Abstract
Members of the Fgd (faciogenital dysplasia) gene family encode a group of critical guanine nucleotide exchange factors (GEFs), which, by specifically activating Cdc42, control cytoskeleton-dependent membrane rearrangements. In its first characterization, we find that FGD2 is expressed in antigen-presenting cells, including B lymphocytes, macrophages, and dendritic cells. In the B lymphocyte lineage, FGD2 levels change with developmental stage. In both mature splenic B cells and immature bone marrow B cells, FGD2 expression is suppressed upon activation through the B cell antigen receptor. FGD2 has a complex intracellular localization, with concentrations found in membrane ruffles and early endosomes. Although endosomal localization of FGD2 is dependent on a conserved FYVE domain, its C-terminal pleckstrin homology domain mediates recruitment to membrane ruffles. FGD2 overexpression promotes the activation of Cdc42 and leads to elevated JNK1 activity in a Cdc42- but not Rac1-dependent fashion. These findings are consistent with a role of FGD2 in leukocyte signaling and vesicle trafficking in cells specialized to present antigen in the immune system.
Fgd2 (faciogenital dysplasia 2) is a novel gene discovered by PCR amplification using primer sequences based on the Fgd1 gene (1) and is a member of a small subfamily of predicted RhoGEFs (including Fgd1, Fgd3, Fgd4 (Frabin), Fgd5, and Fgd6) with C-terminal amino acid homology but highly divergent N-terminal extensions. The Fgd subfamily is part of a larger family of 69 genes with homology to Dbl, a transforming gene identified in diffuse B cell lymphomas (2). RhoGEFs are activators of the Rho (Ras homology) subfamily of GTPases, such as Rac1 to -3, RhoA to -G, and Cdc42, because they are GDP/GTP exchange factors (GEFs).4 As such, they catalyze the release of bound GDP from the GTPases, resulting in the formation of GTP-bound active proteins able to interact with downstream effectors (2). Depending on their subcellular localization and interaction with specific GTPases, they can thus control disparate biological activities, including adhesion, mitosis, cellular polarization, and vesicle trafficking (3).
All Dbl RhoGEF family members carry a signature active site motif composed of tandem Dbl homology (DH) and pleckstrin homology (PH) domains. Substrate specificity is dictated by a “specificity patch” within the DH domain (4), with some RhoGEFs demonstrating specificity for particular GTPases (e.g. TRIO (Rac1) (5) and intersectin (Cdc42) (4)), whereas others, such as the VAV proteins and Dbl, have broad activity for multiple GTPases (6–8). Further domains that influence function, molecular interactions, and cellular localization distinguish RhoGEFs from one another. Among Dbl family proteins, Fgd family members are unique in carrying a FYVE motif (shared in Fab1, YotB, Vac1p, and EEA1) and an additional PH domain at the C terminus.
FYVE domains are zinc finger lipid binding motifs that often target proteins to vesicles by interaction with phosphatidylinositol 3-phosphate (9–13), a lipid that is enriched on endosomal membranes (12). PH domain-containing proteins often bind to phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3), leading to phosphatidylinositol 3-kinase class I-dependent plasma membrane recruitment and possible protein conformational changes (14).
Among the Fgd family members, FGD1, FGD3, and FGD4 have been characterized as proteins. Fgd1 was identified as the gene defective in the Aarskog-Scott syndrome, an X-chromosome-linked disorder involving multiple developmental defects, including bone and body malformations (15). Studies have demonstrated that FGD1 is a guanine nucleotide exchange factor for the GTPase Cdc42, which plays important roles in cytoskeletal regulation and the organization of actin filaments (16, 17). FGD3 and FGD4 have also been shown to modulate the actin cytoskeleton and to be Cdc42-specific exchange factors (18, 19). Interestingly, mutations of Fgd4 (Frabin) are associated with the peripheral nerve disease, Charcot-Marie-Tooth disease (20, 21). Although annotated in genetic data bases, Fgd5 and Fgd6 are so far uncharacterized. Apart from inferences based on sequence homology, little is known about the protein function of FGD2. A recent study indicates that a mutant form of Fgd2 contributes to t-complex ratio distortion, a phenomenon found in some wild strains of mice where decreased motility of sperm is caused by the actions of several genes located in a variant region of chromosome 17 (22). The Fgd2 locus also maps to Idd23, a region of chromosome 17 where the C57BL/6 allele is implicated in the protection from diabetes in the NOD mouse model of diabetes (23).
In this paper, we present the first biochemical characterization of FGD2, including an assessment of its predominant expression in leukocyte subsets, regulation in B cells by B cell receptor (BCR) signaling, enzymatic activity, and subcellular localization. We show that FGD2 is the sole member of the characterized Fgd family that can localize to early endocytic vesicles.
EXPERIMENTAL PROCEDURES
Cloning Mouse Fgd2—Total RNA was harvested from mouse spleen (strain B10.D2/SnJ) using TRIzol (Life Technologies), and cDNA was generated using the SuperScript first strand synthesis system for reverse transcription-polymerase chain reaction (Invitrogen) according to the manufacturer's instructions. Fgd2 cDNA was amplified using PLATINUM Pfx DNA polymerase enzyme (Invitrogen) according to the manufacturer's instructions with the following primers: 5′-TACTCAAGCTTAGGATGGAGCGAGCCTGTGAG and 5′-TGATCACTCGAGATTTCATGATCCAGGGATA. The sequence for mouse Fgd2 from B10.D2 mice had several changes from the published sequence (1) and concurred with the C57BL/6 genomic sequence and cDNA sequences AK042260 and AY301264 (NCBI GenBank™) with two exceptions. The predicted amino acids Ser39 and Glu543 of C57BL/6J origin were changed to isoleucine and aspartic acid, respectively. The B10.D2/SnJ cDNA sequence has been deposited in GenBank™ (DQ344523). Fgd2 cDNA was subcloned into pCMVTag2b and pEGFP-C3 for expression analysis with FLAG and EGFP epitope tags, respectively. A QuikChange kit (Stratagene) was used to generate two FGD2 point mutants. FYVEKT contained full-length FGD2 with mutations in the FYVE domain (Gln454 → Lys, Trp455 → Thr), and GEFAA contained mutations in the DH domain (Ser28 → Ala and Asn288 → Ala). In addition, truncation constructs of FGD2 containing only the DH and PH domains (DHPH) of FGD2 (residues 99–418) and a mutant lacking the C-terminal PH domain (FGD2ΔPH2) of FGD2 (residues 1–526) were also generated. A schematic diagram showing the mutants used in this study is provided in Fig. 5C.
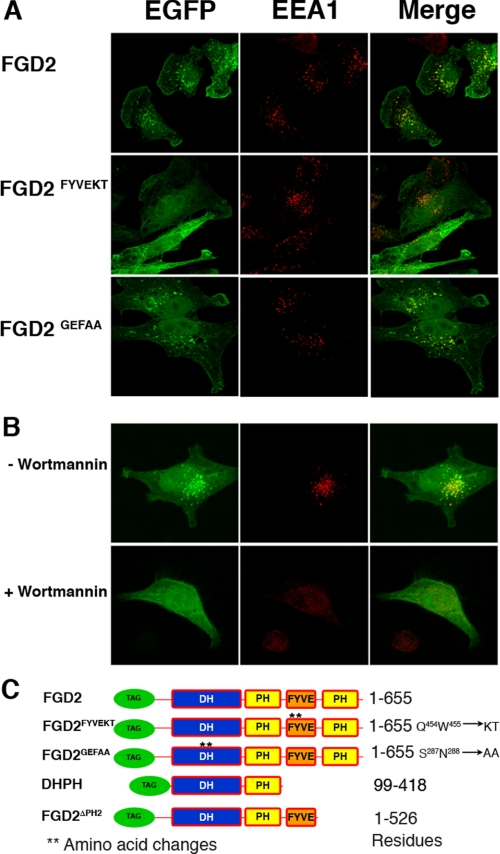
FIGURE 5.
Effects of FYVE domain mutation and wortmannin treatment on the localization of FGD2. A, HeLa cells expressing either wild type EGFP-FGD2 (top), EGFP-FGD2FYVEKT (middle), or EGFP-FGD2GEFAA (bottom) werefixed, permeabilized, and stained with antibody against EEA1 (red). B, HeLa cells expressing EGFP-FGD2 were cultured for 30 min with either 50 nm wortmannin (bottom) or the solvent DMSO alone (top) before fixation and staining for EEA1. C, schematic depictions of FGD2 domains and mutations used in this study. **, mutated residues and modified residues that are listed to the right. TAG, either EGFP or FLAG epitope tags used in the biochemical and microscopy studies.
