5 are adjacent, there is no case of rearrangement of G2A gene segments to those downstream at the G5 locus; the G2A VH contains a distinctive insertion in FR1 (11) and the last codon in G5 JH is a unique ACA (Fig. 2). Among G4 JH, it is only possible to differentiate G4A/E from G4C/G, but in all 58 cDNA clones, both the VH and the JH belonged to G4 genes.
This is the first time that the absence of intercluster Ig rearrangement has been formally demonstrated in sharks and skates. If such events did occur in B cells, it would be at <0.7% frequency.
Evolution of the isotypes
Alignment was straightforward for all but the Cμ2, which were very heterogeneous in the 5′ end and where we had to remove the first 8–14 positions because of the in/dels (http://www.downstate.edu/pharmacology/faculty/hsu_pdf/ji2008.pdf, Fig. 1). GC contents were very similar among segments across loci. The VH and Cμ4 segments were best fit by the GTR+G, and all other segments were best fit by the simpler K2P+G model.
The bootstrap consensus topologies (Fig. 7) are alternative resolutions of the same underlying topology for 4/5 domains: only the data for the Cμ1 segment suggests that it underwent a separate history. However, bootstrap proportions are generally low for most clades, signifying a lack of signal for hierarchical structure. The SH tests confirm this; most pairs of domain data sets do not reciprocally reject candidate bootstrap trees in favor of the best-supporting bootstrap tree. The Cμ2 data do reject the Cμ4 topology (p = 0.042), but the Cμ4 data do not reject Cμ2 topology (p = 0.53); the reciprocal tests for Cμ1 and Cμ4 data sets do come close (p = 0.033 for the fit of the Cμ4 topology to the Cμ1 data set and p = 0.078 for the fit of Cμ1 topology to the Cμ4 data set), but this does not approach significance under corrections for 16 multiple tests. In conclusion, the data sets offer no compelling evidence that segments within loci have evolved with independent histories of duplication/gene conversion.
FIGURE 7.
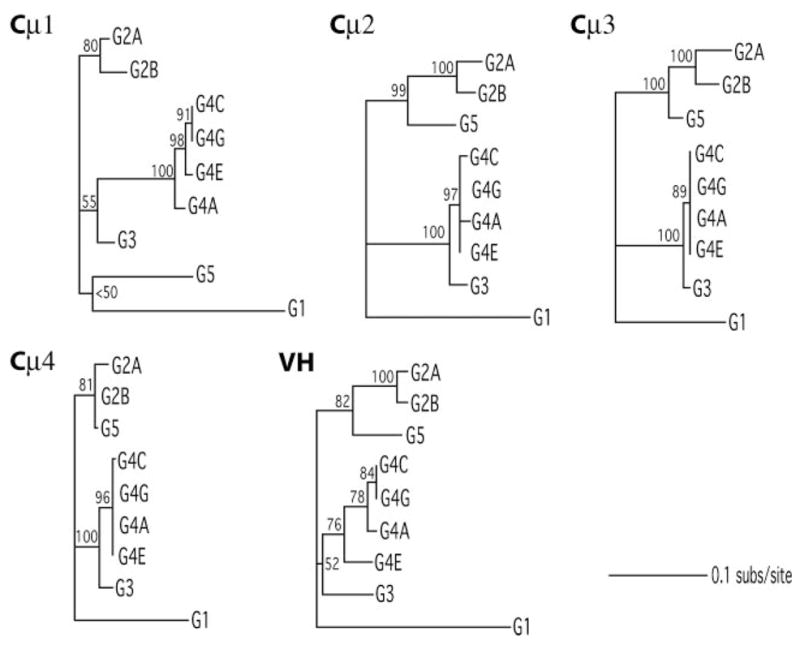
Phylogenetic trees for nine active loci for each of five Igμ domains. All unrooted trees are presented to scale, with bootstrap proportions above branch or at the node defining clade.
