Abstract
NOD.Idd3/5 congenic mice have insulin dependent diabetes (Idd) regions on chromosomes one (Idd5) and three (Idd3) respectively derived from the non-diabetic strains B10 and B6. NOD.Idd3/5 mice are almost completely protected from type 1 diabetes (T1D) but the genes within Idd3 and Idd5 responsible for the disease-altering phenotype have been only partially characterized. To test the hypothesis that candidate Idd genes could be identified by differential gene expression between the diabetes-susceptible NOD strain and a protected NOD congenic strain, genome-wide microarray expression analysis using an empirical Bayes method was applied to RNA purified from activated NOD and NOD.Idd3/5 CD4+ T cells. Remarkably, the top ten most differentially expressed genes were all from the Idd5 region on chromosome one, validating our central hypothesis. The two genes with the greatest differential RNA expression were those encoding decay-accelerating factor (DAF, also known as CD55) and acetyl-Coenzyme A dehydrogenase, long-chain (ACADL), which are located in the Idd5.4 and Idd5.3 regions, respectively. Neither gene has been implicated previously in the pathogenesis of T1D. In the case of DAF, differential expression of mRNA was extended to the protein level; NOD CD4+ T cells expressed higher levels of cell-surface DAF compared with NOD.Idd3/5 CD4+ T cells following activation with anti-CD3 and anti-CD28. DAF upregulation was shown to be IL-4 dependent and blocked under Th1 conditions. These results validate the approach of using congenic mice together with genome-wide analysis of tissue-specific gene expression to identify novel candidate genes in T1D.
Introduction
Identifying genes involved in the pathogenesis of complex, multigenic autoimmune disease is a challenging goal. Pathogenic alleles at loci influencing disease susceptibility are not overtly deleterious to the immune system and thus are not easily identified, although a small number of loci, in addition to the MHC (1), have been identified in human autoimmune diseases (2-7) and in animal models such as the nonobese diabetic (NOD) mouse which spontaneously develops type 1 diabetes (T1D) (8,9). In diseases like T1D, it is likely that dozens of allelic variants interact to promote or discourage disease manifestation, and that no particular susceptibility allele is necessary or sufficient to mediate spontaneous T1D (10). The strongest disease associated gene, the MHC class II molecule, for example, is not sufficient to mediate disease, as shown by the lack of diabetes in B6 mice with homozygous expression of the NOD MHC class II molecule, I-Ag7 (11).
The genetic complexity of T1D in NOD mice has led investigators to use a congenic strain mapping strategy in order to positionally clone causative Idd genes (10). This approach begins with a genome-wide scan of a population of mice segregating the T1D phenotype (developed by crossing the NOD strain with a T1D-resistant strain) with polymorphic genetic markers to identify genome regions conferring resistance and susceptibility to disease (12). The confirmation that these regions contain genes altering T1D susceptibility comes if an altered frequency of T1D is observed in a congenic strain having the region linked with disease introgressed onto the NOD background. The next step, fine-mapping, involves continued backcrossing of the congenic mice to the NOD strain with genetic screening at each generation to detect recombination events occurring in the introgressed interval. Mice with smaller intervals created by such recombination events are used to develop new congenic strains that are then tested for the frequency of T1D to determine if the gene mediating an effect on disease is retained in the smaller segment or if it has been lost. If the smaller interval preserves the disease phenotype produced by the larger region, the number of candidate genes is decreased. If the smaller interval loses its effect, this can be due to the fact that the newly defined interval does not contain the gene, or, alternatively, that there are two genes in the original segment required for the effect on disease and that the recombination event has separated them (13). This approach has been very successful in excluding candidate genes and narrowing the list of possible candidate genes controlling complex diseases (3,8,14-20).
There is a practical limit, however, to fine-mapping, since recombination in the genome is not completely random and hundreds of meioses must be screened to discover a recombination event occurring within a region of one Mb. This makes the development of a congenic strain having only one introgressed gene nearly impossible since one Mb of DNA can contain dozens of genes. When the introgressed regions reach a lower limit, other methods must be employed to prioritize candidate genes in the interval for further biological studies. One approach (the “candidate gene” approach) is to consider the functions of all known genes in the interval and develop hypotheses as to which ones might be relevant to T1D, perform comparative sequencing, and design biological tests based on the mostly likely causative polymorphisms discovered by sequencing (expression studies, searching for splice isoforms, structure/function studies if an amino acid variation is found, etc) to determine if evidence supporting the candidacy of a particular candidate gene can be gathered. The limitation of this approach is that it usually presumes a certain etiologic model, which may overlook novel genes involved in unexpected pathways.
Another method to prioritizing candidate genes in the congenic interval is to first assay each gene in the region for differential expression. The hypothesis is that if cells from congenic mice having disease susceptibility and resistance alleles are compared, they would show significantly different RNA expression patterns under experimentally relevant conditions. There are several experimental considerations and limitations associated with this approach. First, a method for testing all genes in the interval is needed. Second, if the strains are not identical by descent in the chromosome region in question, each interval might have many variant genes unrelated to that causing the T1D phenotype that are differentially expressed, a possibility supported by a recent report (21). Third, it is possible that differential RNA expression is not the molecular mechanism by which variation at the disease-causing gene within the Idd region alters disease susceptibility.
A technique that can address the first limitation is genome-wide microarray expression analysis (22,23). Given microarray chips with sufficient gene substrate, a high proportion of genes in any interval can be tested, although not all splice variants of a gene may be detected efficiently using most microarray chips that are currently available. The combination of microarray analysis with congenic strain fine-mapping has the potential to focus attention on genes within an interval that have no prior functional data in the literature supporting the hypothesis that they are high priority candidate genes. This approach has had variable success in genetic mouse models of autoimmunity. In two different lupus mouse models, microarray analysis identified strong genetic candidates (19,20). In T1D, however, an earlier attempt at this analysis was not successful when applied to expression in whole, naive spleen (24). The authors concluded that analyzing expression in a noninduced whole organ was not informative, implying that selecting specific cell subsets may be more productive.
In this report, we use the Affymetrix microarray expression system to analyze differential gene expression in purified, activated CD4+ T cells from the NOD, NOD.Idd3/5, and B6.G7 (a NOD MHC congenic strain) strains. A large literature supports a pathogenic role for CD4+ T cells in NOD mice, suggesting that some Idd loci may act at the level of the CD4+ T cell. The NOD.Idd3/5 congenic mouse, with the Idd3 interval on chromosome 3 (c3), and the Idd5 interval on chromosome one (c1), is almost completely protected from diabetes (1-2% incidence at seven months of age for NOD.Idd3/5 females compared with 80% for NOD females) (25). The genetic basis of T1D protection from diabetes in NOD.Idd3/5 mice has been partially characterized. The Idd3 region is 780 kb and the prime candidate genes are those encoding IL-2 and IL-21 (16). As detailed in the accompanying paper (26), there are four subregions within the larger Idd5 region: Idd5.1, Idd5.2, Idd5.3, and Idd5.4. The genes accounting for Idd5.1 and Idd5.2 are most likely Ctla4 (3,14,15) and Nramp1 (8,15), respectively. To further characterize the genetic basis of T1D-resistance in NOD.Idd3/5 mice, we formulated two hypotheses: 1) CD4+ T cells from NOD and NOD.Idd3/5 should show differential expression only in the congenic intervals on c1 and c3; conversely, the common NOD genome outside regions on c1 and c3 should be equivalently expressed. Exception: Differential expression outside the congenic intervals could represent downstream effects of genes found in the congenic intervals. 2) NOD.Idd3/5 and B6.G7 mice should show identical expression of genes in the intervals on c1 and c3. Exception: Gene interactions occurring between the non-NOD alleles on chromosomes 1 and 3 in NOD.Idd3/5 mice and the NOD alleles throughout the remainder of the genome could alter expression from that seen in B6.G7 cells in which B6 alleles are present in the remainder of the genome, although expression would be distinguishable from that in cells having the NOD alleles on chromosomes 1 and 3 together with the NOD background. Notably, all non-T cell related genes should serve as internal controls insofar as they would not be expressed in the activated CD4+ T cells. To minimize the skewing effect that low-expressed genes have, due to their low variance, on the estimation of differential expression, we used an empirical bayesian method for statistical analysis of differential expression. We confirmed the microarray results by measuring protein expression using fluorescence-activated cell analysis and/or assessing mRNA levels with reverse transcription of RNA followed by quantitative PCR. Our results show the powerful potential of this approach. Out of over 22,000 genes analyzed on the chip, the top 23 genes found to be differentially expressed between NOD and NOD.Idd3/5 were almost exclusively contained in the B10- and B6-derived regions introgressed onto the NOD background. Two of the most differentially expressed genes, Cd55 (formerly Daf1) and Acadl, have not been implicated in T1D pathogenesis and are novel candidate genes for Idd5.4 and Idd5.3, respectively. The identification of Cd55 and Acadl illustrates that an unbiased genetic approach to gene identification utilizing congenic mouse strains, relevant cell populations, and genome-wide microarray analysis can provide valuable insights into the biological processes underlying T1D.
