Abstract
Background
The recent determination of the complete nucleotide sequence of several Mycobacterium tuberculosis (MTB) genomes allows the use of comparative genomics as a tool for dissecting the nature and consequence of genetic variability within this species. The multiple alignment of the genomes of clinical strains (CDC1551, F11, Haarlem and C), along with the genomes of laboratory strains (H37Rv and H37Ra), provides new insights on the mechanisms of adaptation of this bacterium to the human host.
Findings
The genetic variation found in six M. tuberculosis strains does not involve significant genomic rearrangements. Most of the variation results from deletion and transposition events preferentially associated with insertion sequences and genes of the PE/PPE family but not with genes implicated in virulence. Using a Perl-based software islandsanalyser, which creates a representation of the genetic variation in the genome, we identified differences in the patterns of distribution and frequency of the polymorphisms across the genome. The identification of genes displaying strain-specific polymorphisms and the extrapolation of the number of strain-specific polymorphisms to an unlimited number of genomes indicates that the different strains contain a limited number of unique polymorphisms.
Conclusion
The comparison of multiple genomes demonstrates that the M. tuberculosis genome is currently undergoing an active process of gene decay, analogous to the adaptation process of obligate bacterial symbionts. This observation opens new perspectives into the evolution and the understanding of the pathogenesis of this bacterium.
Background
The genome of Mycobacterium tuberculosis (MTB), considered to be mosaic in structure, has been proposed to be the result of ancient horizontal DNA exchange and subsequent clonal expansion [1]. The MTB genome is highly conserved and is considered a "closed pan-genome" due to the bacterium's restricted niche that limits access to the global microbial gene pool [2]. Despite the limited genetic variation, there is a high degree of phenotypic variability among MTB isolates, including differences in clinical outcome and epidemiological behavior that involve host and environmental factors, as well as bacterial determinants [3,4].
The genome sequences of six MTB strains are available and include the laboratory reference H37Rv [5] and avirulent H37Ra strains and clinical isolates. Strain CDC1551 caused an outbreak with extensive transmission of tuberculosis that is related to increased virulence [6]. The F11 strain represents the largest proportion of all isolates from tuberculosis patients in the Western Cape of South Africa [7]. The C strain is a drug-susceptible strain resistant to reactive nitrogen intermediates that spread among intravenous drug users [8]. The Haarlem strain belongs to a widely distributed genotype with reduced virulence in the murine model [9]. Genome-wide analyses of genetic variation among different MTB strains can help to unveil characteristics regarding the virulence and adaptation of this successful human pathogen.
In this study we analyzed the variation dynamics of the MTB genome by carrying out a multiple genome alignment of the six sequenced strains. The variations identified were then examined using a combinatorial approach that allows simultaneous comparison of all strains, rather than pair-wise comparisons of each to a reference genome, thus providing information about unexplored features of the MTB genome. This study gives insight into the evolution of the MTB genome and the mechanisms of adaptation of this pathogen to its human host.
Methods
Multiple genome alignments were carried out with the freeware MAUVE v1.3 [10] using the genomes of MTB strains H37Rv (NC_000962), H37Ra (NC_009525), CDC1551 (NC_002755), F11 (NC_009565), C (NZ_AAKR00000000) and Haarlem (NZ_AASN00000000), the latter two from the Broad Institute http://www.broad.mit.edu (See Additional File 1 for detailed Methods). A Perl-based package called islandsanalyser (Additional File 2) was designed to analyze the distribution and frequency of the LSPs identified in the alignment.
Results and discussion
Genome structure and genetic variation
The multiple genome alignment of the six MTB genomes showed extensive collinearity and the absence of widespread major rearrangement events. The limited number of rearrangements identified involved mobile genetic elements located at different sites along the genome. These variations were considered InDels, and not true rearrangements, when absent from some of the genomes in the dataset. This is exemplified by the phiRv1 phage, which was absent in the F11 and C strains and had a location that differed in strains H37Rv and H37Ra with respect to strains CDC1551 and Haarlem. Differential insertions of mobile genetic elements such as IS6110 were also detected and, again, considered as InDels when not present in the same location in all strains analyzed. This analysis indicated great stability in genomic structure and organization, characteristics that may be a consequence of the intracellular life-style of the tubercle bacilli, which limits the interaction with other microbial gene pools.
