SUMMARY
Protein N of bacteriophage λ activates the lytic phase of phage development in infected E. coli cells by suppressing the activity of transcriptional terminators that prevent the synthesis of essential phage proteins. N binds tightly to the boxB RNA hairpin located near the 5′ end of the nascent pL and pR transcripts and induces an antitermination response in the RNA polymerase (RNAP) of elongation complexes located at terminators far downstream. We here test a RNA looping model for this N-dependent ‘action at a distance’ by cleaving the nascent transcript between boxB and RNAP during transcript elongation. Cleavage decreases antitermination, showing that an intact RNA transcript is required to stabilize the interaction of boxB-bound N with RNAP during transcription. In contrast, an antitermination complex that also contains Nus factors retains N-dependent activity after transcript cleavage, suggesting that these host factors further stabilize the N-RNAP interaction. Thus the binding of N alone to RNAP is controlled by an RNA looping equilibrium, but after formation of the initial RNA loop and in the presence of Nus factors the system no longer equilibrates on the transcription time-scale, meaning that the ‘range’ of antitermination activity along the template in the full antitermination system is kinetically controlled by the dissociation rate of the stabilized N-RNAP complex. Theoretical calculations of nucleic acid end-to-end contact probabilities are used to estimate the local concentrations of boxB-bound N at elongation complexes poised at terminators, and are combined with N activity measurements at various boxB-to-terminator distances to obtain an intrinsic affinity (Kd) of ~ 2 × 105 M for the N-RNAP interaction. This RNA looping approach is extended to include the effects of N binding at nonspecific RNA sites on the transcript and the implications for transcription control in other regulatory systems are discussed.
Keywords: Transcription, transcript elongation, transcription regulation, enhancers, protein-RNA interactions, looping, nonspecific binding, antitermination
INTRODUCTION
Specific gene transcription is often controlled by ‘action at a distance’ effects that reflect processes in which regulatory proteins bind to specific nucleic acid sequences at one position on the DNA genome and modulate transcription at another. These processes generally involve nucleic acid looping mechanisms that facilitate interaction between a bound regulatory protein assembly and a functional transcription complex.1–3 Looping-facilitated interactions are essential for proper transcription complex assembly and function and have been observed in a number of biological contexts, including intiation and elongation of prokaryotic and eukaryotic transcription,1–6 replication,7 RNA splicing,8 recombination,9,10 and telomere maintenance.11 Despite the ubiquity of these processes, many of the mechanisms that control interactions facilitated by cis-looping are poorly understood.
A minimal model of regulation by nucleic acid looping involves four steps: (i) binding of regulatory protein factor(s) to specific sequences on the DNA or RNA; (ii) looping of the nucleic acid ‘tether’ to bring the bound protein(s) to the (usually DNA-bound) target complex that is to be regulated; (iii) binding of the regulatory proteins to the target complex in cis, with an affinity that depends both on the looping-dependent local protein concentration at the target and on the intrinsic affinity of the protein(s) for the target; and (iv) interaction of the protein(s) with the target complex to achieve regulation. The effectiveness of each of these steps depends on those preceding; thus protein binding in trans to the nucleic acid binding site determines the fraction of protein-nucleic acid complexes available for looping-facilitated binding of protein to the target site, which in turn determines the fraction of target-bound complexes that can modulate the regulatory response. Determination of the relationship between regulatory protein binding and changes in transcriptional activity requires measurement of the efficiency of all four steps in a single experimental system. Such experiments allow determination of the affinity of regulatory proteins for their targets, identify regulatory mechanisms that arise from the cumulative effect of multiple mechanistic steps, and isolate key parameters whose modulation by accessory factors changes regulatory output.
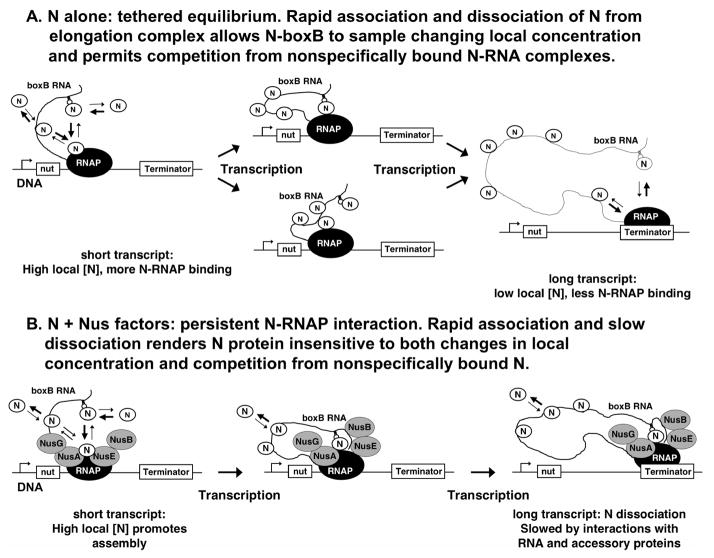
The antitermination function of the N protein of bacteriophage λ provides a simple model system for the study of such interactions. N protein activates the transcription of developmental genes of phage λ by contacting the RNA polymerase (RNAP) of transcriptional complexes of the E. coli host and increasing the read-through of intrinsic and Rho-dependent terminators that otherwise prevent synthesis of transcripts required in the lytic pathway of phage development.12–14 This antitermination process occurs only at terminators located within the pR and pL operons of λ and requires the presence of nut DNA sequences that are located just downstream of the pL and pR promoters.15,16 These nut sequences, functioning in their RNA forms, are present on the transcripts that originate from promoters pL and pR17–19 and contain the boxA and boxB RNA sequences that act as binding sites for regulatory factors involved in antitermination.20,21 N binds to the boxB RNA hairpin sequence element20,22 and contacts (by cis RNA looping) transcription elongation complexes paused at terminators located as far as thousands of bps downstream of the boxB binding site.23–25 In vitro, in the absence of boxB, N protein alone can cause high levels of terminator readthrough by binding nonspecifically to the transcript RNA.26,37
The ability of N to act ‘at a distance’ (via cis RNA looping) to suppress transcript termination at terminators located downstream from the nut-encoded boxB binding site is inversely related to the distance along the template DNA between the nut site and the transcriptional terminators of the pL and pR operons. In vivo, DNA insertions in this region decrease, and DNA deletions increase, the expression of genes located as much as 10,000 bp from nut.23 The effective ‘range’ of the N-dependent antitermination effect in vitro is smaller, with N and the E. coli NusA protein cooperating to suppress terminators located 300 bp from nut with >95% efficiency,26 but failing to suppress terminators positioned >500 bps from nut.25 The diminished activity of N at terminators located significantly downstream of the nut site can be restored in vitro by the addition of the E. coli host factors NusA, NusB, NusE (also called ribosomal protein S10) and NusG.24,25 These host protein factors do not facilitate antitermination in the absence of N, but they do increase the range of in vitro N-dependent antitermination to >3 kbp from the nut site. The Nus factors bind to the boxA RNA sequence located adjacent to boxB and also to N, to RNAP and to one another,21,27–30 suggesting that they work together to stabilize the interaction of N protein with the RNAP. However, little is known about how the Nus factors increase the stability of the N-RNAP complex in the context of RNA looping.
A generally accepted model for action at a distance in the N system proposes that the interaction of N protein with the RNAP of the elongation complex is weak and requires an elevated local concentration of boxB-bound N protein at the elongation complex to permit binding. This elevated local concentration is provided by RNA looping, as outlined above.3,18,19,22 The local concentration of transcript-bound N protein at RNAPs positioned at the target terminators is high for nut sites close to the terminator, because the volume through which the tethered N can diffuse (and therefore its local concentration) is controlled by the length of the RNA transcript. The longer RNA tether that results when the nut site is far from the terminator increases the volume of solution around the RNAP that is available to the boxB-tethered N protein, thus lowering the effective local concentration of N at the target. Thus, as elongation proceeds, extension of the RNA transcript results in a ‘reverse titration’ of RNAP with N protein, with the local concentration of N protein bound ‘in cis’ at the boxB hairpin decreasing. Eventually, at terminators for which the length of the RNA ‘tether’ between boxB and the terminator exceeds an upper limit, the local concentration of boxB-tethered N protein becomes too low to permit effective equilibrium binding of N to the RNAP of the transcription complex.
The local concentration of N protein at the elongation complex has been calculated for various nut to terminator distances using quantitative models of RNA looping. This approach uses a freely jointed chain model and empirical measurements of RNA flexibility to calculate the probability that two proteins bound to the same transcript will make contact.3,31–34 This probability is expressed as the local molar concentration of one protein in the vicinity of another bound at a defined position on the nucleic acid chain as a function of the separation of the two binding sites (e.g., boxB and RNAP at the terminator) along the template. The basic premises of this model have been verified experimentally by monitoring the kinetics of ligase-catalyzed loop closure,35 enhancer-dependent RNA splicing,8 FLP recombinase-dependent DNA looping36 and other looping processes.
In this work we have estimated the looping-dependent binding affinity of N protein for RNAP by comparing the fraction of RNA transcripts that read through transcription termination signals at various boxB-to-terminator distances with the calculated local concentrations of N protein at these same distances. To do this we first verified that N protein acts via a RNA looping mechanism and then showed that the looping-facilitated interaction of N protein with RNAP achieves equilibrium on the time scale of the transcription process. We have sought to verify this second point experimentally because transcript elongation increases the size of the RNA loop even while the N protein is bound to RNAP. This raises the question of how and whether the RNAP -- bound to N protein via an elongating RNA tether -- senses the changing local concentration of N. A simple interpretation of the local concentration model postulates that N protein binds to and dissociates from the polymerase of the elongation complex on a time scale that is faster than the rate of RNA synthesis from the nut site to the terminator, allowing the binding interaction to ‘sample’ the changing (equilibrium) local concentration of N provided by the looping of the growing RNA chain. This model requires the presence of an intact RNA during elongation. An alternative model proposes that, after the initial looping event, N forms a persistent complex with the RNAP of the elongation complex, with dissociation of N from RNAP being slower than transcript elongation. This scenario requires an intact loop only during the initial N binding phase and means that the range of the N effect should depend on the dissociation rate of the complex rather than on the equilibrium binding constant of N to RNAP.
We here test these versions of the local concentration model by cleaving the RNA transcript between boxB and the terminator while the intervening template region is being transcribed. In this way we can ask whether the binding interaction ‘samples’ the changing (equilibrium) local concentration of N due to the elongation of the RNA chain, or whether the tethered N is persistently bound on this time scale. We then use a RNA-looping-dependent in vitro antitermination assay to quantitate the decrease in N activity at distal terminators, and compare this distance-dependent difference in termination levels with the quantitative predictions of the local concentration model. We note that N protein binds to (and is active at) both the boxB site and nonspecific RNA binding sites on the nascent transcript 26,37, requiring that we extend our quantitative theoretical models of nucleic acid looping to include also the local concentrations of proteins bound at nonspecific sites. Our results suggest that, in the presence of an intact RNA chain, the boxB-bound N protein is involved in a rapid cis-looping equilibrium with the transcribing RNAP. In contrast, addition of the full complement of Nus host factors appears to stabilize the binding of N protein to RNAP sufficiently so that the rate of dissociation of the tethered N from the RNAP becomes slow relative to the rate of the transcript elongation process. As a consequence the N-dependent antitermination interaction transitions from an equilibrium system to one controlled by the kinetics of N dissociation from the Nus-factor-containing elongation complex.
These studies show that the kinetics of tethered association and dissociation are critical parameters in the mechanism of N-dependent antitermination in particular, and in ‘action at a distance’ regulatory systems in general. Additionally this work suggests that nonspecific binding of proteins to long nucleic acid loops is likely to occur even when the difference in protein binding affinity between specific and nonspecific nucleic acid binding sites is large. Thus extended looping models must take into account the looping interactions of both specifically and nonspecifically bound protein, and a complete picture of looping-facilitated regulation must include the relationship between the interaction kinetics of enhancers and repressors with their targets, and the rates of the molecular processes that these binding events control.