Methods
Mice
NOD.B6 Idd3 B10 Idd5 mice, (reference 25, hereafter referred to as NOD.Idd3/5 mice), NOD and B6.H2g7 (hereafter called B6.G7) mice were bred and housed under specific pathogen free conditions and all procedures were conducted according to approved protocols of the University of Pittsburgh School of Medicine Animal Care and Use Committee. Idd5 congenic strains 974, 1092, 1595, 2574, 1094 and 2193 were obtained from the Taconic Emerging Models program.
Preparation, purification and stimulation of splenocytes
The spleen from each mouse was removed aseptically and minced. After lysing red blood cells, the cells were washed three times with PBS. To purify CD4-positive splenocytes, splenocytes were prepared by magnetic separation using a MiniMACS system (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. Purified CD4-positive splenocytes were suspended in RPMI1640 medium (Gibco-BRL, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum (Gibco-BRL) and 1 mM L-alanyl-glutamine (Life Technologies, Grand Island, NY), 100 U/ml penicillin, 100 μg/ml streptomycin (Life Technologies), 1 mM sodium pyruvate (Life Technologies), and 50 μM 2-ME. The CD4-positive splenocytes (1×106) were transferred to each well of a 24-well plate precoated with anti-CD3 antibody and anti-CD28 antibody was added (1 μm/ml). The cells were cultured for the indicated period and harvested. Th2 conditions were: recombinant mouse IL-4 (10 ng/ml) and/or anti-mouse IFN-γ antibodies (10 μg/ml), whereas Th1 conditions were: recombinant mouse IL-12 (5 ng/ml) and/or anti-mouse IL-4 antibodies. In some studies, anti-mouse IL-4 receptor antibodies alone were added (BD Pharmingen, San Diego, CA).
Flow cytometry
After culture, cells were incubated with Fc blocker (BD Pharmingen, San Diego, CA) and stained with labeled antibodies for 20 min at 4°C. Samples were analyzed on an FACS Caliber (BD bioscience, Miami, FL). Anti-CD4 and anti-CD55 antibodies were purchased from BD bioscience (San Diego, CA).
RNA extraction
Total RNA was extracted from cultured cells using the RNeasy mini kit (Qiagen, Valencia, CA). The RNA was redissolved in RNase-free water and yield estimated by spectrophotometry; equal quantities of RNA were used for analysis. Samples were hybridized to the Affymetrix mouse chips (see below) at the Genomics and Proteomics Core Laboratory at the University of Pittsburgh.
Real time RT PCR analysis of DAF mRNA expression
CD4+ T cell RNA was reverse-transcribed using an oligo-dT primer and Reverse Transcription System (Promega, WI, USA) according to the manufacturer’s instructions. Real time PCR was carried out for DAF and GAPDH (internal control) in an ABI Prism 7300 sequence detector (PE Applied Biosystems). All reactions were performed using TaqMan Universal MasterMix; primer/probe sets were purchased from Applied Biosystems. (PE Applied Biosystems). The obtained mRNA level was expressed relative to the GAPDH PCR product amplified from the same sample; DAF value=2^((Ct of GAPDH) -(Ct of DAF)).
Real time RT PCR analysis of ACADL mRNA expression
RNA was extracted from purified CD4+ T cells in TRIZOL® (Invitrogen) and 1000 ng of total RNA was used in a cDNA synthesis reaction with Superscript II reverse transcriptase (Invitrogen). cDNA was used as template in a TaqMan PCR reaction (prepared with TaqMan Universal PCR Master Mix, Applied Biosystems) with the following primers and probes designed to detect ACADL mRNA: forward GATTTATCAAGGGCCGGAAG, reverse GAAATCGCCAACTCAGCAAT and probe Fam-TGTCCGATTGCCAGCTAATGCC-Tam. β2-microglobulin was used to normalize expression levels as described previously (3).
Microarray techniques
MOE430A Affymetrix high-density oligonucleotide array chips containing 506,944 oligonucleotide probes for 22,690 genes were used in the analysis. Total RNA was converted to ds cDNA according to standard methods and purified using an Affymetrix cDNA clean-up column. An aliquot of the ds-cDNA equivalent to 5-7 μg of starting RNA is added as template to an in vitro transcription reaction as per the ENZO BioArray high efficiency RNA transcript labeling kit, and the resulting biotinylated cRNA purified using an Affymetrix RNA clean-up column. After elution the cRNA is quantified by spectrophotometry and 20 μg of cRNA is incubated at 94°C in fragmentation buffer (40 mM Tris-Acetate pH 8.1, 100 mM KOAc, 30 mM MgOAc) for 35 minutes. A 1 μl aliquot of the sample is run on an Agilent Bioanalyzer to verify that fragmentation has resulted in RNA of the desired size distribution.
Fifteen micrograms of the fragmented RNA is added to a final volume of 300 μl hybridization cocktail, applied to the GeneChip® of interest and incubated overnight at 45°C with rotation. Following hybridization the sample is removed and the GeneChip cassette filled with non-stringent wash buffer. The chip is loaded onto an Affymetrix Fluidics station for wash and stain. The GeneChips® are then stained for ten minutes in streptavidin-phycoerythrin (SAPE) solution (1X MES stain buffer, 2 mg/ml acetylated BSA, 10 μg/ml SAPE; 1X MES stain buffer contains 100 mM MES, 1M [Na+], 0.05% Tween 20). Non-stringent buffer is used to wash off the first stain solution. Signal amplification is achieved by ten minutes incubation with biotinylated anti-streptavidin (1X MES stain buffer, 2 mg/ml acetylated BSA, 0.1 mg/ml Normal Goat IgG, 3 μg/ml biotinylated anti-streptavidin) followed by a second ten minute incubation with SAPE. The chip is washed and filled with non-stringent wash buffer before being removed from the fluidics station and scanned using the GeneArray® scanner.
Data analysis
Gene expression for activated CD4+ T cells from NOD (four samples), B6.G7 (four samples), and NOD.Idd3/5 (three samples) mice were used. Preprocessing of the data consisted of Robust Multichip Average (RMA) background correction, quantile normalization and RMA expression summarization as described by Irizarry et al. (27). Preprocessing was implemented using the affy library of the bioconductor package of R (28). The advantages of this preprocessing procedure over other methods, for instance the stock Affymetrix MAS5.0, are described in Bolstad et al. (29). Correlations, Boxplots and MVA plots were used to verify that the preprocessing successfully reduced the variability of the expression measures between chips.
For statistical analysis of protein expression, Mann-Whitney test and paired t tests were performed in GraphPad Prism and JMP-IN software. Physical location of genes was established using Ensembl V34 October 2005.
