Abstract
Galectin-1 (Gal1) and galectin-3 (Gal3) are two members of a family of carbohydrate-binding proteins that are found in the nucleus and that participate in pre-mRNA splicing assayed in a cell-free system. When nuclear extracts (NE) of HeLa cells were subjected to adsorption on a fusion protein containing glutathione S-transferase (GST) and Gal3, the general transcription factor II-I (TFII-I) was identified by mass spectrometry as one of the polypeptides specifically bound. Lactose and other saccharide ligands of the galectins inhibited GST-Gal3 pull-down of TFII-I while non-binding carbohydrates failed to yield the same effect. Similar results were also obtained using GST-Gal1. Site-directed mutants of Gal1, expressed and purified as GST fusion proteins, were compared with the wild-type (WT) in three assays: (a) binding to asialofetuin-Sepharose as a measure of the carbohydrate-binding activity; (b) pull-down of TFII-I from NE; and (c) reconstitution of splicing in NE depleted of galectins as a test of the in vitro splicing activity. The binding of GST-Gal1(N46D) to asialofetuin-Sepharose was less than 10% of that observed for GST-Gal1(WT), indicating that the mutant was deficient in carbohydrate-binding activity. In contrast, both GST-Gal1(WT) and GST-Gal1(N46D) were equally efficient in pull-down of TFII-I and in reconstitution of splicing activity in the galectin-depleted NE. Moreover, while the splicing activity of the wild-type protein can be inhibited by saccharide ligands, the carbohydrate-binding deficient mutant was insensitive to such inhibition. Together, all of the results suggest that the carbohydrate-binding and the splicing activities of Gal1 can be dissociated and therefore, saccharide-binding, per se, is not required for the splicing activity.
Keywords: lectins, carbohydrate-binding proteins, RNA processing, general transcription factor
Introduction
Galectins are a family of widely distributed proteins that: (a) bind to β-galactoside containing glycoconjugates; and (b) contain characteristic amino acid sequences in the carbohydrate recognition domain (CRD) of the polypeptides [1, 2]. Most members of the galectin family are predominantly intracellular proteins and observations of their localization in the cell nucleus have been reported for 12 of the 15 galectins known to date [3]. In previous studies, we had reported the nuclear localization of galectin-1 (Gal1) and galectin-3 (Gal3) in the form of a ribonucleoprotein complex [4–6]. Several key findings suggest that Gal1 and Gal3 are two of the many proteins involved in the splicing of pre-mRNA, assayed in a cell-free system [7–9]: (a) nuclear extracts (NE) derived from HeLa cells, capable of carrying out splicing of pre-mRNA, contained both Gal1 and Gal3; (b) depletion of both galectins from NE, either by lactose (Lac) affinity chromatography or by antibody adsorption, resulted in the concomitant loss of splicing activity; (c) either recombinant Gal1 or recombinant Gal3 was able to reconstitute splicing activity in a galectin-depleted extract; (d) saccharides which bind to Gal1 and Gal3 with high affinity, such as Lac and thiodigalactoside (TDG), inhibited the splicing reaction when added to a complete NE whereas non-binding saccharides such as cellobiose failed to have any effect; and (e) when a splicing reaction containing 32P-labeled pre-mRNA is subjected to immunoprecipitation with either anti-Gal1 or anti-Gal3, radiolabeled RNA species corresponding to the starting substrate, intermediates of the splicing reaction, and mature mRNA products of active spliceosomes are all co-precipitated with the specific galectin. All of these results suggest that Gal1 and Gal3 are associated with spliceosomal complexes throughout the splicing pathway.
The polypeptide of Gal1 consists of a single domain, the CRD. On the other hand, the polypeptide of Gal3 can be delineated into two distinct domains: (a) an NH2-terminal domain (ND) containing multiple repeats of a nine-residue motif, PGAYPGXXX; and (b) a COOH-terminal CRD that shows sequence similarity with the corresponding CRDs of other members of the galectin family [1, 2]. Previous experiments have documented that the CRD alone (Gal1 or the carboxyl-terminal domain of Gal3) was necessary and sufficient for reconstitution of splicing activity in a galectin-depleted NE [8]. The availability of site-directed mutants of Gal1, devoid of carbohydrate-binding activity [10, 11], provided the key reagents to test whether saccharide-binding by the CRD, per se, is necessary for splicing activity.
In the present studies, we report that using a fusion protein containing glutathione S-transferase (GST) and Gal1 (GST-Gal1) or Gal3 (GST-Gal3), we can pull-down from NE the general transcription factor II-I (TFII-I), a protein previously identified as a spliceosomal component by proteomic analysis [12]. This association between TFII-I and the galectins occurs via the CRD and is sensitive to inhibition by saccharide ligands. We have taken advantage of this interaction to document that site-directed mutants of Gal1, devoid of carbohydrate-binding activity, retained the association with TFII-I as well as the cell-free splicing activity, thereby dissociating the saccharide-binding of the protein from its spliceosomal function.
Materials and Methods
GST fusion proteins and pull-down assay
The cDNAs for wild-type (WT) and three mutants (N46D, C60S, and E71Q) of human Gal1 have been described [10, 11]. Each cDNA was subcloned into the BamHI restriction site of the pGEX-2T vector (Pharmacia) to produce a fusion protein between GST and Gal1. The plasmid pWJ31 containing the cDNA for murine Gal3 [13] was subcloned into the EcoRI restriction site of the vector pGEX-5X-1 to produce GST-Gal3. Agrwal et al. [14] had described site-directed mutagenesis of the cDNA for murine Gal3 in which Gly 138 and Gly139 were replaced by two stop codons. Thus, the translation open reading frame stops at Pro137, giving an ND covering residues 1–137. This mutant cDNA of Gal3 was subcloned into the expression vector pGEX-5X-1 to express GST-Gal3ND. Each of the constructs was subjected to DNA sequencing to verify: (a) the juncture of the fusion protein between GST and either Gal1 or Gal3; and (b) the wild-type and mutant amino acid at the mutagenized residue.
GST fusion proteins were expressed in E. coli BL-21 codon plus (DE3) cells (Stratagene) by induction with 100 µM isopropyl-β-D-galactopyranoside for 2–3 hours at 30 °C. Cells were pelleted and stored at −70 °C. Thawed bacterial pellets were suspended in PBS containing protease inhibitors (4 µg/ml aprotinin, 5 µg/ml leupeptin, 0.2 µg/ml pepstatin A, and 1 mM Pefabloc (Roche)) and sonicated using a microtip probe. Triton X-100 was added to a final concentration of 0.1%. After rocking for 1 hour at 4 °C, cell debris was removed by centrifugation at 12,000 × g for 10 minutes at 4 °C. The supernatant was purified on the basis of GST binding to glutathione-agarose beads (Pierce).
For GST pull-down experiments, ~10 µg of each GST fusion protein were incubated with 20 µl of packed glutathione beads in the presence of 60% buffer D (20 mM Hepes-KOH, pH 7.9, 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol (DTT)), either at room temperature for 1.5 hours or at 4 °C for ~14 hours. Unbound material was removed and the beads were washed three times with 400 µl of 60% buffer D. The beads were then incubated with 36 µl of NE (~200 µg total protein) along with 24 µl of 60% buffer D, with 14.7 mM creatine phosphate, 2.4 mM MgCl2, and 0.4 mM ATP (final concentrations). In experiments to test the effect of saccharides on the pull-down assay, they were included in this addition at a concentration of 100 mM. The incubation was carried out at 4 °C for 12 hours. After removal of unbound material, the beads were washed four times with 200 µl of 60% buffer D. The material bound to the beads was then eluted by incubation with glutathione elution buffer (16 mM glutathione, 60 mM HEPES-KOH, pH 7.9, 11.4% glycerol, 57 mM KCl, and 0.114 mM EDTA) at 31 °C for 30 minutes, followed by incubation at room temperature for one hour. The eluted material was then subjected to SDS-PAGE analysis.
Antibody reagents
For antibodies directed against TFII-I, we used two affinity purified preparations purchased from Bethyl Labs. Antibody #557 was derived from serum of rabbits immunized with a peptide sequence contained in exons 27 and 28 of TFII-I; antibody #558 was generated in a similar fashion using a peptide sequence in exons 32 and 33. Human autoimmune serum reactive against the Sm epitopes (anti-Sm) found on the core polypeptides of snRNPs was purchased from The Binding Site. For antibodies directed against the Survival of Motor Neuron Protein (SMN), we used a mouse monoclonal antibody (directed against residues 14–174 of the SMN polypeptide) purchased from BD Transduction Labs.