We obtained the estimated total length of each tree to compare the rates of evolution among the VH and CH sets. The results in Table III show that VH is not the fastest evolving set, and that the rate of change is comparable among the domains. A comparison of synonymous vs nonsynonymous substitutions was done for each data set. Even though the test we performed has limited power (18), the domains exhibit signatures of differing selection regimes (Table III). Although ds differed among segments by ~70%, dn differed >three-fold: Cμ 4 is under strong stabilizing selection (dn/ds = 0.171) whereas both VH (dn/ds = 0.974) and Cμ 2 (dn/ds = 0.991) show evidence for quite strong selection among loci. Given these relatively high dn/ds, we ran branch-specific dn/ds models for these two gene segments (model = 1 in PAML’s codem1). Maximum likelihood estimates of dn/ds were >1.0 for 10/15 branches in the VH tree, and for 9/15 branches in the Cμ2 tree. This is the signature of strong diversifying selection among the nine segments for each of these domains.
Table III.
Substitution patterns in nurse shark VH and CH gene segments across nine loci
| Domain | Length (bp) | Treelengtha | dnb | Dsb | dn/ds |
|---|---|---|---|---|---|
| VH1 | 303 | 0.50 | 0.0303 | 0.0311 | 0.974 |
| CH1 | 300 | 0.52 | 0.0240 | 0.0472 | 0.508 |
| CH2 | 300 | 0.47 | 0.0330 | 0.0333 | 0.991 |
| CH3 | 306 | 0.35 | 0.0201 | 0.0389 | 0.517 |
| CH4 | 390 | 0.33 | 0.0070 | 0.0410 | 0.171 |
Expected substitutions/site.
Averaged across all branches of the tree.
Discussion
This is the first report on the entire complement of IgM H chain genes generating the Ab repertoire of an elasmobranch fish. The genomic DNA from two genetically unrelated individuals, shark-33 and shark-Y, were respectively cloned into bacteriophage and BAC vectors. Except for certain G4 members, all H chain genes we deem functional were present in both animals. These five Groups were originally defined in neonatal pups (19); given their presence in unrelated animals these Groups represent isotypes, or IgM subclasses.
Transcription of pseudogenes has been reported in other elasmobranch systems (24), and we have found that at least one of them (G2C) in nurse shark can undergo somatic rearrangement as well. Thus, it is only possible to distinguish the active genes by obtaining a defect-free complete sequence whose rearranged, mutated products show evidence of selection on the cell producing the protein. This is how we have defined nine functional genes in the shark-33 genomic and cDNA libraries. The IgH miniloci are located at least 120 kb apart and >99% of V(D)J recombination in B cells takes place among gene segments within the locus; all nine shark-33 loci can be targeted for somatic hypermutation.
Having established the nature of IgH organization and expression in nurse shark, we discuss this information in the context of our current understanding of Ig gene diversification and regulation in a representative of the earliest jawed vertebrate. The most important aspect of generating a wide repertoire of Ab specificities appears to be junctional diversification. The VH may have evolved to accommodate varying and longer CDR3. It was pointed out in the sandbar shark, whose VH are very similar to nurse shark, that their CDR2 loops are predicted to be more regular and compact to allow for space at the domain interface (25). Certainly, there is no variation in VL-VH CDR1/CDR2 sizes among the nurse shark VH, unlike in mammals.
Rearrangement requirement for the IgM repertoire
In contrast to other elasmobranch species, every gene in nurse shark must undergo recombination to be expressed. This means that the primary IgM repertoire of the nurse shark is generated by intracluster rearrangement in nine μ genes. As discussed elsewhere (11, 26), the extensive trimming and N region addition that occurs during three rearrangement events creates highly diversified CDR3. Thus, while the combinatorial diversification that exists in tetrapods is absent in sharks, rearrangement nonetheless plays the major role in generating the B cell repertoire in all vertebrates.