Results
Empirical Bayes analysis of normalized expression datasets and algorithmic queries to generate candidate gene lists
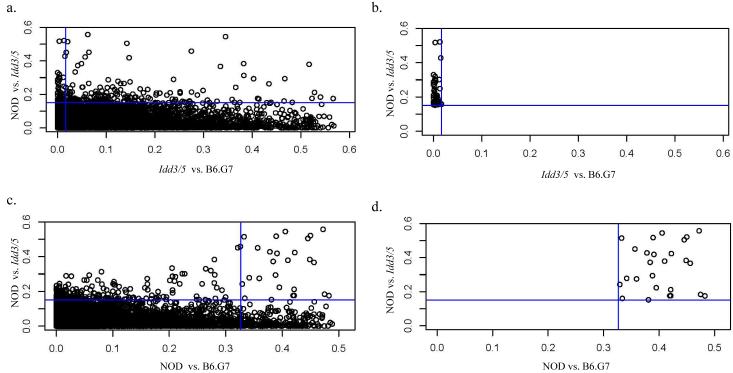
After the normalization of gene array data (see methods), we used an empirical Bayes method to generate an estimated posterior logarithm of odds for differential gene expression. The Bayesian approach was chosen because it is robust to anomalies caused by small variance estimates in low-expression genes (30). This produced a log odd score for each gene for comparing expression in CD4+ T cells between each pair of strains: NOD vs. NOD.Idd3/5, NOD vs. B6.G7, and B6.G7 vs. NOD.Idd3/5. Since the log odd statistic is appropriate for ranking genes according to differential expression and does not have an associated P-value, statistical significance was assessed using the usual t statistic. To generate lists of candidate genes from this overall dataset, we queried the dataset with algorithms designed to test central biological hypotheses. This consisted of generating rules expressing the biological hypothesis and applying the rules to the data set. The first algorithm tested our central hypothesis that NOD and NOD.Idd3/5 gene expression will differ throughout the introgressed regions, intersected with a second condition that NOD.Idd3/5 and B6.G7 expression will be similar across the interval. This algorithm can be represented as: [(NOD neq Idd3/5) AND (Idd3/5 = B6.G7)]. To implement algorithm one, we produced a list of genes satisfying the condition of differential expression of NOD and NOD.Idd3/5 CD4+ T cells genes at the 96th percentile of log odds (Note that the percentile chosen is arbitrary and has no effect on the top of the lists, only the bottom cut-off) and intersected it with the list of genes at the 50th percentile of the NOD.Idd3/5 versus B6.G7 log odds. This resulted in a list of 227 genes (1% of the original dataset) ordered by log odds score, as displayed in Figure one. Figure 1a shows the complete set of genes; the vertical and horizontal lines represent the conditions imposed by the algorithm. Figure 1b depicts the set of genes meeting the algorithmic criteria. Next, we selected the most differentially expressed genes on the list, as ranked by their odds ratio, and determined their chromosomal location. Table One demonstrates the first eleven most differentially expressed genes, as ordered by log odds of differential expression. Remarkably all of the genes were from the congenic NOD.Idd3/5 B10 genetic intervals on chromosome 1. Two probe sets with high differential expression did not yield a gene product in Ensembl (data not shown). This list was not “selected” from the larger list; i.e. it shows the first 11 genes on the larger list. Note that the cut-off is arbitrary; however the further one goes down the list of 227 genes meeting our criteria, the more one begins to find smaller log odd scores, nonsignificant P-values, and the emergence of genes located in non-introgressed chromosomal regions. However, it is important to emphasize the possibility that the candidate gene that will be proven to be the Idd gene at some point in the future may well not be at or near the top of the list. Moreover, the biological hypothesis embedded in algorithm one does not address the possibility that differential expression of genes in the Idd3/5 regions cause the differential expression of genes outside the introgressed regions (see below).
Figure one.
Dot plot representation of whole gene chip data set queries by hypothesis driven algorithms. Shown is the distribution of genes representing the whole data set of NOD, NOD.Idd3/5, and B6.G7 gene chips, analyzed using algorithmic queries represented on the x and y axes (see text). a): Genes meeting the criteria (NOD neq NOD.Idd3/5) (vertical line) intersected with (Idd3/5 = B6.G7) (horizontal line). “b” shows the subset of genes meeting the criteria in “a”. c: Genes meeting the criteria (NOD neq NOD.Idd3/5) (vertical line) intersected with genes meeting the criteria (NOD neq NOD.Idd3/5) (horizontal line). “d” shows the subset of genes resulting from the criteria applied in “c”.
Table One.
| Gene Symbol | NOD vs. Idd3/5 Odds | NOD vs. Idd3/5 Max | NOD vs. Idd3/5 P-value | Idd3/5 vs. B6.G7 Odds | Chromosome location (Mb) |
|---|---|---|---|---|---|
| Cd55 (formerly Daf1) | 0.5210 | NOD | 0.0008 | 0.0134 | c1 (130.2) |
| Acadl | 0.5176 | IDD | 0.0002 | 0.0036 | c1 (67.1) |
| Npl | 0.4278 | NOD | 0.0057 | 0.0152 | c1 (153.4) |
| Rnpep | 0.3300 | IDD | 0.0255 | 0.0001 | c1 (135.1) |
| Hspe1 | 0.3195 | NOD | 0.0301 | 0.0034 | c1 (55.4) |
| Glrp1 | 0.3017 | IDD | 0.0231 | 0.0121 | c1 (88.3) |
| Zc3h11a | 0.3016 | NOD | 0.0099 | 0.0027 | c1(133.4) |
| Rgs16 | 0.2871 | NOD | 0.0726 | 0.0001 | c1 (153.6) |
| Csrp1 | 0.2844 | IDD | 0.0448 | 0.0033 | c1 (135.6) |
| 6430706D22Rik | 0.2730 | IDD | 0.0381 | 0.0001 | c1 (88.2) |
| Insig2 | 0.2372 | NOD | 0.0822 | 0.0013 | c1 (121.1) |
Gene expression is ordered by odds ratio (from Empirical Bayes analysis). The “Max” column indicates which strain showed the higher expression. Chromosomal location was determined by Ensembl (V34)query. Genes represented by more than one probe set are shown only once.
The second algorithm we employed to query the dataset is expressed as: [(NOD neq Idd3/5) AND (NOD neq B6.G7)]. This algorithm intersected the set of genes showing differential expression between NOD and NOD.Idd3/5 with the set of genes differentially expressed between NOD and B6.G7. The 96th percentile of the log odds was the cut-off for each comparison and generated a list containing n=229 genes. One might expect that if NOD and NOD.Idd3/5 gene expression varied at the introgressed regions, NOD and B6.G7 gene expression should also vary at the same regions, producing a list identical to algorithm one. As shown in Figure 1c, however, this algorithm clearly produced a different set of genes than algorithm one. Applying the (NOD neq G7) condition to the (NOD neq Idd3/5) condition intersects a set of genes, depicted in figure 1d, that includes 10 of the 11 genes in Table one, and 12 additional genes (Table two). Table two displays the first 22 most differentially expressed genes, as ordered by log odds of differential expression, resulting from the second algorithm. The robustness of our approach was again confirmed since we observed that 19 of the 22 differentially expressed genes are located in the introgressed regions on chromosomes one and three (Table two). Finally, using the Idd5 subregion boundaries as defined by the T1D frequencies of Idd5 congenic strains (Fig. 3 and ref. 26), 17 of the 22 differentially expressed genes are still included within the boundaries of 3 of the 4 known Idd5 subregions: Icos in Idd5.1, Acadl in Idd5.3 and 15 genes, including Daf1, in Idd5.4. It is notable that the differential expression of ICOS has been reported previously in studies comparing activated T cells from Idd5 congenic mice (15, 31). In these studies it was hypothesized that the differential expression of ICOS is caused by allelic variants at Ctla4.
Table Two.