The rat monoclonal antibody designated as anti-Mac-2 [15, 16] was used as antibody directed against Gal3. Affinity purified polyclonal rabbit anti-Gal1 and anti-GST antibodies were prepared using the immunogen GST-Gal1(WT), purified on the basis of binding to two columns: (a) glutathione-agarose and elution with glutathione; and (b) Lac-agarose and elution with Lac. Approximately 70 ml of antisera, pooled from four bleeds of rabbit #55, were subjected to ammonium sulfate fractionation (50% of saturation). The immunoglobulin-containing precipitated fraction was solubilized in, and dialyzed against, phosphate-buffered saline (PBS) and passed over a 5 ml column of GST-agarose. The unbound (flow-through) fraction was immediately loaded over the same column (six passes over the same column to insure binding). The bound fraction was eluted with 0.1 M glycine-HCl (pH 2.2) and this was dialyzed immediately against PBS to neutralize the pH. The bound and eluted material from the GST affinity column is designated as affinity purified anti-GST. The immunogen, purified GSTGal1( WT), was bound to glutathione-agarose and covalently cross-linked with dimethylpimelimidate (20 mM; Pierce). The reaction was carried out in 0.2 M sodium borate (pH 9) for 1 hour at room temperature; the cross-linked beads were washed twice with 0.2 M ethanolamine, pH 8, followed by a 2-hour incubation at room temperature in the same buffer to block unreacted groups. The unbound fractions of the antisera, depleted of anti-GST antibodies, were passed over this GST-Gal1(WT) affinity column (six passes to insure binding). The bound and eluted material from this column is designated as affinity purified rabbit anti-Gal1. At 1:2000, 1:5000, and 1:10,000 dilutions, this affinity purified anti-Gal1 blots Gal1 in NE of HeLa cells and purified GST-Gal1(WT) but does not blot GST.
NE and splicing reactions
Human HeLa S3 cells were grown in suspension culture by the National Cell Culture Center (Minneapolis, MN) and were shipped as a cell pellet on wet ice. NE was prepared in buffer D as described by Dignam et al. [17]. NEs were frozen as aliquots in a dry ice-ethanol bath and stored at −80 °C. Protein concentrations were determined by the Bradford assay [18]. In this study, the protein concentration of NE was 4–6 mg/ml. Splicing reaction mixtures, in a total volume of 12 µl, contained dialyzed NE sample (10 µl), [32P]MINX pre-mRNA [19], 2.5 mM MgCl2, 1.5 mM ATP, 20 mM creatine phosphate, 0.5 mM DTT, and 20 U RNasin (Promega). Splicing reactions were incubated at 30 °C for 45 minutes. The assay was stopped by addition of proteinase K and SDS to final concentrations of 4 mg/ml and 0.1%, respectively. RNA was extracted and then subjected to electrophoresis in 13% polyacrylamide-8.3 M urea gels, followed by autoradiography [8]. Quantitation of product formation was carried out by exposing the gel to a Storage Phosphor Screen (Amersham Biosciences), scanning on a Storm 860 scanner (Molecular Dynamics), and using the program Image Quant (Molecular Dynamics) to determine the percentage of radioactivity in specific bands in each lane.
Splicing reaction mixtures were also carried out in a total volume of 50 µl, containing 30 µl of NE and [32P]MINX. These splicing reactions were incubated at 30 °C for 40 minutes and then subjected to immunoprecipitation. Antibodies were bound to protein A-Sepharose beads (Pharmacia) in 60% buffer D containing 0.05% Triton X-100. After removal of unbound material and washing, the splicing reaction (containing 106 cpm of 32P-labeled RNA in 50 µl) was added to the antibody beads in 200 µl of 60% buffer D containing 0.05% Triton X-100. Incubation was carried out at 4 °C for 1.5 hours. The unbound material was removed and the beads were washed three times, each with 0.5 ml 60% buffer D containing 0.05% Triton X-100. SDS solution and proteinase K were added to a final concentration of 0.1% and 4 mg/ml, respectively, and the samples were incubated at 37 °C for 20 minutes. The RNA components of the precipitated fraction were extracted and analyzed as described above.
NEs were depleted of Gal1 and Gal3 by adsorption on beads covalently coupled with rabbit anti-Gal1 and rat anti-Mac-2. The beads (150 µl) were washed with 20 mM Hepes, pH 7.9, 0.5 M NaCl; 30 µl NE were added and incubated on ice for 20 minutes in disposable spin columns (Millipore). The unbound fraction was removed and the beads were washed with 12 µl of 60% buffer D adjusted to 0.42 M NaCl; this wash was combined with the unbound fraction. Aliquots of nondepleted and depleted NE were dialyzed against 60% buffer D for 40 minutes at 4 °C [8]. In reconstitution experiments, GST or GST-Gal1 (wild-type and mutant proteins) was added to the depleted NE prior to dialysis. The dialyzed fractions were then assayed for splicing activity.
Assay of Carbohydrate-binding Activity
The preparation of the affinity beads, asialofetuin (ASF)-Sepharose 4B, has been described [20]. Approximately 180 nmoles of ASF were coupled per ml of beads. GST proteins (350 ng each of GST, GST-Gal1 (wild-type and three mutants)) were incubated with 35 µl of ASF-Sepharose for 2 hours at 4 °C. The incubations were carried out in 60% buffer D containing 0.1% NP-40 (Pierce) in the presence and absence of Lac (100 mM). The material not bound to the beads was removed by centrifugation (1000 × g); after resuspension, the beads were washed four times in 60% buffer D containing 0.1% NP-40. The GST proteins bound to the beads were eluted and subjected to SDS-PAGE and immunoblotting with anti-GST and anti-Gal1 antibodies.
The chemiluminescent signal provided by horseradish peroxidase conjugated to the secondary antibody was detected using the Western Lightning reagent (Perkin Elmer Life Sciences). This signal was quantitated with a BioRad model GS505 Molecular imager system and associated software. Known amounts of GST and GST-Gal1 were used to establish standard curves. The intensity of the immunoblotted band derived from the incubation carried out in the presence of Lac represented non-specific binding not inhibitable by Lac; this accounted for about 3% of the total binding for both the wild-type and mutant proteins. Lac-inhibitable specific binding was calculated by subtracting this value from the total binding.
SDS gel electrophoresis, silver staining, and immunoblotting
Samples were subjected to SDS-PAGE as described by Laemmli [21]. Proteins were visualized by silver staining as described by Merril et al. [22]. For immunoblotting, samples were electrophoretically transferred onto Hybond Nitrocellulose membrane (Amersham Biosciences) in the presence of buffer containing 25 mM Tris, 193 mM glycine, and 10% methanol, pH 8.3. Following transfer, membranes were incubated overnight in 10% nonfat dry milk in Tris-buffered saline containing Tween 20 (10 mM Tris, 0.5 M NaCl, 0.05% Tween 20; T-TBS). Antibodies for immunoblotting were diluted in T-TBS containing 1% nonfat dry milk and incubated with membranes for one hour at room temperature. This was followed by four washes (15 minutes each) in T-TBS. The membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for one hour and washed four times. Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse immunoglobulin (Bio-Rad) and goat anti-rat immunoglobulin (Roche) were each used at a dilution of 1:10,000. The proteins were visualized using chemiluminescence.
Mass spectrometric analysis of selected gel slices derived from GST pull-downs
After SDS-PAGE, the gel was stained with Coomassie Brilliant Blue G (Brilliant Blue G-colloidal concentrate; Sigma) [23]. Gel slices corresponding to the ~40 kD and ~135 kD regions were washed with 100 mM NH4HCO3 (100 µl per 1 mm3 slice) for 5 minutes and the wash was discarded. Each gel slice was then subjected to two rounds of dehydration in acetonitrile (50 µl; 15 minutes). The gel slices were then rehydrated and digested with trypsin (Promega) following the protocol of Shevchenko et al [24]. The trypsin digests were fractionated by reverse-phase high pressure liquid chromatography, followed by electrospray ionization mass spectrometry (LC-MS/MS). The mass spectrometry and the subsequent MS/MS ion search (Mascot) were carried out by the Proteomics Core of the Research Technology Support Facility at Michigan State University.
