Abstract
Lysyl oxidase (LOX), an amine oxidase critical for the initiation of collagen and elastin cross-linking, has recently been shown to regulate cellular activities possibly by modulating the functions of growth factors. In this study, we investigated the interaction between LOX and transforming growth factor-β1 (TGF-β1), a potent growth factor abundant in bone, the effect of LOX on TGF-β1 signaling, and its potential mechanism. The specific binding between mature LOX and mature TGF-β1 was demonstrated by immunoprecipitation and glutathione S-transferase pulldown assay in vitro. Both proteins were colocalized in the extracellular matrix in an osteoblastic cell culture system, and the binding complex was identified in the mineral-associated fraction of bone matrix. Furthermore, LOX suppressed TGF-β1-induced Smad3 phosphorylation likely through its amine oxidase activity. The data indicate that LOX binds to mature TGF-β1 and enzymatically regulates its signaling in bone and thus may play an important role in bone maintenance and remodeling.
Lysyl oxidase (LOX)2 is a copper-dependent amine oxidase that initiates the process of covalent intra- and intermolecular cross-linking in collagen and elastin (1). The critical role of LOX in tissue stability is well exemplified by “lathyrism,” the condition where deleterious effects in connective tissues are caused by lathyrogens such as β-aminopropionitrile (BAPN) (2). In lathyritic animals, bone is one of the most severely affected tissues revealing kyphoscoliosis, bone deformities, weakening of tendons and ligament attachments, dislocation of joints, impaired bone fracture healing, and ectopic bone exostoses (3, 4). BAPN is a potent and irreversible inhibitor of LOX catalytic activity and thus prevents cross-linking of immature collagen and elastin into mature, stable, and insoluble fibers. Therefore, it has been thought that the phenotypes seen in lathyritic animals are due primarily to the lack of collagen/elastin cross-linking.
Recent reports, however, have revealed novel functions for LOX, including the regulation of gene transcription and cellular functions. Although the mechanisms are still not clear, those functions could be associated with its ability to oxidize substrates, other than collagen and elastin, such as basic fibroblast growth factor (5) as well as histone H1 and H2 (6, 7). Thus, lathyritic phenotypes could be due in part to the loss of LOX control of cellular functions. Indeed, several studies have reported that collagen synthesis/expression significantly increased when osteoblasts or chondrocytes were cultured in the presence of BAPN (8–11), which is suggestive of such functions.
In bone, there are several major growth factors, including transforming growth factor-β (TGF-β), bone morphogenetic proteins (BMPs), insulin-like growth factor and platelet-derived growth factor, tumor necrosis factor-α, and basic fibroblast growth factor. Among those factors, TGF-βs and BMPs are a group of growth factors that are basic in nature (theoretical pI > 8.5) and up-regulate collagen expression (12–16). TGF-β1 is one of the most potent growth factors enriched in bone matrix modulating many aspects of bone physiology (see a review by Janssens et al. (12)). This growth factor is secreted and stored as a small or large latent complex in bone matrix, but it can be released and activated by the action of osteoclasts (17, 18) and other mechanisms (18, 19). Numerous studies, although not always consistent, have shown that TGF-β1 stimulates the recruitment and proliferation of osteoblast progenitors (20, 21) and stimulates matrix protein production, including collagen, but inhibits the late stage of osteoblast differentiation and matrix mineralization (22, 23) and osteocalcin expression (24, 25). It is also involved in the modulation of osteoclast differentiation (26). The abundance of this growth factor with such potent effects on cells in a dynamic environment of bone clearly requires the need for tight regulation of its biological activities.
In this study, we investigated the potential interaction between LOX and TGF-β1 in vitro and in bone matrix, LOX control of TGF-β1 signaling, and its potential mechanism. The results indicated that LOX may play a pivotal role in regulation of TGF-β1 activity that is critical for bone physiology and pathology.
EXPERIMENTAL PROCEDURES
Antibodies and Proteins—The following antibodies were used in this study: anti-V5 antibody (Invitrogen), anti-hemagglutinin (HA) antibody (Roche Diagnostics), anti-phospho-Smad3 antibody (BIOSOURCE), anti-glutathione S-transferase (GST) antibody (Sigma), anti-Smad3, anti-phospho-Smad1/5/8 and anti-β-actin antibodies (Cell Signaling Technologies), and anti-TGF-β1 antibody (R & D Systems). Two types of polyclonal anti-LOX antibodies were used, one purchased from Imgenex (anti-LOXi) and another reported in previous studies (anti-LOXh) (27, 28). Recombinant human TGF-β1 protein (rhTGF-β1) was purchased from R&D Systems.
Cell Lines and Culture Conditions—Human embryonic kidney 293 cells were purchased from Clontech and maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biologicals), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen) in a 5%CO2 atmosphere at 37 °C. The medium was changed twice a week. The mouse calvaria-derived MC3T3-E1 subclone 4 (MC) cells were purchased from American Type Culture Collection (CRL-2593) and maintained in α-minimum essential medium (Invitrogen) with the same supplements as described above. The medium was changed twice a week.
Molecular Cloning of Mouse LOX cDNA—Total RNA was isolated from MC cells using TRIzol (Invitrogen). Two μg of total RNA was used for reverse transcription (RT), and the cDNA was synthesized using the Omniscript RT kit (Qiagen). The cDNA containing the coding region of the mouse LOX was generated by PCR using Hotstar Taq polymerase (Qiagen). The sequences of the primers were designed as follows: forward primer, 5′-CCCGGTCTTCCTTTTTCTCCTAGCC-3′, and reverse primer, 5′-ATACGGTGAAATTGTGCAGCCTGA-3′. The PCR product was then ligated into the pcDNA3.1/V5-His-TOPO mammalian expression vector (Invitrogen) and sequenced at the University of North Carolina, Chapel Hill, DNA sequencing facility (University of North Carolina, Chapel Hill, NC), and the plasmid containing LOX cDNA in a sense orientation (pcDNA3.1/LOX/V5-His) was generated. To obtain pcDNA3/LOXdm/V5-His (LOXdm, LOX with double mutations converting lysine 314 to alanine and tyrosine 349 to phenylalanine resulting in an inactive LOX) (29), the following two additional sets of primers were used: forward primer, 5′-GCTGAAGGCCACGCAGCAAGCTTCTGT-3′, and reverse primer, 5′-ACAGAAGCTTGCTGCGTGGCCTTCAGC-3′, and forward primer 5′-TGTTATGACACCTTTGCGGCAGACATA-3′, and reverse primer, 5′-TATGTCTGCCGCAAAGGTGTCATAACA-3′. The PCR product was ligated into the pcDNA3.1/V5-His-TOPO mammalian expression vector and sequenced at the University of North Carolina, Chapel Hill, sequencing facility.
To investigate the binding domain of LOX to TGF-β1, several HA-tagged LOX constructs were generated, i.e. full-length LOX (LOX-HA), mature LOX (mLOX-HA, LOX without the propeptide), LOX with double mutations (LOXdm-HA), and LOX propeptide (LOXPP-HA). The coding sequence of LOX was subcloned from pcDNA3.1/LOX/V5-His by PCR and the primers were designed as follows: for LOX and LOXdm, forward primer, 5′-GCGGATCCATGCGTTTCGCCTGGGCTGTGCTC-3′, and reverse primer, 5′-GCCTCGAGATACGGTGAAATTGTGCAGCCTGAGGC-3′; for mLOX, the same primers used for the full-length LOX and the additional primers forward primer, 5′-CTTCTCCGCTGCGACGACCCCTACAATCCCTAC-3′, and reverse primer, 5′-GTAGGGATTGTAGGGGTCGTCGCAGCGGAGAAG-3′;, and for LOXPP, forward primer, 5′-GCGGATCCATGCGTTTCGCCTGGGCTGTGCTC-3′, and reverse primer, 5′-GCCTCGAGGCCCACCATGCGATCTATGTGGCT-3′. The PCR products were digested with BamHI and XhoI, ligated into pcDNA3/HA mammalian expression vector (30), and sequenced at the University of North Carolina, Chapel Hill, DNA sequencing facility.