| Gene Symbol | NOD vs. Idd3/5 Odds | NOD vs. Idd3/5 Max | NOD vs. Idd3/5 P-val | NOD vs. B6.G7 Odds | NOD vs. B6.G7 Max | NOD vs. B6.G7 P-val | Chromosome Location | Idd Region |
|---|---|---|---|---|---|---|---|---|
| Suclg2 | 0.557 | IDD | 0.00006 | 0.472 | B6.G7 | 0.00002 | c6 95.9Mb | Idd6 |
| CD55 (formerly Daf1) | 0.521 | NOD | 0.00076 | 0.45 | NOD | 0.00014 | c1 130.2 | Idd5.4 |
| Acadl | 0.518 | IDD | 0.00018 | 0.389 | B6.G7 | 0.00145 | c1 67.1 Mb | Idd5.3 |
| Lad1 | 0.514 | IDD | 0.00106 | 0.332 | B6.G7 | 0.01168 | c1 135.7 Mb | Idd5.4 |
| Tnni1 | 0.458 | IDD | 0.00332 | 0.326 | B6.G7 | 0.00498 | c1 135.6 Mb | Idd5.4 |
| Exosc9 | 0.451 | IDD | 0.00446 | 0.321 | B6.G7 | 0.00935 | c3 36.0 Mb | **NONE |
| Ptprv | 0.451 | NOD | 0.00257 | 0.357 | NOD | 0.00206 | c1 135.0 Mb | Idd5.4 |
| Npl | 0.428 | NOD | 0.00565 | 0.378 | NOD | 0.00159 | c1 153.4 Mb | Idd5.4 |
| Stk25 | 0.423 | IDD | 0.01131 | 0.422 | B6.G7 | 0.00069 | c1 93.4 Mb | Idd5.4 |
| D15Wsu75e | 0.419 | IDD | 0.00386 | 0.391 | B6.G7 | 0.00099 | c15 82.0 Mb | NONE |
| Ramp1 | 0.383 | NOD | 0.0169 | 0.28 | NOD | 0.01706 | c1 91.0 Mb | Idd5.4 |
| Gzmd | 0.379 | IDD | 0.02119 | 0.41 | NOD | 0.00058 | c14 50.7 Mb | Idd8 |
| Ugt1a10 | 0.334 | IDD | 0.0026 | 0.205 | B6.G7 | 0.00444 | c1 87.9 Mb | Idd5.4 |
| Rnpep | 0.33 | IDD | 0.0255 | 0.265 | B6.G7 | 0.02056 | c1 135.1 | Idd5.4 |
| Hspe1 | 0.319 | NOD | 0.03005 | 0.274 | NOD | 0.0183 | c1 55.4 | **NONE |
| Glrp1 | 0.302 | IDD | 0.02314 | 0.223 | B6.G7 | 0.0178 | c1 88.3 | Idd5.4 |
| Zc3h11a | 0.302 | NOD | 0.00988 | 0.227 | NOD | 0.02069 | c1 133.4 Mb | Idd5.4 |
| Rgs16 | 0.287 | NOD | 0.0726 | 0.206 | NOD | 0.08094 | c1 153.6 Mb | Idd5.4 |
| Csrp1 | 0.284 | IDD | 0.04475 | 0.211 | B6.G7 | 0.04412 | c1 135.6 | Idd5.4 |
| Icos | 0.278 | NOD | 0.07522 | 0.342 | NOD | 0.00909 | c1 61.3 | Idd5.1 |
| Ralb | 0.275 | IDD | 0.02153 | 0.359 | B6.G7 | 0.00066 | c1 119.2 | Idd5.4 |
| 6430706D22Rik | 0.273 | IDD | 0.03812 | 0.161 | B6.G7 | 0.09667 | c1 88.1 | Idd5.4 |
Gene expression is ordered by odds ratio (from Empirical Bayes analysis). The “Max” columns indicate which strain showed the higher expression. Chromosomal location was determined by Ensembl (V34) query. Genes represented by more than one probe set are shown only once.
Indicates that the gene is not within the boundaries of an Idd region but is within the boundaries of the introgressed genetic interval.
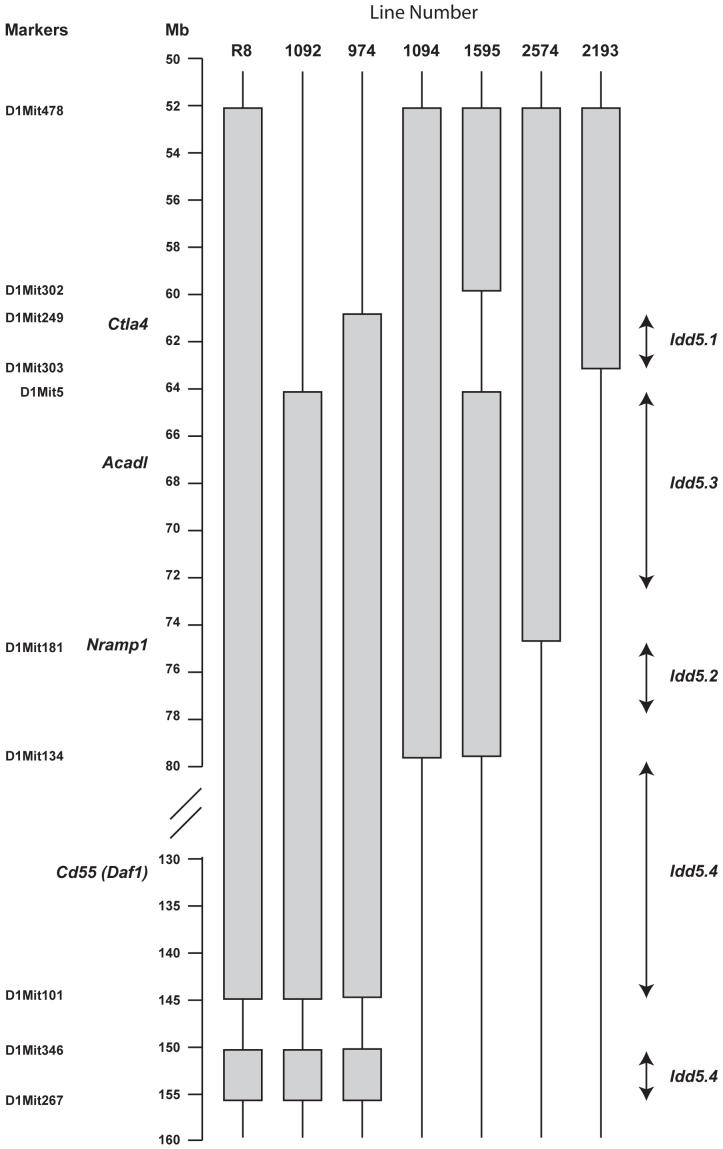
Figure three.
Genetic map of NOD Idd5 congenic strains. The shaded regions represent the B10-derived regions introgressed onto the NOD background.
Comparison of the results of algorithms one and two (Tables one and two) shows that substitution of the condition (NOD neq B6.G7) for (Idd3/5 = B6.G7) has the effect of adding a set of genes to the list which had been excluded by the condition (Idd3/5 = B6.G7). Moreover, the additional genes included at least three that are not from the Idd3/5 introgressed chromosomal regions, including the most differentially expressed gene in the data set, SucIg2 on chromosome 6. This raises the biological question of how NOD and Idd3/5 CD4+ T cells can differ in expression of a gene, SucIg2 (succinate-Coenzyme A ligase, GDP-forming, beta subunit), which is of NOD origin in both strains. The simplest explanation is that algorithm two permits the inclusion of genes subject to gene interactions; i.e. downstream effects of differential expression of genes in the introgressed region on genes outside the region, or visa versa (see discussion).
Confirming differential expression of candidate genes Cd55 and Acadl
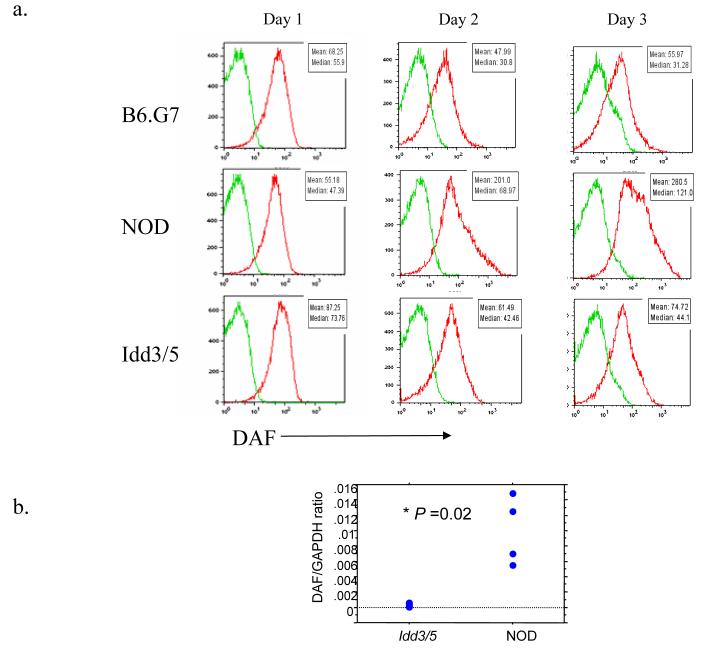
The list of 23 novel candidate genes shown in Tables one and two represent about 0.1 percent of all genes on the gene chip. Nonetheless, in experimental terms, investigating this number of genes represents an enormous effort; moreover the results are complicated by the likelihood that gene interactions account for some of the differences in the genes listed in Tables one and two. We therefore decided to initially focus our investigation on the most differentially expressed gene in the Idd5.4 and Idd5.3 regions identified by algorithm 1 (Table one), Cd55 and Acadl. Cd55 encodes the protein DAF (decay-accelerating factor), also known as CD55. Cd55 is located in the distal segment of the B10-derived Idd5 region present in the NOD.Idd3/5 mouse. First, we evaluated DAF protein expression on purified CD4+ T cells (cultured under the same conditions as the gene chip analysis) from NOD, NOD.Idd3/5, and B6.G7 mice. As shown in Figure 2a, DAF was significantly upregulated on the cell surface of NOD CD4+ T cells compared with NOD.Idd3/5 or B6.G7 CD4+ T cells, thereby confirming the gene chip results at the protein level. Reverse transcription of RNA obtained from the CD4+ T cells followed by quantitative PCR (TaqMan methodology) analyses also confirmed the results obtained in the microarray experiments; there was increased expression of DAF RNA in activated NOD CD4+ T cells compared with similarly activated cells from NOD.Idd3/5 and B6.G7 mice (figure 2b).