We had postulated that RAG was selected as V(D)J recombinase in evolution because of its unprecedented role in generating diverse sequence, not only of varying amino acid sequence but also, uniquely, loop length differences (23, 27). Other diversification mechanisms as somatic mutation or gene conversion would be not be able to create this type of sequence heterogeneity, and not reliably in the same position (between main chain structures) that could tolerate such a wide spectrum of changes. In the nurse shark it is clear that the generation of junctional diversification in itself is sufficient a role for RAG. We suggest that the significant contribution by junctional diversification reflects the primary novelty introduced by RAG in the early vertebrate ancestor, whereas combinatorial diversity coevolved with the complex tetrapod and bony fish translocon organization.
However, cartilaginous fishes are well known for VH sequences that are already recombined, some in-frame and potentially functional, in the germline (7, 8). The role of these genes in the Ab repertoire is still unknown, and none has yet been identified as encoding a particular specificity. The nurse shark does not carry germline-rearranged IgM genes per se. Because IgM1gj probably exists only as a secreted protein (28) it would neither compete with, nor pre-empt, rearrangement and expression of any IgM H chain gene in developing B cells.
There are some germline-joined V regions among the nurse shark L chain genes, which are otherwise organized as VL-JL-CL miniloci. Of the >70 IgL, there are pre-rearranged VJ present in three out of the four L chain isotypes (28, 29), and some have been found in both pup and adults (NS3, NS5–16). We propose that, given a large gene pool, such as the 70 IgL in nurse shark or the 100–200 IgH in horn shark and skate, there is a greater allowance for germline changes. An occurrence of Ig joining in the germ cells, like a VL to JL, may be selected like any other mutant. For example, the germline-joined CDR3 of L chain NS5–16 is only six amino acids long, although other members of NS5 carry CDR3 ranging 9–14 amino acids (27). We proposed that this L chain may have a role in creating a subpopulation of Ag-combining sites, like the anti-phosphorylcholine specificity in mouse Ab that is generated only in the absence of terminal transferase (30).
We conclude from their absence in the nurse shark that germ-line-joined μ-chain specificities are not absolutely necessary for elasmobranch Ab repertoire, but that there may be species-specific adaptations that were selected for.
H chain exclusion
Given the large number of Ig loci in elasmobranchs, it was not clear for many years whether there was in fact clonal expression of the Ag receptor on B cells. Recent work in clearnose skate by Eason and coworkers (31) found that few μ cDNA sequences containing CDR3 could be amplified from single lymphocytes. They concluded that only one gene out of the 100–200 IgH was selected at a time for rearrangement and expression, likening the process to that of the allelically excluded odorant receptors (32). They reasoned that if more IgH were activated, the many germline-joined VDJ genes present in the clearnose skate genome would have been transcribed in every cell. However, this was a preliminary study in which most germline IgH in clearnose skate had not been characterized.
For this question the nurse shark is a more straightforward experimental system because every IgH must rearrange to be expressed. We found in single, surface Ig-positive B cells, 1–3 rearranged VDJ whereas the other IgH were in germline configuration (48). In almost all cells only one VDJ is functionally rearranged. Together with the organizational and expression information presented in this study, we conclude that the shark H chain rearrangement process does not involve long-range effects or differential chromatin activation, as it does in mammals and that shark H chain exclusion results from restricted activation of IgH genes.
In mammalian precursor B cells, the formation of an IgH holo-complex, consisting of nuclear proteins that bind and cooperatively engage the promoter and enhancer, recruits the chromatin remodeling and DNA modifications that render the gene segments open to RAG action (33). The fact that there are few VDJ per shark lymphocyte and an absence of intercluster rearrangement suggests that the B cell-specific chromatin remodeling is localized to discrete nurse shark IgH. Moreover, we did not observe any relationship between recombination events at the G2A and G5 genes, found to be adjacent in this study (W. Feng and E. Hsu, unpublished observations).