Antibodies—Polyclonal antiserum to FGD2 was generated by immunizing rabbits with recombinant FGD2 fused to thioredoxin (TRX-FGD2) and a poly-His sequence using the pET32 expression vector (Novagen). Anti-FLAG M2 antibody-coupled agarose (Sigma), anti-FLAG M2 antibody horseradish peroxidase (Sigma), anti-Myc (clone 4G6; Upstate Biotechnology), anti-EGFP (JL-8, Clontech), goat anti-mouse IgG-peroxidase (horseradish peroxidase), goat anti-rabbit IgG-horseradish peroxidase, and goat F(ab′)2 anti-mouse IgM, goat anti-mouse IgG-horseradish peroxidase (Jackson Immunochemicals), anti-glyceraldehyde-3-phosphate dehydrogenase (Chemicon International), S23 (anti-3-83 BCR)5, and Y3 (anti-H-2Kb) (24) were also used. For confocal microscopy stains, EEA1 (clone 14/EEA1; BD Biosciences), GM130 (clone 35/GM130; BD Biosciences), LAMP2 (H4B4; Developmental Studies Hybridoma Bank), cortactin (clone 4F11; Upstate Biotechnology), Cdc42 (sc-87; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), Rac1 (23A8; Upstate Biotechnology), RhoA-C (06-770; Upstate Biotechnology), rhodamine, or Alexa 647-labeled phalloidin (Invitrogen) and goat anti-mouse Alexa 568 (Invitrogen) were used.
Cell Culture—HEK293, COS-7, and HeLa cells were cultured in Iscove's modified Dulbecco's medium with 10% fetal bovine serum, 25 mm HEPES, 1 mm sodium pyruvate, 55 μm 2-mercaptoethanol, penicillin, and streptomycin (Invitrogen). Bone marrow-derived macrophages were cultured using L cell-conditioned medium essentially as described (25), except that Iscove's modified Dulbecco's medium was used instead of RPMI 1640. The principle method for generating bone marrow-derived dendritic cells with GM-CSF was as described (26). Following 10 days of culture in 200 units/ml GM-CSF, nonadherent bone marrow cells were then cultured a further 2 days in GM-CSF (100 units/ml) and 1 μg/ml lipopolysaccharide (055: B5) (LPS; Sigma).
Western Analysis of FGD2 Expression—Whole cell lysates from thymocytes, pooled lymph nodes, total bone marrow, bone marrow-derived macrophages, bone marrow-derived dendritic cells, splenocytes, and splenocytes either B220-enriched or -depleted using magnetic beads (Miltenyi Biotec) were generated from B10.D2 mice using 0.5% Nonidet P-40 lysis buffer containing 10 mm Tris, 150 mm NaCl, 0.1 mm EDTA, and 1 mm phenylmethylsulfonyl fluoride and Complete™ protease inhibitors (Roche Applied Science). Insoluble nuclei were removed by centrifugation, and lysates were subjected to SDS-PAGE under reducing conditions, followed by Western blotting and detection using a polyclonal rabbit anti-FGD2 antiserum or glyceraldehyde-3-phosphate dehydrogenase antibody. A mouse tissue INSTA-Blot (Imgenex) containing immobilized brain, heart, small intestine, kidney, liver, lung, muscle, stomach, spleen, ovary, and testis lysates was also probed with the polyclonal rabbit anti-FGD2 antiserum, according to the manufacturer's instructions.
Expression with Stimulation—Immature B cells from 3-83 BCR Tg mice were expanded from bone marrow using interleukin-7 cultures as described (27). One day after interleukin-7 removal, 20 μg/ml either anti-BCR antibody (S23) or control (Y3) antibody was added to the cells for the indicated times. Mature B cells were purified from spleen and lymph nodes from B10.D2 mice using negative selection with CD43 MACS beads and columns (Miltenyi Biotec). CD43-B cells were cultured at 1 × 107 cells/ml with either 10 μg/ml goat F(ab)2 anti-mouse IgM (Jackson Immunochemicals) or LPS (1 μg/ml) and incubated at 37 °C for various time points prior to lysis and analysis by Western blotting.
Real Time PCR Analyses of mRNA Expression—Pro-B cells were isolated by cell sorting from bone marrow cells of RAG1–/– mice, where B220+CD43+ were positively selected. Pre-B cells from bone marrow of B10.D2SnJ mice were sorted using the markers B220+ CD43– IgM– IgD–. Immature B cells were isolated from B10.D2 mice carrying the 3-83 BCR transgene (28) and were sorted for B220+ IgM+ IgD– CD43– marked cells. Immature B cells undergoing receptor editing were isolated from B10.Br 3-83 transgenic bone marrow, where B10.Br mice express the BCR antigen H-2k in the bone marrow. Cells were sorted for B220+ IgD– CD43– IgM–. B220+ cells from spleen and pooled lymph nodes were isolated using B220 magnetic bead separation (Miltenyi). Total RNA was purified from sorted B cell subsets using TRIzol (Invitrogen), and contaminating genomic DNA was removed by DNase I digestion. First strand cDNA was generated using oligo(dT) and the Superscript III first strand synthesis system (Invitrogen) before Fgd2 and β-actin were amplified. A 158-bp amplicon for actin was amplified using primers 5′-gcattgctgacaggatgcag and 5′-CCTGCTTGCTGATCCACATC annealing at 57 °C. Fgd2 was amplified using primers 5′-AGGAACCTGAGGAGAAGAGGGTC and 5′-gcgtggaaacgatagatggaggag, annealing at 57 °C, yielding a 189-bp product. The PCRs were performed in triplicate using the 7900 HT Applied Biosystems machine and quantified using the Sybr green PCR kit (Qiagen). Ct values for each reaction were generated using the SDS software (Applied Biosystems). Fgd2 Ct values were subtracted from actin, and mean results were expressed as the exponential product ratio.
Northern Analysis of Fgd2 Expression—20 μg of total RNA isolated from immature B cells cultured with or without anti-BCR antibody for 24 h was electrophoresed under denaturing conditions. After photographing the RNA gel stained with ethidium bromide, the RNA was transferred to a nylon Zeta probe membrane (Bio-Rad) before UV cross-linking and probing with an Fgd2 cDNA probe generated using the primers 5′-CTTCCAGGAGGTGGTCACCCG and 5′-AACCAGGTAGCGTTCCATTG.
Cdc42 Activation Assay—COS-7 cells were transiently transfected by Lipofectamine2000 (Invitrogen) mixed with cDNA encoding Myc-tagged Cdc42, EGFP, EGFP-tagged Fgd2, or EGFP-tagged Fgd2 mutants. The dominant active mutant EGFP-Cdc42Q61L and the dominant negative mutant, EGFP-Cdc42T17N, were used as positive and negative controls, respectively. Active Cdc42 was assessed by a pull-down assay, as described (29). Briefly, 24 h post-transfection, cells were lysed with a buffer containing 50 mm Tris, pH 7.8, 200 mm NaCl, 5 mm MgCl2, 1 mm dithiothreitol, 1% Nonidet P-40, 10% glycerol, and protease inhibitors (Roche Applied Science). Lysates were precleared with GST bound to glutathione beads for 10 min, prior to incubation for 1 h with ACK1 (activated Cdc42-associated kinase 1)-Crib (Cdc42- and Rac1-interacting binding) (amino acids 439–502)-GST fusion protein bound to glutathione beads. After washing three times with wash buffer (25 mm Tris, 40 mm NaCl, 30 mm MgCl2, 1 mm dithiothreitol, 1% Nonidet P-40), beads were subjected to SDS-PAGE and Western blotting. Bead-associated active Cdc42 was detected using anti-Myc and anti-EGFP antibodies. Rac1 and RhoA activity assay kits containing PAK-Crib GST and Rhotekin-RBD GST beads, respectively, were purchased from Upstate Biotechnology (now Millipore) and used according to the manufacturer's instructions.
JNK1 Activity Assays—HEK293 or COS7 cells were transiently transfected with FLAG-JNK1 together with EGFP or various EGFP-FGD2 mutants in the presence of either Cdc42 or Rac1. JNK1 activity assays were performed as described (30) by immunoprecipitation of JNK1 with anti-FLAG beads (Sigma) and incubation of [γ-32P]ATP with a GST-ATF-2 fusion protein as substrate. Phosphorylated ATF-2 was determined by autoradiography, and the amounts of expressed proteins were determined by Western blotting of total cellular lysates.