Results
GST-Gal3 pull-down of TFII-I from NE
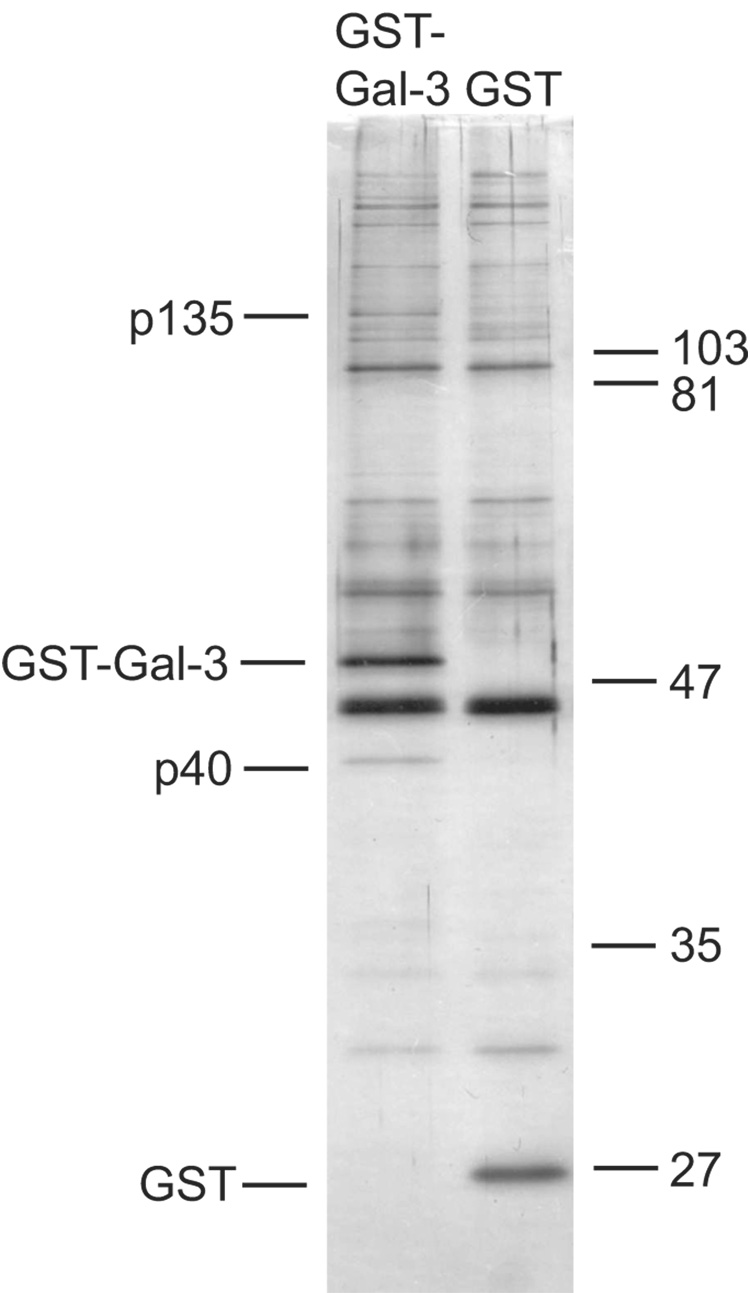
HeLa cell NE was subjected to adsorption onto glutathione beads containing GST-Gal3 and GST. The material bound to the beads was eluted with soluble glutathione and analyzed by SDS-PAGE. Comparison of the silver-stained gels revealed two major bands present in the material bound to the GST-Gal3 beads but not to GST; these were designated as p40 and p135, corresponding to their approximate molecular weights (Fig. 1). Gel slices containing p40 and p135 were analyzed by mass spectrometry. The MS/MS ion search on the analysis of the p135 gel slice revealed twelve matches, representing five distinct tryptic peptides (each with a carboxyl terminal lysine or arginine), with the amino acid sequence of human TFII-I. In contrast, the corresponding gel slice from parallel SDS-PAGE of the material derived from the GST beads did not yield any of these TFII-I peptides. The MS/MS ion search on the analysis of the p40 gel slice yielded four matches representing three peptides of actin.
Figure 1. Comparison of the polypeptides bound to GST-Gal3 and GST.
NE was subjected to GST pull-down in 60% buffer D containing 14.7 mM creatine phosphate, 2.4 mM MgCl2, and 0.4 mM ATP. The material bound to the beads was eluted with 16 mM glutathione and subjected to SDS-PAGE. Polypeptides were revealed by silver staining. The positions of migration of p135, GST-Gal3, p40, and GST are highlighted on the left; the positions of migration of molecular weight markers are indicated on the right.
Although initially identified as a general transcription factor [25], TFII-I has actually been studied under a wide variety of contexts and therefore, the same polypeptide has acquired a number of different names: (a) Serum Responsive Factor-Phox 1 Interacting Protein (SPIN) [26]; and (b) Bruton's tyrosine kinase-associated protein of Mr ~135,000 (BAP135) [27]. Inasmuch as we had previously documented that Gal1 and Gal3 are factors involved in pre-mRNA splicing [7–9], it was of particular interest that a proteomic analysis of the spliceosome identified TFII-I as one of its proteins [12]. On this basis, we have studied the association of TFII-I with Gal3 (and Gal1) as reported below; on the other hand, we have not pursued the pull-down of actin any further.
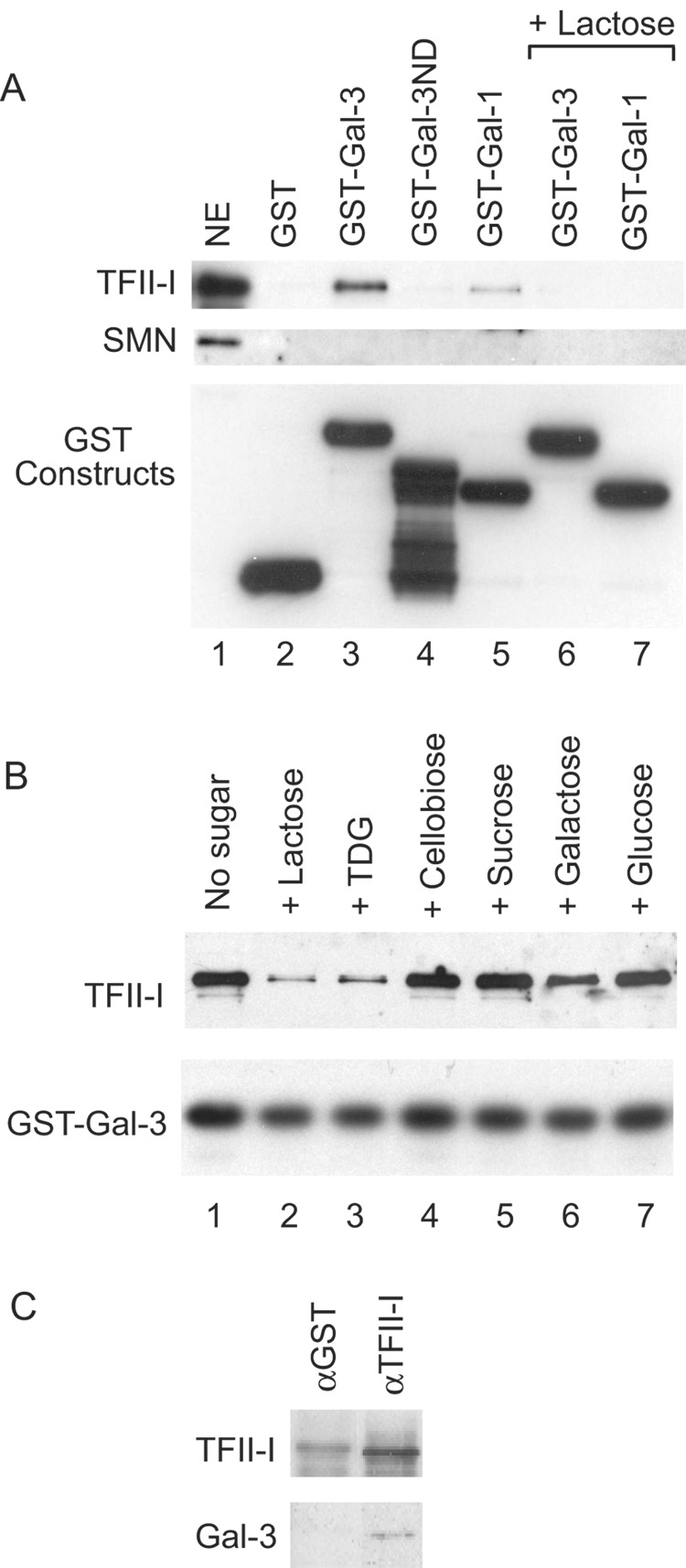
The identification of TFII-I was confirmed by immunoblot analysis. Using antibodies directed against TFII-I (# 558), a positive reaction was observed in the material bound to GST-Gal3 but not in the material bound to GST (Fig. 2A, lanes 2 and 3). In contrast, antibodies directed against the SMN protein failed to yield a positive immunoblot in the GST-Gal3 pull-down material (Fig. 2A). Using anti-GST antibodies, we ascertained that approximately equal amounts of GST proteins were used in the pull-down assay. These results suggest that the GST-Gal3 pull-down of TFII-I represented a specific association, either directly or indirectly, between the two proteins.