Transfection, Immunoprecipitation, and Western Blot Analysis—To obtain a plasmid containing TGF-β1 coding sequences (pcDNA3.1/TGF-β1/V5-His), PCR products were amplified using the normal mouse kidney cDNA (BD Biosciences) as a cDNA template, purified, ligated into the pcDNA3.1/V5-His-TOPO mammalian expression vector, and sequenced. The sequences of the primers were designed as follows: forward primer, 5′-CATGCCGCCCTCGGGGCTG-3′, and reverse primer, 5′-GCTGCACTTGCAGGAGCGC-3′. 293 cells were co-transfected with pcDNA3/LOX/HA vector and pcDNA3.1/TGF-β1/V5-His or pcDNA3.1/V5-His vector harboring the coding sequences of bone morphogenetic proteins-2, -4, -6, or -7 (BMP-2, -4, -6, and -7) that are available in our laboratory (31) using a FuGENE 6 transfection reagent (Roche Diagnostics) according to the manufacturer's instructions. After co-transfection, the cultured media were collected and immunoprecipitated with either anti-V5 or anti-HA antibody. The samples were then incubated with protein A-Sepharose 4B conjugate beads (Zymed Laboratories Inc.) for 30 min, and the beads were washed twice with lysis buffer containing 150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 10 mm EDTA, 1% Triton X-100, 1% deoxycholate, 1.5% aprotinin, and 1 mm phenymethylsulfonyl fluoride. The protein bound to the beads was dissolved in SDS sample buffer (100 mm Tris-HCl, pH 8.8, 0.01% bromphenol blue, 36% glycerol, and 4% SDS) in the presence of 10 mm dithiothreitol, applied to 4–12% gradient SDS-PAGE, transferred onto a polyvinylidene fluoride membrane (Immobilon-P, Millipore), and subjected to Western blot (WB) analysis with anti-V5 or anti-HA antibody. The immunoreactivity was visualized by an alkaline phosphatase conjugate substrate kit (Bio-Rad). To determine whether LOX binds to the propeptide of TGF-β1 (latency-associated peptide, LAP) or mature TGF-β1, pcDNA3.1/LAP/V5-His was subcloned from pcDNA3.1/TGF-β1/V5-His by PCR. The primers designed were as follows: forward primer, 5′-GCTGCACTTGCAGGAGCGC-3′, and reverse primer, 5′-TCTCCGGTGCCGTGAGCTGTG-3′. The PCR product was then ligated into the pcDNA3.1/V5-His-TOPO mammalian expression vector and sequenced, and the plasmid containing LAP cDNA in a sense orientation (pcDNA3.1/LAP/V5-His) was obtained. 293 cells were transiently transfected with the pcDNA3/LOX/HA and either pcDNA3.1/TGF-β1/V5-His or pcDNA3.1/LAP/V5-His, using FuGENE 6 transfection reagent. The total amounts of plasmid were kept constant (2.5 μg) by supplementing pcDNA3.1/V5-HisA (empty vector; Invitrogen). The media were collected and subjected to immunoprecipitation and Western blot (IP-WB) analysis in the same manner as described above.
To investigate the binding domain of LOX to mature TGF-β1, 293 cells were transiently transfected with pcDNA3.1/TGF-β1/V5-His and pcDNA3/LOX/HA, pcDNA3/mLOX/HA, pcDNA3/LOXdm/HA, or pcDNA3/LOXPP/HA, and the media were collected and subjected to IP-WB analysis in the same manner as described above.
Generation of 293 Cell-derived Stable Clones Overexpressing LOX—293 cells were transfected with pcDNA3.1/LOX/V5-His construct as described above. After transfection, cells were cultured in the presence of 400 μg/ml G418 (Invitrogen) for 3–4 weeks to select stable clones. Positive clones derived from single G418-resistant cells were then isolated by cloning rings and further grown in the same conditions. As a control, 293 cells were also transfected with empty vector, and the stable clones (EV clones) were generated in the same manner. The cultured medium from each clone was subjected to IP-WB analysis with anti-V5 antibody to verify the LOX-V5/His protein (LOX-V5).
Purification and Identification of LOX-V5/His Fusion Protein—The 293-derived clones that synthesized the highest level of LOX-V5 were cultured onto 15-cm plates for 6 days, and the culture media were collected in the presence of protease inhibitor mixtures (Sigma). LOX-V5 was purified using a nickel-nitrilotriacetic acid-agarose resin (Qiagen) at 4 °C, and the purified proteins were pooled, dialyzed against 0.2 m sodium borate, pH 8.2, and kept at -80 °C until used. The protein concentrations were measured by a DC protein assay kit (Bio-Rad). To assess the purity of LOX-V5, aliquots of the sample were dissolved in SDS sample buffer containing 10 mm dithiothreitol, separated by 4–12% SDS-PAGE, and subjected to Coomassie Brilliant Blue R-250 staining or WB analysis. For the latter analysis, two anti-LOX antibodies (anti-LOXi and anti-LOXh) and anti-V5 antibody were used. The major Coomassie Brilliant Blue-stained protein bands on the gel were cut and subjected to matrix-assisted laser desorption ionization-mass spectrometric analysis (MALDI-MS) at the University of North Carolina, Chapel Hill, Proteomics Facility to confirm the identity of LOX.
LOX Activity Assay—The LOX enzyme activity was measured using the Amplex Ultra Red fluorescence assay (32). Five or 10 μg of LOX-V5 with or without 500 μm BAPN was suspended in 2 ml of 0.1 m sodium borate, pH 8.2, containing, 1.2 m urea, 1 unit/ml horseradish peroxidase (Biochemika), 10 μm Amplex Red (Molecular Probes), 10 mm 1,5-diaminopentane dihydrochloride (Sigma). The mixture was incubated for 30 min at 37 °C, and the fluorescence intensities were measured with excitation and emission wavelength at 563 and 587 nm, respectively, using F2000 spectrofluorometer (Hitachi).
GST Pulldown Assay—The sequences of the primers were designed to amplify the mature form of TGF-β1 (residues 279–390) and they were as follows: forward primer, 5′-GCGAATTCGCCCTGGATACCAACTATTGCTTC-3′, and reverse primer, 5′-GCCTCGAGTCAGCTGCACTTGCAGGAGCGCAC-3′. The PCR products were amplified using the normal mouse kidney cDNA as a template, purified, and ligated into a pGEX4T-1 vector (GE Healthcare) and transformed into BL21 strain of Escherichia coli (Stratagene). An empty pGEX4T-1 vector was also transformed into the bacterial cells to produce GST protein alone. After DNA purification, plasmids were analyzed by restriction enzyme digestion and sequenced, and the plasmid harboring the mature form of GST-TGF-β1 (pGEX4T-1-TGF-β1) was obtained. After the bacteria transformed with pGEX4T-1-TGF-β1 or pGEX4T-1 were cultured for several hours at 37 °C, 250 μm of isopropyl d-1-thiogalactopyranoside (Sigma) was added to induce the synthesis of GST-TGF-β1 or GST protein. After incubating for 24 h at 20 °C, the cultures were centrifuged, lysed in a buffer containing PBS and 1% Triton X-100, and sonicated for 20 s three times with an interval of 3 min on ice. The lysates were collected by centrifugation, incubated with glutathione-Sepharose beads (GE Healthcare) overnight at 4 °C, and the beads were extensively washed with PBS. The recombinant GST-TGF-β1 and GST proteins were then released from the beads with the elution buffer (10 mm glutathione, 50 mm Tris-HCl, pH 8.0) at 4 °C. The protein concentrations were measured by a DC protein assay kit. The purity of the recombinant proteins was assessed by 4–12% SDS-PAGE.
GST pulldown was then performed in the following manner. Five μg of GST or purified GST-TGF-β1 fusion proteins (2.5 or 5 μg) were incubated with 10 μg of LOX-V5 in 50 mm Tris-HCl, pH 8.0, for 1 h at 4 °C. Glutathione-Sepharose beads (GE Healthcare) were then added and further incubated for 30 min at 4 °C. The beads were then washed three times with 50 mm Tris-buffered saline, including 0.02% Tween 20 (TBST), and the proteins bound were released by boiling for 5 min in SDS sample buffer containing 10 mm dithiothreitol and subjected to WB analysis with anti-V5 antibody. The immunoreactivity was visualized by alkaline phosphatase conjugate substrate kit.
Direct Binding of LOX-V5 and rhTGF-β1 by Immunoprecipitation—Two hundred ng of rhTGF-β1 were incubated with 10 μg of LOX-V5 in 50 mm Tris-HCl, pH 8.0, for 1 h at 4 °C. The complex was immunoprecipitated with anti-V5 antibody, and the protein G-Sepharose 4B conjugate beads (Zymed Laboratories Inc.) were then added to the solutions and further incubated for 30 min. The samples were treated in the same manner as described above and subjected to WB analysis with anti-V5 antibody.
Laser-scanning Confocal Microscopy—The co-localization of LOX and TGF-β1 was investigated in a MC cell culture system by laser-scanning confocal microscopy. MC cells were cultured for 3 weeks as described above, washed with PBS, and fixed with 10% formaldehyde for 10 min. Cell/matrix layer was then cut into 1 × 1-cm pieces and placed on a glass slide. The slides were then immersed in PBS and treated with 20 μg/ml proteinase K (Roche Applied Science) for 10 min. Samples were incubated with two primary antibodies, i.e. anti-TGF-β1 and anti-LOXi antibodies, in PBS containing 1.5% goat serum for 30 min, washed with PBS, and incubated with species-specific fluorescence-labeled secondary antibodies, mouse Alexa Fluor 594 and rabbit Alexa Fluor 488 (Invitrogen), for 30 min each. After washing with PBS, the specimens were mounted, and the immunofluorescence was observed under a Zeiss LSM5 Pascal at University of North Carolina, Chapel Hill, microscopy services laboratory.
