Abstract
Misfolded proteins in the endoplasmic reticulum (ER) are exported to the cytosol for degradation by the proteasome in a process known as ER-associated degradation (ERAD). CPY* is a well characterized ERAD substrate whose degradation is dependent upon the Hrd1 complex. However, although the functions of some of the components of this complex are known, the nature of the protein dislocation channel remains obscure. Sec61p has been suggested as an obvious candidate because of its role as a protein-conducting channel through which polypeptides are initially translocated into the ER. However, it has not yet been possible to functionally dissect any role for Sec61p in dislocation from its essential function in translocation. By changing the translocation properties of a series of novel ERAD substrates, we are able to separate these two events and find that functional Sec61p is essential for the ERAD-L pathway.
Perturbations in protein biogenesis in the ER2 can lead to the accumulation of misfolded proteins with potentially catastrophic cytotoxic consequences. The ERAD quality control system identifies aberrant proteins and targets them for destruction. This disposal mechanism involves misfolded proteins being “dislocated” across the ER membrane to the cytosol where they are ubiquitinated before being delivered to the proteasome for degradation (1).
Three distinct ERAD pathways can be distinguished according to the topology of the misfolded lesion. ER lumenal proteins are degraded via the ERAD-L pathway, as are integral membrane proteins with lesions in a lumenal domain. Membrane proteins with cytosolic lesions are degraded by the ERAD-C pathway (2), whereas those with misfolded transmembrane domains are degraded by the ERAD-M pathway (3, 4). All three pathways require the cytosolic Cdc48p-Ufd1p-Npl4p complex, which delivers ubiquitinated substrates to the proteasome. ERAD-L involves recognition of aberrant domains by a lumenal surveillance complex, comprising Yos9p and Kar2p, which maintains the substrate in an ERAD-competent conformation. The substrate is then delivered to the membrane-associated Hrd1 complex comprising the E3 ubiquitin ligase Hrd1p and its co-factors Hrd3p, Usa1p, and Der1p (3, 4). At this stage the substrate must be dislocated across the ER membrane where it is ubiquitinated by the Hrd1p ubiquitin ligase in combination with the E2 ubiquitin-conjugating enzyme Ubc7p and its membrane anchor, Cue1p. The ubiquitinated substrate is then passed to the Cdc48p-Ufd1p-Npl4p complex, which is itself anchored to the membrane by the Ubx2p receptor (1).
The identity of the protein dislocation channel has been the subject of considerable debate. Derlin-1, a recently identified human homologue of yeast Der1p has been suggested to form a pore in the ER membrane through which unfolded ERAD substrates are exported for degradation (5, 6). Yeast Der1p is an integral membrane protein required for the degradation of ERAD-L substrates including CPY*, KHN, and KWW (2, 7), but the molecular function of Der1p/Derlin-1 is not yet known. In yeast, Der1p is not essential for viability, but its widespread conservation does suggest an important role in eukaryotes.
Another candidate for the dislocation channel is Sec61p, which is a core component of the translocation channel through which proteins are imported into the ER (8). A number of observations support a role for Sec61p in ERAD including its association with a variety of ERAD substrates (9–11) plus the intriguing observation that proteasomes interact with the Sec61 complex both in vivo and in vitro (12). Studies in yeast have shown that the degradation of an unfolded mutant form of alpha factor is reduced in a cell free assay using microsomes from various sec61 mutant strains (13), whereas the degradation of CPY* is delayed in sec61-2 mutant cells (14). However, the interpretation of these data is complicated by the fact that the sec61 mutant alleles examined were also defective in the initial translocation of both alpha factor and CPY*.
In this paper we employ the sec61-3 mutant, which has a cold-sensitive defect in the signal recognition particle (SRP)-dependent co-translational translocation pathway. We therefore engineered two novel SRP-dependent derivatives of CPY*, one integral membrane form and one soluble, and examined their translocation and ERAD properties. We demonstrate that Sec61p is required for ERAD of these novel substrates in a manner that is independent of any effect on translocation.