Figure 2. Immunoblotting analysis of various GST pull-down and immunopreciptation experiments.
(A) NE (lane 1), representing 15% of the material subjected to pull-down by various GST constructs: GST (lane 2), GST-Gal3 (lane 3), GST-Gal3ND (lane 4), GST-Gal1 (lane 5), GSTGal3 in the presence of 100 mM Lac (lane 6), and GST-Gal1 in the presence of 100 mM Lac (lane 7). Top panel: immunoblotting by anti-TFII-I (#558); middle panel: immunoblotting by anti-SMN; and bottom panel: immunoblotting by anti-GST to monitor the amount of GST fusion proteins bound to the glutathione beads.
(B) NE was subjected to GST-Gal3 pull-down in the absence of any carbohydrate (lane 1) or in the presence of 100 mM of Lac (lane 2), TDG (lane 3), cellobiose (lane 4), sucrose (lane 5), galactose (lane 6), and glucose (lane 7). Top panel: immunoblotting by anti-TFII-I (#558); lower panel: immunoblotting by anti-GST to monitor the amount of GST-Gal3 fusion protein bound to the glutathione beads.
(C) NE was subjected to immunoprecipitation by anti-TFII-I (#557), a rabbit antiserum affinity purified over the immunogen. Anti-GST, an antiserum that went through the same affinity purification procedure, was used as a negative control. The immunoprecipitate was subjected to blotting with anti-TFII-I and anti-Mac-2, a rat monoclonal antibody directed against Gal3.
Effect of the saccharide ligands and involvement of the CRD
Such a conclusion is supported by the observation that Lac, a specific saccharide ligand of Gal3, inhibited the GST-Gal3 pull-down of TFII-I (Fig. 2A, lane 6). A limited survey was therefore carried out to determine the effects of various carbohydrates on the GST-Gal3 pull-down. The disaccharide ligands, Lac and thiodigalactoside (TDG), both inhibited the pull-down (Fig. 2B, lanes 2 and 3). In contrast, cellobiose and sucrose, two disaccharides that do not bind to Gal3 [28], failed to yield the same effect (Fig. 2B, lanes 4 and 5). The galectins exhibit a much lower affinity for the monosaccharide ligand galactose and indeed, the latter showed only a weak inhibitory effect (Fig. 2B, lane 6). Finally, glucose does not bind to the galectins and showed no effect on the GST-Gal3 pull-down of TFII-I (Fig. 2B, lane 7). These saccharide-specific effects suggest that the CRD of the Gal3 polypeptide may be involved in the TFII-I pull-down.
The amino acid sequence of the Gal3 polypeptide can be delineated into two domains (a) an ND containing multiple repeats of a 9-residue motif, PGAYPGXXX; and (b) a COOH-terminal CRD that shows sequence similarity with the corresponding CRD of other members of the galectin family. While full-length GST-Gal3 interacted with TFII-I, GST-Gal3ND failed to yield TFII-I in the pull-down (Fig. 2A, lanes 3 and 4). On the other hand, GST-Gal1, which contains a single CRD, interacted with TFII-I and this interaction was also sensitive to Lac inhibition (Fig. 2A, lanes 5 and 7). Therefore, it appears that the site responsible for the TFII-I pull-down resides within the CRD of either Gal1 or Gal3.
Finally, we tested for the presence of Gal3 in the immunoprecipitates of anti-TFII-I as a reciprocal of the GST-galectin pull-down experiments. Indeed, anti-TFII-I immunoprecipitated not only its own cognate antigen but Gal3 as well (Fig. 2C). The anti-TFII-I antibody (#557) is a polyclonal rabbit antiserum affinity purified over the peptide immunogen. For a negative control, we used an anti-GST antiserum that went through the same affinity purification procedure. No Gal3 was observed in the control precipitate (Fig. 2C).
Immunoprecipitation by anti-TFII-I of 32P-labeled RNA in the splicing reaction
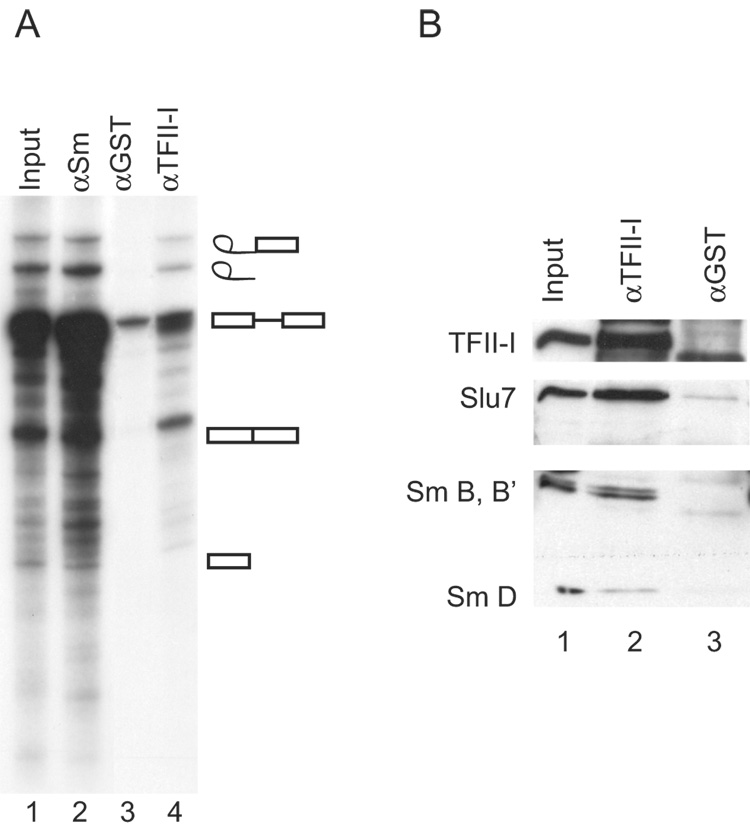
When a splicing reaction containing [32P]pre-mRNA substrate was subjected to immunoprecipitation with antibodies directed against either Gal1 or Gal3, spliceosomal complex(es) were coprecipitated along with the cognate antigen, a conclusion based on finding 32P-labeled RNA species that are produced on the spliceosome during the splicing reaction [9]. We therefore used the same strategy to test for an association of TFII-I with the spliceosome. Indeed, the starting pre-mRNA substrate, the products of the splicing reaction, as well as the intermediates (exon 1 and lariat-exon 2) were all observed in the anti-TFII-I precipitate (Fig. 3A, lane 4). In contrast, much less radioactivity, corresponding only to the pre-mRNA substrate species, was observed in the precipitate of the control anti-GST antibody (Fig. 3A, lane 3). The profile of RNA precipitation observed with anti-TFII-I was also found with human autoimmune serum reactive against the Sm epitopes of the core polypeptides of small nuclear ribonucleoprotein complexes (snRNPs), which served as a positive control (Fig. 3A, lane 2).
Figure 3. Analysis of spliceosomal RNA species and proteins immunoprecipitated by various antisera.
Splicing reactions incubated for 40 minutes with 32P-labeled MINX were subjected to antibody adsorption. (A) The bound RNA was analyzed by electrophoresis through a 13% polyacrylamide- 8.3 M urea gel system, followed by autoradiography. Lane 1 - the splicing reaction mixture (4%) that was subjected to immunoprecipitation; lane 2 - immunoprecipitate of anti-Sm; lane 3 – immunoprecipitate of affinity-purified anti-GST; lane 4 - immunoprecipiate of anti-TFII-I (#558). (B) Following a splicing reaction, the sample (input, lane 1) was subjected to immunoprecipitation by anti-TFII-I (lane 2) or anti-GST (lane 3). Top panel: immunoblotting by anti-TFII-I (#558); middle panel: immunoblotting by anti-Slu7; bottom panel: immunoblotting by anti-Sm.
Along with the analysis for 32P-labeled RNA species, parallel samples of the immunoprecipitate were subjected to SDS-PAGE and immunoblotting (Fig. 3B). In addition to its own antigen, the anti-TFII-I precipitate also yielded positive reactions with: (a) the Sm epitopes of snRNPs, and (b) Slu7, a factor required for the second trans-esterification reaction during splicing [29], representing a late stage splicing complex (Fig. 3B, lane 2). On the basis of analysis of both RNA and proteins species, therefore, it appears that TFII-I is a bona fide component of spliceosomes, as the proteomic study of Rappsilber et al [12] had implicated.