Figure two.
DAF protein and mRNA expression analyses on NOD, NOD.Idd3/5, and B6.G7 CD4+ T cells. CD4+ T cells were purified and stimulated with anti-CD3/CD28 for three days and were then analyzed by flow cytometry for cell surface expression of the DAF protein (a) and quantitative PCR following reverse transcription for mRNA expression (b).
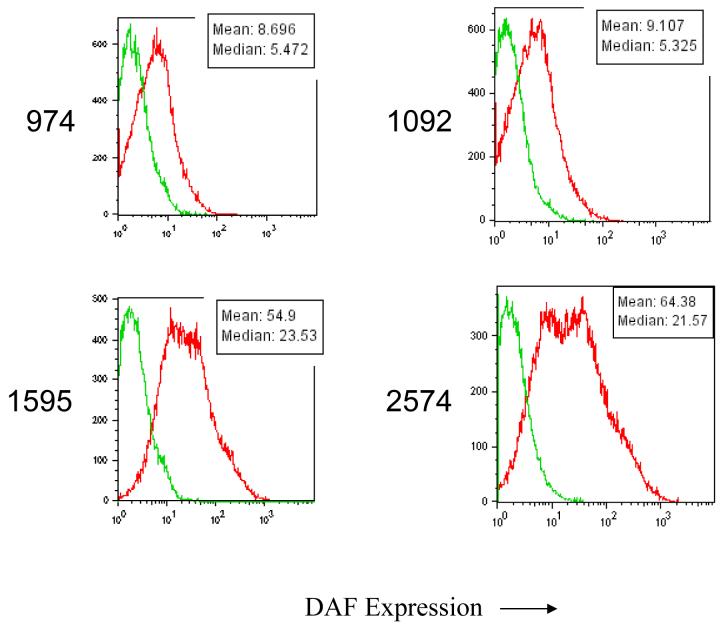
Having confirmed the gene chip results for Cd55, we examined additional Idd5 congenic strains of mice with progressively smaller B10-derived chromosome one regions, some with NOD alleles at Cd55 and some with B10 alleles, for DAF protein expression (Figure 3, genetic map, and figure 4, expression studies). In each case, CD4+ T cells from mice with B10 alleles at Cd55 (lines 1092 and 974) showed decreased DAF expression, while the CD4+ T cells from mice with the NOD allele (lines 1595 and 2574) showed relatively higher expression (Figure 4). These results identify Cd55 as a candidate gene for Idd5.4, although further dissection of this region by making additional Idd5.4 congenic strains is required to substantiate this candidacy since the B10-derived genetic intervals comprising Idd5.4 are large, approximately 70 Mb (Figure 3). Hundreds of genes are located in these 70 Mb of introgressed DNA including Cd55 and several other differentially expressed genes listed on Tables 1 and 2.
Figure four.
Analysis of DAF expression on anti-CD3/CD28 stimulated CD4+ T cells from NOD.Idd5 congenic strains: increased DAF expression is observed only on cells having the NOD DAF allele. CD4+ T cells were purified and stimulated with anti-CD3/CD28 as in Figure two.
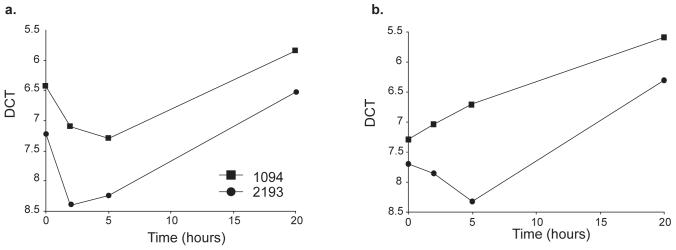
In contrast to the very large Idd5.4 region, Acadl is located in the much smaller (9.3 Mb) Idd5.3 region (Figure 3 and ref. 26) and is the only gene within Idd5.3 detected in the microarray analysis to be listed as one of the most differentially expressed genes. We confirmed and extended the gene chip results of increased expression of the B10 ACADL allele by assessing ACADL mRNA expression by quantitative PCR following reverse transcription in activated CD4+ T cells from congenic mice having smaller B10-derived intervals in the Idd5 region than the NOD.Idd3/5 congenic strain (Figures 3, 5). Line 1094 CD4+ T cells, which have the B10 ACADL allele, had increased expression of ACADL mRNA compared with cells from line 2193 mice having the NOD allele (figure 5). These observations localize the differential expression of Acadl to a 16 Mb congenic interval that overlaps the 9.3 Mb Idd5.3 region making Acadl a major candidate gene for mediating the disease-causing phenotype localized to the Idd5.3 region (26).
Figure five.
Differential expression of ACADL. Expression of ACADL was assessed in RNA isolated from CD4+ T cells stimulated in vitro with CD3 and CD28 for 0, 5 and 24 hours. Two strains were compared for ACADL expression, line 1094 (filled squares) and line 2193 (filled circles). Line 1094 has B10 alleles at Idd5.3 whereas line 2193 has NOD alleles at Idd5.3. Data from two experiments of four performed are shown (a and b). All four experiments had similar results. Comparisons of the delta Ct values of ACADL mRNA obtained from CD4 T cells from lines 1094 and 2193 demonstrated significant differences at the 5 and 24 hour time points (P = 0.06, 0.01, and 0.02 using a paired t test at 0, 5, and 24 h, respectively).