We proposed that the full accessibility of shark H chain genes is dependent on the formation of the IgH holocomplex, which occurs at low frequency due to limiting amounts of B cell lineage nuclear factors, an idea along the same lines as Liang et al., (34) who suggested that κ-chain exclusion is due to probabilistic enhancer activation. Once a viable IgM receptor is expressed, ongoing rearrangement will cease in that cell. Whether there exist 15 or 200 IgH per genome, the result is that likely only one H chain protein will be expressed per shark B cell.
Multiple cluster evolution
Most of the H chain sequence information available for cartilaginous fishes has been for VH of IgM or IgW. Using VH from horn shark, nurse shark, sandbar shark, little skate, and ratfish, Ota and coworkers (35) generated a phylogenetic tree showing species clustering of VH sequence, which suggests that, within each species, the gene segments evolved by continuous and gradual turnover.
The phylogenetic trees generated for the VH and CH domains of the functional nurse shark genes showed that the clusters evolved independently, without exchange of domains between different Groups. Overall, G3 and G4 probably derived from a common ancestor, as did G2 and G5. The adjacent positioning of G2A and G5 is suggestive of a tandem duplication of the entire locus at a distant point back in time, and there are differences in all five domains. Although the G4 genes must be located no closer to each other than are G2/G5, exchange of information apparently has taken place between the four defined G4A, G4E, G4C, and G4G by gene conversion or unequal crossing over. The uneven sharing of identity across domains argues against their being the result of recent whole-locus duplication events.
To compare the rates of evolution among the VH and CH sets, the estimated substitution rates per nucleotide was obtained per domain. The results in Table III show that none of the domains is evolving significantly faster than the others. There exists much literature on the evolutionary diversification of VH gene segments, and it might have been expected that the VH diverged faster than C, in accordance with the Ag-binding role played by this portion of the Ig protein. Given our actual result, we went on to look for differences in the nature of the substitutions in each domain.
As anticipated, the VH sequences showed a greater rate of replacement changes (dn/ds of 0.974) than Cμ4, which had a stabilizing dn/ds of 0.171. We expected that VH would be selected for diversification and Cμ4 for conservation, because this is what can be observed from a comparison of μ sequences from different species (36) and would conform to their perceived functions. However, unexpectedly, dn/ds in Cμ2 equaled that of the VH and this is inspite of the removal of the most diverse positions in the 5′ end. Branch-specific dn/ds models were run for these two domains, and as dn/ds >1 for the majority of branches, it appears that Cμ2 as well as VH are under positive selection for increased sequence diversity.
Ig domain evolution
We aligned the five domains of the nine H chains and marked homologous positions where there existed three or more amino acid substitutions (http://www.downstate.edu/pharmacology/faculty/hsu_pdf/ji2008.pdf, Fig. 1b). For VH, 10/13 positions were located in or adjacent to CDR1, CDR2, and potential CDR3, showing good correlation between selection for heterogeneity and the Ag-combining site. In Cμ1 (14 positions) and Cμ2 (26 positions) the sequence variability is largely part of loops. Overall, the Cμ2 substitutions tend to be on the solvent-exposed y face (37), and in the loops connecting the y segments. Whether this could be the result of positive selection or the contrary, relaxation of purifying selection, is difficult to say (38) because, unlike the VH, less is known of C region activities and Cμ2 in particular. There is little Cμ2 structural information other than that its N-terminal region allows flexibility for the Fab, enabling the Ag-bound arms to rotate out of the planar pentamer structure (39). The sequence of shark Cμ2 in this region is the most diverse of the C region, containing in/dels and, together with the loops pointing to the Cμ2 domain, forms a cleft with diverse sequence among the different H chains. We suggest these may be sites with differing affinities to receptors or effector molecules, possibly in the monomeric form, where they may be better exposed.