Comparison of wild-type and mutant Gal1 proteins in carbohydrate-binding and TFII-I pull-down assays
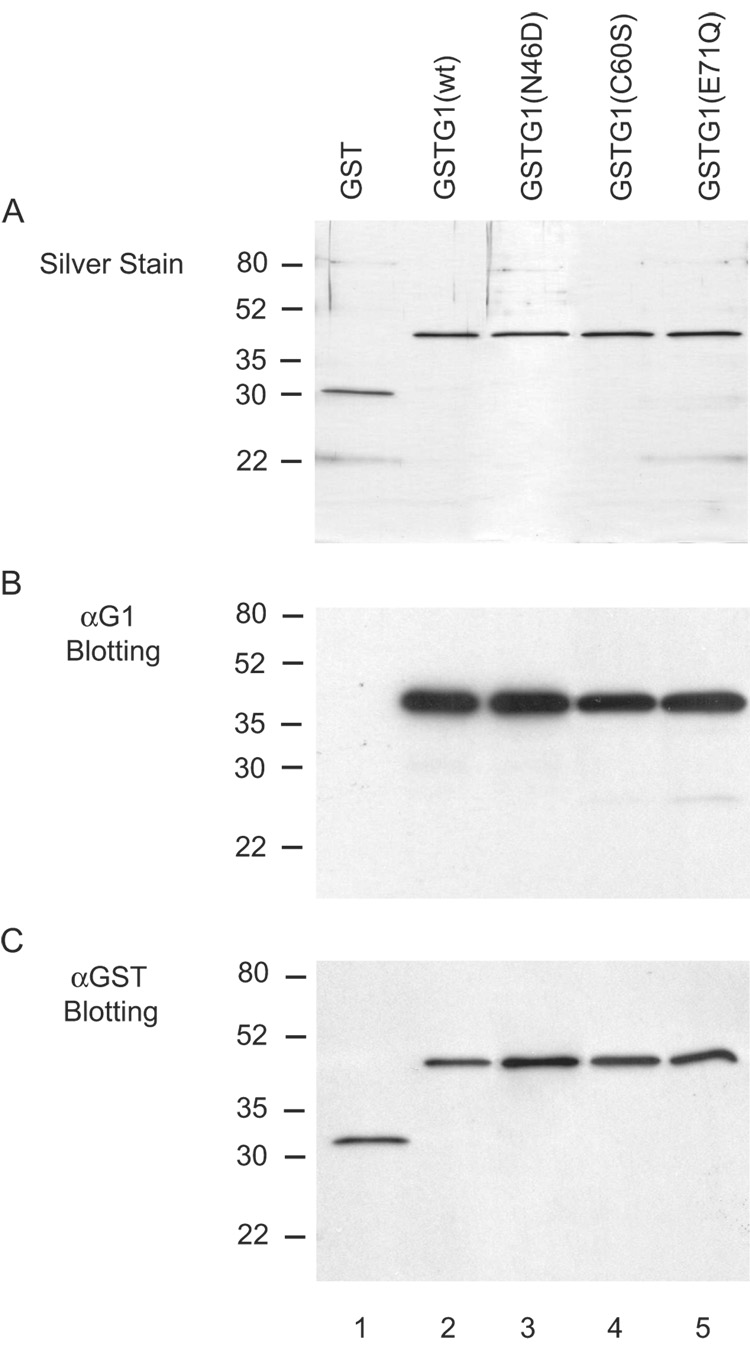
Hirabayashi and Kasai [10, 11] had demonstrated that substitutions at highly conserved residues of the CRD of human Gal1, such as Asn 46 and Glu 71, resulted in loss of saccharide-binding activity. The availability of such site-directed mutants provided the key reagents to test whether saccharide-binding, per se, is necessary for the splicing activity. The cDNAs corresponding to WT and the N46D and E71Q mutants were each subcloned to produce a fusion protein between GST and Gal1. As a control, we also carried out the same analysis on another mutant, C60S, whose substitution did not abolish the carbohydrate-binding activity [10, 11]. On SDS-PAGE and silver staining (Fig.4A), each of the purified GST proteins yielded a single band with a mobility corresponding to the expected molecular weights: GST, ~27 kD; and GST-Gal1 (wild-type and mutants), ~42 kD. Immunoblotting with affinity purified anti-Gal1 antibodies yielded a single band at the same molecular weight for GST-Gal1 (wild-type and mutant proteins) (Fig. 4B, lanes 2–5). No reaction was observed between anti-Gal1 and GST (Fig. 4B, lane 1). Finally, immunoblotting of the respective GST protein preparations with affinity purified anti-GST antibodies yielded the same single band patterns as were observed by silver staining (Fig. 4C). All of these results establish the purity of the protein reagents to be compared in the functional assays below.
Figure 4. Characterization of the preparations of fusion proteins containing wild-type or mutant Gal1 by SDS-PAGE.
Lane 1: GST; lane 2, GST-Gal1(WT); lane 3, GST-Gal1(N46D); lane 4, GST-Gal1(C60S); and lane 5, GST-Gal1 (E71Q). The proteins (~30 ng in each lane) were electrophoresed through 12.5% acrylamide gels. Panel A: silver staining; Panel B: immunoblotting with affinity purified anti-Gal1 antibodies (#55); and Panel C: immnoblotting with affinity purified anti-GST antibodies. The positions of migration of molecular weight standards are indicated on the left.
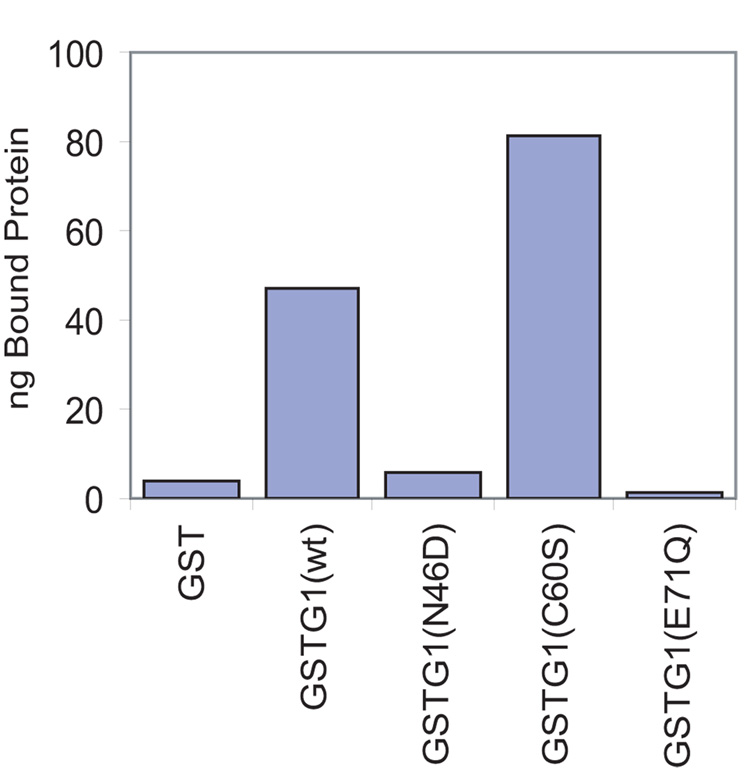
Purified GST-Gal1(WT) was compared to the mutant counterparts in terms of their binding to carbohydrate structures on the glycoprotein asialofetuin, which formed the basis for the original purification of the galectins [20]. The material bound to the ASF-beads was subjected to SDS-PAGE and immunoblotting with anti-GST antibodies. Using known amounts of GST-Gal1 to establish standard curves, we quantitated the Lac-inhibitable binding, as well as the binding not inhibitable by Lac. The latter accounted for about 3% of the total binding observed for both the wild-type and mutant proteins. In terms of Lac-inhibitable binding, 10–15% of the GST-Gal1(WT) added to the assay was bound specifically; the level of binding for GST-Gal1(C60S) was even higher (~23% of the added protein bound specifically). In contrast, less than 1% of the GST-Gal1(N46D) and GST-Gal1(E71Q) added to the assay was bound. The binding of these two mutants was drastically reduced relative to the wild-type protein and was essentially the same as that observed for GST alone (Fig. 5).
Figure 5. Comparison of the carbohydrate-binding activity of GST-Gal1(WT), GSTGal1(N46D), GST-Gal1(C60S), and GST-Gal1(E71Q).