Confocal Microscopy Imaging of Fgd2 Localization—Transiently transfected HeLa cells that had adhered to polylysine- or fibronectin-coated glass coverslips were fixed with 3.4% paraformaldehyde. In some cases, transfected HeLa cells were incubated with either 50 nm wortmannin (Calbiochem) in DMSO or DMSO alone for 30 min prior to fixation and staining. For actin stains, the cells were permeabilized with 0.2% Triton X-100 (phalloidin and cortactin stains). For intracellular compartment stains, the cells were permeabilized with 0.05% saponin prior to staining. After the samples were mounted with Immuno-Fluore mounting medium (MP Biomedicals Inc.), coverslips were visualized by fluorescence microscopy with an MC1024 (Bio-Rad) confocal laser microscope. The EGFP-FGD2 colocalization with intracellular stained compartments was determined using LSM examiner software (Zeiss).
Phospholipid Binding Membranes—Recombinant His-tagged thioredoxin-FGD2 (pET32 expression vector (Novagen)) or fusion partner thioredoxin alone were expressed in Escherichia coli BL21 (DE3) and purified using nickel affinity chromatography. The recombinant FGD2 protein was insoluble and was solubilized with 8 m urea and gradually dialyzed into a final buffer of 25 mm Tris, pH 8, 150 mm NaCl, 10 mm 2-mercaptoethanol, and 0.1% Triton X-100 over 3 days. Purified proteins were incubated with PIP Array membranes (Molecular Probes and Invitrogen) at 0.5 μg/ml for 2 h according to the manufacturer's instructions. After washing, bound proteins were detected with an anti-His epitope antibody (Sigma) and chemiluminescence.
RESULTS
Fgd2 Is Expressed in Primary Lymphoid Cells—When immature B cells encounter self-antigen, they can undergo secondary recombination of the κ light chain locus, to produce an innocuous surface antigen receptor, a tolerance process known as receptor editing (31). To generate large numbers of immature B cells with uniform receptors, we cultured bone marrow from 3-83 BCR transgenic mice in which immature B cells express a prerearranged receptor against H-2Kk antigen (32). In order to identify genes that were differentially expressed between immature B cells and immature B cells forced to undergo receptor editing by exposure to anti-BCR antibodies in vitro, we performed a PCR-based total gene expression analysis screen (33). In this screen, we identified Fgd2 as a candidate RNA that was expressed in immature B cells but was poorly expressed in editing B cells. These findings were also confirmed by Affymetrix chip analysis of U94B arrays with the probe set 115754_at (data not shown). Perusal of the Symatlas data base (34) indicated that Fgd2 mRNA expression was concentrated in lymphoid tissues rich in B cells.
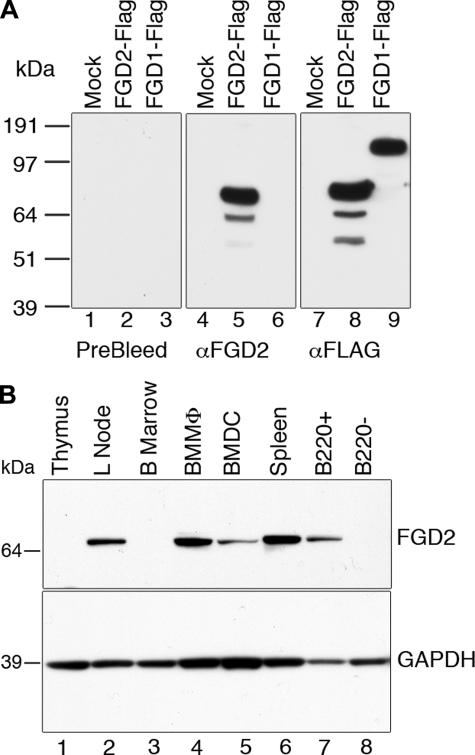
We therefore wanted to assess FGD2 protein expression in lymphoid tissues or isolated leukocyte subsets. To this end, an FGD2-specific rabbit antiserum was prepared. The antiserum's specificity for FGD2 was clear, because, although it could bind to recombinant epitope-tagged FGD2 expressed in HEK293 cells in Western blots, it failed to detect the related FGD1 protein (Fig. 1A, lanes 5 and 6) and FGD3 (not shown). By contrast, both FGD1 and FGD2 could be detected when probed with antibodies directed to their N-terminal FLAG epitope tag (Fig. 1A, lanes 8 and 9). Furthermore, FGD2 has a distinctly lower predicted molecular weight (Mr 74,600) than all other Fgd family members, including FGD1 (Mr 105,000) (Fig. 1A, lanes 8 and 9). In addition, the preimmune sera did not detect any proteins in the HEK293 lysates (Fig. 1A, lanes 1–3).
FIGURE 1.
FGD2 protein expression in lymphoid organs and leukocyte subsets. A, specificity of anti-FGD2 serum. FGD2-FLAG (lane 2) or FGD1-FLAG (lane 3) was expressed in HEK293 cells, cell lysates were prepared, and expression was detected by immunoblot using either preimmune serum (left), anti-FGD2 polyclonal serum (center), or FLAG antibodies (right). As a negative control, empty vector was also transiently transfected and cell lysate prepared (Mock lane 1). B, lymphoid tissues and cell lysates were electrophoresed, and Western blotted membranes were probed with FGD2 antiserum (top) or glyceraldehyde-3-phosphate dehydrogenase antibody (bottom). Macrophages (BMMΦ) were generated in vitro by expansion from bone marrow in macrophage colony-stimulating factor (M-CSF; lane 4). Dendritic cells (BMDC) were similarly generated by expansion with GM-CSF and matured with LPS (lane 5). Splenocytes were fractionated into B220 enriched (B220+) or B220-depleted (B220–) samples by magnetic beads prior to lysis.
This rabbit-derived antiserum was used in Western blotting of whole cell lysates prepared from mouse thymus, pooled lymph nodes, bone marrow, and spleen and magnetically separated CD45R+ (B220+) and B220– splenic cells. No reactivity was detected in lysates of unfractionated bone marrow or thymus (Fig. 1B, lanes 1 and 3), whereas high level expression was evident in lymph nodes and spleen (Fig. 1B, lanes 2 and 6), consistent with the high levels of mRNA expressed in these latter tissues. B220+ splenic cells were enriched, whereas B220– splenic cells were depleted of immunoreactive FGD2 protein (Fig. 1B, lanes 7 and 8), indicating that B cells are the major source of FGD2 in the spleen. In addition, in lysates prepared from macrophages and dendritic cells that were expanded from bone marrow by stimulation with granulocyte colony-stimulating factor or GM-CSF, respectively, FGD2 expression was detectable (Fig. 1B, lanes 4 and 5). The expression of FGD2 in nonleukocyte tissues was tested by probing an INSTA-Blot mouse tissue membrane (Imgenex). We were unable to detect FGD2 protein in brain, liver, kidney, testis, muscle, ovary, or small intestine but did confirm splenic expression in this assay (data not shown). These experiments indicate that leukocytes express FGD2, and that in lymphoid tissues, B lymphocytes and myeloid cells, in particular macrophages, also express the protein.
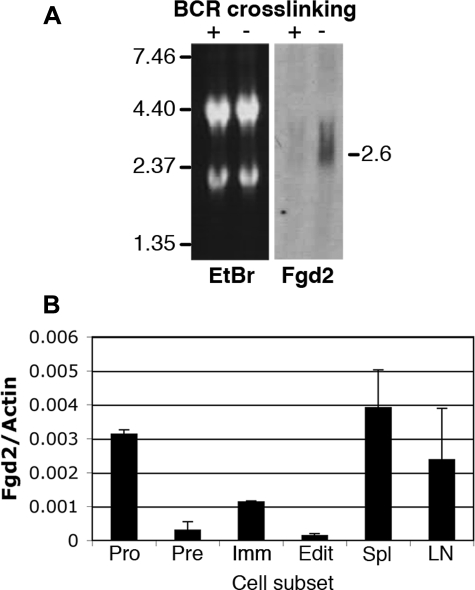
Fgd2 mRNA Expression and Regulation in B Lymphocyte Development—Because we were initially interested in identifying genes that were regulated during B lymphocyte development and selection, and Western blotting and array analysis indicated that Fgd2 expression was particularly high in B cells, we carried out a more detailed analysis of the developmental regulation and response to antigen receptor signaling of its RNA expression. A Northern blot assay confirmed that Fgd2 is differentially expressed in interleukin-7-expanded primary immature B lymphocytes. Fig. 2A shows that immature B cells treated with anti-BCR antibodies (+) for 24 h in vitro had low level expression of Fgd2 mRNA relative to control IgG-treated cells (–). To determine if BCR signaling affected Fgd2 expression in vivo, immature bone marrow B cells were directly sorted from 3-83 BCR transgenic mice that expressed or lacked cognate autoantigen (32) (Fig. 2B). Immature bone marrow B cells differed in their levels of Fgd2 mRNA depending on whether cognate self-antigen was absent (Fig. 2B, Imm) or present (Edit). These data confirmed in ex vivo cells that BCR ligation by self-antigen suppressed Fgd2 expression in immature B cells. We also compared Fgd2 levels in other B lineage subsets. Sorted pre-B cells (CD43–B220+sIg–) expressed low levels of Fgd2, whereas pro-B cells (CD43+B220+sIg–) expressed high levels. Mature B lymphocytes from spleen and lymph nodes expressed high levels of Fgd2 mRNA (Fig. 2B). We conclude that Fgd2 expression changes strikingly with preimmune developmental progression and that BCR ligation can alter Fgd2 expression in the bone marrow, probably by inducing developmental arrest.