Identification of LOX-TGF-β1 Complex in Bone Matrix—Femurs obtained from 0 to 1-year-old bovine animals were purchased from Aries Scientific (Dallas, TX) and kept at -80 °C until use. Both femoral heads were removed and the mid-shafts were longitudinally cut. After the bone marrow was removed and washed with cold PBS, bones were cut into small pieces, defatted with methylene chloride and methanol solution (2:1) overnight at 4 °C, washed with cold distilled water, and lyophilized. The bone fragments were then pulverized to a fine powder in liquid nitrogen using a freezer mill (Spex Certiprep), washed with cold distilled water, lyophilized, and subjected to sequential extraction by the method reported previously (33, 34) with some modifications (35). Briefly, ∼1 g of bone powder was first extracted with 5 ml of 6 m guanidine-HCl (GH), pH 7.4, for 2 days at 4 °C, and the supernatant was separated by centrifugation at 15,000 × g for 30 min, exhaustively dialyzed against cold distilled water, and lyophilized (G1 representing the matrix molecules that are not associated with mineral). The residue (mineral-associated matrix) was then demineralized with 0.5 m EDTA, pH 7.4, for 2 weeks at 4 °C with several changes of EDTA, and the supernatants were separated by centrifugation as described above, pooled, dialyzed against cold distilled water, and lyophilized (E representing soluble matrix molecules associated with mineral). The residue was further extracted with 5 ml of 6 m GH, pH 7.4, for 2 days at 4 °C, and the extract was collected as described above (G2 including mineral-associated, insoluble matrix). All fractions were weighed, dissolved in lysis buffer, and centrifuged. The protein concentrations in the supernatants were determined by a DC protein assay kit. Fifteen μg of proteins in each fraction was dissolved in SDS sample buffer and subjected to WB analysis with either anti-LOXi or anti-TGF-β1 antibody. The immunoreactivity was visualized by alkaline phosphatase-conjugated substrate kit. To confirm the specific binding, various amounts of E fraction (500, 1000, and 2000 μg of protein) dissolved in lysis buffer were immunoprecipitated with either 5 μl of anti-LOXi or anti-LOXh antibodies or 5 μl of normal rabbit serum (negative control) overnight at 4 °C. Then protein G-Sepharose 4B conjugate beads were added and incubated for 15 min at 4 °C, and the beads were washed three times with lysis buffer. The immunocomplex was then released from the beads as described above and subjected to WB analysis with anti-TGF-β1 antibody. The immunoreactivity was visualized by alkaline phosphatase conjugate substrate kit.
Effect of LOX on Smad Phosphorylation—To determine the effect of LOX on the TGF-β1 activity, MC cells were plated onto 35-mm culture dishes at a density of 2.0 × 105/dish in duplicate and cultured. MC cells were transiently transfected with 1, 2.5, and 5 μg of the pcDNA3.1/LOX/V5-His using FuGENE 6 transfection reagent. After 48 h, cells were treated with rhTGF-β1 (5 ng/ml) for 30 min in the presence or absence of 300 μm BAPN or 200 units/ml of catalase (Worthington). Another set of cells was transiently transfected with 5 μg of pcDNA3.1/LOXdm/V5-His and treated with rhTGF-β1 in the presence or absence of BAPN. The cells were lysed with 400 μl of RIPA buffer containing 150 mm NaCl, 50 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 0.1% SDS, 0.5% deoxycholate, 1% aprotinin, and 1 mm phenylmethylsulfonyl fluoride for 1 h at 4 °C. After centrifugation, the supernatants collected were then subjected to WB analysis with anti-phospho-Smad3 (BIOSOURCE) and anti-Smad3 antibodies (Cell Signaling). The intensity of phosphorylated Smad3 (pSmad3) protein from each sample was normalized to that of total Smad3 protein using Scion Image software (Scion Corp.). The expression level of LOX in the cultured medium was quantified by IP-WB analysis with anti-LOXi antibody.
The effect of LOX on BMP signaling was also examined. MC cells were plated and cultured as described above and transiently transfected with 1, 2.5, and 5 μg of the pcDNA3.1/LOX/V5-His using FuGENE 6 transfection reagent. After 48 h, cells were treated with rhBMP-2 (100 ng/ml) for 30 min in the presence or absence of 300 μm BAPN. Cell lysates were prepared and subjected to WB analysis with anti-phospho-Smad1/5/8 and β-actin antibodies (Cell Signaling). The intensity of phosphorylated Smad1/5/8 (pSmad1/5/8) protein from each sample was normalized to that of β-actin protein using Scion Image software. The expression level of LOX in the cultured medium was evaluated by IP-WB analysis with anti-V5 antibody.
In another set of experiments, MC cells were cultured as described above. On the following day, cells were treated with 2.5 or 5 μg of LOX-V5, 5 μg of LOX-V5 with 300 μm BAPN, or 300 μm BAPN alone. Cells were then treated with 5 ng/ml rhTGF-β1 for 30 min, and the phosphorylation of Smad3 was evaluated in the same manner as described above.
RNA Interference—MC cells were plated onto 35-mm culture dishes at a density of 5.0 × 104/dish in duplicate and cultured in the same manner as described above. On the following day, cells were transfected with 3.75 μg of LOX siRNA ID 156159, 156160, 156161 or Silencer negative control AM4611 (Ambion) using siPORT amine transfection agent (Ambion). After 48 h, the cells were treated with 5 ng/ml rhTGF-β1 for 30 min, and Smad3 phosphorylation was examined in the same manner as described above. The suppression of LOX protein in the cultured medium was verified by IP-WB analysis with anti-LOXi antibody.
Statistical Analysis—For statistical analysis, three independent experiments were performed, and Kruskal-Wallis one-way analysis of variance and multiple comparison were used. The data were presented as mean ± S.D., and a p value less than 0.05 was considered significant.
RESULTS
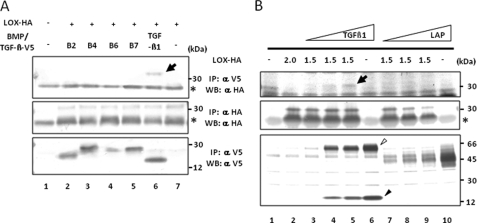
Mature LOX Binds to Mature TGF-β1—The binding of LOX to TGF-β1 and BMPs (BMP-2, -4, -6, and -7) was investigated by co-expressing those proteins with two types of tag, HA (∼1 kDa) for LOX and V5 (∼5 kDa) for other proteins followed by IP-WB analysis (Fig. 1A). When IP and WB were performed with either anti-V5 or -HA antibody alone, V5-tagged BMPs (Fig. 1A, lower panel, lanes 2–5), V5-tagged TGF-β1 (Fig. 1A, lower panel, lane 6), or HA-tagged mature (m)LOX (Fig. 1A, middle panel, lanes 2–7) was detected at the expected molecular weight of each protein. Of the proteins tested, only TGF-β1 was shown to bind mLOX (Fig. 1A, upper panel, lane 6). None of the BMPs tested showed binding (Fig. 1A, upper panel, lanes 2–5). Even when a higher level of BMP2 (2-fold) was expressed, no binding was observed (data not shown). Then the binding of mLOX to TGF-β1 or its propeptide (LAP) was evaluated. By overexpressing full-length TGF-β1-V5 (∼66 kDa), both full-length (∼66 kDa) and mature TGF-β1-V5 (∼17 kDa) were secreted (Fig. 1B, lower panel, lanes 4–6) as reported previously (36). Results showed that the binding of mLOX to full-length/mature TGF-β1 occurred in a dose-dependent manner (Fig. 1B, upper panel, lanes 3–5), whereas LAP (propeptide of TGF-β1) alone did not bind to LOX at any doses (Fig. 1B, upper panel, lanes 7–9), indicating that LOX bound specifically to mature TGF-β1. Some faint, indistinct bands observed in the upper panel of Fig. 1B, lanes 7–9, are most likely unrelated to the specific binding as none of them changed with the increased levels of LAP expression. Although not shown in Fig. 1, full-length LOX-HA was also expressed and bound to TGF-β1 in this system (see Fig. 2).
FIGURE 1.