The GST proteins were incubated with ASF-Sepharose for 2 hours at 4 °C in 60% buffer D containing 0.1% NP-40. Parallel incubations were carried out in the presence and absence of 100 mM Lac. Proteins bound to the ASF-beads were quantitated by immunoblotting with anti-GST. The values shown represent Lac inhibitable specific binding.
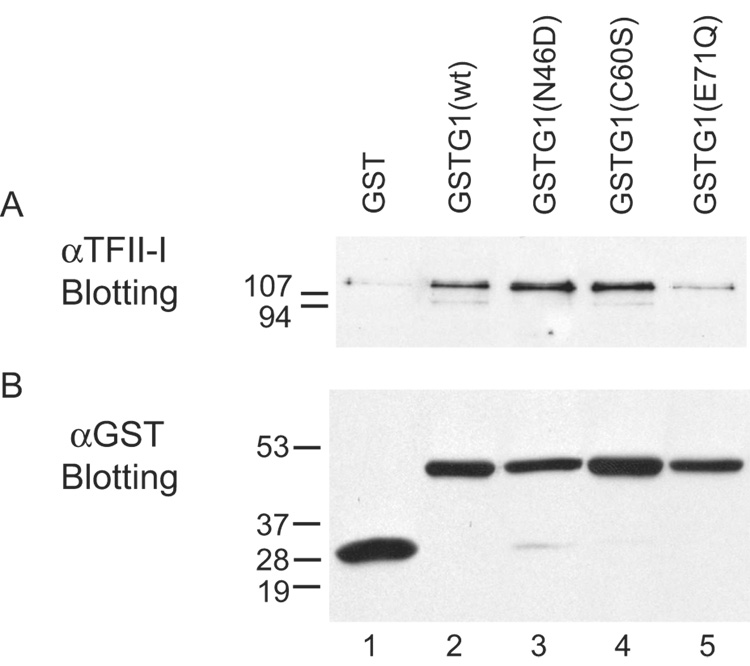
In contrast to the carbohydrate-binding assay, GST-Gal1(N46D), as well as GST-Gal1(WT) and GST-Gal1(C60S), yielded good signals for TFII-I in the pull-down assay (Fig. 6A, lanes 2–4). The signal for TFII-I in the GST-Gal1(E71Q) pull-down was substantially weaker; however, it was nevertheless still above the background level observed for GST alone (Fig. 6A, lanes 5 versus 1). Control immunoblots with anti-GST showed that approximately equal amounts of GST proteins were bound to the glutathione beads in the pull-down assay (Fig. 6B). Thus, it appears that GST-Gal1(C60S) retained both carbohydrate and TFII-I binding activities observed with GST-Gal1(WT). For GST-Gal1(N46D), on the other hand, the mutant polypeptide must have retained sufficient structure to preserve the association with TFII-I while the saccharide-binding activity was compromised. This argues against the possibility that the single amino acid substitution resulted in gross misfolding of the polypeptide. Finally, it appears that the mutation in GST-Gal1(E71Q) reduced both the carbohydrate-binding activity and its association with TFII-I, possibly reflecting some misfolding of the polypeptide.
Figure 6. Comparison of the GST pull-down of TFII-I in NE by wild-type and mutant Gal1 polypeptides.
The GST proteins were incubated overnight with glutathione beads at 4 °C in 60% buffer D. After washing to remove the unbound material, the beads were then incubated with NE for 12 hours at 4 °C. Material bound to the various beads was eluted with glutathione (16 mM) and analyzed by SDS-PAGE and immunoblotting. Panel A: immunoblotting with anti-TFII-I (# 558). Panel B:immunoblotting with anti-GST to ascertain that approximately equal amounts of GST proteins were bound to the beads in the pull-down assay.
Reconstitution of splicing in galectin-depleted NE by GST-Gal1(WT) and GST-Gal1(N46D)
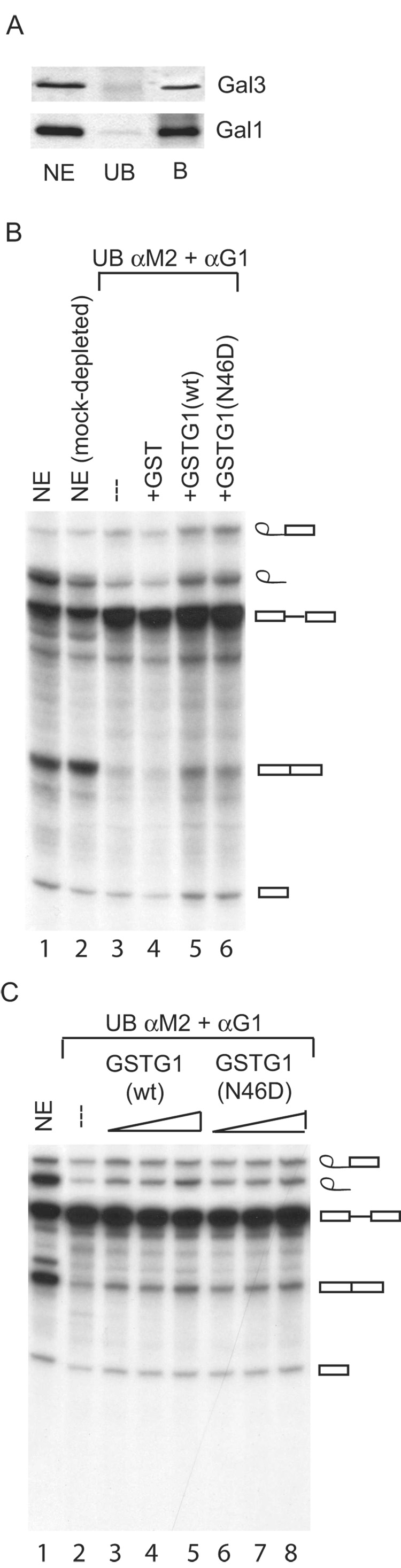
NE was prepared in buffer C, which contained 0.42 M NaCl to dissociate splicing complexes. This NE was incubated with beads covalently coupled with anti-Gal1 and anti-Gal3. Western blotting analysis documented that both proteins were present in the NE and in the bound fraction of the antibody beads. Only trace amounts of either Gal1 or Gal3 remained in the unbound fraction (Fig. 7A).
Figure 7. Comparison of the splicing activities of NE, NE after depletion of Gal1 and Gal3, and depleted NE reconstituted with GST, GST-Gal1(WT), and GST-Gal1(N46D).
NE was depleted of Gal1 and Gal3 by adsorption on beads covalently coupled with anti-Gal1 and anti-M2 (rat anti-Mac-2). Panel A: Immunoblotting for Gal1 and Gal3 in NE, the unbound (UB) fraction of the double antibody adsorption, and the bound (B) fraction. The amount of material electrophoresed in the NE and UB lanes represents ~40% of the amount electrophoresed in the B lane. Panel B: Splicing of 32P-labled MINX pre-mRNA. Lane 1, the complete (non-depleted) NE; lane 2, mock-depleted NE; lane 3, the unbound (UB) fraction of the double antibody adsorption; lanes 4–6, depleted extract reconstituted with GST, GST-Gal1(WT), and GST-Gal1(N46D), respectively. The concentration of the GST proteins was 6.5 µM. Panel C: Dose-response of the reconstitution of splicing activity by GST-Gal1(WT) and GST-Gal1(N46D). Lane 1, the complete NE; lane 2, the unbound (UB) fraction of the double antibody adsorption; lanes 3–5, reconstitution with 1 µM, 6.5 µM, and 13 µM GST-Gal1(WT); and lane 6–8, reconstitution with 1 µM, 6.5 µM, and 13 µM GST-Gal1(N46D). In panels B and C, products of the splicing reaction were analyzed by electrophoresis through a 13% polyacrylamide-urea gel and autoradiography. The positions of migration of the pre-mRNA substrate, the splicing intermediates (exon 1 and lariat-exon 2), and RNA products (ligated exon 1-exon 2 and intron lariat) are indicated on the right.