FIGURE 2.
Fgd2 mRNA expression is regulated during B cell development and is suppressed by BCR ligation in vitro and in vivo. A, Northern blot detection of Fgd2 message in total RNA of immature bone marrow B cells stimulated with (+) or without (–) anti-BCR antibodies for 24 h. Left, ethidium bromide stained gel revealing 18 and 28 S ribosomal RNA bands; right, Northern blot detection of Fgd2. B, quantitative analysis of Fgd2 mRNA expression in B cell subsets in vivo. CDNA was generated from total RNA harvested from sorted ex vivo B cell subsets: pro (CD43+B220+sIg–), pre (CD43–B220+sIg–), immature B (3-83 BCR Tg with innocuous receptor; i.e. H-2d background (Imm)), editing immature B cells (3-83 BCR Tg with in vivo autoantigen; H-2k (Edit)), spleen (Spl), and lymph nodes (LN). Real time PCR was performed, and the mean and S.D. of Fgd2 and actin amplicons were calculated from triplicate samples.
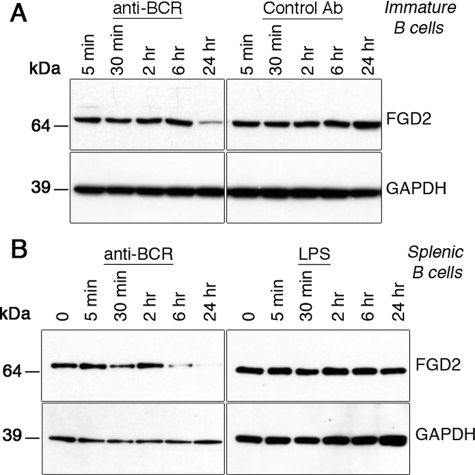
FGD2 Protein Expression in B Cells Is Regulated by a BCR-specific Signal—Since we found Fgd2 mRNA to be repressed in immature B cells undergoing tolerance-induced receptor editing, we were interested in whether this was also true for the protein level in both immature and mature B cells. We therefore used Western blotting with FGD2 antiserum to assess FGD2 protein expression and regulation in primary mouse B cells stimulated with anti-BCR antibody. Immunoblotting analysis carried out with lysates from both immature (Fig. 3A) and mature splenic B cells (Fig. 3B) cultured up to 24 h with or without anti-BCR antibodies revealed that anti-BCR-treated cells had strikingly lower FGD2 protein expression (Fig. 3, A and B), which correlated with the changes observed in RNA expression (Fig. 2A). FGD2 protein expression in immature B cells was suppressed after 24 h of BCR stimulation (Fig. 3A, left panels) but was unaffected by the presence of a control antibody (Fig. 3A, right panels). Similarly, mature splenic B cells down-regulated FGD2 protein expression upon BCR stimulation, albeit with faster kinetics (Fig. 3B, left panels). To test if the repression of FGD2 was specific to a BCR-mediated signal or could also be mediated by a different mitogenic signal, such as signals mediated by Toll-like receptors, we stimulated splenic B cells with the mitogen LPS. This treatment failed to down-regulate FGD2 expression (Fig. 3B, right panels), and we therefore conclude that FGD2 protein down-regulation is the result of a BCR-specific signal.
FIGURE 3.
FGD2 is regulated by a BCR-specific signal. A, immature B cells generated in interleukin-7 culture of 3-83 BCR transgenic bone marrow were challenged with 20 μg/ml idiotypic anti-BCR or isotype control antibody for the indicated times, and protein expression was analyzed by Western blotting. B, splenic purified B cells were stimulated with anti-BCR antibody (goat F(ab)2 anti-IgM 10 μg/ml) or LPS (1 μg/ml) for the times indicated, and FGD2 (top) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH; bottom) protein levels were determined by Western blotting.
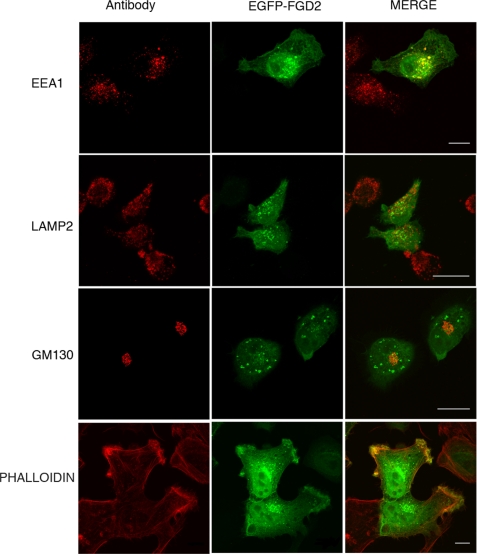
Dual Subcellular Localization of FGD2—Rho family GTPases, such as Rho, Rac, and Cdc42, can control many processes within cells, depending on localized regulation by their cognate RhoGEFs, GDP dissociation inhibitors, or GTPase-activating proteins. For example, Rho promotes contractile actin stress fibers and focal adhesions (35); Cdc42 induces filipodia and foci (36); and Rac regulates formation of lamellipodia and membrane ruffles (37). To follow the subcellular localization of FGD2, an EGFP-FGD2 fusion protein was transiently expressed in HeLa cells. As shown in Fig. 4, EGFP-FGD2 was present in the cytoplasm and nucleus, with a proportion of FGD2 concentrated to vesicular structures and areas of membrane ruffling. Because FGD2 contains a FYVE domain, and the binding of phospholipids via FYVE domains is associated with the endosomal localization of some proteins, we compared the intracellular localization of FGD2 with the co-staining of intracellular compartment markers. Imaging experiments in transfected HeLa cells indicated that about half of EGFP-FGD2 present on vesicular structures associated with early endosomes, as judged by EEA1 colocalization (Fig. 4 upper right panel). In contrast, little colocalization of FGD2 with either LAMP2 or GM130 staining was observed, indicating poor co-localization of FGD2 with lysosomes or the cis-Golgi, respectively (Fig. 4, middle and lower panels). The same intracellular localization of FGD2 was observed when linked to a different tag (FLAG epitope) and stained for FLAG and EEA1 (data not shown).
FIGURE 4.
Colocalization of FGD2 with organelle markers reveals partial localization to early endosomes and membrane ruffles. Shown are confocal fluorescent microscopy images of HeLa cells transfected with EGFP-FGD2 prior to fixation, permeabilization, and staining with antibodies against the endosomal marker EEA1 (upper panels), the lysosomal marker LAMP2 (middle panels), the cis-Golgi marker GM130 (lower panels) and phalloidin, a stain for F-actin (bottom panels). Bound antibody was detected with anti-mouse IgG Alexa 568 (red). A 10-μm scale is shown as white bars.
We also observed concentration of FGD2 on membrane areas that appeared to be ruffles. We co-stained EGFP-FGD2-expressing cells with phalloidin to highlight the areas of actin-rich membrane ruffles. Strong co-localization of EGFP-FGD2 with phalloidin was observed (Fig. 4, bottom right panel). In HeLa cells overexpressing FGD2, we also observed increases in the numbers of membrane ruffles at steady state, but this was not consistent across many experiments.
Localization of FDG2 to Early Endosomes Is FYVE Domain-dependent—To determine if the conserved FYVE domain directed the vesicular staining pattern of FGD2, we assessed the endosomal association of FGD2 and mutant molecules in which the FYVE domain had been mutated (FGD2FYVEKT; Q454K/W455T) Although wild type EGFP-FGD2 still demonstrated a vesicular pattern co-staining with EEA1 (Fig. 5A, upper panel), mutation of the FYVE domain abrogated the co-localization with this early endosomal marker (Fig. 5A, middle panel). However, a functional DH domain is not required for endosomal localization as the FGD2GEFAA DH domain mutant co-localized with EEA1 (Fig. 5A, lower panel). Mutants used in this study are depicted in a diagram (Fig. 5C).