Binding of LOX to TGF-β1 by IP-WB analysis. A, binding of mLOX to TGF-β1 and BMPs. To identify the binding, LOX-HA and TGF-β1-V5 or BMP-2, -4, -6, or -7-V5 (B2, B4, B6, and B7) were co-expressed and immunoprecipitated with anti-(α) V5 antibody, and the binding was detected by WB analysis with α HA antibody. The binding of mLOX to TGF-β1(upper panel, lane 6, LOX-HA is indicated by an arrow) but not to any BMPs (upper panel, lanes 2–5) was clearly observed. The expression levels of mLOX-HA and BMPs/TGF-β1-V5 were verified by IP-WB analyses with α HA antibody (middle panel, mLOX-HA is detected at 32 kDa) and with α V5 antibody (lower panel), respectively. An asterisk indicates IgG light chain. B, binding of mLOX to TGF-β1 and its propeptide LAP. The binding was detected by using mLOX-HA and TGF-β1-V5 or LAP-V5. Dose-dependent binding of mLOX-HA to a mixture of full-length (∼62 kDa indicated by an open triangle) and mature TGF-β1-V5 (∼17 kDa indicated by a closed triangle) was observed (upper panel, lanes 3–5, LOX-HA is indicated by an arrow) but not LAP with any doses tested (upper panel, lanes 7–9). The expression levels of mLOX-HA, TGF-β1-V5, and LAP-V5 are shown in the middle and lower panels, respectively. An asterisk indicates IgG light chain. Molecular masses are shown on the right.
FIGURE 2.
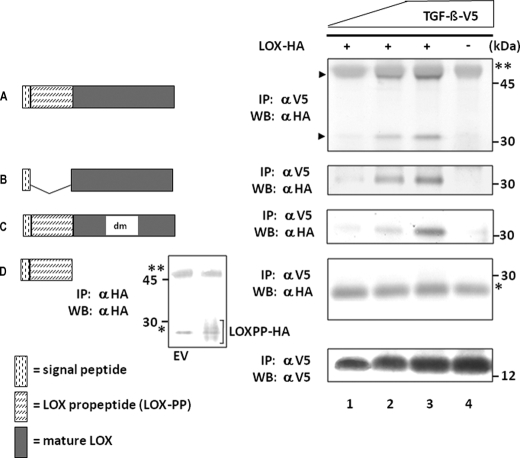
LOX constructs and their binding to TGF-β1 by IP-WB analysis. Four LOX-HA constructs generated, i.e. LOX-HA (A), mature LOX-HA (B), LOX with double mutations (LOXdm-HA) (C), and LOX propeptide (LOXPP-HA) (D), are shown on the left. The binding of each construct to TGF-β1-V5 was analyzed by IP with anti-(α) V5 antibody followed by WB with α HA antibody and shown on right. Expression levels of TGF-β1-V5 are shown by WB with α V5 antibody at the bottom panel on the right. When LOX-HA was expressed, both full-length (50 kDa) and mature LOX-HA (33 kDa) were synthesized (indicated by arrowheads). Note that full-length, mature, and dmLOX-HA showed binding to TGF-β1-V5 in a dose-dependent manner (a–c, lanes 1–3), but no binding was observed for LOXPP-HA (d). The presence of LOXPP-HA (∼28 kDa) in the medium was confirmed by IP-WB analysis with α HA antibody in comparison with the negative control (EV) and is shown in a small inset left to d. LOXPP-HA is indicated by a bracket. IgG heavy (50 kDa) and light (25 kDa) chains are indicated by two and one asterisks, respectively.
To further characterize the binding of LOX to TGF-β1, we generated HA-tagged full-length LOX (residue 1–411, LOX-HA) (Fig. 2a), mature LOX (signal peptide and mature LOX: residues 1–16 and 162–411, mLOX-HA) (Fig. 2b), LOXdm (LOX with double mutations converting lysine 314 to alanine and tyrosine 349 to phenylalanine resulting in an inactive LOX, LOXdm-HA) (Fig. 2c), and LOX propeptide (signal peptide and propeptide: residues 1–161, LOXPP-HA) (Fig. 2d). One of these proteins and TGF-β1-V5 were then transiently co-expressed in 293 cells, and the binding was assessed by IP-WB analysis in the same manner as described above. Three different levels of TGF-β1 were expressed for the binding assay (Fig. 2, bottom panel). The dose-dependent binding was detected for LOX-HA (Fig. 2a), mLOX-HA (Fig. 2b), and LOXdm-HA (Fig. 2c), but the binding to LOXPP-HA was not detected at any dose (Fig. 2d). The presence of LOXPP in the medium was confirmed by IP-WB analysis with anti-HA antibody in comparison with the negative control (EV). The immunopositive band was observed as a smear band at ∼28 kDa, as reported (37), that partially overlapped with the IgG light chain (shown in an inset in Fig. 2). When LOX-HA was expressed, both full-length (55 kDa) and mature forms (35 kDa) of LOX were present in 293 cells (indicated by arrowheads in of Fig. 2a). Although the full-length LOX-HA was partially overlapped with the IgG heavy chain, the dose-dependent binding was readily observed for both full-length and mature LOX. These results indicate that the binding of LOX to TGF-β1 occurs via the mature form of LOX and not its propeptide domain.
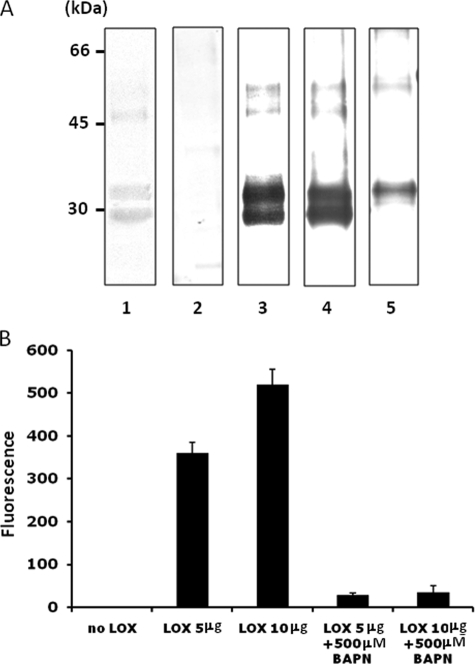
Characterization of Recombinant LOX-V5 Protein—The purified LOX-V5 protein was first characterized by SDS-PAGE and WB analyses. When stained with Coomassie Brilliant Blue, two major protein bands were observed at ∼30 and 35 kDa, respectively. Additional minor bands at ∼48 and 55 kDa were also observed. Those proteins were further analyzed by WB analysis with anti-V5 antibody (Fig. 3A, lane 3) and two anti-LOX antibodies (anti-LOXi and anti-LOXh) (Fig. 3A, lanes 4 and 5). The 30-kDa band was immunoreactive to two antibodies (anti-V5 and anti-LOXi antibodies) and the 35-kDa band to all three antibodies (anti-V5, anti-LOXh, and anti-LOXi antibodies). Similarly, the minor components at a high molecular weight region also showed immunoreactivities to those antibodies in a similar manner. The 48-kDa band was immunopositive to the two antibodies and the 55 kDa to all three antibodies. When WB analysis was performed with normal rabbit serum (NRS), no immunoreactivity was detected (Fig. 3A, lane 2). The 55- and 48-kDa proteins are likely full-length LOX with and without glycosylation as reported previously (38). The major protein bands migrated at 30 and 35 kDa were cut from the gel and subjected to protein identification using MALDI-MS at University of North Carolina, Chapel Hill, Proteomics Facility. Four tryptic peptides separated were all identified as peptides of LOX (residues 225–231, 246–254, 263–277, and 372–391, respectively) (GenBank™ accession protein NP_034858.1), thus confirming that both proteins are indeed LOX. Possibly, the 30-kDa LOX is a product of 35-kDa LOX through additional proteolytic processing.
FIGURE 3.
Characterization of recombinant LOX-V5 protein. A, SDS-PAGE and WB analysis. Purified LOX-V5 protein was subjected to SDS-PAGE analysis and stained with Coomassie Brilliant Blue (lane 1) and WB analysis with NRS (lane 2), anti-(α) V5 antibody (lane 3), α LOXi antibody (Imgenex) (lane 4), and α LOXh antibody that was previously reported (27, 28) (lane 5). Two major Coomassie Brilliant Blue-stained bands were observed at ∼30- and ∼35-kDa bands. Both were immunoreactive to α V5 (lane 3) and α LOXi (lane 4) antibodies and the 35 kDa to α LOXh antibody (lane 5). Both proteins were identified as LOX by MALDI-MS (see text). The minor Coomassie Brilliant Blue-stained bands at higher molecular weight region (∼48 and ∼50 kDa) also showed immunoreactivities to those antibodies in a similar manner. No immunoreactivity was found with NRS. B, amine oxidase activity of LOX-V5 protein. Note that the amine oxidase activity of LOX-V5 protein was retained and increased in a dose-dependent manner, and the activity was nullified by 500 μm BAPN. Error bars indicate mean ± S.D. of three independent experiments.
Purified LOX-V5, including full-length and mature LOX-V5, was then subjected to the amine oxidase assay by the method reported by Palamakumbura and Trackman (32). The activity was readily detected and increased with the dose, but it was nullified by the addition of 500 μm BAPN. These results demonstrate that the recombinant LOX-V5 generated is active as an amine oxidase (Fig. 3B). Purified LOXdm-V5 (LOX with double mutations converting lysine 314 to alanine and tyrosine 349 to phenylalanine resulting in an inactive LOX, see under “Experimental Procedures”) was also subjected to this assay and showed no activity (data not shown).