RESULTS
N protein and the boxB RNA hairpin suffice to demonstrate distance-dependent antitermination in vitro
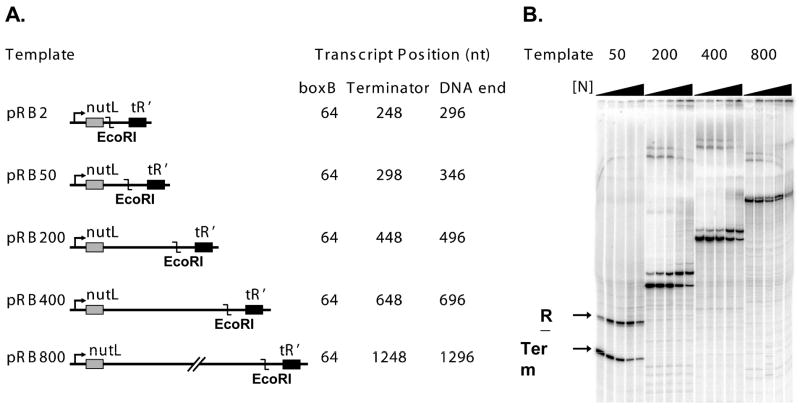
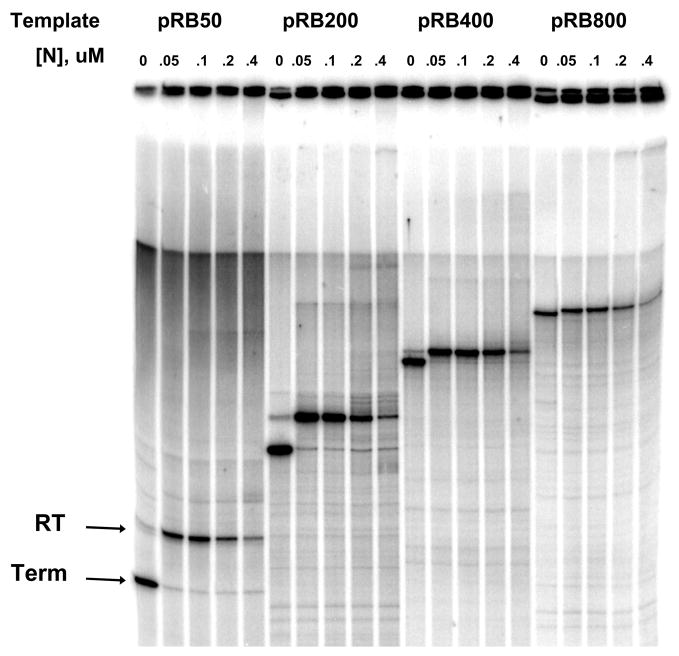
The requirement for RNA looping in N-dependent antitermination has not been unambiguously established. In addition, previous measurements of the N-dependent antitermination ‘range effect’ were performed in the presence of NusA protein, which binds to both N and RNA polymerase and thus provides a potential RNA looping-independent pathway for recruitment of N to RNAP. To establish the minimal requirements for the N range effect, we measured terminator read-through in reactions containing only N, RNAP and a series of templates containing different lengths of DNA between the nut and terminator sequences (Figure 1A). Transcription of each template produced two RNAs in each gel lane of Figure 1B, corresponding to the terminated and the read-through transcripts, respectively. Addition of antitermination protein N increased the fraction of transcripts that read through the terminators; this fraction was calculated by measuring the intensities of the terminated and readthrough bands.
Figure 1. Transcription templates and reactions.
(A) Transcription templates used to measure the effect on N-dependent antitermination activity of the boxB-to-terminator RNA length. All templates contain pL promoter, nutL DNA encoding the boxB RNA sequence (grey box) and the λ tR′ terminator (black box); template sequences are identical except for insertions between nut and tR′. Insertions share the same 5′ sequence and differ in length of the 3′ end. (B) Read-through (RT) and terminated (Term) RNA bands for transcription reactions performed with templates pRB50, pRB200, pRB400 and pRB800 in the presence of increasing concentrations of λN protein.
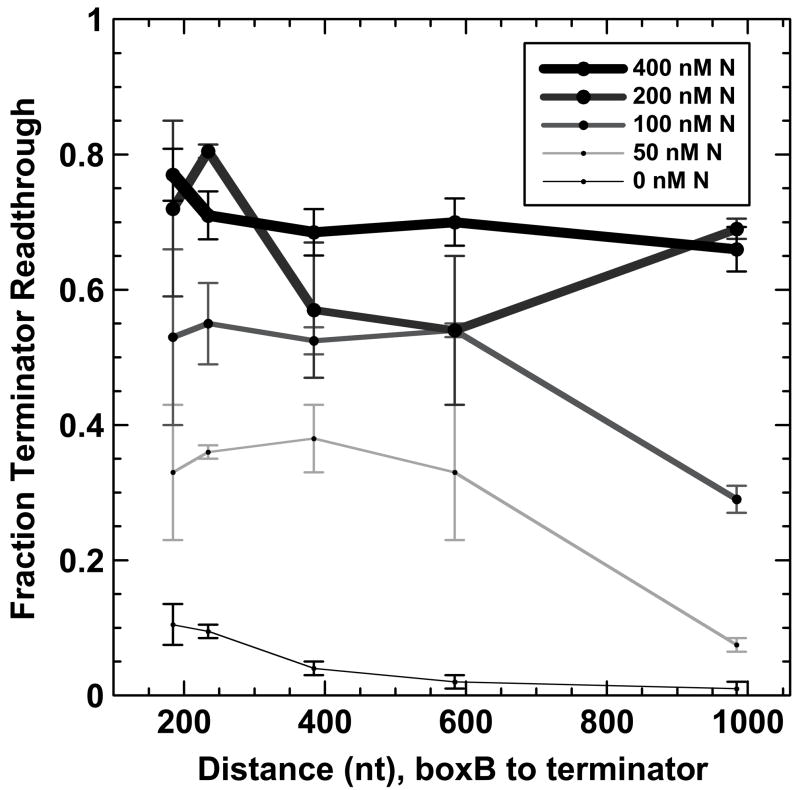
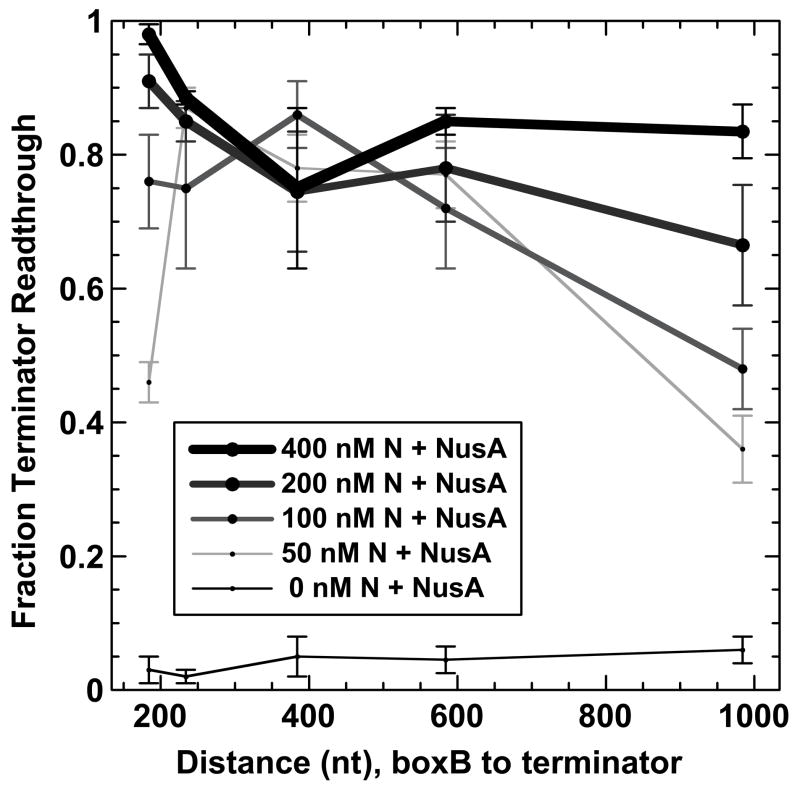
Figure 2 shows the fraction of terminator read-through as a function of increasing RNA (and DNA) distance in nts from the boxB RNA sequence to the termination position within intrinsic terminator tR′ at four different total N protein concentrations. At high N concentrations (200 and 400 nM) read-through is high at all boxB-to-terminator distances, presumably due to the nonspecific binding of N protein to the transcript RNA.26,37 At lower N levels (100 and 50 nM) antitermination is observed at short (200–600 nt) distances and decreases as the boxB-to-terminator distance surpasses 500 nts. This distance-dependent decrease in antitermination at low N concentrations shows that N-dependent ‘action at a distance’ does not require NusA. In addition, because the transcription complexes generated in these experiments differ only in the lengths of the DNA templates (and therefore the lengths of the RNA transcripts) the observed decrease in N activity at distal terminators reflects a lattice-length-dependence of N-dependent antitermination activity that is consistent with an equilibrium RNA looping mechanism for this minimal system.
Figure 2. Effect of template distance between the boxB-N protein binding site and the terminator on N protein-dependent antitermination activity in the absence of accessory proteins.
The fraction of full-length RNA transcripts produced by transcriptional readthrough of terminator tR′ for each template in Figure 1 at the indicated concentrations of N protein are plotted as a function of transcript distance (in nts) between the N binding site (boxB RNA) and the target terminator.
A continuous RNA transcript is required for N-boxB-dependent antitermination activity
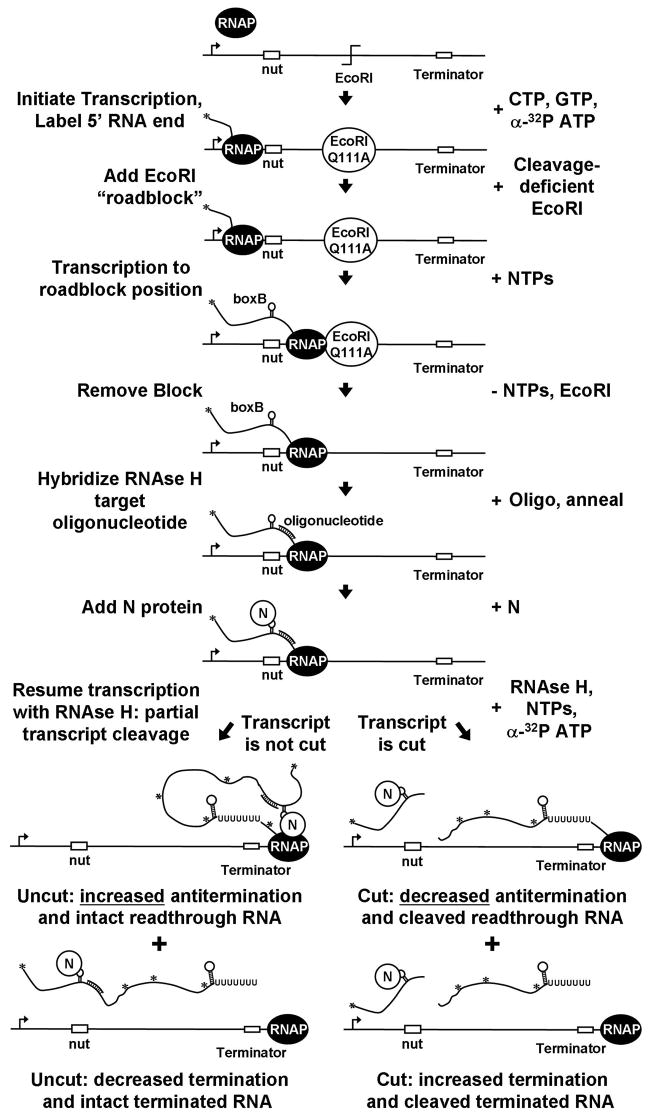
We used this minimal N-dependent antitermination system to test the requirement for a continuous RNA tether between the N-boxB complex and a transcription complex poised at the terminator by enzymatically cleaving the nascent RNA with RNAse H during transcript elongation from boxB to the terminator. The experimental steps are outlined in Figure 3. Because direct N binding to RNAP does not occur at the concentrations of N used in these experiments,26 we reasoned that if the looping-dependent interaction of N-boxB complexes with RNAP equilibrated rapidly, cleavage of the RNA transcript would result in irreversible dissociation of N-boxB complexes from RNAP and loss of N-dependent antitermination activity. Alternatively, if the N-boxB complex were stably bound to the RNAP of the elongation complex during the time required for transcription from boxB to the terminator, the cleaved N-boxB complex should remain bound to the RNAP and retain its antitermination activity. The assay thus offers a rough estimate of the kinetics of the RNA-looping-dependent binding of the N-boxB complex to RNAP located at the terminator.†
Figure 3. Schematic of transcript cleavage experiments.
Procedure used to form stalled antitermination complexes and simultaneously re-extend and cleave RNA transcripts during transcription between boxB and the terminator. Asterisks indicate positions of α-32P ATP incorporation in the RNA transcript. Re-extension of transcripts and cleavage with RNAse H produces five radiolableled RNAs, corresponding to the 5′ cleavage product and pairs of cut and uncut terminated and readthrough transcripts.
Antitermination complexes were formed after ‘stalling’ elongation complexes on the DNA template with a ‘road-block’ protein (the cleavage-deficient restriction enzyme mutant EcoRI Gln111A). These experiments were initiated in the absence of N, and the ‘roadblock’ EcoRI mutant was then removed from the DNA template by the addition of salt and sequestered by a competing dsDNA oligonucleotide carrying the EcoRI sequence. N protein was added after the desalting step to form functional antitermination complexes and a ssDNA oligomer was hybridized to the transcript between boxB and the terminator to provide a cleavage target for RNAse H.