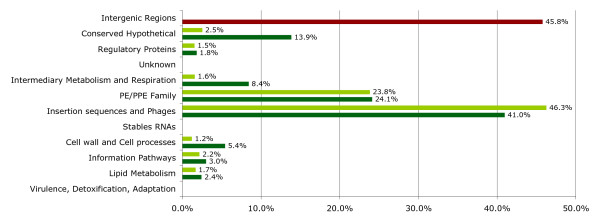
Genetic variation in MTB is considered to occur through single nucleotide polymorphisms (SNPs) [11] and irreversible genetic events, such as large sequence polymorphisms (LSPs), which are believed to be the main source of phenotypic variability [12]. To determine the extent of genetic variation caused by LSPs, the polymorphisms identified in the multiple alignment were classified into different functional categories according to the genes affected (Figure 1). LSPs from 50 bp (the smallest size reported) to 12 kb were found to involve 120 genes. We did not find LSPs in genes for stable RNAs or in genes previously implicated in virulence, adaptation or detoxification. Two functional categories represented 65% of the total genes containing LSPs: 1) mobile genetic elements such as insertion sequences and phages and 2) the PE/PPE family of genes that contain repetitive sequences. This is consistent with the notion that these elements are known to drive the generation of small genetic polymorphisms [13]. The remaining LSPs were found in categories where it is common to find genes with redundant functions and suggests that MTB allows gene loss while avoiding deleterious effects. Finally, LSPs were detected in three genes annotated as transcriptional regulators (Rv0823c, Rv0792c and Rv1353c) that, although not essential for MTB survival [14], may be important in controlling gene expression under certain conditions.
Figure 1.
Functional category classification of the LSPs found in the multiple alignments. Dark green bars represent the variation in each category respect to the total of LSPs. The percentages are given with respect to a total of 166 matches because there are genes that are classified in more than one functional category. Light green bars represent the percentage of LSPs found respect to the total number of genes in each category. The percentage of the intergenic region category (red bar) represents the LSPs that involved a non-coding region.
The extent of LSPs present in intergenic regions had never before been determined for various genomes. We found that 45.8% of the total LSPs identified were present (complete or partially) in intergenic regions, indicating that variation is not targeted solely to coding sequences. This multiple alignment also identified LSPs in more genes, 120 versus 95, than would have been identified using microarrays to compare these same MTB strains against the H37Rv reference genome. This difference is due to the fact that our analysis was combinatorial and not limited to the identification of deletions with respect to a single reference genome. Thus this approach identified differential IS6110 integration sites, LSPs as small as 50 bp and LSPs in intergenic regions and genes of the PE/PPE family, which had not been investigated before [12]. These results, in addition to the SNP and microsatellite polymorphisms previously reported [15], indicate that the genetic variation within MTB is greater than previously expected.