Direct Binding of LOX-V5 to TGF-β1—To determine whether LOX directly binds to TGF-β, we performed a GST pulldown assay by using GST-fused mature TGF-β1 (GST-TGF-β1) and active LOX-V5 protein (see above). LOX-V5 protein was confirmed by WB with anti-V5 antibody (Fig. 4A, lane 1). Although the binding was not detected between LOX-V5 and GST alone (Fig. 4A, lanes 2), it was readily observed between LOX-V5 and GST-TGF-β1 in a dose-dependent manner (Fig. 4A, lanes 3 and 4).
FIGURE 4.
Direct binding of LOX-V5 to mature TGF-β1. A, GST pulldown assay. Ten μg of recombinant LOX-V5 protein (see Fig. 3) was incubated with 2.5 or 5 μg of GST-TGF-β1 or 5 μg of GST and subjected to GST pulldown assay, and the binding was detected by WB analysis with anti-(α) V5 antibody (upper panel). LOX-V5 without pulldown was used as positive control (upper panel, lane 1). No binding was detected to GST alone (lane 2). The binding of GST-TGF-β1 to LOX-V5 was clearly detected in a dose-dependent manner (lanes 3 and 4). WB analysis with α GST for GST and GST-TGF-β1 used (input) are shown in the lower panel. B, direct binding between LOX-V5 and rhTGF-β1 by IP-WB. Twenty ng of rhTGF-β1 was incubated with and without 10 μg of LOX-V5, immunoprecipitated with α V5 antibody, and detected with α TGF-β1 antibody (upper panel). An immunoreactive band was detected when LOX-V5 was incubated with rhTGF-β1(upper panel, lane 2) but absent when LOX-V5 protein was not added (upper panel, lane 1). WB analysis with α TGF-β1 for rhTGF-β1 used (input) is shown in the lower panel.
The direct binding between LOX and TGF-β1 was further confirmed by IP-WB analysis using LOX-V5 and rhTGF-β1. When those two proteins were incubated, immunoprecipitated with anti-V5 antibody, and subjected to WB analysis with anti-TGF-β1 antibody, the TGF-β1 was detected (Fig. 4B) indicating the direct binding between those two proteins.
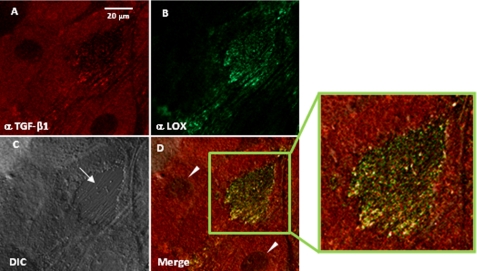
Co-localization of LOX and TGF-β in an Osteoblast Culture System—To investigate the endogenous association of LOX with TGF-β1 in an osteoblastic MC cell culture system, the localization of both proteins was assessed by laser-scanning fluorescence microscopy (Fig. 5). A fibrous extracellular matrix (ECM) structure between cell bodies was identified in the culture and confirmed by differential interference contrast image (Fig. 5C). TGF-β1 was shown in red (Fig. 5A), LOX in green (Fig. 5B), and co-localizalization of the two in yellow in the merged image (Fig. 5D). The results demonstrated that both proteins were co-localized in the ECM of the MC culture (Fig. 5D) indicating their close association in this culture system (Fig. 5D, inset).
FIGURE 5.
Co-localization of LOX and TGF-β1 in an MC cell culture system. After 3 weeks of culture, immunofluorescence staining for LOX and TGF-β1 was performed and observed under laser-scanning confocal microscopy. A, TGF-β1 is shown in red; B, LOX is shown in green; D, merged image of A and B showing colocalization of the two molecules in yellow. An enlarged image (inset) of the merged ECM is shown on the right. C, differential interference contrast (DIC) image confirming the fibrous ECM (indicated by an arrow). An arrowhead indicates a nucleus.
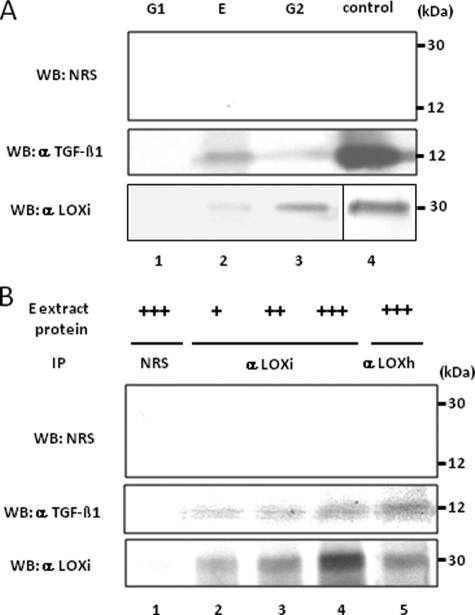
Identification of a LOX-TGF-β1 Complex in Bone Matrix— The close association of LOX with TGF-β1 was further assessed in bone matrix that was fractionated into G1, E, and G2 by sequential extraction. An equal amount of protein from each fraction was subjected to WB analysis with anti-LOXi and anti-TGF-β1 antibodies. Both LOX and TGF-β1 in bone matrix were identified in mineral-associated matrix fractions (E and G2) but not in the G1 fraction, although their relative distribution in each fraction was different (Fig. 6A). This suggested that LOX and TGF-β1 were both closely associated with mineral in bone. Then various amounts of E fraction (500, 1000, and 2000 μg) were subjected to IP with anti-LOXi and WB with anti-TGF-β1 or anti-LOXi antibody to investigate their potential endogenous binding in bone matrix. The results shown in Fig. 6B demonstrated that the endogenous binding between LOX and TGF-β1 occurs in a dose-dependent manner in bone matrix.
FIGURE 6.
Binding of LOX and TGF-β1 in bone matrix. A, presence of LOX and TGF-β1 proteins in bone matrix extracts. WB analyses were performed with anti-(α) LOXi antibody (lower panel), α TGF-β1 antibody (middle panel), and NRS (upper panel). The immunoreactive bands for LOX and TGF-β1 were detected at the expected molecular weight in E and G2 fraction of bone (lanes 2 and 3) but not in G1 fraction (lane 1). No immunoreactive bands were detected with NRS (upper panel). G, guanidine-HCl; G1, first G extract; E, EDTA extract; G2, second G extract. LOX isolated from bovine aorta by the method reported (75) and rhTGF-β1 were used as positive controls (lane 4). B, LOX-TGF-β1 binding complex in bone E extract. Various amounts of E extract were subjected to IP-WB analysis in combination of α LOXi, α LOXh, α TGF-β1 antibody, or NRS as indicated. Note that immunopositive bands of TGF-β1 are detected in a dose-dependent manner (middle panel, lanes 2–4) when IP was performed with α LOXi. An immunopositive band was also observed when IP was performed with α LOXh antibody (lane 5). No immunoreactivity was detected when NRS was used (upper panel, lanes 1–5, middle/lower panel, lane 1). +, ++, +++, 500, 1000, and 2000 μg, respectively, of E extract protein.
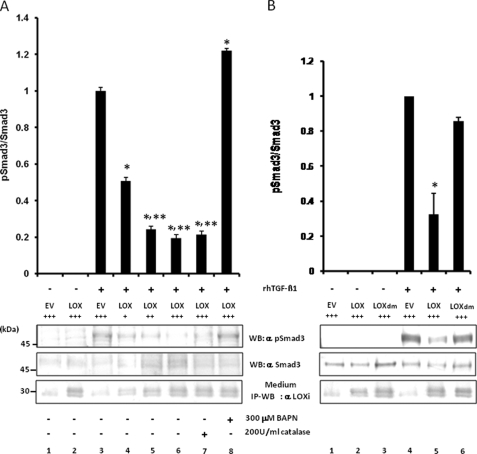
Effect of LOX Enzyme Activity on TGF-β Signaling—Because the binding of LOX to TGF-β1 both in vitro and in vivo was confirmed, we then examined the effect of LOX on TGF-β1 signaling and its potential mechanism. The TGF-β1 signaling was measured as the ratio of pSmad3 protein to the total Smad3 protein. MC cells were transiently transfected with empty vector (EV) or various amounts of pcDNA3.1/LOX/V5-His vector in the presence or absence of 300 μm BAPN or 200 units/ml catalase. The LOX-V5 protein level in each treatment group was evaluated by IP-WB analysis with anti-LOX antibody (Fig. 7, A and B, lower panel). Then rhTGF-β1 was added to those groups, and the TGF-β1 signaling (pSmad3/total Smad3) was evaluated. Without the addition of rhTGF-β1, cells transfected with EV or pcDNA3.1/LOX/V5-His vector did not induce Smad3 phosphorylation (Fig. 7A, lanes 1 and 2). Endogenous LOX expression was detected in cells transfected with EV (Fig. 7A, lanes 1 and 3). When rhTGF-β1 was added to EV, Smad3 phosphorylation was induced (Fig. 7A, lane 3). However, the TGF-β1-induced Smad3 phosphorylation was suppressed when LOX-V5 protein was expressed (Fig. 7A, lane 4), and the suppression occurred in a dose-dependent manner (Fig. 7A, lanes 4–6). The LOX-mediated inhibition was not affected in the presence of catalase indicating that the effect is not because of the H2O2, a by-product of the LOX-mediated oxidation (Fig. 7A, lane 7). The LOX-induced suppression was completely rescued in the presence of BAPN (Fig. 7A, lane 8). A slight increase in TGF-β1 signaling in this group compared with EV control (Fig. 7A, lane 3) could be due to the BAPN effect on endogenous LOX. We further determined whether the LOX catalytic activity was critical for the suppression by the use of LOXdm (inactive LOX, see under “Experimental Procedures”) by employing the same approach. When MC cells were transfected with the same dose of EV, pcDNA3.1/LOX/V5-His, or pcDNA3.1/LOXdm/V5-His, in the absence of rhTGF-β1, the Smad3 phosphorylation was not observed (Fig. 7B, lanes 1–3). The TGF-β1-induced Smad3 phosphorylation (Fig. 7B, lane 4) was significantly suppressed when LOX-V5 was expressed (Fig. 7B, lane 5), thus confirming the results shown in Fig. 7A. However, no significant suppression was observed when LOXdm was expressed (Fig. 7B, lane 6). The slight suppression by LOXdm, although statistically not significant, could be due to the physical binding of LOXdm to TGF-β1 (Fig. 2c, lanes 1–3). These results clearly indicate that LOX-mediated suppression of TGF-β1 signaling is due primarily to the amine oxidase activity of LOX.