Two competing reactions were then launched simultaneously. One, involving elongation of RNAP through the termination position, was initiated by the addition of NTPs (containing α-32P ATP for labeling of the 3′ end of the RNA) to the reaction. The other, involving cleavage during transcript elongation of the transcript at a defined RNA site located upstream of the terminator, was initiated by also adding RNAse H. Simultaneous addition of NTPs and RNAse H resulted in cleavage of approximately 50% of the RNA transcripts generated by NTP addition, allowing the antitermination efficiency of both the cut and the uncut complexes to be measured in one reaction.
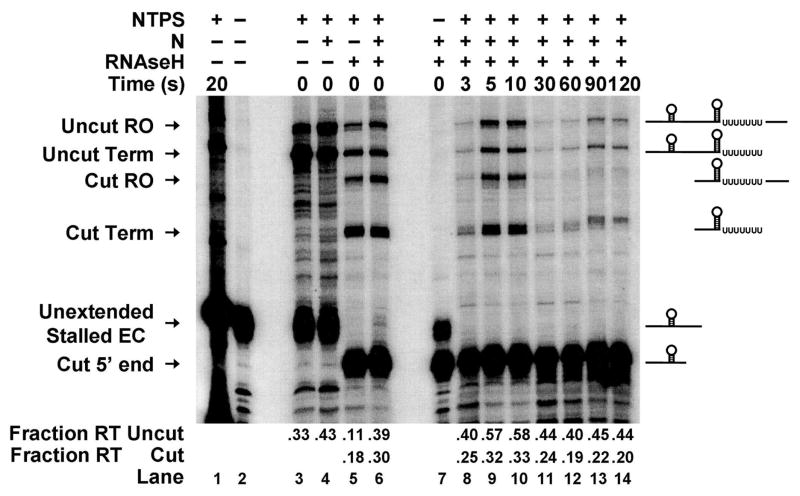
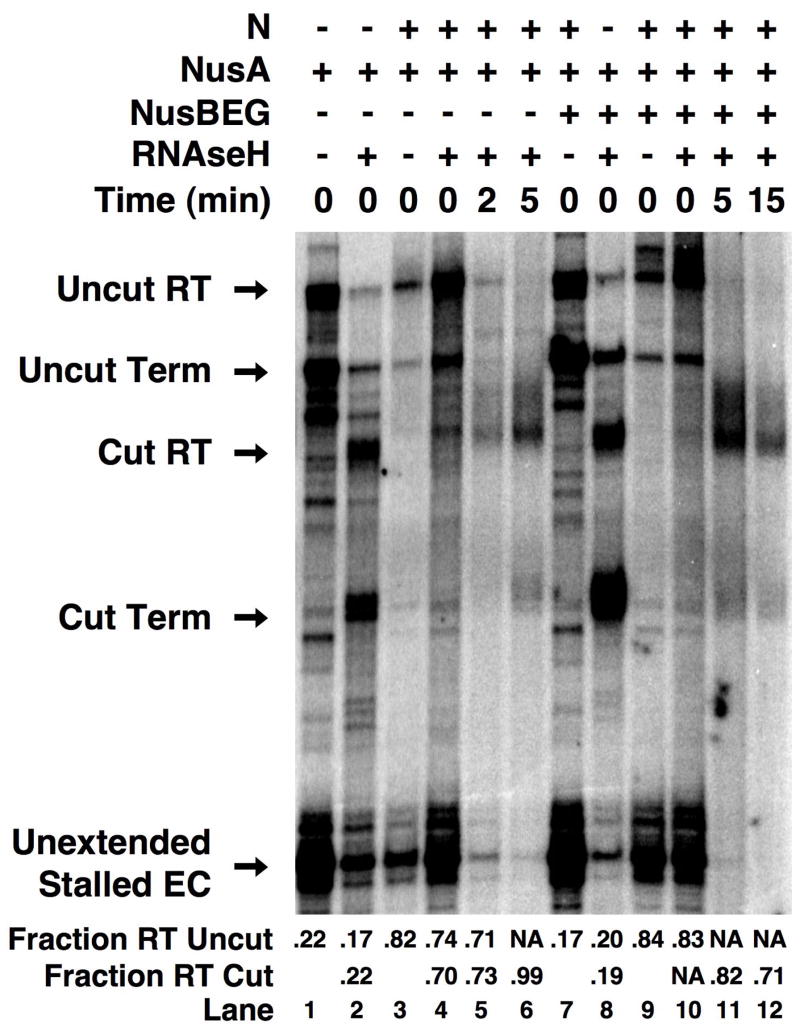
Figure 4 shows the results of transcript cleavage on antitermination function. In these reactions a fraction of stalled elongation complexes failed to resume transcription upon addition of NTPs (lane 3, “Unextended stalled ECs”), due to the well-known tendency of stalled elongation complexes to dissociate from the 3′ end of the transcript. The remaining fraction of elongation complexes resumed transcription upon NTP addition, and extended the RNA to produce two transcripts that correspond to terminated (lane 3, “Uncut Term”) and full-length readthrough (lane 3, “Uncut RT”) transcripts. Simultaneous addition of RNAse H and NTPs to stalled elongation complexes (lane 5) resulted in both extension and cleavage of RNA transcripts. Cleavage by RNAse H during the brief (~15 second) extension reaction is incomplete, so that the final reaction product contains both RNase H-cut and uncut transcripts. The transcripts that are cut by RNAse H during elongation from the stall position to the terminator continue to elongate, and produce terminated (lane 5, “Cut Term”) and full-length readthrough (lane 5, “Cut RT) RNAs. Transcripts that are not cleaved by RNAse H during elongation also extend through the terminator to produce terminated (lane 5, “Uncut Term”) and full-length readthrough (lane 5, “Uncut RT”) RNAs.
Figure 4. A continuous RNA tether is required for functional interaction of N with RNA polymerase.
RNAse H cleavage of the transcript between boxB and the terminator site decreases N-dependent antitermination activity. Elongation complexes stalled between boxB and terminator sequences were hybridized to a ssDNA oligonucleotide, supplemented with N protein (200 nM) and re-extended with NTPs in the presence of RNAse H. Partial RNAse H cleavage of the elongating transcripts yielded four RNA chains corresponding to terminated (Term) and full-length readthrough (RT) products of the RNAse H-cut and uncut transcripts, and two shorter RNAs corresponding to unextended stalled complexes (Unextended Stalled EC) and the 5′-product generated by RNAse H cleavage of the transcript (Cut 5′ end). The fraction of RNAse H-cleaved transcripts (Fraction RT, cut) that read through terminators and the fraction of intact transcripts that read through terminators (Fraction RT, uncut) are indicated at the bottom. The times between addition of RNAse H and the NTP-extension mix are indicated at the top of the gel.
The observed antitermination efficiency was low in all the reactions (lanes 4 and 6), presumably because the lower concentration of NTPs required for 3′-labeling of the transcript results in increased termination. Nevertheless antitermination was observed in the intact complexes (fraction read-through = 0.39, lane 6). In contrast, antitermination in complexes for which the RNA transcript had been cut was decreased (fraction read-through = 0.30, lane 6). To test whether N protein, given enough time, would completely dissociate from RNAP, we pre-incubated the antitermination complexes with RNAse H for varying amounts of time prior to addition of NTPs and extension through the terminator. Under these conditions the total amount of RNA decreased, presumably due to degradation of transcripts by contaminating RNAses in the RNAse H preparation. However, terminator readthrough decreased still further, approaching levels observed in the absence of N (compare lanes 14 and 5). Thus these experiments show directly that N protein requires an intact RNA between the boxB binding site and RNAP to permit antitermination, and suggest that RNA looping promotes re-association at terminators of dissociated N/boxB complexes in a tethered equilibrium with the functioning elongation complex.
Effect of Nus host factors on antitermination ‘range’
The NusA protein of E. coli is an essential transcription elongation factor that increases termination in the absence of N, and increases antitermination in the presence of N.27,38 Several lines of evidence suggest that NusA may stabilize the cis interaction of N protein and boxB with RNAP. NusA confers resistance to increased salt concentration and to the addition of competitor nucleic acids in antitermination reactions.26 NusA also binds both N and core RNAP, although it has no effect on the binding of N to boxB in isolation.28,41,42 NusA, RNAP and boxB together stabilize the interaction of N with the elongation complex, possibly via autoregulatory conformational changes.43 In sum, these observations predict that complexes of NusA and RNAP will bind the tethered N and boxB more tightly, thus slowing dissociation of the N-boxB assembly from the elongation complex and thus increasing the antitermination range.
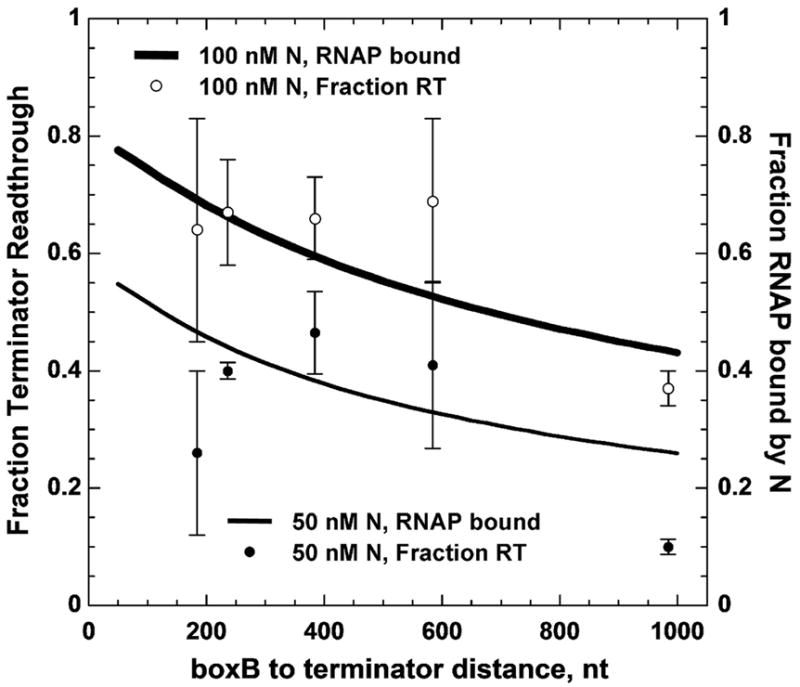
To test this interpretation we repeated in vitro measurements of the distance dependence of antitermination in the presence of NusA (Figure 5). Antitermination levels were higher in the presence of NusA than in its absence at all boxB-to-terminator distances and at all concentrations of N protein (compare with Figure 2). High N concentrations in the presence of NusA induced small decreases in antitermination at distal terminators, again reflecting nonspecific binding of N to the RNA transcript. At lower N concentrations, distance-dependent decreases were observed that were greater than in the absence of NusA. NusA inhibits antitermination by nonspecifically bound N protein, thus the observed decrease likely reflects a higher occupancy of RNAP by boxB-bound N proteins located farther from the terminator than the majority of nonspecific binding sites. The increase in the observed overall antitermination activity of N at downstream terminators, as well as the increase relative to reactions at the same N concentration in the absence of NusA, suggests that a greater fraction of N-boxB assemblies were bound to elongation complexes in the presence of NusA than in its absence.
Figure 5. Effect of template distance between the boxB-N protein binding site and the terminator on N-dependent antitermination activity in the presence of NusA.
The fraction of full-length RNA transcripts produced by transcriptional readthrough of terminator tR′ for each template in Figure 1 at the indicated concentrations of N protein in the presence of 120 nM NusA protein are plotted as a function of transcript distance between boxB and the terminator.
To further examine the relationship between antitermination complex stability and antitermination range we repeated the above measurements in the presence of NusA and NusB, NusE, and NusG (Figure 6). In these reactions a significant portion of the RNAs remained in the wells of the polyacrylamide gels used to measure fraction read-through; control reactions showed that these complexes consisted mostly of full-length RNAs that had read through terminators (see Materials and Methods). For the remaining RNA transcripts, addition of these factors abolished the distance dependence of antitermination at all concentrations and distances, such that the lowest levels of antitermination were in excess of 95%. These data are consistent with previous reports that show that antitermination in the presence of the full complement of Nus factors can persist for thousands of nts downstream from boxB23,24 and suggest that almost all elongation complexes are bound by N protein at downstream terminators in the presence of Nus factors.
Figure 6. Antitermination 'range' in the presence of N, NusA, NusB, NusE, NusG and transcript-encoded boxB.
Products of transcription reactions performed with templates pRB250, pRB200, pRB400 and pRB800 in the presence of indicated concentrations of N and NusA (120 nM), NusB, NusE and NusG proteins (250nM each). In all cases, the fraction of transcripts that ‘read through’ the terminator was >95%.