Unlike in mammals, shark IgM is secreted as pentamer and monomer, a switch that takes place during an Ab response (40, 41). This event suggests that the shark monomer must be adapted for at least a similarly effective level of biological activities as the polymeric form, unlike in mammals (42). There is little information on shark IgM effector functions. All the IgM genes must generate receptor and secretory forms, and such structural requirements and those directing polymer assembly are located in the Cμ3/Cμ4 domains (43, 44, and references therein) which are thus very well conserved. As such, only the shark Cμ1 and Cμ2 domains are available to coevolve with (different) Fc receptors. Ab-dependent opsonization by neutrophils has been demonstrated in nurse shark (45), and we suggest, for instance, that it is possible that only some IgM subclasses bind the neutrophil Fc receptor. Fc receptor recognition can involve CH2; for instance, the binding of FcεRII with human IgE requires CH2 as well as CH3 (46).
The four IgG subclasses share 66–79% amino acid identity in CH and 55–64% in CH3; they mediate distinct biological activities via preferential binding to activating or inhibitory Fc receptors (47). The nurse shark groups share 66–91% identity in Cμ2; the 66% identity between G1 and G2 or the 70% identity between G2 and G3/G4 might manifest itself in different effector functions. Interestingly, the CH2 exon is deleted from the nurse shark IgM1gj gene, and we speculate that this perhaps obviates competition with IgM effector function because high levels of IgM1gj are present in neonatal serum (21). IgM1gj is but one example of the plasticity of the IgH system of cartilaginous fishes. It has evolved with changes in both VH and CH and probably no longer serves an Ig function, either for binding conventional Ags or for Ag disposal. In Hydrolagus, there is a family of IgH genes where the Cμ1 has been replaced with a Cμ2-like domain (8), which may affect L chain interaction or polymerization. The existence of such genes further demonstrates selection for variant C regions among IgM clusters and the capacity of duplicated Ig genes to diverge and evolve new roles.
Acknowledgments
We thank David Redding and Stevan Springer for technical help with the phylogenetics. We thank Martin Flajnik for critical reading of this manuscript and Jonathan Rast for comments on Hydrolagus.
Footnotes
This work was supported in part by grants from the National Institutes of Health (GM068095) and Natural Sciences and Engineering Research Council Canada. A.M. and E.H. were fellows of the Wissenschaftskolleg zu Berlin, 2006–2007.
Abbreviation used in this paper: BAC, bacterial artificial chromosome.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Flajnik MF, Du Pasquier L. Evolution of innate and adaptive immunity: can we draw a line? Trends Immunol. 2004;25:640–644. doi: 10.1016/j.it.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Rast JP, Litman GW. Towards understanding the evolutionary origins and early diversification of rearranging antigen receptors. Immunol Rev. 1998;166:79–86. doi: 10.1111/j.1600-065x.1998.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 3.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 4.Stavnezer J, Amemiya CT. Evolution of isotype switching. Semin Immunol. 2004;16:257–275. doi: 10.1016/j.smim.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Hinds KR, Litman GW. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 6.Litman GW, Rast JP, Shamblott MJ, Haire RN, Hulst M, Roess W, Litman RT, Hinds-Frey KR, Zilch A, Amemiya CT. Phylogenetic diversification of immunoglobulin genes and the antibody repertoire. Mol Biol Evol. 1993;10:60–72. doi: 10.1093/oxfordjournals.molbev.a040000. [DOI] [PubMed] [Google Scholar]