FIGURE 7.
Effect of LOX as an amine oxidase on TGF-β signaling in MC cells. A, effect of LOX overexpression and its amine oxidase activity on TGF-β signaling. TGF-β1 signaling was measured as phospho(p)-Smad3 relative to total Smad3 (pSmad3/Smad3). Without the treatment with rhTGF-β1, Smad3 phosphorylation was not induced in MC cells transfected with EV (lane 1) or those overexpressing LOX-V5 (lane 2). When rhTGF-β1(5 ng/ml) was added to EV for 30 min, Smad3 phosphorylation was induced (lane 3); however, the rhTGF-β1-induced phosphorylation level was decreased when LOX-V5 was expressed in a dose-dependent manner (lanes 4–6). The LOX inhibition of TGF-β1-induced Smad3 phosphorylation was not affected in the presence of 200 units/ml catalase (lane 7) but completely rescued in the presence of 300 μm BAPN (lane 8). Error bars indicate mean ± S.D. of three independent experiments. *, significantly different (p < 0.05) from EV. * and **, significantly different (p < 0.05) from EV and LOX+. B, effects of enzymatically active and inactive LOX on TGF-β1-induced Smad3 phosphorylation. Without TGF-β1, no phosphorylation was observed in EV (lane 1), those expressing LOX-V5 (lane 2), or LOXdm-V5 (lane 3). Smad3 phosphorylation was induced upon the treatment with rhTGF-β1 (5 ng/ml) for 30 min in EV (lane 4); however, it was significantly down-regulated by active LOX-V5 (lane 5) but almost unaffected by inactive LOXdm (lane 6). The level of LOX expression was determined by IP-WB withα LOXi antibody (lower panel). Error bars indicate mean ± S.D. of three independent experiments. *, significantly different (p < 0.05) from EV. +, ++, and +++ indicate the relative amount of EV or LOX expression vectors transfected.
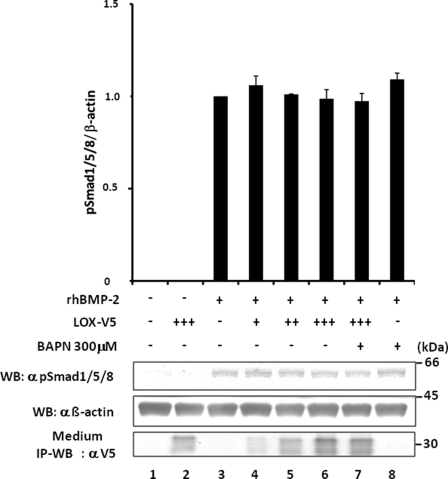
By employing the same approach, the effect of LOX and its enzyme activity on BMP-2 signaling was examined. The BMP-2 signaling was measured as the ratio of pSmad1/5/8 protein to the level of β-actin protein. Without the addition of rhBMP-2, cells transfected with EV or pcDNA3.1/LOX/V5-His vector did not induce Smad1/5/8 phosphorylation (Fig. 8, lanes 1 and 2). The phosphorylation was induced by BMP-2 (Fig. 8, lane 3), and the phosphorylation level did not change with the addition of various doses of LOX-V5 (Fig. 8, lanes 4–6) in the presence (Fig. 8, lanes 7 and 8) or absence (Fig. 8, lanes 3–6) of BAPN. This further confirms the specificity of the LOX-mediated suppression of TGF-β signaling.
FIGURE 8.
Effect of LOX overexpression on BMP-2 signaling in MC cells. The BMP-2-induced signaling was calculated as phospho-Smad1/5/8 relative to β-actin. Without rhBMP-2 treatment, Smad 1/5/8 phosphorylation was not induced in cells transfected by EV (lane 1) and those overexpressing LOX-V5 (lane 2). Upon rhBMP-2 treatment (100 ng/ml) for 30 min, the phosphorylation was induced in EV (lane 3). The level of BMP-2-induced Smad1/5/8 phosphorylation was not affected with the presence of various levels of LOX-V5 (lanes 4–6). The presence of BAPN did not affect the phosphorylation levels (lanes 7 and 8). The level of LOX-V5 protein was determined by IP-WB with α V5 antibody (lower panel). +, ++, and +++ indicate the relative amount of EV or LOX expression vectors transfected.
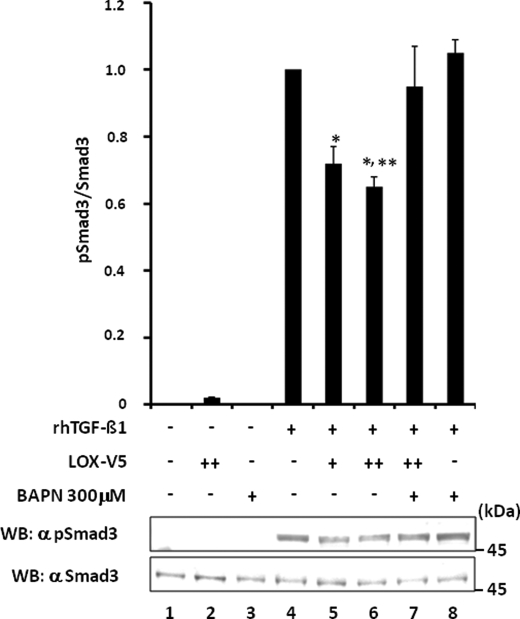
In another set of experiments, the effect of exogenous addition of LOX-V5 protein with or without BAPN on TGF-β1 signaling in MC cells was evaluated. The results are shown in Fig. 9. Without rhTGF-β1 treatment, Smad3 phosphorylation was not observed (Fig. 9, lanes 1–3). The phosphorylation was induced by the treatment with TGF-β1 (Fig. 9, lane 4), but it was significantly diminished with the addition of LOX-V5 in a dose-dependent manner (Fig. 9, lanes 5 and 6). However, this LOX-V5-mediated inhibition was rescued in the presence of BAPN (Fig. 9, lane 7). When BAPN and rhTGF-β1 were added to MC cells in the absence of exogenous LOX-V5, Smad3 phosphorylation was induced to the level comparable with that of TGF-β1 alone (Fig. 9, lane 8 versus 4). A slightly higher level of the TGF-β1 signaling of this group compared with EV control (lane 4), although statistically not significant, may reflect the BAPN effect on endogenous LOX.
FIGURE 9.
Effect of exogenous addition of LOX-V5 on TGF-β signaling in MC cells. Without rhTGF-β1, Smad3 phosphorylation was not induced in MC cells (lane 1) and those treated with LOX-V5 (lane 2) or BAPN (lane 3). The phosphorylation was induced in EV upon 5 ng/ml rhTGF-β1 treatment for 30 min (lane 4), but the phosphorylation level (calculated as pSmad3/Smad3) decreased with the addition of LOX-V5 in a dose-dependent manner (lanes 5 and 6). The inhibition of TGF-β1-induced Smad3 phosphorylation by LOX was rescued by the presence of 300 μm BAPN (lane 7). With the presence of BAPN (lane 8), rhTGF-β1-induced Smad3 phosphorylation was comparable with that of control (lane 4). Error bars indicate mean ± S.D. of three independent experiments. *, significantly different (p < 0.05) from lane 4. * and **, significantly different (p < 0.05) from lanes 4 and 5.