DAF upregulation on CD4+ T cells is promoted by IL-4 and suppressed in Th1 conditions
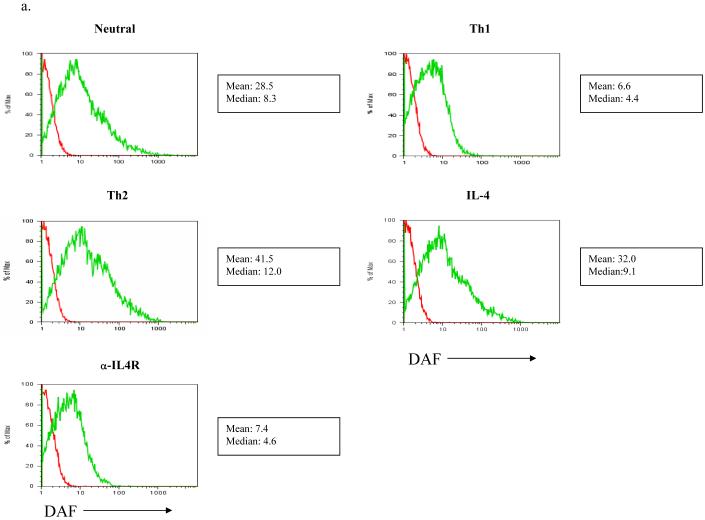
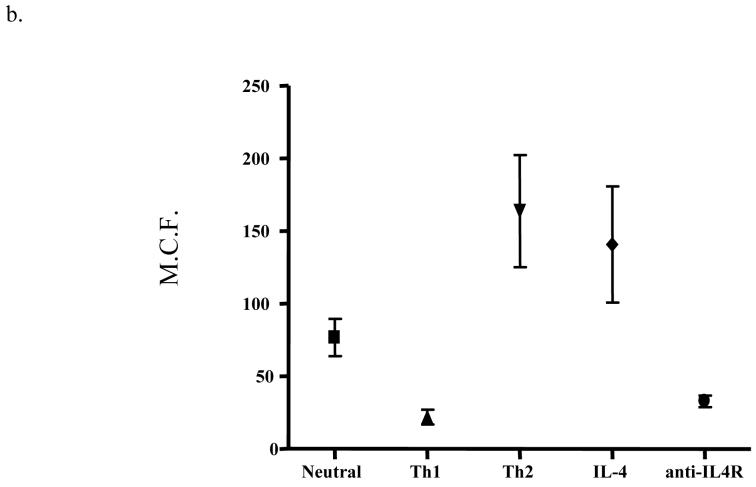
Our discovery of a variation in DAF expression that localizes to a chromosome region that includes Cd55 is of interest since the knockout of the DAF/CD55 gene increases T cell activity and susceptibility to EAE (see discussion). We thus hypothesized that the differential expression of DAF by the B10 and NOD alleles would have functional consequences on the immune response, and initiated studies to investigate the regulation of DAF expressed at the cell surface of CD4+ T cells in different cytokine environments (Figure 6 and Table three). As noted above, NOD CD4+ T cells upregulate DAF under neutral conditions. However, as shown in Figure 6 a and b, Th1 conditions prevented activation-induced DAF upregulation on NOD CD4+ T cells whereas Th2 conditions strongly enhanced DAF upregulation. Th2 conditions did not, however, increase DAF expression on NOD.Idd3/5 CD4+ T cells (not shown). Next, we asked which components of the Th2 culture conditions were sufficient for upregulation of DAF. We found that IL-4 alone added to culture supported strong upregulation of DAF on NOD CD4+ T cells (Figure six a and b, Table three); the expression levels of DAF with both Th2 and IL-4 alone were significantly different from neutral conditions (P= 0.026 and P = 0.05), but expression under Th2 conditions did not differ from IL-4 alone. Moreover, anti-IL4 receptor antibodies alone added to culture with CD3 and CD28 stimulation completely prevented DAF upregulation; the expression levels of DAF with anti-IL-4R antibody were significantly different from neutral conditions (P = 0.049), whereas expression under Th1 conditions was not significantly different from anti-IL4R alone. The strong effect of IL-4 on DAF upregulation raises interesting issues related to T1D pathogenesis in NOD mice.
Figure six.
DAF expression on NOD CD4+ T cells is upregulated by both Th2 conditions and IL-4, and downregulated by both Th1 conditions and anti-IL-4R. NOD CD4+ T cells were purified and stimulated with anti-CD3/CD28 as in Figure two, under neutral, Th1, and Th2 conditions (see methods) or with only IL-4 or anti-IL-4 receptor antibody. a) One representative of seven experiments is shown. b) The mean channel fluorescence from all experiments plus SEM is shown for each condition. The complete data set is shown in Table three. Neutral condition DAF expression was significantly different from DAF expression using Th1, Th2, IL-4 and anti-IL-4R conditions (P = 0.012, 0.026, 0.05, and 0.049 respectively), and Th1 DAF expression was significantly different from that under Th2 conditions (P = 0.015). Th1 condition DAF expression was not significantly different from that using the anti-IL4R condition nor was Th2 condition DAF expression significantly different from that using the IL-4 condition. All significance testing was performed using the paired t test.
Table Three.
| Exp | Neutral | Th1 | Th2 | IL-4 | α-IL-4R |
|---|---|---|---|---|---|
| 1 | 111.9 | 38.3 | 238 | n.d. | n.d |
| 2 | 39.5 | 18.3 | 155.8 | 92.7 | 48.6 |
| 3 | 28.5 | 6.6 | 41.5 | 32.0 | 7.4 |
| 4 | 71.6 | 22.8 | 129.6 | 134.7 | 22.9 |
| 5 | 76.3 | 23.6 | 253.6 | 116.5 | 22.4 |
| 6 | 118.5 | n.d. | n.d. | 296.2 | 38.1 |
| 7 | 89.9 | n.d. | n.d. | 196.6 | 32.3 |
n.d.= not done. All experiments are after 3d culture of NOD CD4+ T cells under the stated conditions. See figure 7 for p value calculations.
Discussion
Identification of candidate genes in complex autoimmune diseases, even when they are confined to a defined genetic interval, remains a challenging scientific problem. Hypothesis driven approaches (e.g. investigating immunological mediators in T1D, or investigating possible autoantigens) will overlook genes in novel and unexpected pathways whereas a “hypothesis free” whole genome approach can miss candidate genes by investigating too many variables simultaneously. Indeed, a prior published attempt to discover causative genes in NOD congenic mice analyzed expression in whole, unstimulated spleen preparations and did not detect compelling candidate genes in the Idd intervals (24). In the current study, we have reapproached the search for candidate loci by focusing on gene expression in activated CD4+ T cells from both NOD and NOD congenic mice. Our approach, which combines a whole genome analysis with hypotheses based on the importance of CD4+ T cells in T1D pathogenesis, is expressed in our algorithms stating that we expect to find a subset of genes differentially expressed in the introgressed congenic intervals.
As summarized in Tables one and two, this approach was highly successful; 20 of 23 genes identified were located in the introgressed regions on chromosomes one and three of the NOD.Idd3/5 mouse. Notably, these genes were not selected from the larger list arbitrarily, but are the top most differentially expressed genes as determined by our Empirical Bayesian approach. Remarkably, two of the most differentially expressed genes, Daf1 and Acadl, were determined to be located within the boundaries of the Idd5.4 and Idd5.3 loci, respectively. Moreover, mRNA and protein expression analyses confirmed the gene chip results and thereby established Daf1 and Acadl as candidate genes.
Nonetheless, as exemplified in several entries in Table two, our approach produced more complicated results as well. By changing the parameters of the algorithmic query, we generated additional candidate genes; moreover, several of the candidate genes in Table two were not located in the introgressed regions on chromosomes one and three. SucIg2, for example, emerges as the most differentially expressed gene on both lists, but it is located on chromosome six. NOD and NOD.Idd3/5 mice have identical alleles on chromosome six, which raises the question of how and why the NOD and NOD.Idd3/5 T cells differ so dramatically in expression of genes that are identical. While we do not explore the mechanism of this differential expression here, the simplest explanation is that it represents a downstream effect of one of the Idd3/5 genes in the introgressed intervals. In other words, differential regulation of a gene in the introgressed Idd3 or 5 regions in NOD.Idd3/5 vs. NOD CD4+ T cells affected the expression of SucIg2 differentially in NOD and NOD.Idd3/5 mice, leading to upregulation of SucIg2 in NOD.Idd3/5 CD4+ T cells. For example, it is possible that the increased expression of Acadl in Idd3/5 and B6.G7 CD4+ T cells leads to greater energy production that in turn upregulates Suclg2, which encodes an enzyme important in the citric acid cycle. The same hypothesis applies to Gzmd, which is located on Chromosome 14 and therefore has the same allele in the NOD and NOD.Idd3/5 strains. Gzmd is overexpressed in NOD.Idd3/5 CD4+ T cells compared with NOD CD4+ T cells, which in turn express higher levels than B6.G7 CD4+ T cells. In this case, the non-NOD allele in the NOD.Idd3/5 intervals appears to act on or upstream of Gzmd to upregulate its response, and the amount of upregulation is modified by one or more genes that differ between NOD and B6 in other areas of the genome. The highlighting of such downstream gene amplification events by microarray analysis should provide significant insights into disease pathogenesis. Since the combined activity of protective alleles at Idd3 and Idd5 provide more disease protection than would be expected if the alleles were acting in a multiplicative fashion (32), the discovery of compelling differential gene expression events in activated Idd3/Idd5 CD4+ T cells could reveal how the combination of particular protective alleles in an “autoimmune” pathway can significantly alter the balance between health and disease.