The requirement for a continuous RNA tether is relieved by the presence of Nus factors
To test whether the increased ability of N protein to suppress termination at downstream terminators in the presence of accessory factors might be due to a Nus-factor-dependent reduction in the dissociation rate of N protein from elongation complexes, we repeated our RNA transcript cleavage experiments in the presence of both N and NusA, or of N and NusA, B, E and G (Figure 7). In the absence of N and accessory factors, fractional readthrough of the terminator was 0.25 (data not shown). In the presence of NusA (lane 2) or NusA, B, E and G (lane 8), but no N, both cut and uncut transcripts showed increased termination, as expected for the well-known effect of these factors acting in trans. In the presence of N, reactions with NusA (lane 4) and NusA, B, E and G (lane 10) showed resistance to cleavage of transcripts by RNAse H (consistent with previous RNA footprinting experiments showing stable antitermination complexes in the presence of accessory factors18) and weak signals for terminated and runoff products of cut transcripts. Consequently, reactions were incubated with RNAse H for several minutes prior to addition of NTPs and resumption of transcription, resulting in near-complete cleavage of the RNA transcripts. Reactions with NusA (lanes 5–6) showed strong readthrough on both RNAse H-cleaved and intact transcripts. Reactions with NusA, B, E, and G (lanes 11–12) also showed strong read-through of the terminator on both cleaved and intact transcripts. We conclude that the presence of the full set of Nus host factors makes the interaction of N and RNAP sufficiently persistent to eliminate its dependence on the presence of an intact RNA tether during transcription, although the tether is clearly required to permit the complex to form initially.
Figure 7. A continuous RNA tether is not required for functional interaction of N with RNA polymerase in the presence of accessory factors.
RNAse H cleavage of the transcript between boxB and the terminator site does not decrease N-dependent antitermination activity in the presence of transcription accessory factors. Experiments were performed as in Figure 3; i.e., elongation complexes stalled between boxB and terminator sequences were hybridized to a ssDNA oligonucleotide, supplemented with N protein (200 nM), NusA (120 nM), and NusB, E and G (250 nM each), and incubated with RNAse H for the indicated times before NTP addition and resumption of transcription. Reactions yielded four RNA chains corresponding to terminated (Term) and full-length readthrough (RT) products of the RNAse H-cut and uncut transcripts, and two shorter RNAs corresponding to unextended stalled complexes (Unextended Stalled EC) and the 5′-product generated by RNAse H cleavage of the transcript (not shown). The fractions of full-length RNA transcripts produced by transcriptional readthrough of terminator tR′ of RNAse H-cleaved (Fraction RT Cut) and intact (Fraction RT Uncut) transcripts, and the times between addition of RNAse H and the NTP-extension mix, are indicated above and below the relevant lanes.
Comparison of measured and predicted RNA looping efficiencies
We next estimate the binding affinity of N for the RNAP of the elongation complex in the tethered equilibrium observed in the absence of Nus factors. In this system binding of a single N protein to RNAP results in antitermination.37 We therefore used measurements of terminator read-through at various nut-to-terminator distances (Figure 2) to determine the fraction of elongation complexes bound by N. We then calculated the binding of N protein in trans to RNA transcripts present in antitermination assays, and estimated the local concentration of transcript-bound N protein at the nut-to-terminator distances of the in vitro antitermination assays to obtain an apparent tethered equilibrium binding affinity of N for RNAP in this system.
The extent of N protein binding in trans to the nascent RNA transcripts produced by the in vitro antitermination assays was estimated using the ‘exact’ method described in the Appendix, which incorporates the effects of nonspecific binding site overlap on N binding. Under the conditions used in the experiments of Figure 2, N protein binds to the boxB RNA site and also to nonspecific RNA and DNA binding sites, resulting in the involvement of a number of different protein binding configurations in these transcription systems. We calculated the partition of N protein onto the available boxB, RNA, and DNA sites using measured binding affinities of N for boxB, nonspecific RNA, and nonspecific DNA, and estimations of the number of potential nonspecific sites with the elongation complex located at the terminator. Complexes of N and DNA are inactive for antitermination,37 presumably because the DNA is too stiff to allow looping. Therefore we considered only elongation complexes with one or more molecules of N protein bound to the RNA transcript to be capable of binding to the RNAP of the elongation complex and bring about antitermination. The fraction of transcripts bound by N at boxB and/or nonspecific RNA sites was then determined using the ‘exact’ method described in the Appendix.
Having determined the distribution of N protein-RNA and N-boxB complexes on the RNA transcripts contained in our transcription reactions, we next calculated the looping-facilitated binding of tethered N protein to RNAP bound at the terminator. We determined the local concentration lc of each protein-RNA complex tethered at a distance of nt nucleotide residues from the RNAP of the elongation complex using the relation:31
| (1) |
The looping-facilitated binding of the N-boxB and N-RNA complexes on the transcript to RNAP poised at termination positions was then calculated as:
| (2) |
where Kd N-RNAP is the equilibrium constant for N binding to RNAP in the tethered looping equilibrium, and lck(1),lck(2)…lck(i) are the local concentration of the 1st, 2nd, …, ith bound N protein on the transcript (see Appendix for details of derivations and treatment of transcripts bound with regulatory proteins at multiple sites).
Figure 8 compares the predicted fraction of RNAP molecules bound to N protein with experimental measurements of terminator read-through on the five templates shown in Figure 1 at 50 and 100 nM concentrations of N. To permit comparison of the fractional occupancy of N protein on RNAP with experimentally measured terminator readthrough, the experimental data were normalized to total change in antitermination due to the presence of N protein (see Figure legend). Regression analysis was used (Figure 8) to estimate the binding constant for the interaction of transcript-bound N protein with RNAP within the elongation complex. For experiments performed at 50 nM and 100 nM N concentration, the best fit values using the ‘exact’ method (see Appendix) were Kd = 2.8 × 10−5 and Kd = 1.7 × 10−5 M, respectively. Regression of the combined data using the ‘exact’ method yielded a best-fit value of Kd = 2 × 10−5 M for the tethered interaction of N with RNAP. This value is in good agreement with previous estimates (Kd = ~5 × 10−6 M) obtained from experimental studies of transcript elongation kinetics.39
Figure 8. Comparison of experimental and predicted values for terminator readthrough produced by the addition of N protein as a function of RNA length.
Experimental measurements from Figure 2 at input concentrations of 50 (black circles) and 100 (white circles) nM N protein were normalized to the total change in terminator read-through due to the addition of N protein; thus the maximum terminator readthrough at 400 nM N (0.77) in Figure 2 corresponds to a Fraction Terminator Readthrough (right y-axis) of 1, and the terminator readthrough in the absence of N (0.11) in Figure 2 corresponds to a Fraction Terminator Readthrough of 0. Predictions of the fraction of elongation complexes that are bound in cis by transcript-tethered N protein (Fraction RNAP bound by N, left y-axis) at 50 (thin solid lines) and 100 (thick solid lines) nM N protein as a function of increasing RNA loop length were generated using the ‘exact’ model described in the Appendix and a tethered equilibrium constant (Kd) of 2 × 10−5 M. The Kd for the binding of N to boxB was 0.5 nM at the salt concentrations used in the transcription reactions; apparent Kd values for nonspecific binding of N to ssRNA and dsDNA were 100 nM and 135 nM, respectively. Binding constants were measured and nucleic acid concentrations were estimated as described in Materials and Methods.
The fit of the model to the data for transcript lengths of 100–800 bps (from boxB to the terminator) appears to describe adequately the binding of N to RNAP in the minimal antitermination system. We note that at the longest distance (terminator 984 bps from boxB) the experimentally measured terminator read-through values are somewhat smaller than predicted by the model for both concentrations of N protein. This discrepancy likely stems from the tendency of the long RNA transcripts to aggregate in the wells of acrylamide gels used to measure fractional readthrough (see Materials and Methods). Alternatively the experimental RNA may be stiffer than the RNA used to generate the model. To test the latter possibility, Kuhn length values and equilibrium association constants for N-RNAP binding were subjected to simultaneous regression. This procedure failed to improve the fit of the model to the experimental data. Nevertheless, the ability of the model to predict the majority of antitermination events more accurately than models that ignore the effects of nonspecific binding (see Discussion) emphasizes the important role of such binding in the control of regulatory nucleic acid looping processes.
DISCUSSION
In this work we provide evidence that the N protein of phage λ, when bound to the boxB hairpin of the nascent RNA transcript, uses RNA looping to bind to the polymerase of the E. coli elongation complex during active transcription. We show that in the minimal antitermination system (containing only the binary N-boxB complex) N interacts transiently with the RNAP and ‘range control’ of the antitermination effect is exerted through a RNA looping equilibrium. In contrast, in the presence of the full complement of Nus factors, N binding to the RNAP of the elongation complex via cis looping becomes persistent on the transcription time scale, resulting in kinetic (dissociation-dependent) regulation of the ‘range’ of the antitermination effect. The observation that looping involves a persistent complex in this simple transcriptional regulatory system implies that more complicated systems may rely on persistent complexes as well, especially in vivo where macromolecular crowding may force looping-facilitated interactions to operate under kinetic control.40 These results show that nucleic acid looping models for transcriptional regulation must include the effects of tethered protein association and dissociation rates on target occupancy.
Previous experimental studies of range effects in N-dependent antitermination included NusA protein which, in addition to stabilizing the cis (via RNA looping) N binding pathway could also -- in principle -- provide a second (trans) pathway for the direct association of N with RNAP as a NusA-N dimer. To test this latter possibility we used a minimal in vitro assay containing only N, RNAP and variable length boxB-encoding DNA templates, and showed that decreases in N activity as the transcript is extended are manifested in the presence of N protein alone. The elongation complexes generated by these reactions vary only in the lengths of the RNA transcript and DNA template, and we have shown that when both the RNA transcript and the DNA template are increased by the same length, as they are in these experiments, the binding of N protein in trans to nonspecific sites on the RNA and DNA remains proportionate and does not significantly alter antitermination levels ((37); see also the Appendix). We conclude that antitermination range in this minimal antitermination system is controlled by RNA-length-dependent interactions of N protein with the RNAP of the elongation complex.
We then asked directly, using an RNA-cleavage methodology, whether N-dependent ‘action at a distance’ occurs via a cis RNA looping mechanism during active transcription. In the minimal N-boxB-RNAP reaction, cleavage of the intervening transcript strongly reduced antitermination. This observation, taken alone, could have a number of mechanistic interpretations. However, based on the results that we have presented here and in earlier studies by our lab and others,17,26 it seems most likely that the N protein (together with the transcript segment to which it is bound) dissociates from the RNAP shortly after transcript cleavage and is unable to rebind directly (now in trans) in the absence of the local concentration increase provided by an intact RNA tether. The cis RNA looping interpretation is also consistent with the lack of results supporting a non-tethering role for boxB and the intervening RNA in stabilizing antitermination complexes. Assuming that the decrease in activity observed with antitermination complexes after transcript cleavage is indeed due to dissociation of the N-boxB complex from the RNAP, we can assert that this dissociation process is fast compared to transcription (the transcription reactions in our cleavage assays go to completion in ~15 sec). Fast dissociation of N-boxB complexes from RNAP is consistent with the weak binding of N to RNAP calculated from local concentration models and functional assays, and explains our inability to measure directly the binding of N to RNAP in trans (37; S.E.W., Marc Van Gilst and C.R.C., unpublished results).
We show here that complexes of N and boxB are rendered inactive within ~15 sec following transcript cleavage. During this time the elongation complex will have transcribed over ~500 nts of template DNA, which represents a small fraction of the nut-to-terminator distances along the genome that manifest diminished N activity in our range assays. If N dissociates from the RNAP during the transcription process, substantial reassociation must occur in order to explain the higher antitermination levels observed in the absence of transcript cleavage. We thus conclude that N and boxB exist in a RNA looping equilibrium with RNAP on the time scale of transcription, and that this equilibrium is modulated by increasing the length of the RNA tether linking boxB and the RNAP located at the terminator. Such an equilibrium has been proposed for N-dependent antitermination,3,18,22 and appears to regulate the behavior of other ssRNA loop-dependent control systems as well.8,44
The present results can also be used to estimate the association constant for the binding of tethered N protein to RNAP located at terminator positions. To this end the antitermination assay results shown in Figure 2 were used to calculate the in trans binding constant of N protein to the elongation complex.22,37 We then used theoretical models to estimate the local concentration of N protein near RNAP when the elongation complex is poised at the terminator. Combining these local concentration estimates with an expression for the equilibrium association constant for N protein binding to RNAP produced an expression for the fraction of RNAP bound by N protein in our antitermination assays, and allowed us to employ regression analysis to estimate a value of Kd = 2 × 10−5 M for the N-RNAP interaction. This value is consistent with previous estimates of this parameter.39 The quantitative approximations used in the model to generate the local concentration values of N are discussed in the Appendix.