Distribution and frequency of polymorphisms
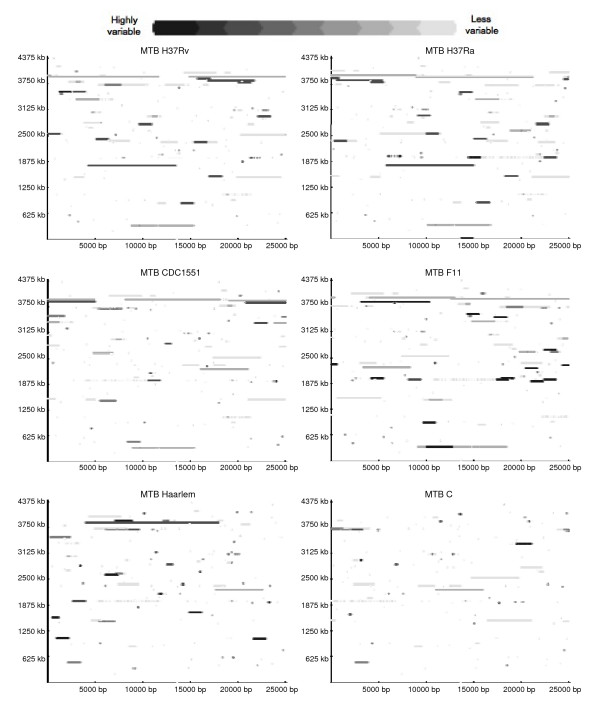
The observed skewed distribution of LSPs in the different functional categories could be the result of traits intrinsic to the MTB genome that constrain the accumulation of polymorphisms to specific regions of the chromosome. In this scenario, all the MTB strains should display similar patterns of LSP distribution. We therefore developed a tool (Additional file 2) to evaluate the presence and frequency of polymorphisms using a sliding window that identifies and represents variable regions as a continuous string, rather than as a step-wise count, for the presence of LSPs (as in a histogram). This avoids the over representation of small polymorphisms and the under representation of large polymorphisms that result from using discrete window presence/absence counts. The graphical output, which represents both LSP size and variability among strains, showed that the polymorphisms were spread across the entire genome and differed among strains, indicating that they were not constrained to the same genomic regions (Figure 2). As expected, highly variable regions corresponded to IS6110 insertion sites and the presence of the phiRv1 phage. This differential distribution and frequency of polymorphisms raised the possibility that there should be a limited number of polymorphisms unique to each strain.
Figure 2.
Distribution and frequency of LSPs in strains H37Rv, H37Ra, CDC1551, F11, Haarlem and C. The picture depicts LSP sites observed for each of the six strains (MTB H37Rv, H37Ra, CDC1551, F11, Haarlem and C) with respect to the other strains. The frequency of the LSP can be inferred from the color scale. Background color represents invariable positions. The base of the square is 25,000 bp and the sizes of the LSPs are proportionately scaled.
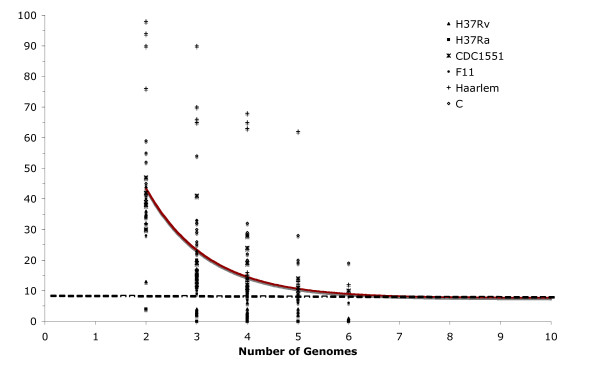
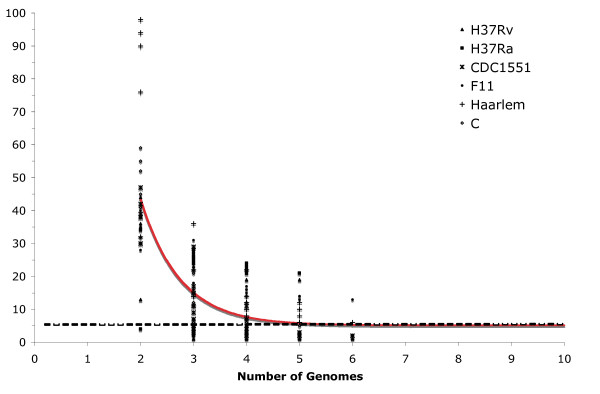
To test this hypothesis we determined the number of strain-specific polymorphisms, independently of strain order, by the sequential addition and comparison of new MTB genomes. Strain-specific polymorphisms were separated into the following categories: i) specific deletions (SD), defined as a region absent in a particular strain and present in the rest of the dataset when n-1 genomes are compared, where n is the number of genomes compared; and ii) specific insertions (SI), a region unique to a particular strain and absent in the rest of the dataset when n-1 genomes are compared. The results obtained for the number of strain-specific deletions and insertions for all combinations of comparisons in a collection length of n = 2, 3, ...6, of the six genomes in the dataset were fitted to an exponential decay function to determine the number of specific deletion and specific insertions when the nth genome is included (Figures 3 and 4).
Figure 3.