EXPERIMENTAL PROCEDURES
Yeast Strains—The strains used in this study are listed in Table 1. Yeast strains were grown in either YPD medium (2% peptone, 1% yeast extract, 2% glucose) or minimal medium (0.67% yeast nitrogen base, 2% glucose, appropriate supplements) at temperatures required for individual experiments (17, 24, 30, or 37 °C). Analysis of plasmid-borne forms of CPY*, DPY*, and OPY* were performed in strains lacking any endogenous CPY (prc1::KANMX). To make strains of the required genotypes, we re-engineered the sec61-3 allele in target strains in the following way. The sec61-3 mutation was introduced into pBW11 (15) by site-directed mutagenesis with Primers 1 and 2 generating plasmid pCW11 Table 2. A 2.4-kb KpnI-PstI fragment containing the mutagenized sec61-3 allele was cloned into KpnI-PstI of YIp352, generating an integrative vector pCW14, which was subsequently linearized with Xba1 and transformed into yeast. Loss of the integrative vector part was selected for on media containing 5-fluo-orotic acid, and the resulting single copy genomic copy of sec61-3 was confirmed by PCR and DNA sequencing.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| MWY61 | Mataprc1::KanMX leu2 his3 trp1 ura3 der1::KanMX | This study |
| MWY63 | Mataprc1::KanMX leu2 his3 ura3 ade2 pep4-3 sec61-3 | This study |
| MWY64 | Mataprc1::KanMX leu2 ura3 ade2 pep4-3 | This study |
| MWY71 | Mataprc1::KanMX leu2 his3 trp1 ura3 ade2 sec65-1 | This study |
| MWY77 | Mataprc1::KanMX leu2 his3 ura3 doa10::KanMX | This study |
| MWY82 | Mataprc1::KanMX leu2 his3 trp1 ura3 ade2 sec62-1 | This study |
| W303-cd3 | Mata leu2 his3 trp1 ura3 ade2 der3 prc1-1 | This study |
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| prc1-1_F | CCGAGCTCGCGATGTTGGTCATCCAGTACACTTCGGTAGC |
| prc1-1_R | GGAATCAGCACATAGTTCTTGAACCAGCTCAG |
| CPY_Nde1_F | CTACTCAACTTAAAGTATACATACCATATGAAAGCATTCACCAGTTTACTATG |
| CPY_Nde1_R | CATAGTAAACTGGTGAATGCTTTCATATGGTATGTATACTTTAAGTTGAGTAG |
| DPAPB_F2 | GAGGACATATGAGAGTCGGTATTATCTTCGTTTTGTTGATTTGGGGTACTGTTT |
| DPAPB_R2 | GTGCCGTAGGCCTTCTATACTCTTCAACAACAAAACAGTACCCAAAATCAACAA |
| OST2F | TTCTTTGAGCTCCGAGGCAGCAGTCTCTGTGGAGGGGGTACC |
| OST2R | CCGGATATCGGCTCGTATTGGGCAGCAGAAGACACGTTGAAAAAACATAGG |
| Primer 1 | CTCAAATGGCCTTGAGCGAGTTGGCCTACTACATCCAACC |
| Primer 2 | GGTTGGATGTAGTAGGCCAACTCGCTCAAGGCCATTTGAG |
Plasmid Constructions—Plasmids encoding CPY* (pMW319), DPY* (pMW339), and OPY* (pMW342) were generated as follows. The CPY* encoding allele of PRC1 (prc1-1) was amplified from genomic DNA with primers prc1-1_F and prc1-1_R. A 2379-bp HindIII-SacI DNA fragment was cloned into pRS315 and pRS316 to generate pMW319 and pMW320, respectively. A 645-bp BamHI-SacI fragment from pMW320 was cloned into pBLUESCRIPT KS+ to generate pMW321. A NdeI restriction site was introduced by site-directed mutagenesis with primers CPY_NdeI_F and CPY_NdeI_R to generate pMW325. Signal sequence coding sequence of DPAP B was amplified with primers DPAPB_F2 and DPAPB_R2 and following digestion with NdeI and StuI cloned into pMW325 to generate pMW330. Finally, a 748-bp SacI-BamHI fragment from was cloned from pMW330 into pMW319 to generate pMW339. An OST1 sequence was PCR-amplified with primers OST2F and OST2R, digested with SacI and EcoRV, and cloned into SacI-StuI of pMW319 to generate pMW342. To be able to make strains of the required genotypes, we decided to regenerate the sec61-3 allele in target strains in the following way: The sec61-3 mutation was introduced into pBW11(22) by site-directed mutagenesis with Primers 1 and 2 generating plasmid pCW11. A 2.4-kb KpnI-PstI fragment was cloned into YIp352, generating pCW14.