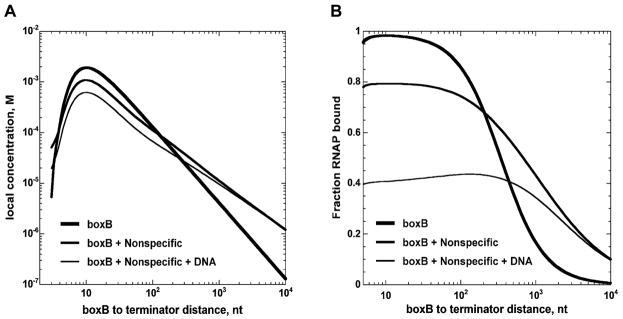
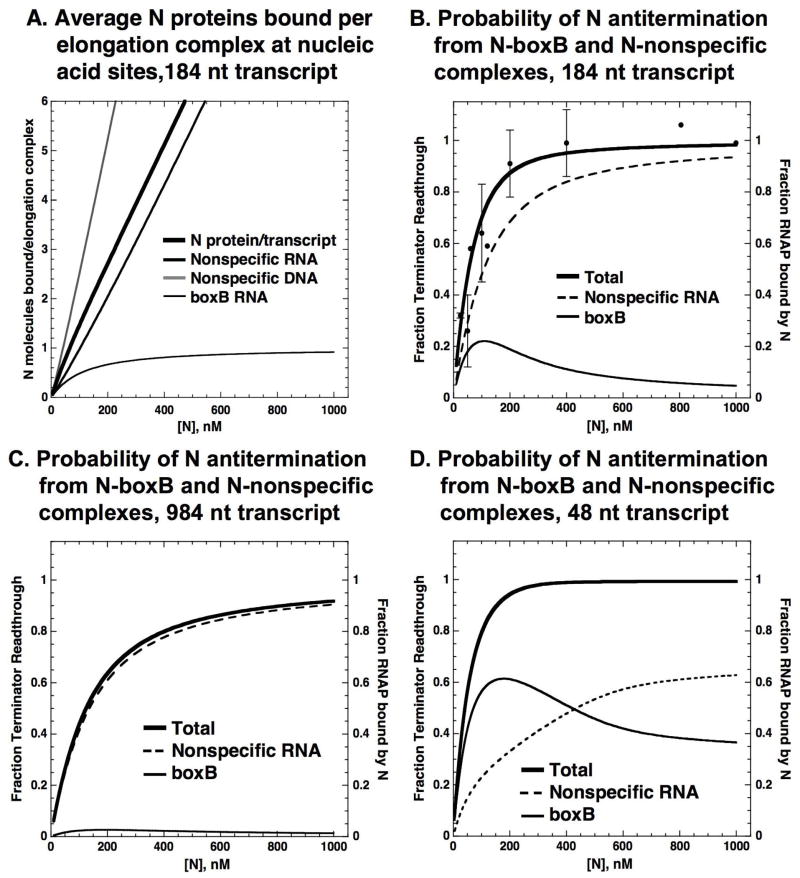
Because N protein is active in the absence of boxB (i.e., when bound to nonspecific RNA binding sites on the transcript26,37) we extended the looping model to include the effects of nonspecific binding of the regulatory protein, which occurs for N at significant levels despite an ~1000-fold affinity difference for N binding to boxB and to nonspecific RNA sites. Nonspecific binding is likely to occur in other looping contexts as well (especially in in vitro experimental systems that employ ‘naked’ nucleic acid lattices), because the large number of overlapping45 binding sites on lattices used in looping experiments compensates for the lower nonspecific binding affinities.‡ Figure 9A shows the calculated local concentrations of boxB-bound N protein in the presence and absence of nonspecific binding sites on the nucleic acid components of the elongation complex and illustrates how the interplay of N binding to nonspecific sites on the RNA transcript in trans and in cis can alter looping specificity.
Figure 9. Effect of nonspecific binding on the local concentration of N, on N binding to RNAP and on terminator read-through as a function of transcript length. A. Local concentrations of N protein in the presence and absence of nonspecific interactions between N, RNA and DNA.
Thick line: predicted local concentration of N (bound at boxB) at RNAP (bound at the terminator) as a function of distance between boxB and the terminator in the absence of competition from nonspecific sites. At a total N protein concentration of 100 nM, more that 99% of the boxB sites are bound. Medium line: average local concentration when 100 nM N protein is partitioned between boxB and nonspecific nucleic acid binding sites. Thin line: average local concentration when 100 nM N protein is partitioned between boxB and the nonspecific nucleic acid binding sites when the transcription template has an additional 800 bp dsDNA adjacent to the transcribed region. Complexes of N with nonspecific RNA possess antitermination activity, and increase the fraction of RNAP bound by N; complexes of N with DNA are inactive for antitermination (see text), and act as a “sink” for free N protein B. Fraction of RNAP bound by ‘cis-looped’ N in the presence and absence of nonspecific interactions. Thick line: predicted fraction of RNAP bound to ‘cis-looped’ N-boxB complexes in the absence of nonspecific binding to RNA and DNA. Medium line: fraction of RNAP bound to cis-looped N protein tethered at boxB or at a nonspecific binding site. Thin line: fraction of RNAP bound to cis-looped N protein tethered at boxB or at a nonspecific site when the transcription template contains an additional 800 bp of dsDNA adjacent to the transcribed region. The simulations were performed using the ‘exact’ model described in the Appendix with a total N protein concentration of 100 nM and the same Kd values for N binding to RNAP and nucleic acid as in Figure 8. Unless otherwise noted, the templates used in the simulations had the same number of DNA base pairs outside the transcribed region as the experimental templates depicted in Figure 1 and described in the Materials and Methods.
When transcripts are short, nonspecific binding to the nascent transcript can be ignored and the local concentration of N-boxB complexes at the elongation complex will be high. However, when the total effect of nonspecific N binding is taken into account significant amounts of N protein bind to the DNA of the elongation complex, resulting in less binding of N to the RNA transcript and a lower effective local concentration of N protein at the elongation complex. When transcripts are long, both specific and nonspecific binding to the RNA regulate the local concentration of N, but nonspecific N binding will have the greater effect because nonspecific binding of N to the RNA chain can occur closer to the RNAP than specific binding to boxB. The presence of additional DNA adjacent to the transcription unit (as is the case in vivo) further stimulates the decrease in local concentration at short distances. The effect of these changes in effective local concentration on the binding of N to RNAP are depicted in Figure 9B. In the context of transcription, the net effect of nonspecific binding in our minimal N-dependent antitermination experiments is to decrease specificity at both ends of the range of N function, suggesting that transcriptional specificity relies as much on the suppression of nonspecific binding as on the enhancement of specific binding. These ideas are outlined schematically in Figure 10A.
Figure 10. Model for control of antitermination by the tethered interaction of N protein with elongation complexes. A. The minimal in vitro antitermination complex.
N protein binds to the transcript-encoded boxB sequence and loops to contact RNAP; N also binds to nonspecific sites on the transcript. Binding of tethered N-boxB complexes to RNAP equilibrates during transcription, with fast on- and off-rates (arrows) that permit N to sample the changing local concentration provided by the lengthening RNA. The binding of N to RNAP and antitermination activity are controlled by the transcript length-dependent change in local N concentration. B. The 'physiological' antitermination complex consists of N, RNAP, boxB, and the NusA, B, E and G proteins. N binds to boxB and the high local concentration of N provided by the short RNA transcript facilitates initial binding of the N-boxB to the transcription complex. The interaction of the N-boxB complex with RNAP and Nus factors is persistent relative to the time required for transcription between boxB and the terminator. The binding of N to RNAP and the antitermination activity are therefore not sensitive to transcript length, but depend on the slow (relative to transcription) rate of dissociation of Nus-factor-stabilized N from the elongation complex.
In the more general context of nucleic acid looping-dependent processes, the effects of nonspecific binding will depend on the activity parameter used to measure end-to-end looping contacts. For systems resembling N-dependent antitermination, where nonspecific binding of N to RNA (but not to DNA) produces activity, function will be increased for large RNA loops. For systems where nonspecific binding does not produce activity, such binding will decrease the free protein available for specific binding and thus reduce activity by acting as a ‘sink’ for the regulatory protein. In both kinds of systems high levels of nonspecific binding may also change the stiffness and/or the conformational freedom of the nucleic acid loop itself, resulting in either increased or decreased activity.
While the presence of a continous tether during transcription is required for N activity in minimal systems, it does not appear to be required (once the N-RNAP binding interaction has been initiated by cis RNA looping) for range-control of N-dependent antitermination in the more physiologically relevant system that includes the E. coli Nus factors. Reactions in which the transcript was cleaved by RNAse H in the presence of N and NusA, B, E, and G show the same amount of terminator readthrough as do reactions run in the presence of these same Nus factors in which the transcript was intact (compare lanes 9 and 11, Figure 7). Similar results were obtained when only NusA was added to the transcription reactions. Taken together, the results show a pattern of increased read-through in the presence of Nus factors, consistent with existing reports of kinetically-stabilized antitermination complexes.18,30,46
These observations are supported by our measurements of antitermination range in the presence of Nus factors. Inclusion of NusA increases antitermination at low N concentrations, and modestly increases the range of antitermination. Inclusion of NusA, NusB, NusE, and NusG in the antitermination system extends the range of antitermination activity to the point where no activity decrease is observed at RNA lengths up to 1000 nts. This finding is consistent with the participation of these Nus factors in the formation of stable antitermination complexes with N and RNAP in isolation,18,30 increasing both the apparent activity of N on the read-through seen at multiple terminators24,25 and increasing the antitermination range24 (see Figure 10B).
A persistent complex of this sort may increase the specificity of N function by minimizing the effects of nonspecific binding and nuclease activity on antitermination activity, and explains the near-complete antitermination defect of mutants that lack boxB. We have used binding measurements and a simple mass balance model to estimate the binding of N protein in trans to the nucleic acid sites (boxB, nonspecific RNA, and nonspecific DNA) contained in elongation complexes produced by in vitro transcription of the 184 nt template pRB2 (Figure 11). The large number of overlapping nonspecific binding sites present on the DNA template and RNA transcript of these systems results in the binding of multiple N proteins to nonspecific nucleic acid binding sites, while the boxB RNA site can bind only one molecule of N (Figure 11A). N protein bound to nonspecific DNA is inactive in inducing antitermination and thus acts as a ‘sink’ for free N protein. The remaining molecules of N protein, bound to boxB and nonspecific sites on the RNA transcript, compete with one another for RNAP binding. Figure 11B shows that the outcome of such a competition is that the fraction of RNAP molecules bound by N-boxB complexesis small compared to the fraction of the RNAP molecules bound by N protein located at nonspecific RNA sites. Furthermore this nonspecific N binding effect increases as the transcripts increase in length (Figure 11C). Only when the RNA transcript is very short, as it is immediately following transcription of nutL, does the proportion of RNAP molecules bound by N-boxB complexes exceed that bound by N-nonspecific RNA complexes (Figure 11D).
Figure 11. Nonspecific binding of N protein in trans to RNA and DNA controls the fraction of N molecules specifically bound at the boxB RNA site available for RNA looping-facilitated binding of N to RNAP. Effect of nonspecific binding and transcript length on the antitermination activity of N protein bound at the boxB RNA site. A. Number of N molecules bound to the nucleic acid framework of the elongation complex as a function of N concentration.
Predicted average number of N molecules bound to the RNA and DNA binding sites of elongation complexes produced by in vitro transcription of the 184 nt template pRB2 with RNAP at the terminator at a total input N concentration of 100 nM. Thick line: total N molecules bound to transcript RNA. Thin line: N molecules bound to boxB RNA. Grey line: N molecules bound to nonspecific DNA. Medium line: N molecules bound to nonspecific RNA sites. B. Probability of N-RNA and N-boxB complexes binding to RNAP and inducing antitermination. Probability that individual N-RNA and N-boxB complexes from Figure 11A will bind RNAP and induce antitermination. DNA acts as a ‘sink’ for N protein and N molecules bound nonspecifically to DNA do not compete for RNAP binding. Thick line: total fraction of RNAP molecules bound by N-boxB and N-RNA complexes. Dashed line: fraction of RNAP bound by N-RNA complexes. Thin line: fraction of RNAP bound by N-boxB complexes. Circles: experimental fraction of elongation complexes reading through the terminator. C. Probability of N-RNA and N-boxB complexes binding to RNAP on a long transcript containing a single boxB binding site. Predicted N-RNAP binding on the 984 nt pRB800 transcript. Thick line: total fraction RNAP bound by N. Dashed line: fraction RNAP bound by N-RNA complexes. Thin line: fraction RNAP bound by N-boxB complexes. D. Probability of N-RNA and N-boxB complexes binding to RNAP on a short transcript containing a single boxB binding site. Predicted N-RNAP binding on a hypothetical transcript containing 48 nt between boxB and terminator. The template used in this simulation has the same number of DNA bp outside the transcribed region as the experimental templates. Thick line: total fraction RNAP bound by N. Dashed line: fraction RNAP bound by N-RNA complexes. Thin line: fraction RNAP bound by N-boxB complexes. We used the ‘exact’ predictive model (see Appendix) to calculate the partitioning of N protein onto boxB, RNA, and DNA and the fraction of elongation complexes that read through the terminator due to a looping-facilitated interaction between RNAP and N protein bound to boxB and/or nonspecific sites on the transcript. The model assumes that the N-RNAP interaction equilibrates on the time scale of elongation, that the affinity of boxB- and RNA-bound N proteins to RNAP is equal, and that interaction between RNAP and a molecule of N protein bound to boxB or a nonspecific site on the transcript results in antitermination. The same Kd values for N binding to RNAP, boxB, nonspecific ssRNA sites and nonspecific dsDNA sites were used as in Figure 8.