- 7.Kokubu F, Litman R, Shamblott MJ, Hinds K, Litman GW. Diverse organization of immunoglobulin VH gene loci in a primitive vertebrate. EMBO J. 1988;7:3413–3422. doi: 10.1002/j.1460-2075.1988.tb03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rast JP, Amemiya CT, Litman RT, Strong SJ, Litman GW. Distinct patterns of IgH structure and organization in a divergent lineage of chrondrichthyan fishes. Immunogenetics. 1998;47:234–245. doi: 10.1007/s002510050353. [DOI] [PubMed] [Google Scholar]
- 9.Kokubu F, Hinds K, Litman R, Shamblott MJ, Litman GW. Complete structure and organization of immunoglobulin heavy chain constant region genes in a phylogenetically primitive vertebrate. EMBO J. 1988;7:1979–1988. doi: 10.1002/j.1460-2075.1988.tb03036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsu E, Criscitiello MF. Diverse immunoglobulin light chain organization in fish retain potential to revise B cell receptor specificities. J Immunol. 2006;177:2452–2462. doi: 10.4049/jimmunol.177.4.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF, Hsu E. Somatic hypermutation and junctional diversification at Ig heavy chain loci in the nurse shark. J Immunol. 2005;175:8105–8115. doi: 10.4049/jimmunol.175.12.8105. [DOI] [PubMed] [Google Scholar]
- 12.Kokubu F, Hinds K, Litman R, Shamblott MJ, Litman GW. Extensive families of constant region genes in a phylogenetically primitive vertebrate indicate an additional level of immunoglobulin complexity. Proc Natl Acad Sci USA. 1987;84:5868–5872. doi: 10.1073/pnas.84.16.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo M, Kim H, Kudrna D, Sisneros NB, Lee S-J, Mueller C, Collura K, Zuccolo A, Buckingham EB, Grim SM, et al. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics. 2006;7:106. doi: 10.1186/1471-2164-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nylander JAA. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University: 2004. [Google Scholar]
- 15.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, Massachusetts: 2002. Version 4. [Google Scholar]
- 16.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetics inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 17.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 18.Anisimova M, Bielawski JP, Yang Z. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol. 2001;18:1585–1592. doi: 10.1093/oxfordjournals.molbev.a003945. [DOI] [PubMed] [Google Scholar]
- 19.Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004;5:8. doi: 10.1186/1471-2172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brodeur PH, Riblet R. The immunoglobulin heavy chain variable region (Igh-V) locus in the mouse: I. One hundred Igh-V genes comprise seven families of homologous genes. Eur J Immunol. 1984;14:922–930. doi: 10.1002/eji.1830141012. [DOI] [PubMed] [Google Scholar]
- 21.Rumfelt LL, Avila D, Diaz M, Bartl S, McKinney EC, Flajnik MF. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc Natl Acad Sci USA. 2001;98:1775–1780. doi: 10.1073/pnas.98.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825–833. doi: 10.1093/intimm/11.5.825. [DOI] [PubMed] [Google Scholar]
- 23.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity. 2002;16:571–582. doi: 10.1016/s1074-7613(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 24.Miracle AL, Anderson MK, Litman RT, Walsh CJ, Luer CA, Rothenberg EV, Litman GW. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int Immunol. 2001;13:567–580. doi: 10.1093/intimm/13.4.567. [DOI] [PubMed] [Google Scholar]
- 25.Ramsland PA, Kaushik A, Marchalonis JJ, Edmundson AB. Incorporation of long CDR3s into V domains: implications for the structural evolution of the antibody-combining site. Exp Clin Immunogenet. 2001;18:176–198. doi: 10.1159/000049197. [DOI] [PubMed] [Google Scholar]
- 26.Fleurant M, Changchien L, Chen CT, Flajnik MF, Hsu E. Shark Ig light chain junctions are as diverse as in heavy chains. J Immunol. 2004;173:5574–5582. doi: 10.4049/jimmunol.173.9.5574. [DOI] [PubMed] [Google Scholar]
- 27.Lewis SM, Wu GE, Hsu E. The origin of V(D)J diversification. In: Honjo T, Alt FW, Neuberger MS, editors. Molecular Biology of B Cells. Elsevier Academic Press; Amsterdam: 2004. pp. 473–489. [Google Scholar]