Membrane Association Experiments—The microsomes were prepared as previously described (16). The microsomes were treated with 100 mm Na2CO3 at 0 °C for 30 min and spun at 100,000 × g for 60 min. Equal proportions of pellets and supernatants were analyzed by Western blotting with appropriate antibodies. Antibodies against CPY, Kar2p, and Sec61p have previously been described (17–20).
Pulse-Chase Analysis—Temperature shifts were done for 2 h prior to radiolabeling. The cells were grown in minimal medium to A600 = 0.2 and labeled with [35S]methionine for 20 min (5 min for experiments looking at translocation). Chase was performed by the addition of cold methionine and cysteine to final concentrations of 2 mm each. 5 A600 units of cells were taken for each time point, and the cell extracts were prepared for immunoprecipitation as described (17). Radiolabeled proteins were visualized using a Fujifilm FLA3000 phosphorimaging device, and quantitation was performed on signals within the linear range of detection using the Aida Image Analyzer v.3.44 software.
RESULTS
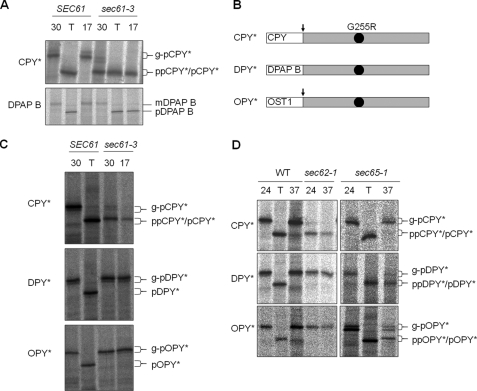
Properties of New ERAD Substrates—A single mutation (G255R) in the yeast vacuolar protease carboxypeptidase Y (CPY) results in a misfolded protein (CPY*), which is a substrate for the ERAD-L pathway (21). PreproCPY* is translated in the cytosol and translocated post-translationally into the ER. We were interested in addressing the role of Sec61p in ERAD using CPY* as a substrate, but all known mutant alleles of sec61 are deficient in post-translational translocation. The sec61-3 mutant allele is no exception but differs in that it also exhibits a cold-sensitive defect in co-translational translocation. This mutant translocates SRP-dependent precursors efficiently at 30 °C but is defective in post-translational translocation at this temperature and is completely deficient in all translocation at 17 °C (Fig. 1A). We therefore reasoned that an SRP-dependent ERAD substrate might translocate normally in sec61-3 cells at 30 °C and might therefore allow us to test specifically for any effect on ERAD following the loss of Sec61p function at 17 °C.
FIGURE 1.
Translocation properties of CPY*, DPY*, and OPY*. A, wild type (SEC61) cells and sec61-3 mutant cells each expressing CPY* were grown at 30 °C and either shifted to 17 °C or incubated with tunicamycin (T) for 2 h prior to radiolabeling as described under “Experimental Procedures.” Immunoprecipitated forms of CPY* and DPAP B were analyzed by SDS-PAGE: translocated glycosylated forms (g-pCPY* and mDPAP B), translocated but unglycosylated forms upon treatment with tunicamycin (pCPY* and pDPAP B), and untranslocated precursor (ppCPY* and pDPAP B). B, schematic representation of the three substrates used in this study. All are based on CPY* and contain the G255R mutation (black dot), which prevents CPY* folding. Predicted signal sequence cleavage sites indicated (arrows), DPY* has the DPAP B signal sequence, and OPY* has the signal sequence from Ost1p. C, translocation of CPY*, DPY*, and OPY* was analyzed in wild type and sec61-3 cells as described in A. D, wild type (WT), sec62-1 cells, or sec65-1 cells expressing either CPY*, DPY*, or OPY* were grown at 24 °C and then either shifted to 37 °C for 1 h or treated with tunicamycin prior to radiolabeling and immunoprecipitation as described above.