The affinity of FYVE domains for endosomal vesicles requires phosphorylated phosphatidylinositol (phosphatidylinositol 3-phosphate) and is regulated by phosphatidylinositol 3-kinase activity (11). We therefore assessed the FGD2 endosomal localization in the presence and absence of the broad phosphatidylinositol 3-kinase inhibitor, wortmannin. As shown in Fig. 5B, the vesicular pattern of staining of EGFP-FGD2 was largely blocked when transfected HeLa cells were cultured for 30 min in the presence of wortmannin (lower panels) but was unchanged by incubation of the cells with the solvent DMSO alone (upper panels). The treatment of cells with wortmannin and other more specific class III phosphatidylinositol 3-kinase inhibitors like 3-methyladenine has been shown to affect the localization of FYVE domain-containing proteins like EEA1, leading to alterations in the morphology of early endosomes but not their disappearance (38, 39). In our cells, the wortmannin treatment had the expected effects on EEA1 localization (Fig. 5B, lower panel). We conclude that a proportion of FGD2 is recruited to endosomal membranes via the FYVE domain and by the presence of phosphatidylinositol 3-phosphate or other phosphatidylinositides generated by phosphatidylinositol 3-kinase activity.
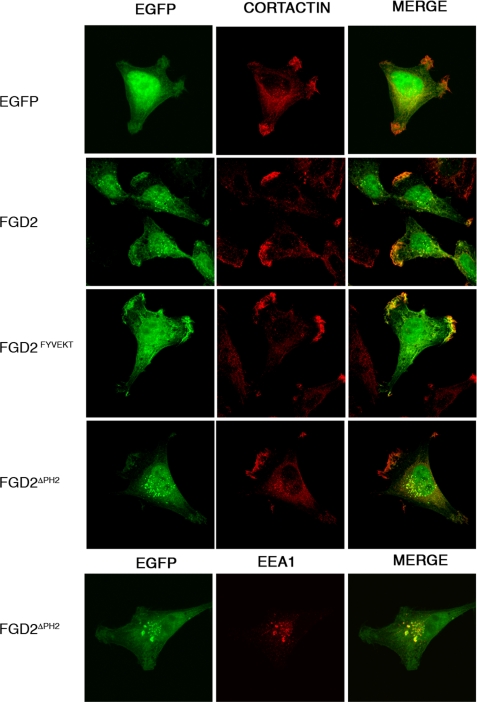
FGD2 Recruitment to Membrane Ruffles is PH Domain-dependent—Both FGD1 and FGD4 (Frabin) are recruited to the actin cytoskeleton within membrane ruffles either directly, by binding actin (40), or indirectly, through interactions with proteins like cortactin (16). Since FGD2 lacks an identifiable actin or cortactin binding domain we speculated that the C-terminal PH domain might mediate its recruitment to membrane ruffles. To test this hypothesis, we expressed an EGFP-FGD2 mutant lacking the C-terminal PH domain (FGD2ΔPH2) in HeLa cells and co-stained these cells for cortactin, a protein enriched in areas of newly polymerized actin filaments. In contrast to wild type FGD2, the FGD2ΔPH2 mutant remained predominantly cytoplasmic and was not concentrated in membrane ruffles (Fig. 6). Its localization to early endosomes was not affected by the deletion of this second PH domain. On the contrary, it appeared as if the vesicular staining pattern was increased in FGD2ΔPH2 expressing cells, and approximately half of these vesicles co-stained with the endosomal marker EEA1. Given that PH domains are known to specifically bind to phosphoinositides, our experiments are consistent with a role of the C-terminal PH domain in the translocation of FGD2 to activated membrane ruffles rich in phosphatidylinositol 4,5-bisphosphate and PI(3,4,5)P3 lipids.
FIGURE 6.
The C-terminal PH domain drives FGD2 co-localization with cortactin in membrane ruffles. HeLa cells expressing either EGFP alone (top panel), EGFP-FGD2 fusion protein (second panel), EGFP-FGD2FYVEKT (third panel), or EGFP-FGD2ΔPH2 (fourth panel) were grown on coverslips prior to fixation and permeabilization with Triton X-100. The samples were stained for cortactin, a protein localizing to newly polymerized actin filaments, using a monoclonal antibody, followed by Alexa 586-goat anti-mouse IgG (red). Cells expressing EGFP-FGD2ΔPH2 were also stained for the endosomal marker EEA1 (bottom).
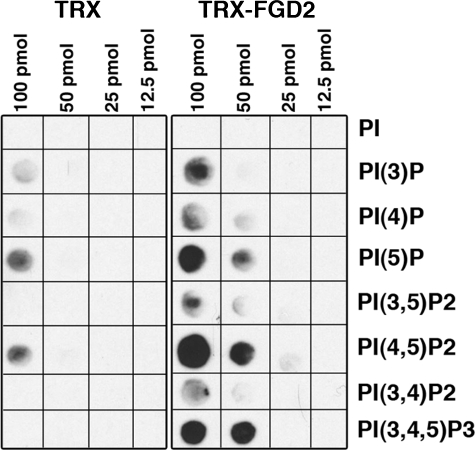
Recombinant FGD2 Binds Phosphatidylinositides Found at Plasma Membranes—The above experiments suggest that the FYVE and the C-terminal PH domain mediate FGD2 recruitment to early endosomes and membrane ruffles, respectively. We therefore assessed if FGD2 could directly bind phosphatidylinositides found at endosomal and plasma membranes. A recombinant fusion protein of TRX-FGD2 bound to multiple phosphatidylinositides spotted onto a membrane, whereas the control TRX protein alone had limited binding under the same experimental conditions (Fig. 7). TRX-FGD2 bound phosphatidylinositol 3-phosphate, as would be predicted from its localization to early endosomes. However, recombinant TRX-FGD2 appeared to have higher affinity for phosphatidylinositol 5-phosphate, phosphatidylinositol 4,5-bisphosphate, and PI(3,4,5)P3, phosphoinositides also found in membrane ruffles. Since PH domains have affinity for phosphatidylinositol 4,5-bisphosphate and PI(3,4,5)P3, we assume that the PH domains of FGD2 are functional and contribute to the lipid binding properties of this protein. Indeed, we have demonstrated that interactions of the C-terminal PH domain with membrane lipids accounts at least in part for the partial co-localization of FGD2 with cortactin at regions of ruffling plasma membranes. The interaction of recombinant FGD2 with these phospholipids was specific, because TRX-FGD2 did not bind to phosphatidic acid, assorted sphingolipids, or phosphatidylinositol alone in this assay (data not shown).
FIGURE 7.
Recombinant FGD2 directly binds to phospholipids. His-thioredoxin protein alone (TRX; left) or His-thioredoxin-FGD2 fusion protein (TRX-FGD2; right) was incubated with membranes spotted with listed concentrations of phospholipids. After washing, bound protein was detected with an anti-His antibody and chemiluminescence. PI, phosphatidylinositol; PI(3)P, phosphatidylinositol-3-phosphate; PI(4)P, phosphatidylinositol-4-phosphate; PI(5)P, phosphatidylinositol-5-phosphate; PI(3,4)P2, phosphatidylinositol 3,4-bisphosphate; PI(3,5)P2, phosphatidylinositol 3,5-bisphosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate.