There are few nonspecific RNA sites available when the boxB RNA sequence is first transcribed, and thus initial N binding is primarily at boxB. Subsequent binding of Nus factors to the boxA element of the RNA and the formation of a stable antitermination complex renders the elongation complex insensitive to competition from N molecules bound at nonspecific sites elsewhere on the growing RNA transcript. Stable complexes are also expected to be insensitive to cleavage of the pL mRNA by RNAse III (which occurs concurrently with transcription47–49), as well as to other events (such as concurrent translation) that may alter the looping properties of the RNA transcript.
These considerations lead us to propose that physiological antitermination involves a series of reaction steps, some that equilibrate during transcription and some that do not. Thus N protein, shortly after it is synthesized, will first bind tightly to the boxB RNA site. The N-boxB complex then binds weakly to the RNAP of the elongation complex in a tethered cis-RNA looping equilibrium. The fractional occupancy of RNAP by N-boxB will then depend on the rates of association and dissociation of the antitermination complex from the RNAP of the elongation complex relative to the rate of transcription. For reactions containing only N-boxB, rapid equilibration affords the N-boxB complex time to explore changes in the local concentration provided by the RNA tether. In contrast, reactions between RNAP and N that involve the full complement of Nus cofactors assembled around N may be relatively slow, and thus initial binding to RNAP of the entire tethered and N-dependent antitermination complex may be slow as well. However, since the interaction of RNAP with the antitermination complex is completely (or at least primarily) through N, the initial binding to the RNAP of the elongation complex will continue to depend on the length of the RNA tether in the same way as does the binding of the N-boxB complex alone.
On the other hand, after binding of the full antitermination complex, the rate of dissociation from the RNAP of the Nus factor-stabilized N-boxB complex will also be slow. Therefore, after transcription has proceeded and the RNA tether is long, a single dissociation event is likely to result in local concentrations of tethered N that are too low to facilitate productive interactions with the elongation complex and antitermination activity will appear to switch to kinetic (dissociation rate) control. Under slow dissociation conditions the most important regulatory parameter then becomes the time required for the elongation complex to transcribe from the nut site to the terminator, which will depend on the length of the template DNA, the presence of pause sites, etc. As a consequence the form of the regulation of this cis-looping system, and possibly the regulation of other systems as well, is likely to depend on the rates of a series of inter-dependent association and dissociation reactions relative to the rate of the regulated process itself.
Numerous studies have correlated the equilibrium binding of N protein in trans to boxB RNA and boxB mutants with antitermination activity, but the effects of RNA looping on this process, the role of N binding in trans to specific and nonspecific sites contained in elongation complexes, and the effect of accessory factors on looping dynamics, all of which regulate N activity, have not been previously examined. Our present work suggests that in vivo antitermination defects of phages carrying mutations in N, boxB, and accessory factors might be divided into two phenotypic classes. One class of such mutations may disrupt assembly of the antitermination complexes, preventing antitermination at terminators located close to boxB. The second proposed class of mutations would allow assembly but also increase the ‘cis’ dissociation rate of N protein from RNAP, thus shortening the observed antitermination range. Mutations that likely lead to assembly defects include NusA124 and boxB G1A (nutL44),46 since both of these mutations decrease antitermination at terminators located close to boxB. In addition, while it is generally true that N- and boxB- mutants that bind well also promote antitermination, there exist mutants that bind but do not antiterminate in vivo or in vitro,20 and at least one mutant that binds poorly, but antiterminates reasonably well in vivo42. These phenotypes may reflect differences in the kinetics of assembly of transcription complexes; alternatively they may be due to differences in the RNA chain dynamics of the cis-looping process that change the occupancy of the RNAP of the elongation complex by N at terminators and thus alter the observed termination efficiency. Study of these mutants in the context of the quantitative antitermination model developed here and in (37) (see also (14)) may further illuminate the control points of this looping-dependent regulatory system.
MATERIALS AND METHODS
Protein and Oligonucleotide Preparation
The N protein of phage lambda,22 E. coli RNA polymerase,28 NusA ((38), with modifications (28)), NusB and E,50 NusG,51 and EcoRI G111A,52 were purified as described. The ssDNA used in the RNAse H experiments (5′-CACACCCCAAAGGCCT-3′) was hybridized to a complementary RNA site generated by transcription of a template sequence located 15 nt downstream from boxB in pRB2 and related plasmids. Concentrations of DNA, RNA, and proteins were determined spectrophotometrically using calculated molar extinction coefficients for oligonucleotides and proteins53 and bulk extinction coefficients (50 μg/O.D.) for transcription templates and calf thymus DNA (Sigma).
Plasmids and Transcription Templates
Plasmid pRB2 was constructed as previously described.39 Plasmid pRB250 was constructed by insertion of an oligonucleotide encoding λ bps16236-16286 and complementary ends for the PstI-EcoRI DNA between nutL and the tR′ terminator of pRB2. Plasmids pRB200, pRB400, pRB800, and pRB1600 were constructed by inserting λ DNA (16236–16436, 16236–16636, and 16236–17036, respectively) at the same site in pRB2. All templates for in vitro transcription were prepared from parent plasmids by PCR with Vent DNA polymerase (New England Biolabs, Beverly, MA); each template includes 139 bps dsDNA upstream of the transcription start site. PCR fragments containing the pL-tR′ transcription unit were isolated by electrophoresis on 1% agarose, purified using Quiagen (Valencia, CA) gel purification spin columns, precipitated with ethanol, and quantitated specrophotometrically with ε260 = 6.6 × 103 M−1 cm−1 nt−1.
In vitro Transcription Reactions
Transcription reactions (10 μl) containing 25nM template dsDNA (Figure 1A) and 25 nM E. coli RNAP in transcription buffer [20 mM Tris-Cl (pH 7.6), 0.1 mM EDTA, 5 mM MgOAc, 50 mM KOAc, 5 mM DTT, and 5% (v/v) glycerol] were incubated at 30°C with 150 μM ApU, 10 μM ATP, CTP, GTP, and 250 nM α-32P ATP to form elongation complexes stalled at the first U position on the transcription templates. Complexes were chased with 1 mM NTPs, 10 μg/ml of rifampicin, and proteins (120 nM NusA, 250 nM NusB, E, and G, and indicated amounts of N protein) for 7 msec per nt of transcript RNA between the transcription start site and the terminator position. Reactions were quenched with 0.25% (w/v) SDS/10mM EDTA and digested with 20 μg/ml of proteinase K for 45 min at 45°C, diluted with an equal volume of formamide loading buffer, heated to 94°C for three minutes, and loaded onto a 7 M urea/7% (w/v) polyacrylamide gel. Fraction readthrough was defined as:
where RNAterm and RNArunoff are amounts of radioactivity in terminated and full-length bands (after subtraction of background determined by quantification of the area between bands) as measured using a Model 60 Phosphimager and accompanying software (Molecular Dynamics, Sunnyvale CA). Where indicated by error bars, activity measurements represent the average of three independent measurements; otherwise they represent a single measurement. Transcription experiments often produced bands that stuck in the wells of acrylamide gels (Figure 1B). To determine whether these bands contained a different proportion of terminated and runoff RNAs than bands that ran into the gels, reactions were processed with and without extended proteinase K digestion that resulted in > 97% of RNAs entering the gel. RNAs liberated from the wells in this way showed a greater proportion of runoff than terminated products, with reactions showing equal amounts of radioactivity in the wells and in gel bands manifesting a maximum increase of 20% in fractional terminator readthrough. Accordingly all reactions were performed with proteinase K digestion of the samples, and we take the 20% value as an upper limit for this potential measurement error.
To facilitate comparison of the predicted fraction of RNAP bound by N with experimental measurements of the fraction of RNAP molecules that read through transcriptional terminators (Figures 8 and 11), experimental data from Figure 2 were normalized to the total terminator readthrough due to addition of N protein. On all transcription templates, the maximum fractional terminator readthrough due to addition of N protein was taken to be 0.77, corresponding to the fractional readthrough of template pRB2 at saturating (1 μM) levels of N protein. Terminator readthough in the absence of N protein varied for each template (see Figure 2); for template pRB2 this value was 0.11; thus the experimentally measured value for fraction RT on template pRB2 at 100 nM N concentration (0.53) corresponds to a normalized value of (0.53–0.11)/(0.77–0.11) = 0.64.
EcoRI Gln111 ‘Roadblock’ Transcript Cleavage Reactions
Elongation complexes stalled at the EcoRI site between nut and termination sequences were prepared by incubating EcoRI Gln111A protein (200 nM) and DNA template pRB2 (100 nM) for 5 min at 30°C in NB buffer [50 mM NaCl, 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2]. RNAP (100 nM) was added and the reaction was incubated for an additional 5 min. Transcripts were then initiated and extended to position +11 by addition of 150 μM ApU, 5 μM each of ATP, CTP, GTP and 150 μM α-32P ATP (3000Ci/mmol). After incubation for 3–5 min the transcripts were further extended to the EcoRI position with 500 μM each of ATP, CTP, GTP, UTP and 10 μg/mL rifampicin. The remaining NTPs were removed by twice transferring reactions to Bio-spin 30 (Bio-Rad, Hercules, CA) columns pre-equilibrated in NB buffer plus 10 μg/mL of rifampicin, and desalting as per Bio-Rad directions.
EcoRI Gln111A protein was removed from the template by addition of 250 mM KCl for 20 min at 40°C in the presence of a competitor dsDNA oligonucleotide containing the EcoRI site (2 μM final concentration), together with 300 nM α-32P ATP to 3′-end-label the transcript. Salt and complexes of DNA oligonucleotide with EcoRIGln111A were removed by two passages through a microcon-50 spin column, followed by two passages through a microcon-100 spin column (Millipore, Bedford, MA); between each spin step the complex-containing solution was diluted 10-fold with transcription buffer plus rifampicin. Following the last desalting step, an oligonucleotide (1 μM) complimentary to a RNA transcript sequence located between the nut site and the terminator (see Figure 1A) was added and allowed to anneal for 20 min at 45°C. Indicated amounts of proteins were then added to form antitermination complexes and the complexes were supplemented with RNAse H (0.25 U), 250 μM NTPs, 300 nM α-32P ATP, and rifampicin (10μg/ml) to simultaneously cleave and extend the transcript to (and through) the termination sequences. Time courses of activity were performed by adding RNAse H at the indicated times prior to NTP addition. Where stated, reactions contained 250 nM N protein, 120 nM NusA, and 250 nM NusA, B, E, and G. Reactions in the presence of accessory factors (Figure 7) were carried out as above, but with 150 nM α-32P ATP added prior to the RNAse H oligo annealing step to label the 3′ end of the blocked transcript.
Measurement of N-boxB, N-RNA, N-DNA binding constants and parameters used for prediction of N looping and N-RNAP binding
Equilibrium binding constants and salt dependencies for the nonspecific binding of N to RNA and DNA oligonucleotides were determined using fluorescence quenching and anisotropy and analyzed as previously described using the McGhee-von Hippel model for ligands binding to a homogenous lattice (N binding shows no cooperativity).37 To account for the effects of the Mg++ present in the transcription reactions on the binding of N protein to boxB RNA, the salt dependence of N protein binding to boxB RNA was measured as previously described,37 but here in transcription buffer containing 20 mM Tris-Cl (pH 7.6), 0.1 mM EDTA, 5 mM Mg(OAc)2 and 190, 210, 230 and 250 mM KOAc. Measured N-boxB association constants as a function of salt concentration were plotted as log Ka versus −log[K+], and linear fits and errors were estimated using least-squares analysis and Kaleidagraph (Synergy Software, Reading, PA), yielding (dlogKa/dlog[K+]) = 4.3 (± 0.1) and logKa = 3.7 (± 0.1) at 1 M K+. Calculations for Figures 8, 9, and 11 were performed with the ‘exact’ model (see Appendix) with 25 nM elongation complexes at the termination position.
Acknowledgments
This work was supported in part by NIH Research Grants GM-15792 and GM-29158 to P.H.v.H., and a Lucille P. Markey Charitable Trust grant to the Institute of Molecular Biology at the University of Oregon. C.R.C. and J.P.G. were trainees on NIH Institutional Predoctoral Training Grant GM-07759. P.H.v.H. is an American Cancer Society Research Professor of Chemistry. We are grateful to our laboratory colleagues for many useful discussions of this work.