NE depleted of the galectins showed drastically reduced splicing activity (Fig. 7B, lane 3) compared to the original or mock-depleted NE (Fig. 7B, lanes 1 and 2). We quantitated the product RNAs (ligated exons and liberated intron) as a percent of the total radioactive RNA species in each assay. Product formation in the depleted extract was less than 5%; the corresponding value for the original NE was 45%. The same preparations of the wild-type and mutant proteins that exhibited drastic differences in saccharide-binding activity (Fig. 5) both reconstituted splicing in the galectin-depleted NE (Fig. 7B, lanes 5 and 6). The level of product formation was 15% for GST-Gal1(WT) and 14% for GST-Gal1(N46D). The concentration of GST proteins used was 6.5 µM. At this concentration, the level of reconstitution was comparable to that achieved previously with recombinant Gal1 [8]. In contrast, GST by itself could not reconstitute the splicing activity in the depleted NE (Fig. 7B, lane 4); product formation was 4.8%. GST-Gal1(WT) and GST-Gal1(N46D) showed similar dose-response curves in reconstituting the splicing activity of a galectin-depleted NE (Fig. 7C). Since GST-Gal1( N46D) has lost its carbohydrate-binding activity, it appears that the splicing activity correlates with its retention of the association with TFII-I.
GST-Gal1(C60S) behaved similarly to GST-Gal1(WT) in the reconstitution of splicing assay. Thus, GST-Gal1(C60S) retained all three activities of the wild-type protein. In contrast, it appears that GST-Gal1(E71Q) has lost all three activities as we were unable to obtain a reproducible reconstitution assay with this mutant (data not shown).
Lack of an effect of saccharide ligands on GST-Gal1(N46D) in the splicing assay
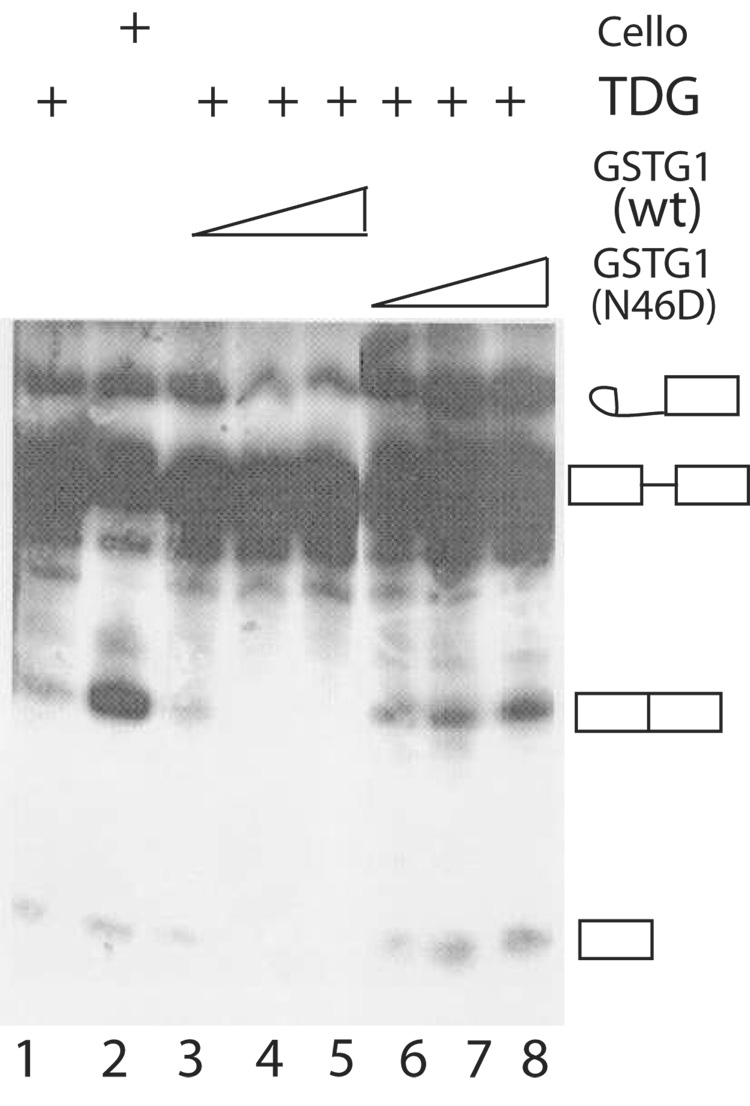
We had previously documented that saccharide ligands such as Lac and TDG inhibited the splicing activity of a complete (non-depleted) NE when added to the cell-free assay [7]. We therefore tested the effect of GST-Gal1(N46D) on NE whose splicing activity is inhibited by TDG. In the present assay, TDG inhibited splicing whereas cellobiose, which does not bind to the galectins, failed to yield the same effect. The level of product formation was much higher in NE containing cellobiose (30%; Fig. 8, lane 2) than the corresponding extract containing TDG (~1%; Fig. 8, lane 1). Addition of GST-Gal1(WT), which can bind to carbohydrate ligands (Fig. 5), did not overcome the inhibitory effect of TDG on splicing (Fig. 8, lanes 3–5). In contrast, addition of the GST-Gal1(N46D) mutant, which was not sensitive to saccharide inhibition, resulted in a dose-dependent increase in splicing activity (Fig. 8, lanes 6–8).
Figure 8. Comparison of the effects of GST-Gal1(WT) and GST-Gal1(N46D) on the splicing activity of NE containing TDG.
Lane 1, TDG addition (150 mM); Lane 2, cellobiose addition (150 mM); Lanes 3–5, TDG (150 mM) and GST-Gal1(WT) (10, 20, and 30 µM); Lanes 6–8, TDG (150 mM) and GST-Gal1( N46D) (10, 20, and 30 µM). Products of the splicing reactions were analyzed by electrophoresis through a 13% polyacrylamide-urea gel and autoradiography. The positions of migration of the pre-mRNA substrate, the splicing intermediates, and the mature RNA product are indicated on the right.
Discussion
The major conclusion derived from the present studies is that the splicing functions of the galectins can be dissociated from their carbohydrate-binding activity. This conclusion is based on several key findings: (a) When NEs of HeLa cells were subjected to pull-down experiments with GST-Gal1 or GST-Gal3, the general transcription factor TFII-I was identified as one of the polypeptides bound. (b) Lac, a saccharide ligand of the galectin family of proteins, inhibited the pull-down of TFII-I from NE. (c) GST-Gal1(N46D), deficient in saccharide-binding activity, retained both its association with TFII-I and the reconstitution of splicing activity in NE depleted of galectins.
An important consideration in the interpretation of these results is that the TFII-I association with Gal1 or Gal3 is linked to spliceosomes. There are now several lines of evidence linking transcription and pre-mRNA splicing (see [30] for a review). Our present demonstration, that anti-TFII-I can coprecipitate RNA species produced during the splicing reaction, complements the chemical studies in implicating transcription factor TFII-I as a part of a functional spliceosome [12]. Moreover, when anti-TFII-I is added to a complete NE during the splicing reaction, a dose-dependent inhibition was observed (R. Gray and J. Wang, unpublished observations).
Like the parent mutant protein Gal1(N46D) [10, 11], our present fusion construct GST-Gal1( N46D) also resulted in a polypeptide deficient in carbohydrate-binding activity. That residue 46 of human Gal1 is critical in saccharide-binding is consistent with the results of X-ray crystallographic analysis of the CRD of galectins, in which the Asn residue at this position serves as an acceptor of a hydrogen bond from the hydroxyl group at C-4 of galactose [31]. This raises the question of how to reconcile the two apparently disparate findings: while carbohydrate-binding is not required for splicing activity and for association with TFII-I, saccharides such as Lac and TDG nevertheless exert an inhibitory effect when added to the splicing assay [7] or to the GST-Gal1 pull-down of TFII-I. One possibility is that the binding site(s) for Lac and for TFII-I overlap and the presence of the saccharide competitively inhibits the binding of the protein ligand. Asn46 in the Gal1 polypeptide is required for hydrogen bonding to the carbohydrate [31] but it does not appear to be required for TFII-I binding. Thus, the N46D mutation perturbs saccharide-binding but not TFII-I association. Alternatively, it is also possible that binding of saccharide ligands to the carbohydrate-binding site results in a conformational change that disrupts the interaction of the galectin polypeptide with a component of the splicing machinery such as TFII-I. Conformational changes in the CRD of galectins have been reported on the basis of NMR analysis [32] and differential scanning calorimetry studies [14].
It should be noted that previous studies had shown that Gal1 forms non-covalently associated dimers that exist in equilibrium with monomeric Gal1 [33–35]. Therefore, it was possible that the observed pull-down of TFII-I by GST-Gal1(N46D) can be accounted for by hybrid dimeric molecules (consisting of one GST-Gal1(N46D) and one wild-type Gal1 derived from NE) tethered to the glutathione beads via GST. Two lines of evidence argue against this possibility. First, while TFII-I pull-down by GST-Gal1(WT) was inhibited by Lac (see Fig. 2A), the corresponding pull-down by GST-Gal1(N46D) was not affected by the disaccharide. If endogenous Gal1 had formed hybrid dimers with GST-Gal1(N46) and were responsible for the observed TFII-I pull-down, then one would expect Lac to inhibit the association. Second, we have compared the level of TFII-I pull-down by GST-Gal1(N46D) using either NE or NE depleted of endogenous Gal1 and Gal3 through adsorption on Lac-agarose beads. We found the same amount of TFII-I in both cases. Therefore, GST-Gal1(N46D) can pull-down TFII-I independent of endogenous Gal1.