- 28.Hsu E, Pulham N, Rumfelt L, Flajnik MF. The plasticity of immunoglobulin gene systems in evolution. Immunol Rev. 2006;210:8–26. doi: 10.1111/j.0105-2896.2006.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Criscitiello MF, Flajnik MF. Four primordial immunoglobulin light chain isotypes, including λ and κ, identified in the most primitive living jawed vertebrates. Eur J Immunol. 2007;37:2683–2694. doi: 10.1002/eji.200737263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benedict CL, Kearney JF. Increased junctional diversity in fetal B cells results ina loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity. 1999;10:607–617. doi: 10.1016/s1074-7613(00)80060-6. [DOI] [PubMed] [Google Scholar]
- 31.Eason DD, Litman RT, Luer CA, Kerr W, Litman GW. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. doi: 10.1002/eji.200425224. [DOI] [PubMed] [Google Scholar]
- 32.Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 33.Oltz EM, Osipovich O. Targeting V(D)J recombinase: putting a PHDS to work. Immunity. 2007;27:539–541. doi: 10.1016/j.immuni.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Liang HE, Hsu LY, Cado D, Schlissel MS. Variegated transcriptional activation of the immunoglobulin κ locus in pre-B contributes to the allelic exclusion of light-chain expression. Cell. 2004;118:19–29. doi: 10.1016/j.cell.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 35.Ota T, Sitnikova T, Nei M. Evolution of vertebrate immunoglobulin variable gene segments. Curr Top Microbiol Immunol. 2000;248:221–245. doi: 10.1007/978-3-642-59674-2_10. [DOI] [PubMed] [Google Scholar]
- 36.Hsu E. The variation in immunoglobulin constant regions in evolution. Semin Immunol. 1994;6:383–391. doi: 10.1006/smim.1994.1048. [DOI] [PubMed] [Google Scholar]
- 37.Beale D, Feinstein A. Structure and function of the constant regions of immunoglobulins. Q Rev Biophys. 1976;9:135–180. doi: 10.1017/s0033583500002390. [DOI] [PubMed] [Google Scholar]
- 38.Hughes AL. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- 39.Perkins SJ, Nealis AS, Sutton BJ, Feinstein A. Solution structure of human and mouse immunoglobulin M by synchrotron X-ray scattering and molecular graphics modeling. J Mol Biol. 1991;221:1345–1366. doi: 10.1016/0022-2836(91)90937-2. [DOI] [PubMed] [Google Scholar]
- 40.Voss EW, Sigel MM. Distribution of 19S and 7S IgM antibodies during the immune response in the nurse shark. J Immunol. 1971;106:1323–1329. [PubMed] [Google Scholar]
- 41.Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol. 2005;35:936–945. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 42.Chen FH, Arya AK, Rinfret A, Isenman DE, Shulman MJ, Painter RH. Domain-switched mouse IgM/IgG2g hybrids indicate individual roles for Cμ2, Cμ3, and Cμ4 domains in the regulation of the interaction of IgM with complement C1q. J Immunol. 1997;159:3354–3363. [PubMed] [Google Scholar]
- 43.Weirsma EJ, Shulman MJ. Assembly of IgM: role of disulfide bonding and noncovalent interactions. J Immunol. 154:5265–5272. [PubMed] [Google Scholar]
- 44.Sorensen V, Rasmussen IB, Sundvold V, Michaelsen TE, Sandlie I. Structural requirements for incorporation of J chain into human IgM and IgA. Int Immunol. 2000;12:19–27. doi: 10.1093/intimm/12.1.19. [DOI] [PubMed] [Google Scholar]
- 45.McKinney EC, Flajnik MF. IgM-mediated opsonization and cytotoxicity in the shark. J Leukocyte Biol. 1997;61:141–146. doi: 10.1002/jlb.61.2.141. [DOI] [PubMed] [Google Scholar]
- 46.Vercelli D, Helm B, Marsh P, Padlan E, Geha R, Gould H. The B-cell binding site on human immunoglobulin E. Nature. 1989;338:649–651. doi: 10.1038/338649a0. [DOI] [PubMed] [Google Scholar]
- 47.Nimmerjahm F, Ravetch JV. Divergent immunoglobulin-G subclass activity through selective Fc receptor binding. Science. 2005;310:1510–1512. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- 48.Malecek K, Lee V, Feng W, Huang J-L, Flajnik MF, Ohta Y, Hsu E. Immunoglobulin heavy chain exclusion in the shark. PLoS Biology. 2008 doi: 10.1371/journal.pbio.0060157. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]