APPENDIX A general method for calculating the activity of transcription regulators that bind specifically or nonspecifically (and functionally or nonfunctionally) to overlapping DNA and RNA sites
Authors: Jim P. Goodarzi, Clarke R. Conant and Peter H. Von Hippel
In this Appendix we calculate the antitermination efficiency of a transcription complex as the probability that the RNAP of a transcription complex poised at a terminator is bound by N protein tethered to the transcript. Our approach is both general and exact, in order to permit others to use these procedures to calculate the binding distribution and thus the regulatory activity of any protein that can bind to both specific and nonspecific binding sites of the nucleic acid components of the cell in either a functional or a non-functional manner. We also compare this complete approach to estimates that are sometimes more easily made by more inexact methods involving various assumptions, because our ‘exact’ approach can also be used to estimate which portions of ‘parameter space’ are most likely to introduce significant errors into the regulatory calculations.
In our analysis of the N-dependent antitermination system we calculate the equilibrium partitioning of N protein between boxB and nonspecific binding sites on the RNA and DNA, as well as the probability that the RNAP of the transcription complex is contacted by the tethered N protein. Since molecules of N protein bound to the DNA or free in solution are functionally inactive, either because they cannot reach the RNAP or because their concentrations are substantially below the dissociation constant of the N-RNAP interaction, we will only consider cis-RNA-looping-facilitated interactions between N and RNAP in calculating antitermination efficiency. If antitermination function does indeed result from tethered N-RNAP interaction when the elongation complex is poised at the terminator, and if this interaction equilibrates on the time scale of elongation, then the probability of antitermination mediated by cis-RNA-looping can be written as
| (A1) |
where fbound[i] is the fraction of RNA transcripts that have i molecules of N protein bound, fN−RNAP[i] is the fraction of complexes that have a N-RNAP contact (given that there are i bound N proteins), and imax is the maximum number of N proteins that can bind to the nascent transcript.
First we will calculate fbound[i], which was done previously for low N protein binding densities by treating every potential binding site on bare ssRNA and ds DNA lattices as an independent species.37 We now extend that model to include situations of higher protein binding density by using a statistical mechanical approach that has been previously used for complexes with more than one ligand binding site54, and to describe the probability of multiple ligand binding events on finite lattices.55
Consider a system of protein and nucleic acid chains where there is only one nonspecific nucleic acid binding site per protein and each bound protein covers n nucleic acid residues. Then, a bare nucleic acid chain of length M will consist of (M − n + 1) overlapping binding sites with equal affinity to the protein. We represent the concentration of a particular unique ligand/lattice configuration with i bound proteins as C*[i]. Then the intrinsic (nonspecific) binding constant of the reaction:
| (A2) |
is given by
| (A3) |
where Lfree is the concentration of free ligand. We note that C*[i + 1] in eqs. (A2) and (A3) is the concentration of a unique ligand/lattice configuration, which is exactly the same as the ligand/lattice configuration of C*[i] except that one additional ligand is bound to a previously free binding site. Using eq. (A3), we can write the concentration of some unique ligand/lattice configuration with i bound proteins as
| (A4) |
Eq. (A4) must be multiplied by the number of ways i proteins, each with a site size of n, can be arranged on a lattice of length M in order to obtain the total concentration of all ligand/lattice configurations with i bound proteins:
| (A5) |
In what follows we drop the star from C*[0] since there is only one ligand/lattice configuration with zero bound proteins (i.e., C[0] = C*[0]).
As described in the text (see also refs. 14 and 37), N protein binds noncooperatively to nonspecific ssRNA and dsDNA binding sites in the transcription complex, and the measured binding of N to ssRNA and dsDNA does not vary significantly with the sequence or composition of the target nucleic acid lattice. Therefore, eq. (A5) is valid for any uninterrupted section of the DNA template or RNA transcript.
RNAP asymmetrically bisects the DNA template when it is poised at the terminator, which results in two uninterrupted sections of dsDNA. Additionally, our analysis of binding experiments of N protein to DNA is consistent with N protein binding simultaneously on opposite sides of the dsDNA lattice37, resembling the situation for E. coli lac repressor, which has been shown to bind nonspecifically to both sides of dsDNA, resulting in a nonspecific binding site size one-half as large as the specific (operator binding) site size.56 Thus for present purposes each uninterrupted section of dsDNA will be treated as two separate sub-lattices. Then, eq. (A5) can be used to write the concentrations of ‘short’ and ‘long’ DNA sub-lattices with i nonspecifically bound N proteins as
| (A6a) |
| (A6b) |
where is the concentration of a short DNA sub-lattice with zero bound proteins, is the concentration of a long DNA sub-lattice with zero bound proteins, is the length of the short DNA sub-lattices, is the length of the long DNA sub-lattices, nDNA is the nonspecific site size of N protein on dsDNA, KDNA is the intrinsic association constant for the nonspecific interaction of N with dsDNA, and Nfree is the concentration of free N protein.
A similar equation can be written for N protein bound to RNA. However, in doing so the higher affinity of N to the boxB site must be taken into account:
| (A7) |
Here CRNA[0] is the concentration of transcripts with zero bound molecules of N protein, is the length of the RNA from the 5′-end of the transcript to a point 18 nts upstream of the 5′-end of the terminator (18 nts of RNA are contained within the RNAP complex and are therefore inaccessible to N37), is the number of nts from the 5′-end of the transcript to the 5′-end of the boxB site, is the number of nts from the 3′ end of the boxB site to a point 18 nts upstream of the 5′ end of the terminator, nRNA is the site size of N protein on RNA, KRNA is the intrinsic association constant for the nonspecific interaction of N with RNA, and KboxB is the association constant for the specific interaction of N with boxB. In eq. (A7), all ligand/lattice configurations with N protein bound nonspecifically to the boxB site are subtracted in the second term, while all ligand/lattice configurations with N protein bound specifically to the boxB site are added in the third term. Accordingly, the combinatorial factors in terms 2 and 3 of eq. (A7) give the number of ways that (i − 1) proteins can be arranged on the transcript, given that there is a protein bound at boxB. This maintains the proper weights of the ligand/lattice configurations with or without specific binding of N to boxB, given that the specific site size of N to boxB (~8 nts from RNase protection experiments20) is equal to nRNA = 11 nts37 and assuming that N cannot bind to the boxB site in a nonspecific manner.
Proceeding from eq. (A7), we can write the concentration of transcripts with i molecules of N protein bound nonspecifically and zero bound to boxB as
| (A8) |
and the concentration of transcripts with (i − 1) molecules of N protein bound nonspecifically and one bound to boxB as
| (A9) |
In order to solve eqs. (A6)–(A9), we require Nfree, which can be obtained from the mass balance equations for the system. The mass balance for N binding to nucleic acid components of the elongation complex is
| (A10) |
where Ntotal is the total concentration of N protein, imax is the maximum number of proteins that can bind to the transcript, jmax is the maximum number of proteins that can bind to the short DNA sub-lattice, and kmax is the maximum number of proteins that can bind to the long DNA sub-lattice. It is also true that
| (A11a) |
| (A11b) |
| (A11c) |
where RNAtotal and DNAtotal are the total concentrations of RNA and DNA lattices in the reaction. To obtain Nfree at any particular Ntotal, eqs. (A11) are solved, respectively, for CRNA[0], , and as a function of Nfree. These equations replace CRNA[0], , and in eq. (A10), and Nfree is adjusted in an iterative fashion until eq. (A10) gives the required value of Ntotal.
Then, the fraction of RNA transcripts that have i bound proteins is
| (A12) |
where CRNA[i] is given by eq. (A7). With eqs. (A8) and (A9), we can also express the fraction of RNA transcripts with i molecules of N protein bound nonspecifically and zero N bound to boxB as well as the fraction of RNA transcripts with (i − 1) molecules of N protein bound nonspecifically and one N bound to boxB:
| (A13a) |
| (A13b) |
We note here that our previous method37 of obtaining fbound[i], where every nucleic acid binding site is treated independently, is formally equivalent to setting
| (A14a) |
| (A14b) |
where kDNA is the number of potential nonspecific binding sites on both sides of a bare dsDNA lattice that is bisected by RNAP poised at the terminator and kRNA is the number of potential nonspecific binding sites on the bare transcript. The combinatorial terms in eqs. (A14) give the number of ways that i proteins can be arranged on a DNA and RNA lattices with kDNA and kRNA independent binding sites, respectively. Since the specific site on the transcript is also treated as an independent species, the concentration of boxB sites bound by N is given by
| (A15) |
where boxBtot is the total concentration of boxB sites. The corresponding mass balance equations for this simplified treatment are
| (A16a) |
| (A16b) |
| (A16c) |
Given the parameters obtained experimentally37, Figure A1 shows that the solution of eqs. (A14)–(A16) is valid as long as the occupancy of the ssRNA or dsDNA lattices does not approach saturation. For our purposes, model calculations using eqs. (A12) or (A13) will be termed ‘exact’, while model calculations using the solution of eqs. (A14)–(A16) will be termed ‘independent’.
Figure A1. Comparison of the ‘exact’ and the ‘independent’ models for N protein binding to nucleic acid components of transcription elongation complexes.
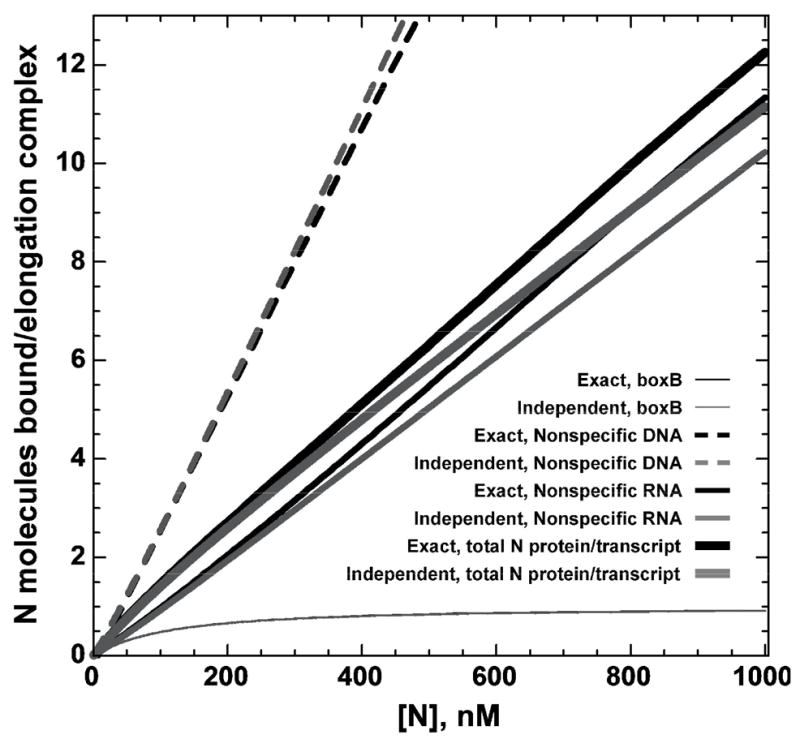
Calculations of the average number of N protein molecules bound to nucleic acid components of a transcription elongation complex as a function of increasing N concentration were made using the ‘exact’ method described in the Appendix (black lines), and the ‘independent’ model, which treats every potential binding site on the bare transcript and template as an independent species (grey lines)37. Thin lines: N protein binding at the boxB RNA site. Medium lines: N protein binding at nonspecific ssRNA sites present in the transcript. Dashed lines: N protein binding at nonspecific dsDNA sites present in the transcription template. Thick lines: N protein binding to the RNA transcript (boxB + nonspecific sites).
In order to determine fN − RNAP[i], we use the concept of local concentration to relate the probability of N-RNAP contact to the length of the RNA ‘tether’. Local concentration (lc) is defined by:
| (A17) |
where is the association equilibrium constant for the tethered reaction and is the association constant for the second order reaction. The local concentration of tethered N protein bound at either boxB or at a nonspecific site on the nascent RNA is given by31
| (A18) |
where nt is length of the RNA tether, the stiffness of the transcript RNA is approximated using empirically determined values for polyr(U) (Kuhn length = 4 nm57). We note that using the parameters for polyr(U) to define the flexibility of ‘real’ mRNA molecules, especially if they carry significant bound protein, may be inappropriate, and that a smaller flexibility parameter may apply. However, polyr(U) is the only RNA for which the Kuhn length has been measured under ‘theta solvent’ conditions, and since the RNA is not fully coated with N protein and the introduction of stacking or secondary structure is likely to effectively shorten the chain, this estimate may not be too unreasonable.
For a single N protein is bound to the transcript, eqs. (A17) and (A18) can be used to obtain the fraction of complexes with an N-RNAP interaction for a unique ligand/lattice configuration with one bound N protein:
| (A19) |
After rearranging, eq. (A19) becomes
| (A20) |
where Kd is the dissociation equilibrium constant of the untethered N-RNAP interaction. Similarly, if there are i molecules of N protein bound to the transcript and only one N protein can interact with RNAP, then the fraction of complexes with an N-RNAP interaction for a unique ligand/lattice configuration with i bound N proteins is
| (A21) |
where lck(x) is the local concentration of the xth bound N protein which has a tether length of (MRNA − k(x) − nRNA).