In addition to TFII-I association and pre-mRNA splicing, Gal1 has been reported to exhibit growth inhibitory activity for cells in culture and this activity also does not appear to depend upon its saccharide-binding activity [36, 37]. Attempts to dissociate the carbohydrate-binding activity from the growth inhibitory activity by site-directed mutagenesis have been confounded, however, by the fact that mutations at residues known to interact with saccharide ligands (e.g. N46D) have not yielded protein products amenable for rigorous tests of growth inhibition (see [38] for a recent review). In this connection, the present study has taken advantage of the availability of binding assays (ASF-binding as a measure of carbohydrate-specific interactions [20] and TFII-I pull-down as a measure of association with the spliceosome) to indicate that saccharide-binding is not essential for splicing activity. Galectins were initially isolated on the basis of their carbohydrate-binding activity and, for the most part, the literature on these proteins has been dominated by studies focused on their activity on the extracellular side, based on their binding to cell surface glycoconjugates and adhesive glycoproteins of the extracellular matrix (see [39] for a recent review). Perhaps attention should also be directed to the intracellular activity that may not depend on carbohydrate recognition. At some point, it would be interesting to address the more general question: did both activities exist in the progenitor of the galectin family or did the two activities arise separately?
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by grants GM-38740 from the National Institutes of Health (JLW), 06-IRGP-858 from the Michigan State University Intramural Research Grant Program (JLW), and MCB-0092919 from the National Science Foundation (RJP)
Abbreviations used: CRD, carbohydrate recognition domain; Gal1, galectin-1; Gal3, galectin-3; NE, nuclear extract; Lac, lactose; TDG, thiodigalactoside; snRNP, small nuclear ribonucleoprotein; TFII-I, general transcription factor II-I; ASF, asialofetuin; GST, glutathione S-transferase; ND, the NH2-terminal domain of Gal3; DTT, dithiothreitol; PBS, phosphatebuffered saline; T-TBS, Tris-buffered saline containing 0.05% Tween 20; SMN, survival of motor neuron protein.
References
- 1.Barondes SH, Castronovo V, Cooper DNW, Cummings RD, Drickamer K, et al. Cell. 1994;76:597–598. doi: 10.1016/0092-8674(94)90498-7. [DOI] [PubMed] [Google Scholar]
- 2.Wang JL, Gray RM, Haudek KC, Patterson RJ. Biochim. Biophys. Acta. 2004;1673:75–93. doi: 10.1016/j.bbagen.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Arnoys EJ, Wang JL. Acta Histochemica. 2007;109:89–110. doi: 10.1016/j.acthis.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Laing JG, Wang JL. Biochemistry. 1998;27:5329–5334. doi: 10.1021/bi00414a057. [DOI] [PubMed] [Google Scholar]
- 5.Hubert M, Wang S-Y, Wang JL, Seve A-P, Hubert J. Exp. Cell Res. 1995;220:397–406. doi: 10.1006/excr.1995.1331. [DOI] [PubMed] [Google Scholar]
- 6.Vyakarnam A, Lenneman AJ, Lakkides KM, Patterson RJ, Wang JL. Exp. Cell Res. 1998;242:419–428. doi: 10.1006/excr.1998.4111. [DOI] [PubMed] [Google Scholar]
- 7.Dagher SF, Wang JL, Patterson RJ. Proc. Natl. Acad. Sci. USA. 1995;92:1213–1217. doi: 10.1073/pnas.92.4.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vyakarnam A, Dagher SF, Wang JL, Patterson RJ. Mol. Cell. Biol. 1997;17:4730–4737. doi: 10.1128/mcb.17.8.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Park JW, Wang JL, Patterson RJ. Nucleic Acids Res. 2006;34:5166–5174. doi: 10.1093/nar/gkl673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirabayashi J, Kasai K. J. Biol. Chem. 1991;266:23648–23653. [PubMed] [Google Scholar]
- 11.Hirabayashi J, Kasai K. Glycoconjugate J. 1994;11:437–442. doi: 10.1007/BF00731280. [DOI] [PubMed] [Google Scholar]
- 12.Rappsilber J, Ryder U, Lamond AI, Mann M. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrwal N, Wang JL, Voss PG. J. Biol. Chem. 1989;264:17236–17242. [PubMed] [Google Scholar]
- 14.Agrwal N, Sun Q, Wang S-Y, Wang JL. J. Biol. Chem. 1993;268:14932–14939. [PubMed] [Google Scholar]
- 15.Ho MK, Springer TA. J. Immunol. 1982;128:1221–1228. [PubMed] [Google Scholar]
- 16.Cherayil BJ, Weiner SJ, Pillai S. J. Exp. Med. 1989;170:1959–1972. doi: 10.1084/jem.170.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.Zillman M, Zapp ML, Berget SM. Mol. Cell Biol. 1988;8:814–821. doi: 10.1128/mcb.8.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roff CF, Wang JL. J. Biol. Chem. 1983;258:10657–10663. [PubMed] [Google Scholar]
- 21.Laemmli UK. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Merril C, Dunau M, Goldman D. Anal. Biochem. 1981;110:201–207. doi: 10.1016/0003-2697(81)90136-6. [DOI] [PubMed] [Google Scholar]
- 23.Rowley A, Choudhary JS, Marzioch M, Ward MA, Weir M, Solari RCE, Blackstock WP. Methods. 2000;20:383–397. doi: 10.1006/meth.2000.0951. [DOI] [PubMed] [Google Scholar]
- 24.Shevchenko A, Wilm M, Vorm O, Mann M. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 25.Roy A, Meisterernst M, Pognonec P, Roeder R. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 26.Grueneberg DA, Henry RW, Bauer A, Novina CD, Cheriyath V, Roy AL, Gilman M. Genes Dev. 1997;11:2482–2493. doi: 10.1101/gad.11.19.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W, Desiderio S. Proc. Natl. Acad. Sci. USA. 1997;94:604–609. doi: 10.1073/pnas.94.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leffler H, Barondes SH. J. Biol. Chem. 1986;261:10119–10126. [PubMed] [Google Scholar]
- 29.Butcher SE, Brow DA. Biochem. Soc. Trans. 2005;33:447–449. doi: 10.1042/BST0330447. [DOI] [PubMed] [Google Scholar]
- 30.Neugebauer KM. J. Cell Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 31.Rini JM, Lobasanov YD. Curr. Opin. Struct. Biol. 1999;9:578–584. doi: 10.1016/s0959-440x(99)00008-1. [DOI] [PubMed] [Google Scholar]
- 32.Umemoto K, Leffler H, Venot A, Valafar H, Prestegard JH. Biochemistry. 2003;42:3688–3695. doi: 10.1021/bi026671m. [DOI] [PubMed] [Google Scholar]
- 33.Briles EB, Gregory W, Fletcher P, Kornfeld S. J. Cell Biol. 1979;81:528–537. doi: 10.1083/jcb.81.3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho M, Cummings RD. J. Biol. Chem. 1995;270:5198–5206. doi: 10.1074/jbc.270.10.5198. [DOI] [PubMed] [Google Scholar]
- 35.Cho M, Cummings RD. Biochemistry. 1996;35:13081–13088. doi: 10.1021/bi961181d. [DOI] [PubMed] [Google Scholar]
- 36.Adams L, Scott GK, Weinberg CS. Biochim. Biophys. Acta. 1996;1312:137–144. doi: 10.1016/0167-4889(96)00031-6. [DOI] [PubMed] [Google Scholar]
- 37.Wells V, Mallucci L. Cell. 1991;64:91–97. doi: 10.1016/0092-8674(91)90211-g. [DOI] [PubMed] [Google Scholar]
- 38.Scott K, Weinberg C. Glycoconjugate J. 2004;19:467–477. doi: 10.1023/B:GLYC.0000014076.43288.89. [DOI] [PubMed] [Google Scholar]
- 39.Camby I, Le Mercier M, Lefrance F, Kiss R. Glycobiology. 2006;16:137R–157R. doi: 10.1093/glycob/cwl025. [DOI] [PubMed] [Google Scholar]