Extrapolation of strain-specific deletions. The number of specific deletions is plotted as a function of the number n of strains. For each n, markers are the 6!/[(n - 1)!·(6 - n)!] values obtained for the different strain combinations. The continuous curve represents the least-squares fit of the function Fs(n) = κs exp [-n/τs] + Ω, where κs = 186.7 ± 44.3, -1/τs = -0.83 ± 0.13 and Ω = 7.6 ± 1.7. The best fit obtained had a correlation of 0.997.
Figure 4.
Extrapolation of strain-specific insertions. The number of specific insertions is plotted as a function of the number n of strains. For each n, markers are the 6!/[(n - 1)!·(6 - n)!] values obtained for the different strain combinations. The continuous curve represents the least-squares fit of the function Fs(n) = κs exp [-n/τs] + Ω where κs = 578.35 ± 198.07, -1/τs = -1.36 ± 0.18 and Ω = 5.1 ± 1.1. The best fit was obtained with a correlation of 0.998.
According to our hypothesis, the number of both types of strain-specific polymorphisms should decrease as more genomes are included. Interestingly, the value of strain-specific polymorphisms did not tend to an asymptotic value of zero but tended towards 7.6 ± 1.7 strain-specific deletions and 5.1 ± 1.1 strain-specific insertions when the nth genome is included. In addition, the variation dynamics differed among strains. For example, the laboratory strains presented fewer strain-specific polymorphisms, 1 SD and 1 SI in H37Rv and 2 SI in H37Ra, than clinical strains isolated during outbreaks. There were 12 strain-specific polymorphisms in CDC1551 (10 SD and 2 SI), 18 in Haarlem (12 SD and 6 SI) and 20 each in F11 (6 SD and 14 SI) and C (19 SD and 1 SI). This indicates that interaction with the human host is important in driving the generation and selection of genomic changes. The tendency observed by extrapolation of the number of genomes to infinite indicated that as additional genomes are included in the analysis only a small number of new strain-specific polymorphisms would be observed. The tendency to accumulate InDels also indicates that the MTB genome is under an active process of gene decay reminiscent of the adaptation process of obligate symbiotic bacterial pathogens [16].
Genomic configuration of MTB strains
The genes involved in strain-specific deletions and insertions are summarized in Tables 1 and 2, respectively. Interestingly, we found that the avirulent laboratory strain H37Ra lacked specific deletions in comparison with the virulent strains, suggesting that loss of virulence in this strain is not a consequence of gene deletion. The larger genome of H37Ra, with respect to H37Rv, also suggests that virulent strains might enhance their fitness by means of gene loss. We also found that strain-specific deletions involving the PE/PPE gene family were identified only in clinical strains, consistent with their proposed function as variable surface antigens and mediators of interactions with host cells [17,18]. Other strain-specific deletions were also found in genes associated with the immune response. For instance, strain F11 lacked an entire gene encoding one of the culture filtrate proteins (CFP10A) known to mediate cellular immune responses [19]. The three LSPs related with transcriptional regulatory proteins correspond to strain-specific deletions of the Haarlem (Rv0823c and Rv1353c) and CDC1551 strains (Rv0792c). In terms of strain-specific insertions, unique insertion sites of the IS6110 element were predominant. Strain F11, for example, had 12 unique insertion sites for this element, suggesting that transposition events are important for variation in this particular genotype [20,21]. Both types of strain-specific polymorphisms were found in intergenic regions, with events such as insertion of transposons and duplication or loss of repeated regions between coding sequences. The identification and analysis of these intergenic regions is very valuable since they may be involved in regulation of adjacent genes or may contain as yet unknown small regulatory RNAs [22].
Table 1.