We identified Cd55 as one of the most differentially expressed genes between NOD and NOD.Idd3/5 T cells. Cd55 is well known as a complement regulatory gene, but recent research has also identified it as a T cell costimulatory molecule that interacts with its ligand, CD97 (33). Splice variants of CD97 produce isoforms having variable numbers of EGF domains that engage DAF with variable affinity (34). DAF likely binds to the first CD97 EGF domain but also requires domains 2 and 5; the alternately spliced variant of CD97 that expresses only domains 1,2, and 5 binds DAF with highest affinity (35). Cd55 knockout mice have been studied in several models of autoimmunity and show worsened experimental glomerulonephritis, experimental myasthenia gravis, and EAE (36-39). Moreover, Cd55-/- mice demonstrated significantly enhanced T cell responses with hypersecretion of IFN-γ (39). Our genetic mapping studies indicate that the NOD allele of Idd5.4 acts as a T1D-resistance allele whereas the B10 allele increases T1D susceptibility (26). Since NOD CD4+ T cells upregulate DAF in response to activation conditions in which NOD.Idd5 CD4+ T cells do not, and DAF knockout mice have increased autoimmunity, our mRNA expression results are consistent with Cd55 as a candidate gene for Idd5.4. Additional congenic strains of mice with the region of Idd5.4 containing Cd55 are currently being developed to test this hypothesis.
Our characterization of DAF regulation clearly showed that NOD DAF is upregulated by Th2 conditions and downregulated by Th1 conditions. We have previously shown that NOD T cells are biased to Th1 expression (40,41), consistent with many other publications suggesting a Th1 bias in NOD mice (42-47). Since Th1 conditions prevent DAF upregulation, it may be that the genetic program generating Th1 conditions in NOD T cells negates the protective effect of DAF by preventing its upregulation. Conversely, many therapeutic interventions associated with the induction of Th2-related phenotypes have prevented T1D (48-50). Given our results, it is possible that one mechanism mediating the protective effect of Th2 conditions on T1D is the upregulation of DAF on NOD CD4+ T cells.
Acknowledgements
The availability of NOD congenic mice through the Taconic Farms Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
WMR is supported by NIH NIDDK 60714 and NIH RFA A102-006
LSW is a Juvenile Diabetes Research Foundation/Wellcome Trust Principal Research Fellow and the research in LSW’s laboratory for this study was also supported by NIH P01 AI039671.
References
- 1.Lambert AP, Gillespie KM, Thomson G, Cordell HJ, Todd JA, Gale EA, Bingley PJ. Absolute risk of childhood-onset type 1 diabetes defined by human leukocyte antigen class II genotype: a population-based study in the United Kingdom. J Clin Endocrinol Metab. 2004;89:4037–4043. doi: 10.1210/jc.2003-032084. [DOI] [PubMed] [Google Scholar]
- 2.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 3.Ueda H, Howson JM, Esposito L, Heward J, Snook H, Chamberlain G, Rainbow DB, Hunter KM, Smith AN, Di Genova G, Herr MH, Dahlman I, Payne F, Smyth D, Lowe C, Twells RC, Howlett S, Healy B, Nutland S, Rance HE, Everett V, Smink LJ, Lam AC, Cordell HJ, Walker NM, Bordin C, Hulme J, Motzo C, Cucca F, Hess JF, Metzker ML, Rogers J, Gregory S, Allahabadia A, Nithiyananthan R, Tuomilehto-Wolf E, Tuomilehto J, Bingley P, Gillespie KM, Undlien DE, Ronningen KS, Guja C, Ionescu-Tirgoviste C, Savage DA, Maxwell AP, Carson DJ, Patterson CC, Franklyn JA, Clayton DG, Peterson LB, Wicker LS, Todd JA, Gough SC. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–511. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 4.Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmune diseases--a general susceptibility gene to autoimmunity? Genes Immun. 2000;1:170–184. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- 5.Barratt BJ, Payne F, Lowe CE, Hermann R, Healy BC, Harold D, Concannon P, Gharani N, McCarthy MI, Olavesen MG, McCormack R, Guja C, Ionescu-Tirgoviste C, Undlien DE, Ronningen KS, Gillespie KM, Tuomilehto-Wolf E, Tuomilehto J, Bennett ST, Clayton DG, Cordell HJ, Todd JA. Remapping the insulin gene/IDDM2 locus in type 1 diabetes. Diabetes. 2004;53:1884–1889. doi: 10.2337/diabetes.53.7.1884. [DOI] [PubMed] [Google Scholar]
- 6.Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, Colle E, Polychronakos C. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nat Genet. 1997;15:289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 7.Pugliese A, Zeller M, Fernandez A, Jr., Zalcberg LJ, Bartlett RJ, Ricordi C, Pietropaolo M, Eisenbarth GS, Bennett ST, Patel DD. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 8.Kissler S, Stern P, Takahashi K, Hunter K, Peterson LB, Wicker LS. In vivo RNA interference demonstrates a role for Nramp1 in modifying susceptibility to type 1 diabetes. Nat Genet. 2006 Apr;38(4):479–83. doi: 10.1038/ng1766. Epub 2006 Mar 19. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton-Williams EE, Serreze DV, Charlton B, Johnson EA, Marron MP, Mullbacher A, Slattery RM. Transgenic rescue implicates beta2-microglobulin as a diabetes susceptibility gene in nonobese diabetic (NOD) mice. Proc Natl Acad Sci U S A. 2001 Sep 25;98(20):11533–8. doi: 10.1073/pnas.191383798. Erratum in: Proc Natl Acad Sci U S A 2001 Nov 6;98(23):13472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wicker LS, Todd JA, Peterson LB. Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol. 1995;13:179–200. doi: 10.1146/annurev.iy.13.040195.001143. [DOI] [PubMed] [Google Scholar]
- 11.Yui MA, Muralidharan K, Moreno-Altamirano B, Perrin G, Chestnut K, Wakeland EK. Production of congenic mouse strains carrying NOD-derived diabetogenic genetic intervals: an approach for the genetic dissection of complex traits. Mamm Genome. 1996;7:331–334. doi: 10.1007/s003359900097. [DOI] [PubMed] [Google Scholar]
- 12.Todd JA, Aitman TJ, Cornall RJ, Ghosh S, Hall JR, Hearne CM, Knight AM, Love JM, McAleer MA, Prins JB, et al. Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature. 1991;351:542–547. doi: 10.1038/351542a0. [DOI] [PubMed] [Google Scholar]
- 13.Wicker LS, Todd JA, Prins JB, Podolin PL, Renjilian RJ, Peterson LB. Resistance alleles at two non-major histocompatibility complex-linked insulin-dependent diabetes loci on chromosome 3, Idd3 and Idd10, protect nonobese diabetic mice from diabetes. J Exp Med. 1994 Nov 1;180(5):1705–13. doi: 10.1084/jem.180.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vijayakrishnan L, Slavik JM, Illes Z, Greenwald RJ, Rainbow D, Greve B, Peterson LB, Hafler DA, Freeman GJ, Sharpe AH, Wicker LS, Kuchroo VK. An autoimmune disease-associated CTLA-4 splice variant lacking the B7 binding domain signals negatively in T cells. Immunity. 2004;20:563–575. doi: 10.1016/s1074-7613(04)00110-4. [DOI] [PubMed] [Google Scholar]
- 15.Wicker LS, Chamberlain G, Hunter K, Rainbow D, Howlett S, Tiffen P, Clark J, Gonzalez-Munoz A, Cumiskey AM, Rosa RL, Howson JM, Smink LJ, Kingsnorth A, Lyons PA, Gregory S, Rogers J, Todd JA, Peterson LB. Fine mapping, gene content, comparative sequencing, and expression analyses support Ctla4 and Nramp1 as candidates for Idd5.1 and Idd5.2 in the nonobese diabetic mouse. J Immunol. 2004;173:164–173. doi: 10.4049/jimmunol.173.1.164. [DOI] [PubMed] [Google Scholar]
- 16.Lyons PA, Armitage N, Argentina F, Denny P, Hill NJ, Lord CJ, Wilusz MB, Peterson LB, Wicker LS, Todd JA. Congenic mapping of the type 1 diabetes locus, Idd3, to a 780-kb region of mouse chromosome 3: identification of a candidate segment of ancestral DNA by haplotype mapping. Genome Res. 2000;10:446–453. doi: 10.1101/gr.10.4.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, Chen SL, Fischer PA, Peterson LB, Wicker LS. The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity. 2000;13:107–115. doi: 10.1016/s1074-7613(00)00012-1. [DOI] [PubMed] [Google Scholar]