The signal sequence of a precursor determines whether it will follow either the Sec62- or SRP-dependent pathway, with more hydrophobic sequences tending to require the latter (22). We therefore sought to create SRP-dependent derivatives of CPY* by replacing its targeting sequence with that of either DPAP-B (DPY*) or Ost1p (OPY*) (Fig. 1B). DPAP-B is a type II integral membrane protein with an amino-terminal signal anchor domain whose targeting is SRP-dependent (23). Ost1p is a type I integral membrane protein with a cleavable signal sequence (24) whose hydrophobicity led us to predict a likely dependence upon SRP.
ER Translocation of DPY* and OPY*—When expressed in wild type cells, both DPY* and OPY* were efficiently translocated and glycosylated, indicating that their respective ER targeting signals are functional. Our hypothesis predicted that DPY* and OPY* would translocate efficiently in sec61-3 cells at 30 °C, and this proved to be the case (Fig. 1C). Interestingly, these proteins also translocate well at 17 °C, suggesting that the SRP-dependent targeting of a polypeptide that is competent for post-translational import can overcome the sec61-3 translocation defect. This finding suggested that the translocation defect in sec61-3 cells might be kinetic in nature, and this has been confirmed by pulse-chase analysis (see supplemental Fig. 1). To confirm that DPY* and OPY* were being targeted via the SRP-dependent pathway, we also examined their translocation in sec65-1 cells, which express a temperature-sensitive form of SRP (23). As expected, preproCPY* translocation is unaffected in sec65-1 cells with only the ER glycosylated form of proCPY* being evident at either 24 or 37 °C. In contrast, DPY* and OPY* translocate efficiently at 24 °C but accumulate as precursor forms at 37 °C (Fig. 1D). These results demonstrate that DPY* and OPY* require functional SRP for their ER targeting. A further characteristic of a genuinely SRP-dependent precursor is that it does not depend on Sec62p for translocation. sec62-1 cells have defects in post-translational translocation at their permissive temperature (24 °C) and are temperature-sensitive for growth at 37 °C (25). We therefore examined translocation of DPY* and OPY* in sec62-1 cells. A profound defect in translocation of CPY* was observed at both 24 and 37 °C, but DPY* and OPY* translocation were unaffected (Fig. 1D). We therefore conclude that unlike CPY*, both DPY* and OPY* translocate via the co-translational SRP-dependent pathway. Most importantly, the sec61-3 mutation has no detectable effect on translocation of these new precursors.
Membrane Association of DPY* and OPY*—The signal peptide of preproCPY* is cleaved during translocation by signal peptidase.
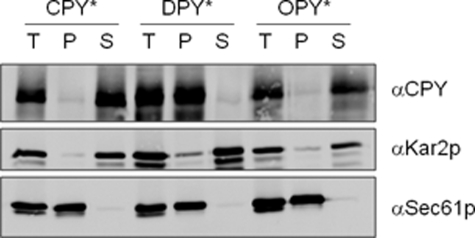
OPY* was predicted to be similarly cleaved, whereas DPY* was predicted to insert into the bilayer as an integral membrane protein. To test these predictions we used carbonate extraction of microsomes to examine the membrane association of the various proteins. We found that DPY* behaved as an integral membrane protein, whereas both CPY* and OPY* were readily extracted by carbonate (Fig. 2). We therefore conclude that OPY* is soluble, whereas DPY* is membrane associated. DPY* and OPY* thus behave entirely differently from CPY* with regards to their mode of translocation and differently from one another with regards to their solubility/membrane association properties.
FIGURE 2.
Membrane association of novel derivatives of CPY*. Microsomes from wild type cells expressing CPY*, DPY*, or OPY* were treated with Na2CO3 as described under “Experimental Procedures.” Total (T), pellet (P), and supernatant (S) fractions were analyzed by Western blots using antibodies against CPY, Kar2p, or Sec61p as indicated.