Genes Involved in strain-specific deletions.
| Position | Size | Genes Involved |
| H37Rv | ||
| 1987985 | 6830 | MT1800 (Glycosyl transferase) – MT1801 (Molybdopterin oxidoredutase) – mmpL14 (Transmembrane transport protein) |
| CDC1551 | ||
| 206981 | 58 | Rv0175 (Probable conserved MCE Associated membrane protein) |
| 888687 | 875 | Rv0792c (GNTR-Family transcriptional regulatory protein) – Rv0793 (Hypothetical protein) – Rv0794c (Putative oxidoreductase) |
| 2161299 | 139 | PPE34 |
| 2349725 | 58 | Intergenic Region between pepE (Xaa-Pro Dipeptidase) and Rv2090 (5'-3' Exonuclease) |
| 2373345 | 58 | C-terminal end of the Rv2112c (Hypothetical protein) |
| 2382265 | 2275 | PPE37 – metH (Methionine synthase) |
| 2701639 | 501 | Rv2406c (Conserved hypothetical protein) – Intergenic region – Rv2407 (Ribonuclease Z) |
| 3727960 | 5284 | PPE54 (Possible recombination event with PE-PGRS49-50 and PPE55) |
| 3732032 | 192 | PPE54 |
| 3948252 | 640 | Rv3519 (Conserved hypothetical protein) |
| F11 | ||
| 374076 | 61 | PPE5 |
| 1509585 | 1101 | Rv1334 (Conserved Hypothetical protein – Rv1335 (CFP10A) – cysM (cysteine synthetase) |
| 1890753 | 172 | Possible Promoter region of Rv1668c (Probable Macrolide ABC transport. ATP binding protein) |
| 2381228 | 2645 | PE22 and Intergenic region |
| 3183106 | 109 | Rv2859c (Hypothetical amidotransferase) |
| 3702663 | 104 | Intergenic region between lpdA (dihydrolipoamide dehydrogenase) and Rv3304 (Conserved hypothetical protein) |
| Haarlem | ||
| 912152 | 92 | Rv0823c (Possible transcriptional regulatory Protein) |
| 965426 | 481 | PE-PGRS15 |
| 1451173 | 115 | Intergenic region betwwen thrB (Homoserine kinase) and thrC (Threonine synthase) |
| 1521604 | 4753 | Rv1353c (Probable transcriptional regulatory protein) – Rv1354c (Possible diguanylate cyclase protein) – moeY (Molibdopterin biosynthesis protein) – Rv1356c (Hypothetical protein) |
| 2060412 | 643 | PE-PGRS33 |
| 2365930 | 1774 | helZ (Helicase) – Rv2102 (Conserved hypothetical protein) |
| 2546849 | 6480 | Rv2271 (Hypothetical protein) – Rv2272 (Probable conserved transmembrane protein) – Rv2273 (Probable conserved transmembrane protein) – Rv2274c (Hypothetical protein) – Rv2275 (Hypothetical protein) – cyp121 (Cytochrome P450) – Rv2277c (Probable glycerolphosphodiesterase) |
| 2792261 | 170 | PE-PGRS42 |
| 3117314 | 439 | Repeat region (DR) Intergenic between Rv2813 and Rv2814 |
| 3192481 | 87 | IS1539 Transposase contained in repeat region |
| 4033587 | 616 | Rv3600c (Conserved hypothetical protein) – panD (Aspartate decarboxylase precursor) |
| 4365135 | 84 | PPE69 and Intergenic region with PE36 |
| C | ||
| 334840 | 6611 | PE-PGRS3-PE-PGRS4-PPE3 (partially absent in Haarlem Strain) |
| 474110 | 125 | Rv0401 (Probable conserved transmembrane protein) |
| 565404 | 113 | Intergenic region between Rv0480c (Probable amidohydrolase) and Rv0481c (Hypothetical protein) |
| 956083 | 54 | Intergenic region between ercc3 (Probable DNA helicase) and Rv0862c (Hypothetical protein) |
| 1984522 | 109 | C-terminal end wag22 (PE-PGRS) |
| 2354142 | 214 | PE-PGRS36 |
| 2456125 | 115 | Intergenic region between qcrB (Ubiquinol cytochrome C reductase) and Rv2197c (Probable conserved transmembrane protein) |
| 2825166 | 53 | Rv2517c (Hypothetical protein) |
| 2914707 | 115 | PE-PGRS44 |
| 3113322 | 736 | Repeat region DR Intergenic between Rv2816c and Rv2817c |
| 3182209 | 54 | Intergenic region between frr (ribosome releasing factor) and pyrH (Uridylate kinase) |
| 3369008 | 113 | PPE47 |
| 3369190 | 81 | PPE48 |
| 3369344 | 62 | PE29 |
| 3369422 | 171 | PE29 |
| 3426378 | 83 | Rv3074 (Hypothetical protein) |
| 3911404 | 11522 | fadD17 (Fatty-acid-CoA Synthase)-PE-PGRS53-PE-PGRS54 |
| 4009833 | 3486 | clpC1 (Probable ATP-dependent protease) – lsr2 (Probable Iron-regulated protein) – lysS (lysyl t-RNA synthetase) |
| 4143780 | 560 | Rv3728 (Probable conserved two domain membrane protein – drug transporter) |
Gene names are according to Tuberculist http://genolist.pasteur.fr/TubercuList/ and for genes absent in the MTB H37Rv genome the names are according to the MTB CDC1551 genome annotation [12]. Position indicated correspond to the site where the deletion has occurred.