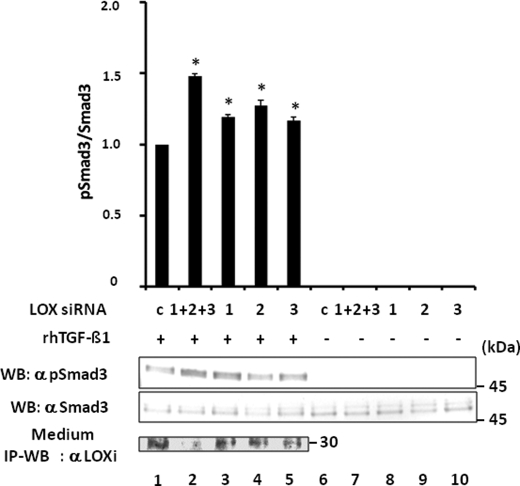
Effect of LOX RNA Interference on TGF-β Signaling—In addition to the gain-of function approaches described above, a loss-of-function experiment was performed using RNA interference technology. MC cells were transiently transfected with Silencer negative control or three different LOX siRNAs constructs. LOX protein levels in the cultured medium were lower in all the siRNA-transfected groups compared with that of negative control (20–80% of the control level; Fig. 10, lane 1 versus lanes 2–5, upper panel). The specificity of RNA silencing construct was evaluated by using LOX and LOX-like protein primers, and all constructs were specific to LOX (data not shown). The LOX level was the lowest when cells were transfected with three siRNA constructs combined (Fig. 10, lane 2). In comparison with the negative control (Fig. 10, lane 1), the TGF-β1-induced Smad3 phosphorylation was significantly increased in all siRNA-transfected groups (Fig. 10, lanes 3–5), but the increased level was highest when cells were transfected with the three siRNA combined (Fig. 10, lane 2). Those cells exhibited the lowest level of LOX protein (Fig. 10, lane 2). Transfection with the LOX siRNA construct alone in the absence of rhTGF-β1 did not induce Smad3 phosphorylation (Fig. 10, lanes 6–10). Therefore, the rhTGF-β1-induced Smad3 phosphorylation was enhanced when the level of endogenous LOX protein was diminished by RNA interference. These results further support the notion that LOX suppresses TGF-β1 signaling.
FIGURE 10.
Effect of LOX suppression by RNA interference on TGF-β signaling in MC cells. When MC cells were transfected with silencer negative control (c) and treated with TGF-β1, Smad3 phosphorylation was induced (lane 1). However, the Smad3 phosphorylation level (pSmad3/Smad3) was further enhanced when LOX expression was suppressed when RNA interference was performed (lanes 2–5). The increase in signaling was enhanced the most in the cells transfected with three siRNA constructs combined (1 + 2 + 3, lane 2) that expressed the lowest level of LOX when compared with those transfected with individual constructs (1, lane 3;2, lane 4, and 3, lane 5). Without TGF-β induction, the phosphorylation was not detected in any of those cell groups (lanes 6–10). The level of LOX expression was determined by IP-WB with α LOXi antibody (bottom panel). Error bars indicate mean ± S.D. of three independent experiments. *, significantly different (p < 0.05) from control (lane 1).
DISCUSSION
Bone organic matrix, primarily composed of type I collagen (∼90%) and noncollagenous component (∼10%), is one of the major storage sites of TGF-βs in the body (39). In bone, TGF-β1, the predominant isoform of TGF-β subfamily, plays pivotal roles in many, if not all, aspects of the tissue development, remodeling, mechanical properties, and aging (12, 24, 40) by controlling cell recruitment and proliferation, cell differentiation, and matrix production. Those multiple effects of TGF-β1 are exerted through its binding to specific transmembranous type I and type II Ser/Thr kinase receptors that propagate the signal to control target genes in various cell types (41). As such, its activity is tightly regulated. In general, the latency of TGF-β is achieved within the cells through its noncovalent association with the propeptide, LAP (small latent complex, SLC), that is further covalently bound to latent TGF-β-binding proteins forming a large latent complex. The secreted large latent complex can then be sequestered within the ECM (18, 42). In order for TGF-β to get activated, it must be released from the complex through several biological as well as physical activation mechanisms (43). It has been reported that bone cells, distinct from other cell types, secrete a significant portion of TGF-β1 as an SLC form in which the growth factor is more readily available and activated (44, 45). Low pH generated by osteoclasts, for instance, can release TGF-β from SLC and activate (18). Thus, the abundance and bioavailability of this highly potent growth factor in bone, a dynamic and constantly remodeling tissue, predict a great need of highly controlled mechanism for its activity.
In bone matrix, there are several TGF-β-binding molecules, including a group of small leucine-rich proteoglycans such as decorin and biglycan (46–57), that may sequester active TGF-β in the ECM, thus hindering the TGF-β binding to its cell receptor (58). Indeed, the absence of these proteoglycans resulted in increased TGF-β activation in bone due likely to the disruption of proper sequestration of this growth factor in ECM, leading to premature apoptosis of bone marrow stromal cells (42). Clearly a proper level of TGF-β-binding molecules in ECM is important to regulate the TGF-β activity during bone formation and remodeling.
In this study, we have demonstrated that LOX, a matrix amine oxidase, directly binds to mature TGF-β1 in vitro, and the binding complex is present in mineralized bone matrix. Furthermore, it has been demonstrated that LOX suppresses TGF-β1 signaling via its amine oxidase activity. This was evident because the suppression was rescued by BAPN, and inactive LOX (LOXdm) was not capable of exerting this effect (Fig. 7B). This effect is not likely due to H2O2, a by-product of the LOX-mediated oxidation reaction, as catalase did not affect the suppression (Fig. 7A). Although we cannot exclude the possibility that LOX binds and regulates other growth factors present in bone, this LOX function appears to be relatively specific as LOX neither binds to osteogenic BMPs (BMP-2, -4, -6, and -7) (Fig. 1) nor affects their signaling (Fig. 8). As stated above, there are a number of mechanisms by which TGF-β function is controlled at the intra- and extracellular levels. However, to the best of our knowledge, the suppression of TGF-β function by enzymatic modification represents a novel mechanism. Likely, LOX binds to mature TGF-β1 first and then oxidizes some of the lysine residues that could be critical for the signal transduction. Although the propeptide domain of LOX is not likely involved in the binding (Fig. 2), at this point it is not clear where the specific binding domain(s) resides within the mature LOX molecule. A further binding study is warranted to identify such TGF-β1 binding domain(s) of LOX by the use of various LOX deletion mutant constructs.
It has recently been proposed that the range of LOX substrates is much broader than speculated in the past and that this acidic enzyme has a strong preference toward basic globular proteins with pI higher than 8 (59). It is of interest to note that mature TGF-β1 is a basic protein (pI 8.59) and that the C terminus of the mature TGF-β (residues 83–112), critical for the binding to its type II receptor (TβRII) to initiate the signaling cascade (60), is enriched in basic amino acids, including several lysine residues. Thus, some of those lysine residues could be oxidatively deaminated by LOX, which diminishes the overall positive charge. The aldehyde produced could then further cross-link to the vicinal ε-amino group of lysine or another LOX-mediated aldehyde derived from mature TGF-β1 and, possibly, TGF-β1-associated proteins in a manner similar to collagen/elastin cross-linking (61). The generation of such cross-links would not only diminish the charge but also covalently stabilize the mature TGF-β molecule(s). In either case, such a modification of TGF-β will likely disrupt the interaction with its receptors, thus diminishing its signaling.
It appears that both LOX and TGF-β1 are present in the mineral-associated fraction of bone matrix as the detection required demineralization. In addition, the two proteins appear to be bound in this fraction. Although the potential effect of the prior GH extraction (G1) on this binding identified in E extract cannot be completely ruled out, this is unlikely as mineral protects the mineralized matrix from denaturation (62). The binding of LOX and TGF-β1 found in vitro where no denaturants were used (Figs. 1, 2, and 4) also support this endogenous binding. The significance of this specific distribution is not clear at this point. It was reported that in dentin, LOX was identified as one of the major matrix proteins in the compartment (G2 fraction in this study) that is associated with insoluble collagen matrix masked by the mineral (63). Because LOX is known to bind type I collagen fibrils (64), LOX together with TGF-β1 are likely incorporated into the collagen matrix through the LOX-collagen interaction that eventually mineralizes. To further define the co-localization and its spatial relationship with collagen fibrils, more detailed analyses such as double-labeling immunoelectron microscopy need to be performed. The potential effect of LOX-TGF-β1 complex on collagen mineralization needs to be further investigated.
A number of studies have demonstrated that LOX expression is up-regulated by TGF-β1 (65–72). Thus, it is noteworthy that mature TGF-β1 and its activity are in turn regulated by LOX. Recently, a potential cross-control of LOX on TGF-β1 effects has been reported, but without any direct role for LOX in the TGF-β1 signaling cascades. The authors demonstrated a decrease of total Smad3 protein in 293T cells overexpressing LOX after TGF-β1 addition for 24 h (73), an effect that was not observed in our study possibly because of the difference in the cell type.