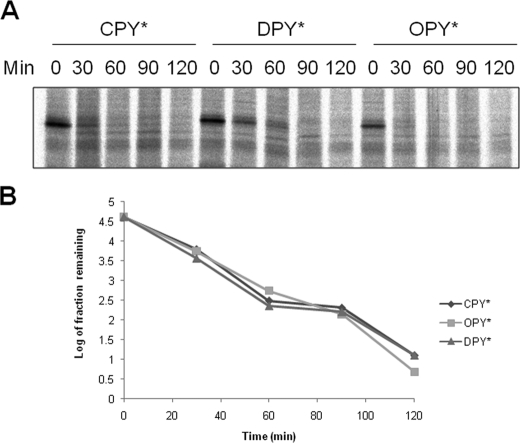
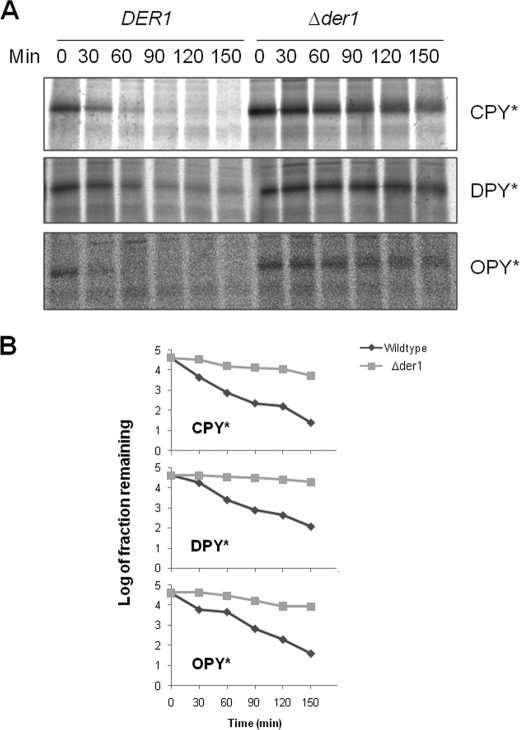
DPY* and OPY* Are Subject to Der1p-dependent ERAD-L—Next we examined whether the translocated forms of DPY* and OPY* were subject to ERAD. Wild type cells expressing CPY*, DPY*, or OPY* were subjected to pulse-chase studies demonstrating that all three substrates degraded with similar kinetics (Fig. 3). Signal cleaved OPY* behaved as a lumenal protein in our carbonate extraction studies, and so one might expect that it would be a substrate for the Der1p-dependent ERAD-L pathway (7). This was confirmed by pulse-chase studies in Δder1 cells in which the rate of OPY* degradation was indistinguishable from that of CPY* (Fig. 4). DPY* shares the same lumenal lesion as CPY*/OPY* but in a membrane-tethered form. Recent studies suggest that ERAD-M can supersede the ERAD-L pathway for substrates that have lesions in both a membrane anchor and a lumenal domain (26, 27); thus any lesion in the DPY* membrane anchor might have led to degradation via the Der1p-independent ERAD-M pathway. However, our data demonstrate that DPY* degradation requires Der1p (Fig. 4) and so conclude that this degradation occurs as a result of the lumenal lesion in this protein. Finally we found that the rates of degradation of DPY* and OPY* were unaffected in Δdoa10 cells (data not shown), confirming that neither was dependent on the ERAD-C pathway (28). We therefore conclude that DPY* and OPY* are efficiently degraded via the ERAD-L pathway.
FIGURE 3.
DPY* and OPY* are substrates for ERAD in wild type cells. A, wild type cells expressing CPY*, DPY*, or OPY* were pulse-labeled with [35S]methionine as described under “Experimental Procedures.” After the addition of cold methionine (time 0), the samples were taken at 30 min intervals, whole cell extracts were subjected to immunoprecipitation with anti-CPY antibodies, and labeled proteins were analyzed by SDS-PAGE. B, quantification of data shown in A.
FIGURE 4.