- 18.Olofsson P, Holmberg J, Tordsson J, Lu S, Akerstrom B, Holmdahl R. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat Genet. 2003;33:25–32. doi: 10.1038/ng1058. [DOI] [PubMed] [Google Scholar]
- 19.Boackle SA, Holers VM, Chen X, Szakonyi G, Karp DR, Wakeland EK, Morel L. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity. 2001;15:775–785. doi: 10.1016/s1074-7613(01)00228-x. [DOI] [PubMed] [Google Scholar]
- 20.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- 21.Cervino AC, Gosink M, Fallahi M, Pascal B, Mader C, Tsinoremas NF. A comprehensive mouse IBD database for the efficient localization of quantitative trait loci. Mamm Genome. 2006 Jun;17(6):565–74. doi: 10.1007/s00335-005-0170-4. Epub 2006 Jun 12. [DOI] [PubMed] [Google Scholar]
- 22.Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci U S A. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 24.Eaves IA, Wicker LS, Ghandour G, Lyons PA, Peterson LB, Todd JA, Glynne RJ. Combining mouse congenic strains and microarray gene expression analyses to study a complex trait: the NOD model of type 1 diabetes. Genome Res. 2002;12:232–243. [PubMed] [Google Scholar]
- 25.Hill NJ, Lyons PA, Armitage N, Todd JA, Wicker LS, Peterson LB. NOD Idd5 locus controls insulitis and diabetes and overlaps the orthologous CTLA4/IDDM12 and NRAMP1 loci in humans. Diabetes. 2000;49:1744–1747. doi: 10.2337/diabetes.49.10.1744. [DOI] [PubMed] [Google Scholar]
- 26.Hunter K, Rainbow D, Todd JA, Peterson LB, Wicker LS. Interactions between Idd5.1/Ctla4 and other type 1 diabetes genes. 2006. Co-submitted. [DOI] [PubMed] [Google Scholar]
- 27.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 28.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 29.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 30.Lin Y, Reynolds P, Feingold E. An empirical Bayesian method for differential expression studies using one-channel microarray data. Statistical Applications in Genetics and Molecular Biology. 2003;2(1) doi: 10.2202/1544-6115.1024. Article 8. [DOI] [PubMed] [Google Scholar]
- 31.Greve B, Vijayakrishnan L, Kubal A, Sobel RA, Peterson LB, Wicker LS, Kuchroo VK. The diabetes susceptibility locus Idd5.1 on mouse chromosome 1 regulates ICOS expression and modulates murine experimental autoimmune encephalomyelitis. J Immunol. 2004;173:157–163. doi: 10.4049/jimmunol.173.1.157. [DOI] [PubMed] [Google Scholar]
- 32.Cordell HJ, Todd JA, Hill NJ, Lord CJ, Lyons PA, Peterson LB, Wicker LS, Clayton DG. Statistical modeling of interlocus interactions in a complex disease: rejection of the multiplicative model of epistasis in type 1 diabetes. Genetics. 2001 May;158(1):357–67. doi: 10.1093/genetics/158.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamann J, Vogel B, van Schijndel GM, van Lier RA. The seven- span transmembrane receptor CD97 has a cellular ligand (CD55, DAF) J Exp Med. 1996;184:1185–1189. doi: 10.1084/jem.184.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray JX, Haino M, Roth MJ, Maguire JE, Jensen PN, Yarme A, Stetler-Stevenson MA, Siebenlist U, Kelly K. CD97 is a processed, seven-transmembrane, heterodimeric receptor associated with inflammation. J Immunol. 1996;157:5438–5447. [PubMed] [Google Scholar]
- 35.Hamann J, Stortelers C, Kiss-Toth E, Vogel B, Eichler W, van Lier RA. Characterization of the CD55 (DAF)-binding site on the seven-span transmembrane receptor CD97. Eur J Immunol. 1998;28:1701–1707. doi: 10.1002/(SICI)1521-4141(199805)28:05<1701::AID-IMMU1701>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 36.Sogabe H, Nangaku M, Ishibashi Y, Wada T, Fujita T, Sun X, Miwa T, Madaio MP, Song WC. Increased susceptibility of decay-accelerating factor deficient mice to anti-glomerular basement membrane glomerulonephritis. J Immunol. 2001;167:2791–2797. doi: 10.4049/jimmunol.167.5.2791. [DOI] [PubMed] [Google Scholar]
- 37.Lin F, Emancipator SN, Salant DJ, Medof ME. Decay-accelerating factor confers protection against complement-mediated podocyte injury in acute nephrotoxic nephritis. Lab Invest. 2002;82:563–569. doi: 10.1038/labinvest.3780451. [DOI] [PubMed] [Google Scholar]
- 38.Lin F, Kaminski HJ, Conti-Fine BM, Wang W, Richmonds C, Medof ME. Markedly enhanced susceptibility to experimental autoimmune myasthenia gravis in the absence of decay-accelerating factor protection. J Clin Invest. 2002;110:1269–1274. doi: 10.1172/JCI16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Miwa T, Hilliard B, Chen Y, Lambris JD, Wells AD, Song WC. The complement inhibitory protein DAF (CD55) suppresses T cell immunity in vivo. J Exp Med. 2005;201:567–577. doi: 10.1084/jem.20040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koarada S, Wu Y, Ridgway WM. Increased entry into the IFN-gamma effector pathway by CD4+ T cells selected by I-Ag7 on a nonobese diabetic versus C57BL/6 genetic background. J Immunol. 2001;167:1693–1702. doi: 10.4049/jimmunol.167.3.1693. [DOI] [PubMed] [Google Scholar]
- 41.Koarada S, Wu Y, Olshansky G, Ridgway WM. Increased nonobese diabetic Th1:Th2 (IFN-gamma:IL-4) ratio is CD4+ T cell intrinsic and independent of APC genetic background. J Immunol. 2002;169:6580–6587. doi: 10.4049/jimmunol.169.11.6580. [DOI] [PubMed] [Google Scholar]
- 42.Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- 43.Trembleau S, Penna G, Bosi E, Mortara A, Gately MK, Adorini L. Interleukin 12 administration induces T helper type 1 cells and accelerates autoimmune diabetes in NOD mice. J Exp Med. 1995;181:817–821. doi: 10.1084/jem.181.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trembleau S, Penna G, Gregori S, Gately MK, Adorini L. Deviation of pancreas-infiltrating cells to Th2 by interleukin-12 antagonist administration inhibits autoimmune diabetes. Eur J Immunol. 1997;27:2330–2339. doi: 10.1002/eji.1830270930. [DOI] [PubMed] [Google Scholar]
- 45.Trembleau S, Penna G, Gregori S, Chapman HD, Serreze DV, Magram J, Adorini L. Pancreas-infiltrating Th1 cells and diabetes develop in IL-12-deficient nonobese diabetic mice. J Immunol. 1999;163:2960–2968. [PubMed] [Google Scholar]
- 46.Rabinovitch A. Immunoregulatory and cytokine imbalances in the pathogenesis of IDDM. Therapeutic intervention by immunostimulation? Diabetes. 1994;43:613–621. doi: 10.2337/diab.43.5.613. [DOI] [PubMed] [Google Scholar]
- 47.Rabinovitch A, Sorensen O, Suarez-Pinzon WL, Power RF, Rajotte RV, Bleackley RC. Analysis of cytokine mRNA expression in syngeneic islet grafts of NOD mice: interleukin 2 and interferon gamma mRNA expression correlate with graft rejection and interleukin 10 with graft survival. Diabetologia. 1994;37:833–837. doi: 10.1007/BF00404341. [DOI] [PubMed] [Google Scholar]
- 48.Rabinovitch A, Suarez-Pinzon WL, Sorensen O, Bleackley RC, Power RF, Rajotte RV. Combined therapy with interleukin-4 and interleukin-10 inhibits autoimmune diabetes recurrence in syngeneic islet-transplanted nonobese diabetic mice. Analysis of cytokine mRNA expression in the graft. Transplantation. 1995;60:368–374. doi: 10.1097/00007890-199508270-00012. [DOI] [PubMed] [Google Scholar]
- 49.Fox CJ, Danska JS. IL-4 expression at the onset of islet inflammation predicts nondestructive insulitis in nonobese diabetic mice. J Immunol. 1997;158:2414–2424. [PubMed] [Google Scholar]
- 50.Adorini L, Trembleau S. Immune deviation towards Th2 inhibits Th1-mediated autoimmune diabetes. Biochem Soc Trans. 1997;25:625–629. doi: 10.1042/bst0250625. [DOI] [PubMed] [Google Scholar]