Table 2.
Genes Involved in strain-specific insertions.
| Position | Size | Genes Involved |
| H37Rv | ||
| 3551227–3552586 | 1359 | Intergenic region between Rv3185 (Probable transposase) and Rv3188 (Hypothetical protein) |
| H37Ra | ||
| 13627–14986 | 1359 | Insertion of the IS6110 element between Rv0010c (Probable conserved membrane protein) and Rv0011c (Hypothetical protein) |
| 1989343–1990797 | 1437 | Insertion of the IS6110 element on the ipl locus |
| CDC1551 | ||
| 804474–804691 | 217 | Intergenic region between rplW (50S ribosomal protein L23) and rplB (50S ribosomal protein L2) |
| 2341782–2341840 | 58 | MT2144 (Hypothetical protein) |
| F11 | ||
| 934662–936021 | 1359 | IS6110 insertion in intergenic region between hypothetical proteins TBFG_10851 and TBFG_10853 |
| 1945992–1947351 | 1359 | IS6110 insertion in Rv1724c (Hypothetical protein) |
| 1992354–1993713 | 1359 | IS6110 insertion in cut1 (Probable cutinase) |
| 1997360–1998719 | 1359 | IS6110 insertion in intergenic region between wag22 (PE-PGRS) and Rv1760 (Hypothetical protein) |
| 2004182–2005541 | 1359 | IS6110 insertion in intergenic region between hypothetical proteins Rv1765c and Rv1766 |
| 2017862–2019221 | 1359 | IS6110 insertion in cyp144 (Cytochome P450) |
| 2270442–2271801 | 1359 | IS6110 insertion in Rv2015c (Hypothetical protein) |
| 2349289–2350648 | 1359 | Intergenic region between Rv2077c (Possible conserved transmembrane protein) – Rv2078 (Hypothetical protein) |
| 2697325–2698684 | 1359 | IS6110 insertion in Rv2390c (Hypothetical protein) |
| 3492136–3493495 | 1359 | IS6110 insertion in Rv3113 (Probable phosphatase) |
| 3564263–3565622 | 1359 | IS6110 insertion in intergenic region between Rv3183 (Possible transcriptional regulatory protein) – TBFG_13208 (Conserved hypothetical protein) |
| 3831596–3831772 | 176 | Intergenic region between Rv3401 (Hypothetical protein) and Rv3402c (Hypothetical protein) |
| 3853062–3860408 | 2394 | TBFG_13461 (Hypothetical protein) – TBFG_13462 (Hypothetical protein) – TBFG_13463 (Hypothetical protein) – TBFG_134614 (Probable Transposase) – Intergenic region |
| Haarlem | ||
| 1071617–1072976 | 1359 | IS6110 insertion in Rv0963c (Conserved hypothetical protein) |
| 1714841–1716200 | 1359 | IS6110 insertion in mmpl12 (Transmembrane protein) |
| 1834511–1834569 | 58 | Rv1637c (Hypothetical protein) |
| 1973210–1973436 | 226 | PPE24 |
| 2606037–2607396 | 1359 | IS6110 insertion in Rv2336 (Hypothetical protein) |
| 4144939–4144997 | 58 | leuA (2-isopropylmalate synthase) |
| C | ||
| 4088473–4088705 | 232 | Rv3680 (Probable anion transporter ATPase) |
Gene names are according to Tuberculist when homologous to MTB H37Rv, if not, genes are named according to the annotation of the respective genome.