ERAD of DPY* and OPY* is Der1p-dependent. A, wild type and Δder1 cells expressing CPY*, DPY*, or OPY* were analyzed as in Fig. 3A. B, quantification of data shown in A.
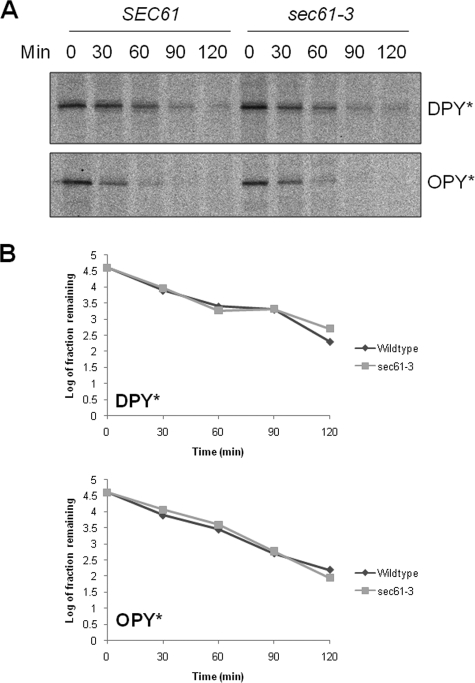
ERAD of DPY* and OPY* Requires Functional Sec61p—Next we sought to examine any requirement for Sec61p. We have earlier shown that DPY* and OPY* can be efficiently translocated into the ER in sec61-3 mutant cells at 30 °C. We therefore tested whether these translocated substrates were competent for ERAD under these same conditions. Wild type or sec61-3 mutant cells expressing either DPY* or OPY* were labeled at 30 °C, and samples taken at various time points during a chase performed at the same temperature. We found the half-life of both DPY* and OPY* in sec61-3 cells to be indistinguishable from that in wild type cells under these conditions (Fig. 5). These results demonstrate that DPY* and OPY* are available for ERAD after translocation at 30 °C in sec61-3 cells and that their degradation occurs with essentially wild type kinetics.
FIGURE 5.
ERAD of DPY* and OPY* is unaffected in sec61-3 cells at 30 °C. A, pulse-chase experiment using wild type and sec61-3 cells at 30 °C as described in Fig. 3A. B, quantification of data shown in A.
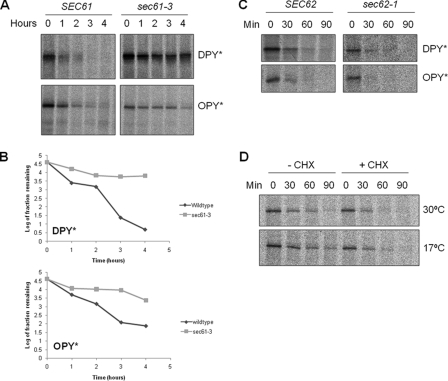
Next we tested for any effect on ERAD following inactivation of the Sec61p-dependent translocase at 17 °C. The ER was preloaded with ERAD-competent substrate by pulse labeling at 30 °C in either wild type or sec61-3 mutant cells. The cells were then shifted to 17 °C and chased in the presence of unlabeled methionine for the times indicated (Fig. 6, A and B). We observed a dramatic increase in the stability of both substrates in sec61-3 cells when compared with wild type controls. Thus functional Sec61p is required for the ER-associated degradation of both DPY* and OPY*.
FIGURE 6.
ERAD of DPY* and OPY is blocked in sec61-3 cells at 17 °C. A, as in Fig. 5 except cells were incubated at 17 °C after the addition of cold methionine, and samples were collected every 60 min for 4 h. B, quantification of data shown in A. C, wild type and sec62-1 cells expressing DPY* or OPY* were analyzed as described in Fig. 3A. D, wild type cells expressing DPY* were analyzed as described in Fig. 3A with or without the addition of 10 μg/ml cycloheximide (CHX).