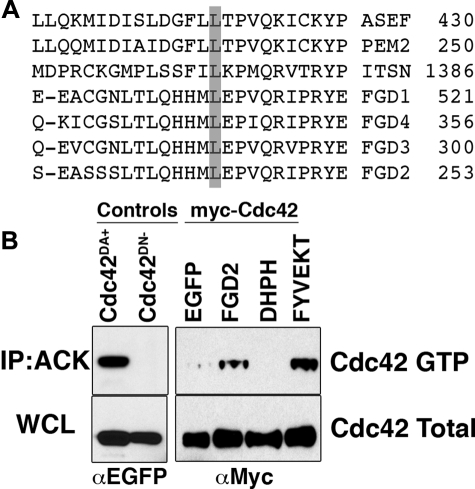
FGD2 Has RhoGEF Activity for Cdc42—Other Fgd family members have been shown to have exchange activity toward Cdc42 directly (FGD1 (11) and FGD4 (17, 18)) and Rac1 indirectly (FGD4 (18)), suggesting that FGD2 might have similar specificity. Alignment of the “specificity patch” region of the DH domain sequences of FGD2 with other Cdc42-specific RhoGEFs (ASEF, h-PEM2, intersectin) further supported this prediction (Fig. 8A); a conserved leucine, which permits effective binding of RhoGEFs to Cdc42 (4), was also conserved in FGD2 and all other Fgd family members (Fig. 8A). To experimentally test this potential regulation of Cdc42 by FGD2, we performed pull-down experiments using lysates of COS-7 cells transiently transfected with Myc-tagged Cdc42 and either EGFP, EGFP-tagged FGD2, or EGFP-tagged FGD2 mutants (Fig. 8B). The Myc-Cdc42 levels were monitored by Western blot in whole cell lysates (WCL) or after pull-down with ACK1-GST beads (IP; ACK). GTP-bound active Cdc42 is specifically pulled down by the ACK1-GST beads in this assay, since an EGFP-tagged constitutively active GTP-bound mutant of Cdc42 (Cdc42DA+) was brought down by the beads, but the GDP-bound mutant of Cdc42 (Cdc42DN–) was not (Fig. 8B, left panels). Compared with the EGFP protein control, EGFP-FGD2 appeared to promote association of the ACK1-GST with Cdc42 (Fig. 8B, right panels), suggesting that it elevates the level of active Cdc42 in the cell lysate. This elevation of Cdc42 GTP levels was also observed when cDNA of an EGFP-tagged FYVE domain mutant of FGD2 (FGD2FYVEKT) was overexpressed (Fig. 8B, right panels). We therefore believe that a functional FYVE domain is not required for the activation of Cdc42 by FGD2. However, an EGFP-tagged truncation mutant of FGD2, consisting of the DH-PH domains only (DHPH), did not appear to activate Cdc42 in this assay (Fig. 8B, right panels), indicating that additional domains are necessary for full FGD2 activity and interaction with Cdc42. Full-length FGD2 was found to mediate Cdc42 activation in several additional experiments (not shown), using either the ACK1-Crib GST or WASP-Crib GST fusion proteins in the pull-down assay. We did not see evidence of endogenous Cdc42 activation in COS-7 cells by recombinant FGD2 expression, indicating that in these cells, the levels of active Cdc42 were too low to be reproducibly measured. In addition, we did not see consistent evidence of activation of either Rac1 or RhoA using PAK1-Crib GST or Rhotekin RBD-GST beads (supplemental Fig. 1). Collectively, these results support the conclusion that when expressed exogenously, FGD2 has RhoGEF activity specific for Cdc42.
FIGURE 8.
FGD2 activates Cdc42. A, the “specificity patch” within the DH domains of RhoGEFs specific for Cdc42 (ASEF, PEM2, intersectin, and FGD1,3,4) are aligned with the corresponding FGD2 sequence. The conserved leucine mediating effective binding to Cdc42 is shaded in gray. B, lysates from COS-7 cells transfected with Myc-Cdc42 and EGFP, EGFP-tagged FGD2, FGD2-DHPH domain (DHPH), or FGD2FYVEKT mutant (FYVEKT) were immunoprecipitated with ACK1-crib-GST beads. Immunoprecipitated beads (IP; Ack, upper panels) or whole cell lysates (WCL; lower panels) were electrophoresed and immunoblotted with anti-Myc (right panels) or anti-EGFP (left panels). Lysates from transfected COS-7 cells expressing dominant active (Cdc42DA+) and negative (Cdc42DN–) EGFP-tagged Cdc42 mutants were used as controls (left panels). This experiment was performed three times with reproducible results.
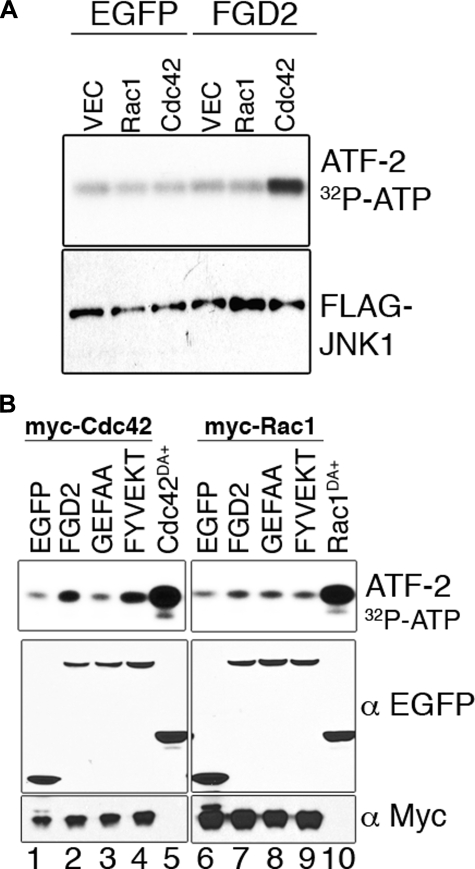
FGD2 Activates JNK1 via Cdc42—Guanine exchange factors can direct downstream signaling of cognate GTPases (41); therefore, we wished to test if FGD2 could activate mitogen-activated protein kinase activity via Cdc42. In co-transfection studies, overexpression of EGFP-tagged FGD2, but not EGFP alone, led to increased JNK1 activity in the presence of Cdc42 but not Rac1 (Fig. 9A). The elevated JNK1 activity, as measured by the phosphorylation of the substrate, GST-ATF-2, was dependent on wild type FGD2 sequence in the DH domain, since a DH mutant of FGD2 (FGD2GEFAA) had a diminished effect on JNK1 activation (Fig. 9B, lane 3). In contrast, mutation of residues in the FYVE domain (FGD2FYVEKT) had no effect on the enhancement of JNK1 activity (Fig. 9B, lane 4). A constitutively active mutant of Cdc42, EGFP-Cdc42Q61L demonstrated the highest activation of JNK. This supports the conclusion that FGD2 utilizes the DH domain to activate JNK1 through Cdc42 but not Rac1, despite the ability of active Rac1 to also mediate JNK1 activation (Fig. 9B, lane 10).
FIGURE 9.
FGD2 enhances JNK1 activity via Cdc42 but not Rac1. A, COS-7 cells were transiently transfected with FLAG-JNK1 cDNA together with EGFP or EGFP-FGD2, in the presence of either empty vector (V), Myc-tagged Rac1 (R), or Myc-tagged Cdc42 (C). JNK1 was immunoprecipitated with anti-FLAG beads and analyzed for kinase activity by incubation with [32P]ATP and the JNK1 substrate ATF-2-GST. Eluates from the FLAG beads were electrophoresed and transferred to membranes; JNK1 activity was determined by the autoradiography of phosphorylated ATF-2. Western blotting from electrophoresed cell lysates confirmed equal expression of transfected JNK1 (lower panel). B, FGD2 requires Cdc42 and an active GEF domain but not an active FYVE domain to stimulate JNK1 activity. JNK1 kinase assays were performed as in A, but COS-7 cells were transfected with Myc-Cdc42 (left panels) or Myc-Rac1 (right panels) and either EGFP alone, EGFP-tagged FGD2, EGFP-tagged FGD2GEFAA mutant, or EGFP-tagged FGD2FYVEKT mutant. EGFP-tagged constitutively active mutants of Cdc42 (lane 5) or Rac1 (lane 10) were used as positive controls. The expression of the various components was confirmed by Western blotting with antibodies against either Myc or EGFP.
DISCUSSION
The present study provides the first characterization of FGD2 expression, cellular localization, and protein function. Our data support the predictions that mouse FGD2 has RhoGEF activity and can regulate Cdc42 and downstream JNK1 activation. Furthermore, we have discovered that Fgd2 has striking developmental and tissue-restricted expression in antigen-presenting cells of the immune system. Particularly, B lymphocytes express high levels of FGD2, but significant expression was also detected in bone marrow macrophages and to lower levels in mature bone marrow dendritic cells. In both immature and mature B cells, FGD2 expression is down-regulated by prior B cell receptor signaling. Another B cell mitogen, LPS, fails to do so, suggesting that FGD2 expression in B cells is regulated specifically by BCR signals. The down-regulation of Fgd2 message after BCR cross-linking does not appear to be permanent, however, since memory B cells also appear to express high levels of Fgd2 mRNA (data from gene expression omnibus accession number GDS1695). These data indicate that Fgd2 mRNA expression remains high in resting B and memory cells but declines upon differentiation into plasma cells. Finally, we show that FGD2 is associated with early endosomes in a phosphatidylinositol 3-kinase-dependent manner and with plasma membrane ruffles, suggesting multiple possible avenues for regulation of activity and cellular function. Since the amino acid sequences of human and mouse FGD2 are 83% identical and are expressed in a similar tissue distribution (based on the Symatlas data base (42)), we predict human FGD2 probably carries out the same biochemical and biological functions as the mouse FGD2 protein studied here. The FGD2-restricted expression pattern in antigen-presenting cells implies that it has a unique role in controlling the biology of these cells. This is underscored by the finding that FGD2 expression is tightly regulated at least during B lymphocyte development. Potential cellular functions that might be modulated by FGD2 include antigen uptake via endocytosis, antigen presentation via regulation of vesicle trafficking, and cell migration via cytoskeletal rearrangements. In accordance with these potential FGD2 functions, we demonstrate that FGD2 is partly localized to early endosomes, through FYVE domain-mediated binding to phospholipids. To our knowledge, FGD2 is the only known RhoGEF family member shown to have a functional FYVE domain and endosomal binding activity. Although both FGD1 and FGD3 have a FYVE domain, sequence analysis shows that FGD1 and FGD3 have species-conserved alterations of the critical tryptophan residue in the WXXD motif of the FYVE domain (see supplemental Fig. 2). Mutation of this residue in EEA1 abrogates endosomal localization, indicating that both FGD1 and FGD3 may have reduced affinity for phosphatidylinositol 3-phosphate and endosomes (43). FGD4, or Frabin, appears to have a conventional FYVE domain motif, but endosomal localization of this protein has not been described, perhaps due to the presence of an F-actin binding domain that may predominantly direct the protein to the actin cytoskeleton (44).