If the boxB site is free, then the concentration of elongation complexes with a unique ligand/lattice configuration of i tethered molecules of N protein and a contact between one of the tethered N proteins and RNAP is
| (A22) |
where CRNA[0](KRNA Nfree)i is the concentration of any unique ligand/lattice configuration with i molecules of N protein bound nonspecifically to the transcript. If the boxB site is bound, then the concentration of elongation complexes with a unique ligand/lattice configuration of i tethered molecules of N protein and a contact between one of the tethered N proteins and RNAP is
| (A23) |
where lcboxB is the local concentration of N protein bound to boxB and CRNA[0](KboxBNfree)(KRNA Nfree)i−1 is the concentration of any unique ligand/lattice configuration with (i − 1) molecules of N protein bound nonspecifically and one N molecule bound specifically to boxB.
The fraction of elongation complexes modified by one of the tethered N proteins, given that there are i proteins bound to the transcript, is obtained by summing eqs. (A22) and (A23) over all possible configurations of i proteins bound to the transcript and dividing CRNA[i], the total concentration of transcripts with i by proteins bound:
| (A24) |
Note that lc Mleft = lcboxB, and the Kronecker Delta δ(Mleft, k(x)) = 1 for any k(x) = Mleft and is equal to zero otherwise. Eq. (A24) preserves the proper weight of ligand/lattice configurations with specific binding of N protein to boxB.
Eqs. (A12) and (A24) can be inserted into eq. (A1) to obtain the probability of antitermination function mediated by cis-RNA-looping. However, the summations of eq. (A24) require large amounts of computation time as the number of bound proteins and the length of nucleic acid lattices are increased. We will therefore use an approximation for the local concentration of nonspecifically bound proteins. If there is no boxB site on the transcript (i.e. KboxB = KRNA), eq. (A24) reduces to
| (A25) |
We evaluate eq. (A25) for one ligand bound nonspecifically and then solve eq. (A26) for lc′, which is the ‘effective’ local concentration for each protein bound nonspecifically to the transcript:
| (A26) |
Then, eqs. (A27) give the approximate fraction of elongation complexes modified by a tethered N protein for RNA transcripts with i molecules of N protein bound nonspecifically and zero bound to boxB and RNA transcripts with (i − 1) molecules of N protein bound nonspecifically and one bound to boxB.
| (A27a) |
| (A27b) |
Using parameters corresponding to the experimental conditions described in the Materials and Methods and the local concentration given by eq. (A18), we have confirmed that the use of eqs. (A27) does not introduce significant error.
Finally, eqs. (A13) and (A27) can be used to determine the approximate probability that tethered N protein contacts RNAP and modifies the activity of the elongation complex at the terminator:
| (A28) |
Conceptually similar experiments, in which RNAse H digestion was targeted to the boxB sequence in the nascent transcript, have been described previously17 and showed that a decrease in antitermination accompanied cleavage. We note that the earlier experimental design was ambiguous in that the observed antitermination decrease could have reflected either the destruction of the boxB binding site or the loss of RNA looping due to transcript cleavage, or both.
In our transcription experiments the Kd for the interaction of N protein with boxB in 50 M monovalent salt is ~0.5 nM, while for N protein binding to nonspecific RNA sites this parameter is ~100 nM; at 0.1 M salt, these values are 1 nM and ~1000 nM, respectively.22 For an elongation reaction that has produced a 1000 nt RNA transcript there will be an ~55,000 nM concentration of overlapping nonspecific binding sites present on both RNA and DNA of the system, providing ample opportunity for competitive (with boxB) nonspecific binding.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bellomy GR, Record MT., Jr Stable DNA loops in vivo and in vitro: roles in gene regulation at a distance and in biophysical characterization of DNA. Prog Nucleic Acid Res Mol Biol. 1990;39:81–128. doi: 10.1016/s0079-6603(08)60624-8. [DOI] [PubMed] [Google Scholar]
- 2.Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 3.Rippe K, von Hippel PH, Langowski J. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem Sci. 1995;20:500–6. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 4.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 5.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14:R101–11. doi: 10.1093/hmg/ddi104. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 6.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–13. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 7.Schnos M, Zahn K, Blattner FR, Inman RB. DNA looping induced by bacteriophage lambda O protein: implications for formation of higher order structures at the lambda origin of replication. Virology. 1989;168:370–7. doi: 10.1016/0042-6822(89)90278-x. [DOI] [PubMed] [Google Scholar]
- 8.Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. Embo J. 1998;17:6747–56. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broach JR. Making the right choice--long-range chromosomal interactions in development. Cell. 2004;119:583–6. doi: 10.1016/j.cell.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 10.Moitoso de Vargas L, Kim S, Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989;244:1457–61. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–68. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Friedman DI. Lytic mode of lambda development. In: Hendix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor Laboratory; New York, NY: 1983. pp. 21–51. [Google Scholar]
- 13.Greenblatt J, Nodwell JR, Mason SW. Transcriptional antitermination. Nature. 1993;364:401–6. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- 14.Van Gilst MR, von Hippel PH. Quantitative dissection of a transcriptional control system: The N-dependent complex of phage 1ambda as a regulatory paradigm. In: Johnson Michael L, Ackers Gary K., editors. Methods in Enzymology, Energetics of Biological Macromolecules), Part C. Vol. 323. 2000. pp. 1–31. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg M, Court D, Shimatake H, Brady C, Wulff DL. The relationship between function and DNA sequence in an intercistronic regulatory region in phage lambda. Nature. 1978;272:414–23. doi: 10.1038/272414a0. [DOI] [PubMed] [Google Scholar]
- 16.Salstrom JS, Szybalski W. Coliphage lambdanutL-: a unique class of mutants defective in the site of gene N product utilization for antitermination of leftward transcription. J Mol Biol. 1978;124:195–221. doi: 10.1016/0022-2836(78)90156-0. [DOI] [PubMed] [Google Scholar]
- 17.Whalen WA, Das A. Action of an RNA site at a distance: role of the nut genetic signal in transcription antitermination by phage-lambda N gene product. New Biol. 1990;2:975–91. [PubMed] [Google Scholar]
- 18.Nodwell JR, Greenblatt J. The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 1991;5:2141–51. doi: 10.1101/gad.5.11.2141. [DOI] [PubMed] [Google Scholar]
- 19.Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992;174:6711–6. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chattopadhyay S, Garcia-Mena J, DeVito J, Wolska K, Das A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc Natl Acad Sci U S A. 1995;92:4061–5. doi: 10.1073/pnas.92.9.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nodwell JR, Greenblatt J. Recognition of boxA antiterminator RNA by the E. coli antitermination factors NusB and ribosomal protein S10. Cell. 1993;72:261–8. doi: 10.1016/0092-8674(93)90665-d. [DOI] [PubMed] [Google Scholar]
- 22.Van Gilst MR, Rees WA, Das A, von Hippel PH. Complexes of N antitermination protein of phage lambda with specific and nonspecific RNA target sites on the nascent transcript. Biochemistry. 1997;36:1514–24. doi: 10.1021/bi961920q. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman ME, Adhya S, Das A. Transcription antitermination by bacteriophage lambda N gene product. J Mol Biol. 1980;140:57–75. doi: 10.1016/0022-2836(80)90356-3. [DOI] [PubMed] [Google Scholar]
- 24.Mason SW, Li J, Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J Biol Chem. 1992;267:19418–26. [PubMed] [Google Scholar]
- 25.DeVito J, Das A. Control of transcription processivity in phage lambda: Nus factors strengthen the termination-resistant state of RNA polymerase induced by N antiterminator. Proc Natl Acad Sci U S A. 1994;91:8660–4. doi: 10.1073/pnas.91.18.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rees WA, Weitzel SE, Yager TD, Das A, von Hippel PH. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc Natl Acad Sci U S A. 1996;93:342–6. doi: 10.1073/pnas.93.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenblatt J, Li J. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J Mol Biol. 1981;147:11–23. doi: 10.1016/0022-2836(81)90076-0. [DOI] [PubMed] [Google Scholar]
- 28.Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991;220:307–24. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 29.Mason SW, Greenblatt J. Assembly of transcription elongation complexes containing the N protein of phage lambda and the Escherichia coli elongation factors NusA, NusB, NusG, and S10. Genes Dev. 1991;5:1504–12. doi: 10.1101/gad.5.8.1504. [DOI] [PubMed] [Google Scholar]
- 30.Mogridge J, Mah TF, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–45. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 31.Rippe K. Making contacts on a nucleic acid polymer. Trends Biochem Sci. 2001;26:733–40. doi: 10.1016/s0968-0004(01)01978-8. [DOI] [PubMed] [Google Scholar]
- 32.Kratky O, Porod G. Röntgenuntersuchung gelöster Fadenmoleküle. Rec Trav Chim Pays-Bas. 1949;68:1106–1123. [Google Scholar]
- 33.Jacobsen H, Stockmeyer WH. Intramolecular Reaction in Polycondensation. I. The Theory of Linear Systems. J Chem Phys. 1950;18:1600–1606. [Google Scholar]
- 34.Flory PJ. Statistical Mechanics of Chain Molecules. Wiley Interscience; New York, NY: 1969. [Google Scholar]
- 35.Shore D, Langowski J, Baldwin RL. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci U S A. 1981;78:4833–7. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ringrose L, Chabanis S, Angrand PO, Woodroofe C, Stewart AF. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. Embo J. 1999;18:6630–41. doi: 10.1093/emboj/18.23.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conant CR, Van Gilst MR, Weitzel SE, Rees WA, von Hippel PH. A quantitative description of the binding states and in vitro function of antitermination protein N of bacteriophage lambda. J Mol Biol. 2005;348:1039–57. doi: 10.1016/j.jmb.2005.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt MC, Chamberlin MJ. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987;195:809–18. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- 39.Rees WA, Weitzel SE, Das A, von Hippel PH. Regulation of the elongation-termination decision at intrinsic terminators by antitermination protein N of phage lambda. J Mol Biol. 1997;273:797–813. doi: 10.1006/jmbi.1997.1327. [DOI] [PubMed] [Google Scholar]
- 40.Marenduzzo D, Finan K, Cook PR. The depletion attraction: an underappreciated force driving cellular organization. J Cell Biol. 2006;175:681–686. doi: 10.1083/jcb.200609066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Gilst MR, von Hippel PH. Assembly of the N-dependent antitermination complex of phage lambda: NusA and RNA bind independently to different unfolded domains of the N protein. J Mol Biol. 1997;274:160–73. doi: 10.1006/jmbi.1997.1389. [DOI] [PubMed] [Google Scholar]
- 42.Xia T, Frankel A, Takahashi TT, Ren J, Roberts RW. Context and conformation dictate function of a transcription antitermination switch. Nat Struct Biol. 2003;10:812–9. doi: 10.1038/nsb983. [DOI] [PubMed] [Google Scholar]
- 43.Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–75. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King RA, Banik-Maiti S, Jin DJ, Weisberg RA. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- 45.McGhee JD, von Hippel PH. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974;86:469–89. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 46.Barik S, Ghosh B, Whalen W, Lazinski D, Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987;50:885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- 47.Lozeron HA, Dahlberg JE, Szybalski W. Processing of the major leftward mRNA of coliphage lambda. Virology. 1976;71:262–77. doi: 10.1016/0042-6822(76)90111-2. [DOI] [PubMed] [Google Scholar]
- 48.Lozeron HA, Anevski PJ, Apirion D. Antitermination and absence of processing of the leftward transcript of coliphage lambda in the RNAase III-deficient host. J Mol Biol. 1977;109:359–65. doi: 10.1016/s0022-2836(77)80039-9. [DOI] [PubMed] [Google Scholar]
- 49.Court DL. RNA processing and degradation by RNase III. In: Belasco J, Brawerman G, editors. Control of mRNA Stability. Academic Press; New York: 1993. pp. 71–116. [Google Scholar]
- 50.Greive SJ, Lins AF, von Hippel PH. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J Biol Chem. 2005;280:36397–408. doi: 10.1074/jbc.M507146200. [DOI] [PubMed] [Google Scholar]
- 51.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–85. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 52.Pavco PA, Steege DA. Elongation by Escherichia coli RNA polymerase is blocked in vitro by a site-specific DNA binding protein. J Biol Chem. 1990;265:9960–9. [PubMed] [Google Scholar]
- 53.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 54.Schellman J. Macromolecular Binding. Biopolymers. 1975;14:999–1018. [Google Scholar]
- 55.Epstein IR. Cooperative and Non-Cooperative Binding of Large Ligands to a Finite One-Dimensional Lattice - Model for Ligand-Oligonucleotide Interactions. Biophys Chem. 1978;8:327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- 56.Revzin A, von Hippel PH. Direct measurements of association constants for the binding of E. coli lac repressor to non-operator DNA. Biochemistry. 1977;16:4769–4776. doi: 10.1021/bi00641a002. [DOI] [PubMed] [Google Scholar]
- 57.Inners LD, Felsenfeld G. Conformation of polyuridylic acid in solution. J Mol Biol. 1970;50:373–339. doi: 10.1016/0022-2836(70)90199-3. [DOI] [PubMed] [Google Scholar]