Strain-specific polymorphisms may also help to explain some of the observed phenotypic differences among MTB strains. This study is therefore hypothesis-generating and functional analyses of the polymorphisms found can be carried out to understand how genetic variation affects the outcome of infection. Our results indicate that circulating strains harbor a higher number of strain-specific polymorphisms (Tables 1 and 2) and suggest that interaction with the human host results in strain-specific genomic polymorphisms that may in turn determine the phenotypic differences among circulating MTB strains. While most deletions tend to be slightly deleterious [12], some strain-specific polymorphisms, such as the Beijing genotype's strain-specific deletion in the pks1/15 region [4,23], may contribute to the pathogen's virulence or its adaptation and the establishment of stable associations with the human host population [24]. The tendency predicted here regarding genetic variation of the MTB genome is based on six strains that represent only a fraction of the global MTB population. Thus it is necessary to further investigate the extent of intra and inter lineage genetic variation for a more accurate prediction of the variation dynamics within the entire MTB population. The inclusion of a larger number of representative strains can provide a broader understanding of how these genetic polymorphisms contribute to the adaptation of certain genotypes to human populations.
Conclusion
The multiple alignment of six MTB genomes identified both frequency and differential distribution patterns of polymorphisms. This study indicated that circulating MTB strains are under active selective pressure that leads to variation by means of gene loss or inactivation and suggests that this species in a process of gene decay. The identification of strain-specific polymorphisms may ultimately translate into molecular markers for strain surveillance and predictors of outbreaks in defined human populations.
Abbreviations
MTB: Mycobacterium tuberculosis; LSP: Large sequence polymorphism; InDels: Insertions and Deletions; SNP: Single nucleotide polymorphism; SD: Specific Deletion; SI: Specific Insertion
Authors' contributions
ACR carried out the multiple alignments and genome comparisons, designed the islandsanalyser, evaluated data, analyzed the results obtained and was in charge of writing the manuscript. MMZ supervised the progress of the work, helped in writing the manuscript and contributed with discussions regarding the overall research and the results obtained. JM carried out the programming of the islandsanalyser.
Supplementary Material
Methods.
Islandanalyser package. The scripts were built using Perl 5. In order to execute them the module Chart::Plot (available at http://search.cpan.org/~smorton/Chart-Plot-0.11/Plot.pm) and the module GD http://stein.cshl.org/WWW/software/GD/ must be installed. It works on any operating system that supports Perl, a list of systems where Perl is available can be obtained at http://www.perl.org/get.html. Contents: genoma.pl, max.pl and Graph.pl and proper documentation.
Acknowledgments
Acknowledgements
This work was supported by Colciencias (project No. 657040820410) and as part of the Colombian Center for Research in Tuberculosis, CCITB (RC No.431-2004). Thanks to Roberto Kolter, Silvia Restrepo, Jaime Robledo, Alejandro Reyes and Juan R. Cubillos for critical reading of the manuscript, Patricia del Portillo and Gabriel Lozano for helpful comments.
Contributor Information
Andrés Cubillos-Ruiz, Email: acubillos@corpogen.org.
Juan Morales, Email: jpmorales@ingenian.com.
María Mercedes Zambrano, Email: mzambrano@corpogen.org.