Although the Rab and Arf families of GTPases are best known for their roles in vesicle budding, docking, fusion, and movement within the mammalian endocytic and secretory pathways (45–47), functions for the Rho family of GTPases in vesicle pathways are being elucidated (48). A role for Cdc42 in intracellular trafficking in yeast has been identified, where Cdc42 is involved in vacuolar membrane docking and fusion (49, 50). Recent studies have also demonstrated that an effector of Cdc42, ACK1, is intimately involved in regulating the endocytosis and degradation of the EGFR (51). In addition, in maturing bone marrow-derived dendritic antigen-presenting cells, which express FGD2, the endocytic capacity correlates with the level of active Cdc42 (52). In this system, as dendritic cells mature, the ability to uptake further antigens by endocytosis is blocked, and mature dendritic cells have reduced levels of active Cdc42 when compared with immature dendritic cells. The intracellular localization of FGD2 at endocytic vesicle membranes would suggest that FGD2 is in a unique position to regulate Cdc42 and possible vesicle formation and trafficking processes in dendritic or other professional antigen-presenting cells.
We also found a portion of FGD2 localizing to membrane ruffles. In contrast to FGD1, which associates with cortical actin via cortactin binding by its Src homology 3 binding domain, we show that FGD2 is recruited to membrane ruffles through binding of phosphoinositides by its C-terminal PH domain. The presence of FGD2 in cortactin-rich membrane ruffles may relate to a role of this protein in the regulation of cell polarization and migration, since membrane ruffles often form at the leading edge of migrating cells. Another possibility is that FGD2 controls macropinocytic endocytosis in antigen-presenting cells. Membrane ruffles are essential parts of the macropinocytic machinery, which controls high volume nonspecific endocytosis. Blockade of Rho family GTPases by toxin B completely inhibits macropinocytosis of immature spleen or bone marrow dendritic cells, as does injection of dominant negative Cdc42 or Rac1 (53). Presumably, FGD2 would exert control over this endocytic pathway via Cdc42, rather than Rac1, since we have not been able to detect direct activation of Rac1 by FGD2. Membrane ruffling is classically viewed as controlled by Rac-specific RhoGEFs. Constitutive ruffling by dendritic cells, which reflects macropinocytosis, appears to be different, however, since previous studies have shown that it is independent of Rac (54).
The recruitment of FGD2 to both endosomal membranes and the actin-rich lamellipodia is driven by two competing signals. In the absence of the C-terminal PH domain, early endosomal recruitment of FGD2ΔPH2 appeared to be enhanced, with a corresponding loss of membrane ruffle localization. In contrast, the FYVE domain mutant FGD2FYVEKT lacked endosomal localization but appeared to have augmented recruitment to the cortactin-rich areas of the cell. We conclude that wild type FGD2 is receptive to both signals and can be recruited to either compartment, depending on the levels of specific phospholipids and phosphatidylinositol 3-kinase activity within cellular compartments.
Our biochemical analysis of FGD2 predicts a specific role of FGD2 in the activation of Cdc42 and not of Rac1. Based on studies of the RhoGEFs Dbs and intersectin, the structure of FGD2 (and indeed all Fgd family members) predicts a role as a Cdc42 activator and seems inconsistent with Rac activation (4). All Fgd members have a conserved leucine residue in the DH domain, which, when changed to an isoleucine in intersectin, leads to a specificity change from Cdc42 to Rac1 (4). Overexpression of FGD2 led to elevated levels of active GTP-bound Cdc42, and FGD2 activated JNK1 activity when expressed with Cdc42, but not Rac1, consistent with Cdc42 specificity. FGD1, FGD3, and FGD4 (Frabin) are known to activate Cdc42 directly (17–19), we presume the same to be true for FGD2, although so far, we have been unable to generate a soluble recombinant FGD2 that contains any exchange activity in vitro.
A recent publication reports that in certain wild strains of mice, overexpression of Fgd2 RNA is associated with an altered allele mapped to the T complex on chromosome 17, leading to altered inheritance of the T complex (22), a phenomenon called t-complex ratio distortion (55). Although normally expressed at very low levels in testes, this study shows that an altered form of Fgd2 RNA is overexpressed in T complex-containing sperm, which in conjunction with other distortion genes can influence the fertilization and thus transmission of the T complex genetic region. The authors propose that FGD2 is acting upstream in the activation of Smok (sperm motility kinase) (22). Cdc42 is expressed in sperm cells, along with other Rho GTPases (56); however it is not clear if the FGD2-mediated distortion effects on sperm fertility in this model are dependent upon Cdc42 function or through a so far uncharacterized regulation of the Smok kinase pathway. Since the Smok kinase described in mice is not conserved across species, and Fgd2 is highly conserved in mammals, we predict that FGD2 will play roles in other Cdc42-dependent signaling pathways. Future studies of the immune response of FGD2-deficient animals may help elucidate this novel protein's function in vivo.
Supplementary Material
Acknowledgments
We thank Dr. Bill Kiosses for helpful advice and microscopy assistance, Dr. David Lo and Digital Gene Technologies for the total gene expression analysis, and Dr. Marianne Martinic for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI033608 and R01AI067403. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: GEF, guanine nucleotide exchange factor; BCR, B cell antigen receptor; PH, pleckstrin homology; DH, Dbl homology; PI(3,4,5)P3, phosphatidylinositol 3,4,5-trisphosphate; TRX, thioredoxin; GM-CSF, granulocyte-monocyte colony-stimulating factor; LPS, lipopolysaccharide; GST, glutathione S-transferase; EGFP, enhanced green fluorescence protein.