It remains formally possible that the severe translocation phenotype associated with sec61-3 at 17 °C might lead to an indirect effect on ERAD, perhaps by blocking the import of some essential factor. To rule this out we first examined ERAD in sec62-1 mutant cells in which post-translational translocation is specifically blocked but found no significant difference in the rate of degradation of either substrate when compared with wild type controls (Fig. 6C). Thus ERAD of these novel substrates does not require ongoing post-translational translocation. Of course sec61-3 cells are also deficient in co-translational translocation at 17 °C. We therefore tested the effects on our ERAD substrates of a complete block in all protein translocation by treating wild type cells with cycloheximide to inhibit protein synthesis. The cells were pulse-labeled at 30 °C and then chased at either 30 or 17 °C in the presence or absence of cycloheximide (Fig. 6D). We found no delay in the degradation of DPY* in the presence of drug. Because protein synthesis is not required for ERAD, it naturally follows that ongoing co-translational import of factors into the ER cannot be required. This is consistent with numerous studies in which ERAD has been observed in cells treated with cycloheximide (29).
DISCUSSION
A variety of studies implicate Der1p in ERAD, but its precise function remains unknown. It is clearly required for the degradation of a range of misfolded luminal proteins including CPY* (7). In contrast, the majority of membrane proteins tested appear to be degraded independently of Der1p (2, 26, 30, 31). One notable exception is the type I integral membrane protein, KWW, whose degradation is defective in der1 mutant cells (2). The misfolded lesion in KWW is located within its luminal domain leading to the suggestion that Der1p is required for the degradation of substrates with misfolded lumenal domains via a pathway now known as ERAD-L (2).
A number of studies have similarly implicated Sec61p in ERAD, but the role of Sec61p in the initial translocation of such substrates into the ER complicates the analysis of degradation kinetics. We have sought to temporally separate the known role of Sec61p in translocation from any subsequent role in ERAD by exploiting the properties of the sec61-3 mutant. We have created two new ERAD substrates, both of which are imported into the ER in an SRP-dependent manner. The first, DPY*, inserts into the ER membrane as a type II integral membrane protein, whereas the second, OPY*, is subject to signal peptide cleavage and is released into the ER lumen. Both DPY* and OPY* are substrates for Der1p-dependent ERAD and so, like CPY*, must be substrates for the ERAD-L pathway. As expected, both DPY* and OPY* were efficiently translocated in sec61-3 cells at the permissive temperature of 30 °C. These translocated forms of DPY* and OPY* were evidently available to the ERAD machinery because both were degraded with kinetics that were indistinguishable from those observed in wild type cells. This allowed us to load the ER in sec61-3 cells with ERAD-competent substrate and to then inactivate Sec61p by shifting cells to 17 °C. Our data demonstrate that the inactivation of sec61-3p at 17 °C results in a rapid and substantial block in the ERAD of both DPY* and OPY*. This cannot be explained by some indirect effect of a block in sec61-3-dependent protein import because neither sec62-1 nor cycloheximide treatment had any similar effect on ERAD.
Our results demonstrate that functional Sec61p is essential for degradation of ERAD-L substrates. This finding supports a model in which Sec61p forms a bi-directional protein-conducting channel for the transport of polypeptide chains both into and out of the lumen of the endoplasmic reticulum. It will be interesting to determine whether or not different accessory factors might engage with Sec61p to determine the directionality of transport.
Supplementary Material
Acknowledgments
We thank Drs. Martin Pool and Barrie Wilkinson for helpful discussions and critical reading of the manuscript and Professors Steve Oliver and Mark Hochstrasser for yeast strains.
Author's Choice—Final version full access.
This work was supported by Wellcome Trust Grant 074149. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; ERAD, ER-associated degradation; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; CPY, carboxypeptidase Y; SRP, signal recognition particle.