References
- Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, Supply P, Vincent V. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–594. doi: 10.1016/j.gde.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, Riley LW. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc Natl Acad Sci USA. 2003;100:15918–15923. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MB, Gagneux S, Deriemer K, Small PM, Barry CE., 3rd The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. J Bacteriol. 2007;189:2583–2589. doi: 10.1128/JB.01670-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, Hoy D, Jones JS, Westmoreland H, Onorato IM. An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med. 1998;338:633–639. doi: 10.1056/NEJM199803053381001. [DOI] [PubMed] [Google Scholar]
- Victor TC, de Haas PE, Jordaan AM, Spuy GD van der, Richardson M, van Soolingen D, van Helden PD, Warren R. Molecular characteristics and global spread of Mycobacterium tuberculosis with a western cape F11 genotype. J Clin Microbiol. 2004;42:769–772. doi: 10.1128/JCM.42.2.769-772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman CR, Quinn GC, Kreiswirth BN, Perlman DC, Salomon N, Schluger N, Lutfey M, Berger J, Poltoratskaia N, Riley LW. Widespread dissemination of a drug-susceptible strain of Mycobacterium tuberculosis. J Infect Dis. 1997;176:478–484. doi: 10.1086/514067. [DOI] [PubMed] [Google Scholar]
- Lopez B, Aguilar D, Orozco H, Burger M, Espitia C, Ritacco V, Barrera L, Kremer K, Hernandez-Pando R, Huygen K, et al. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–37. doi: 10.1046/j.1365-2249.2003.02171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, Cousins DV, Graviss EA, Shashkina E, Kreiswirth BN, Musser JM. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics. 2002;162:1533–1543. doi: 10.1093/genetics/162.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, Hannan M, Goguet de la Salmoniere YO, Aman K, Kato-Maeda M, Small PM. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci USA. 2004;101:4865–4870. doi: 10.1073/pnas.0305634101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Klasson L, Andersson SG. Microbial genome evolution: sources of variability. Curr Opin Microbiol. 2002;5:506–512. doi: 10.1016/S1369-5274(02)00358-2. [DOI] [PubMed] [Google Scholar]
- Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- Sreenu VB, Kumar P, Nagaraju J, Nagarajaram HA. Microsatellite polymorphism across the M. tuberculosis and M. bovis genomes: implications on genome evolution and plasticity. BMC Genomics. 2006;7:78. doi: 10.1186/1471-2164-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Blekhman R, Graur D. The "domino theory" of gene death: gradual and mass gene extinction events in three lineages of obligate symbiotic bacterial pathogens. Mol Biol Evol. 2006;23:310–316. doi: 10.1093/molbev/msj036. [DOI] [PubMed] [Google Scholar]
- Banu S, Honore N, Saint-Joanis B, Philpott D, Prevost MC, Cole ST. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol. 2002;44:9–19. doi: 10.1046/j.1365-2958.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- Delogu G, Brennan MJ. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun. 2001;69:5606–5611. doi: 10.1128/IAI.69.9.5606-5611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly N, Giang PH, Gupta C, Basu SK, Siddiqui I, Salunke DM, Sharma P. Mycobacterium tuberculosis secretory proteins CFP-10, ESAT-6 and the CFP10:ESAT6 complex inhibit lipopolysaccharide-induced NF-kappaB transactivation by downregulation of reactive oxidative species (ROS) production. Immunol Cell Biol. 2007 doi: 10.1038/sj.icb.7100117. [DOI] [PubMed] [Google Scholar]
- Safi H, Barnes PF, Lakey DL, Shams H, Samten B, Vankayalapati R, Howard ST. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol Microbiol. 2004;52:999–1012. doi: 10.1111/j.1365-2958.2004.04037.x. [DOI] [PubMed] [Google Scholar]
- Tanaka MM, Rosenberg NA, Small PM. The control of copy number of IS6110 in Mycobacterium tuberculosis. Mol Biol Evol. 2004;21:2195–2201. doi: 10.1093/molbev/msh234. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Tsolaki AG, Gagneux S, Pym AS, Goguet de la Salmoniere YO, Kreiswirth BN, Van Soolingen D, Small PM. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol. 2005;43:3185–3191. doi: 10.1128/JCM.43.7.3185-3191.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci USA. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methods.
Islandanalyser package. The scripts were built using Perl 5. In order to execute them the module Chart::Plot (available at http://search.cpan.org/~smorton/Chart-Plot-0.11/Plot.pm) and the module GD http://stein.cshl.org/WWW/software/GD/ must be installed. It works on any operating system that supports Perl, a list of systems where Perl is available can be obtained at http://www.perl.org/get.html. Contents: genoma.pl, max.pl and Graph.pl and proper documentation.