D. Nemazee, unpublished results.
References
- 1.Pasteris, N. G., and Gorski, J. L. (1999) Genomics 60 57–66 [DOI] [PubMed] [Google Scholar]
- 2.Rossman, K. L., Der, C. J., and Sondek, J. (2005) Nat. Rev. Mol. Cell Biol. 6 167–180 [DOI] [PubMed] [Google Scholar]
- 3.Buchsbaum, R. J. (2007) J. Cell Sci. 120 1149–1152 [DOI] [PubMed] [Google Scholar]
- 4.Snyder, J. T., Worthylake, D. K., Rossman, K. L., Betts, L., Pruitt, W. M., Siderovski, D. P., Der, C. J., and Sondek, J. (2002) Nat. Struct. Biol. 9 468–475 [DOI] [PubMed] [Google Scholar]
- 5.Liu, X., Wang, H., Eberstadt, M., Schnuchel, A., Olejniczak, E. T., Meadows, R. P., Schkeryantz, J. M., Janowick, D. A., Harlan, J. E., Harris, E. A., Staunton, D. E., and Fesik, S. W. (1998) Cell 95 269–277 [DOI] [PubMed] [Google Scholar]
- 6.Zhu, K., Debreceni, B., Li, R., and Zheng, Y. (2000) J. Biol. Chem. 275 25993–26001 [DOI] [PubMed] [Google Scholar]
- 7.Crespo, P., Schuebel, K. E., Ostrom, A. A., Gutkind, J. S., and Bustelo, X. R. (1997) Nature 385 169–172 [DOI] [PubMed] [Google Scholar]
- 8.Schuebel, K. E., Movilla, N., Rosa, J. L., and Bustelo, X. R. (1998) EMBO J. 17 6608–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patki, V., Virbasius, J., Lane, W. S., Toh, B. H., Shpetner, H. S., and Corvera, S. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7326–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patki, V., Lawe, D. C., Corvera, S., Virbasius, J. V., and Chawla, A. (1998) Nature 394 433–434 [DOI] [PubMed] [Google Scholar]
- 11.Burd, C. G., and Emr, S. D. (1998) Mol. Cell 2 157–162 [DOI] [PubMed] [Google Scholar]
- 12.Gaullier, J. M., Simonsen, A., D'Arrigo, A., Bremnes, B., Stenmark, H., and Aasland, R. (1998) Nature 394 432–433 [DOI] [PubMed] [Google Scholar]
- 13.Stenmark, H., Aasland, R., Toh, B. H., and D'Arrigo, A. (1996) J. Biol. Chem. 271 24048–24054 [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, K. M., Kavran, J. M., Sankaran, V. G., Fournier, E., Isakoff, S. J., Skolnik, E. Y., and Lemmon, M. A. (2000) Mol. Cell 6 373–384 [DOI] [PubMed] [Google Scholar]
- 15.Pasteris, N. G., Cadle, A., Logie, L. J., Porteous, M. E., Schwartz, C. E., Stevenson, R. E., Glover, T. W., Wilroy, R. S., and Gorski, J. L. (1994) Cell 79 669–678 [DOI] [PubMed] [Google Scholar]
- 16.Hou, P., Estrada, L., Kinley, A. W., Parsons, J. T., Vojtek, A. B., and Gorski, J. L. (2003) Hum. Mol. Genet. 12 1981–1993 [DOI] [PubMed] [Google Scholar]
- 17.Zheng, Y., Fischer, D. J., Santos, M. F., Tigyi, G., Pasteris, N. G., Gorski, J. L., and Xu, Y. (1996) J. Biol. Chem. 271 33169–33172 [DOI] [PubMed] [Google Scholar]
- 18.Ono, Y., Nakanishi, H., Nishimura, M., Kakizaki, M., Takahashi, K., Miyahara, M., Satoh-Horikawa, K., Mandai, K., and Takai, Y. (2000) Oncogene 19 3050–3058 [DOI] [PubMed] [Google Scholar]
- 19.Pasteris, N. G., Nagata, K., Hall, A., and Gorski, J. L. (2000) Gene (Amst.) 242 237–247 [DOI] [PubMed] [Google Scholar]
- 20.Delague, V., Jacquier, A., Hamadouche, T., Poitelon, Y., Baudot, C., Boccaccio, I., Chouery, E., Chaouch, M., Kassouri, N., Jabbour, R., Grid, D., Megarbane, A., Haase, G., and Levy, N. (2007) Am. J. Hum. Genet. 81 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stendel, C., Roos, A., Deconinck, T., Pereira, J., Castagner, F., Niemann, A., Kirschner, J., Korinthenberg, R., Ketelsen, U. P., Battaloglu, E., Parman, Y., Nicholson, G., Ouvrier, R., Seeger, J., Jonghe, P. D., Weis, J., Kruttgen, A., Rudnik-Schoneborn, S., Bergmann, C., Suter, U., Zerres, K., Timmerman, V., Relvas, J. B., and Senderek, J. (2007) Am. J. Hum. Genet. 81 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer, H., Veron, N., Willert, J., and Herrmann, B. G. (2007) Genes Dev. 21 143–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deruytter, N., Boulard, O., and Garchon, H. J. (2004) Diabetes 53 3323–3327 [DOI] [PubMed] [Google Scholar]
- 24.Hammerling, G. J., Rusch, E., Tada, N., Kimura, S., and Hammerling, U. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 4737–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vairo, G., and Hamilton, J. A. (1985) Biochem. Biophys. Res. Commun. 132 430–437 [DOI] [PubMed] [Google Scholar]
- 26.Lutz, M. B., Kukutsch, N., Ogilvie, A. L., Rossner, S., Koch, F., Romani, N., and Schuler, G. (1999) J. Immunol. Methods 223 77–92 [DOI] [PubMed] [Google Scholar]
- 27.Verkoczy, L., Ait-Azzouzene, D., Skog, P., Martensson, A., Lang, J., Duong, B., and Nemazee, D. (2005) Immunity 22 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell, D. M., Dembic, Z., Morahan, G., Miller, J. F., Burki, K., and Nemazee, D. (1991) Nature 354 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stofega, M., DerMardirossian, C., and Bokoch, G. M. (2006) Methods Mol. Biol. 332 269–279 [DOI] [PubMed] [Google Scholar]
- 30.Sanna, M. G., Duckett, C. S., Richter, B. W., Thompson, C. B., and Ulevitch, R. J. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6015–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nemazee, D. (2006) Nat. Rev. Immunol. 6 728–740 [DOI] [PubMed] [Google Scholar]
- 32.Nemazee, D. A., and Burki, K. (1989) Nature 337 562–566 [DOI] [PubMed] [Google Scholar]
- 33.Sutcliffe, J. G., Foye, P. E., Erlander, M. G., Hilbush, B. S., Bodzin, L. J., Durham, J. T., and Hasel, K. W. Proc. Natl. Acad. Sci. U. S. A. 97 1976–1981 [DOI] [PMC free article] [PubMed]
- 34.Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., Orth, A. P., Vega, R. G., Sapinoso, L. M., Moqrich, A., Patapoutian, A., Hampton, G. M., Schultz, P. G., and Hogenesch, J. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4465–4470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridley, A. J., and Hall, A. (1992) Cell 70 389–399 [DOI] [PubMed] [Google Scholar]
- 36.Nobes, C. D., and Hall, A. (1995) Cell 81 53–62 [DOI] [PubMed] [Google Scholar]
- 37.Ridley, A. J., Paterson, H. F., Johnston, C. L., Diekmann, D., and Hall, A. (1992) Cell 70 401–410 [DOI] [PubMed] [Google Scholar]
- 38.Kjeken, R., Mousavi, S. A., Brech, A., Griffiths, G., and Berg, T. (2001) Biochem. J. 357 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egami, Y., and Araki, N. (2008) Exp. Cell Res. 314 729–737 [DOI] [PubMed] [Google Scholar]
- 40.Obaishi, H., Nakanishi, H., Mandai, K., Satoh, K., Satoh, A., Takahashi, K., Miyahara, M., Nishioka, H., Takaishi, K., and Takai, Y. (1998) J. Biol. Chem. 273 18697–18700 [DOI] [PubMed] [Google Scholar]
- 41.Zhou, K., Wang, Y., Gorski, J. L., Nomura, N., Collard, J., and Bokoch, G. M. (1998) J. Biol. Chem. 273 16782–16786 [DOI] [PubMed] [Google Scholar]
- 42.Su, A. I., Wiltshire, T., Batalov, S., Lapp, H., Ching, K. A., Block, D., Zhang, J., Soden, R., Hayakawa, M., Kreiman, G., Cooke, M. P., Walker, J. R., and Hogenesch, J. B. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaullier, J. M., Ronning, E., Gillooly, D. J., and Stenmark, H. (2000) J. Biol. Chem. 275 24595–24600 [DOI] [PubMed] [Google Scholar]
- 44.Kim, Y., Ikeda, W., Nakanishi, H., Tanaka, Y., Takekuni, K., Itoh, S., Monden, M., and Takai, Y. (2002) Genes Cells 7 413–420 [DOI] [PubMed] [Google Scholar]
- 45.Christoforidis, S., McBride, H. M., Burgoyne, R. D., and Zerial, M. (1999) Nature 397 621–625 [DOI] [PubMed] [Google Scholar]
- 46.Zerial, M., and McBride, H. (2001) Nat. Rev. 2 107–117 [DOI] [PubMed] [Google Scholar]
- 47.Donaldson, J. G., and Honda, A. (2005) Biochem. Soc. Trans. 33 639–642 [DOI] [PubMed] [Google Scholar]
- 48.Qualmann, B., and Mellor, H. (2003) Biochem. J. 371 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller, O., Johnson, D. I., and Mayer, A. (2001) EMBO J. 20 5657–5665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eitzen, G., Thorngren, N., and Wickner, W. (2001) EMBO J. 20 5650–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen, F., Lin, Q., Gu, Y., Childress, C., and Yang, W. (2007) Mol. Biol. Cell 18 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garrett, W. S., Chen, L. M., Kroschewski, R., Ebersold, M., Turley, S., Trombetta, S., Galan, J. E., and Mellman, I. (2000) Cell 102 325–334 [DOI] [PubMed] [Google Scholar]
- 53.Nobes, C., and Marsh, M. (2000) Curr. Biol. 10 R739–R741 [DOI] [PubMed] [Google Scholar]
- 54.West, M. A., Prescott, A. R., Eskelinen, E. L., Ridley, A. J., and Watts, C. (2000) Curr. Biol. 10 839–848 [DOI] [PubMed] [Google Scholar]
- 55.Lyon, M. F. (2005) Nat. Genet. 37 924–925 [DOI] [PubMed] [Google Scholar]
- 56.Ducummon, C. C., and Berger, T. (2006) Zygote 14 249–257 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.