References
- 1.Meusser, B., Hirsch, C., Jarosch, E., and Sommer, T. (2005) Nat. Cell Biol. 7 766–772 [DOI] [PubMed] [Google Scholar]
- 2.Vashist, S., and Ng, D. T. (2004) J. Cell Biol. 165 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho, P., Goder, V., and Rapoport, T. A. (2006) Cell 126 361–373 [DOI] [PubMed] [Google Scholar]
- 4.Denic, V., Quan, E. M., and Weissman, J. S. (2006) Cell 126 349–359 [DOI] [PubMed] [Google Scholar]
- 5.Lilley, B. N., and Ploegh, H. L. (2004) Nature 429 834–840 [DOI] [PubMed] [Google Scholar]
- 6.Ye, Y., Shibata, Y., Yun, C., Ron, D., and Rapoport, T. A. (2004) Nature 429 841–847 [DOI] [PubMed] [Google Scholar]
- 7.Knop, M., Finger, A., Braun, T., Hellmuth, K., and Wolf, D. H. (1996) EMBO J. 15 753–763 [PMC free article] [PubMed] [Google Scholar]
- 8.Robson, A., and Collinson, I. (2006) EMBO Rep. 7 1099–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Virgilio, M., Weninger, H., and Ivessa, N. E. (1998) J. Biol. Chem. 273 9734–9743 [DOI] [PubMed] [Google Scholar]
- 10.Wiertz, E. J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T. R., Rapoport, T. A., and Ploegh, H. L. (1996) Nature 384 432–438 [DOI] [PubMed] [Google Scholar]
- 11.Scott, D. C., and Schekman, R. (2008) J. Cell Biol. 181 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalies, K. U., Allan, S., Sergeyenko, T., Kroger, H., and Romisch, K. (2005) EMBO J. 24 2284–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilon, M., Romisch, K., Quach, D., and Schekman, R. (1998) Mol. Biol. Cell 9 3455–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plemper, R. K., Bohmler, S., Bordallo, J., Sommer, T., and Wolf, D. H. (1997) Nature 388 891–895 [DOI] [PubMed] [Google Scholar]
- 15.Wilkinson, B. M., Critchley, A. J., and Stirling, C. J. (1996) J. Biol. Chem. 271 25590–25597 [DOI] [PubMed] [Google Scholar]
- 16.Willer, M., Jermy, A. J., Young, B. P., and Stirling, C. J. (2003) Yeast 20 133–148 [DOI] [PubMed] [Google Scholar]
- 17.Stirling, C. J., Rothblatt, J., Hosobuchi, M., Deshaies, R., and Schekman, R. (1992) Mol. Biol. Cell 3 129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson, B. M., Tyson, J. R., Reid, P. J., and Stirling, C. J. (2000) J. Biol. Chem. 275 521–529 [DOI] [PubMed] [Google Scholar]
- 19.Young, B. P., Craven, R. A., Reid, P. J., Willer, M., and Stirling, C. J. (2001) EMBO J. 20 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tyson, J. R., and Stirling, C. J. (2000) EMBO J. 19 6440–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finger, A., Knop, M., and Wolf, D. H. (1993) Eur. J. Biochem. 218 565–574 [DOI] [PubMed] [Google Scholar]
- 22.Ng, D. T., Brown, J. D., and Walter, P. (1996) J. Cell Biol. 134 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stirling, C. J., and Hewitt, E. W. (1992) Nature 356 534–537 [DOI] [PubMed] [Google Scholar]
- 24.Silberstein, S., Collins, P. G., Kelleher, D. J., Rapiejko, P. J., and Gilmore, R. (1995) J. Cell Biol. 128 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deshaies, R. J., and Schekman, R. (1989) J. Cell Biol. 109 2653–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taxis, C., Hitt, R., Park, S. H., Deak, P. M., Kostova, Z., and Wolf, D. H. (2003) J. Biol. Chem. 278 35903–35913 [DOI] [PubMed] [Google Scholar]
- 27.Meusser, B., and Sommer, T. (2004) Mol. Cell 14 247–258 [DOI] [PubMed] [Google Scholar]
- 28.Ravid, T., Kreft, S. G., and Hochstrasser, M. (2006) EMBO J. 25 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hitt, R., and Wolf, D. H. (2004) FEMS Yeast Res. 4 815–820 [DOI] [PubMed] [Google Scholar]
- 30.Hill, K., and Cooper, A. A. (2000) EMBO J. 19 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter, J., Urban, J., Volkwein, C., and Sommer, T. (2001) EMBO J. 20 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.