It has been reported that when chick and murine osteoblasts were cultured in the presence of BAPN, collagen morphology and mineralization were significantly altered (9, 10). Those effects are likely attributable to altered collagen cross-linking as the authors speculated. However, both reports also noted that in the presence of BAPN, collagen synthesis was significantly increased, and the fibrils formed were larger. The similar effect, i.e. increased collagen synthesis by BAPN treatment of cells, was also reported in chondrocyte cultures (8, 11). In addition, up-regulation of collagen mRNA was also seen in lathyritic animals during the early phase of bone fracture healing as well as TGF-β1 mRNA expression (4). Although the potential mechanism of this effect (higher collagen synthesis by BAPN) was not clearly delineated in those reports, it can be explained in part based on our finding. Inactivation of LOX by BAPN likely leads to higher levels of active TGF-β1 that stimulates collagen expression and synthesis. Indeed, in a different set of experiments, we have observed that MC cells stably overexpressing LOX produced significantly less collagen and exhibited lower TGF-β1 signaling (data not shown). We are currently in the process of establishing and characterizing several MC-derived stable clones expressing higher or lower levels of LOX.
Taken together, our results demonstrate, for the first time, that the mature active LOX binds to mature TGF-β1 and suppresses its signaling likely via its amine oxidase activity. This proposed mechanism is distinct from other regulatory mechanisms reported previously such as sequestration of mature TGF-β1 by physical binding (46) and inhibition of TGF-β1 processing (74). This finding may provide a new insight into the control mechanism of bone development and remodeling in which TGF-β1 plays pivotal roles.
Acknowledgments
We thank Drs. Phillip C. Trackman at Boston University and Yuji Mishina at NIEHS, National Institutes of Health, for valuable discussion and suggestions, and Dr. Hidenori Ichijo at University of Tokyo for providing vectors.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DE10489 (to M. Y.) and R21 AR052824 (to M. Y.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: LOX, lysyl oxidase; BAPN, β-aminopropionitrile; TGF-β, transforming growth factor-beta; BMP, bone morphogenetic proteins; HA, hemagglutinin; GST, glutathione S-transferase; MC3T3-E1, MC; RT, reverse transcription; LOXdm, LOX with double mutation; mLOX, mature LOX; LOXPP, LOX propeptide; fluoride; WB, Western blot; IP, immunoprecipitation; MALDI-MS, matrix assisted laser desorption ionizationmass spectrometric analysis; GH, guanidine-HCl; NRS, normal rabbit serum; ECM, extracellular matrix; LAP, latency-associated peptide; SLC, small latent complex; siRNA, short interfering RNA; rh, recombinant human; PBS, phosphate-buffered saline; EV, empty vector.
References
- 1.Kagan, H. M., and Li, W. (2003) J. Cell Biochem. 88 660-672 [DOI] [PubMed] [Google Scholar]
- 2.Levene, C. I., and Gross, J. (1959) J. Exp. Med. 110 771-790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeager, V. L., Buranarugsa, M. W., and Arunatut, O. (1985) J. Exp. Pathol. 2 1-11 [PubMed] [Google Scholar]
- 4.Ekholm, E. C., Ravanti, L., Kahari, V., Paavolainen, P., and Penttinen, R. P. (2000) Bone (Elmsford) 27 551-557 [DOI] [PubMed] [Google Scholar]
- 5.Li, W., Nugent, M. A., Zhao, Y., Chau, A. N., Li, S. J., Chou, I. N., Liu, G., and Kagan, H. M. (2003) J. Cell Biochem. 88 152-164 [DOI] [PubMed] [Google Scholar]
- 6.Giampuzzi, M., Oleggini, R., and Di Donato, A. (2003) Biochim. Biophys. Acta 1647 245-251 [DOI] [PubMed] [Google Scholar]
- 7.Kagan, H. M., Williams, M. A., Calaman, S. D., and Berkowitz, E. M. (1983) Biochem. Biophys. Res. Commun. 115 186-192 [DOI] [PubMed] [Google Scholar]
- 8.Beekman, B., Verzijl, N., Bank, R. A., von der Mark, K., and TeKoppele, J. M. (1997) Exp. Cell Res. 237 135-141 [DOI] [PubMed] [Google Scholar]
- 9.Gerstenfeld, L. C., Riva, A., Hodgens, K., Eyre, D. R., and Landis, W. J. (1993) J. Bone Miner. Res. 8 1031-1043 [DOI] [PubMed] [Google Scholar]
- 10.Hong, H. H., Pischon, N., Santana, R. B., Palamakumbura, A. H., Chase, H. B., Gantz, D., Guo, Y., Uzel, M. I., Ma, D., and Trackman, P. C. (2004) J. Cell. Physiol. 200 53-62 [DOI] [PubMed] [Google Scholar]
- 11.Wong, M., Siegrist, M., Gaschen, V., Park, Y., Graber, W., and Studer, D. (2002) Tissue Eng. 8 979-987 [DOI] [PubMed] [Google Scholar]
- 12.Janssens, K., ten Dijke, P., Janssens, S., and Van Hul, W. (2005) Endocr. Rev. 26 743-774 [DOI] [PubMed] [Google Scholar]
- 13.Yamane, K., Suzuki, H., Ihn, H., Kato, M., Yoshikawa, H., and Tamaki, K. (2005) J. Cell. Physiol. 202 822-830 [DOI] [PubMed] [Google Scholar]
- 14.Kahai, S., Vary, C. P., Gao, Y., and Seth, A. (2004) Matrix Biol. 23 445-455 [DOI] [PubMed] [Google Scholar]
- 15.Takuwa, Y., Ohse, C., Wang, E. A., Wozney, J. M., and Yamashita, K. (1991) Biochem. Biophys. Res. Commun. 174 96-101 [DOI] [PubMed] [Google Scholar]
- 16.Luppen, C. A., Leclerc, N., Noh, T., Barski, A., Khokhar, A., Boskey, A. L., Smith, E., and Frenkel, B. (2003) J. Biol. Chem. 278 44995-45003 [DOI] [PubMed] [Google Scholar]
- 17.Pircher, R., Jullien, P., and Lawrence, D. A. (1986) Biochem. Biophys. Res. Commun. 136 30-37 [DOI] [PubMed] [Google Scholar]
- 18.Annes, J. P., Munger, J. S., and Rifkin, D. B. (2003) J. Cell Sci. 116 217-224 [DOI] [PubMed] [Google Scholar]
- 19.Annes, J. P., Chen, Y., Munger, J. S., and Rifkin, D. B. (2004) J. Cell Biol. 165 723-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeilschifter, J., Pignat, W., Leighton, J., Marki, F., Vosbeck, K., and Alkan, S. (1990) Biochem. J. 270 269-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machwate, M., Jullienne, A., Moukhtar, M., Lomri, A., and Marie, P. J. (1995) Mol. Endocrinol. 9 187-198 [DOI] [PubMed] [Google Scholar]
- 22.Alliston, T., Choy, L., Ducy, P., Karsenty, G., and Derynck, R. (2001) EMBO J. 20 2254-2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeda, S., Hayashi, M., Komiya, S., Imamura, T., and Miyazono, K. (2004) EMBO J. 23 552-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromigue, O., Modrowski, D., and Marie, P. J. (2004) Curr. Pharm. Des. 10 2593-2603 [DOI] [PubMed] [Google Scholar]
- 25.Bonewald, L. F. (2002) J. Musculoskelet. Neuronal Interact. 2 239-241 [PubMed] [Google Scholar]
- 26.Fox, S. W., and Lovibond, A. C. (2005) Mol. Cell. Endocrinol. 243 19-26 [DOI] [PubMed] [Google Scholar]
- 27.Li, P. A., He, Q., Cao, T., Yong, G., Szauter, K. M., Fong, K. S., Karlsson, J., Keep, M. F., and Csiszar, K. (2004) Brain Res. Mol. Brain Res. 120 115-122 [DOI] [PubMed] [Google Scholar]
- 28.Fogelgren, B., Polgar, N., Szauter, K. M., Ujfaludi, Z., Laczko, R., Fong, K. S., and Csiszar, K. (2005) J. Biol. Chem. 280 24690-24697 [DOI] [PubMed] [Google Scholar]
- 29.Wang, S. X., Mure, M., Medzihradszky, K. F., Burlingame, A. L., Brown, D. E., Dooley, D. M., Smith, A. J., Kagan, H. M., and Klinman, J. P. (1996) Science 273 1078-1084 [DOI] [PubMed] [Google Scholar]
- 30.Mochida, Y., Parisuthiman, D., Kaku, M., Hanai, J., Sukhatme, V. P., and Yamauchi, M. (2006) J. Biol. Chem. 281 36044-36051 [DOI] [PubMed] [Google Scholar]
- 31.Mochida, Y., Parisuthiman, D., and Yamauchi, M. (2006) Adv. Exp. Med. Biol. 585 101-113 [DOI] [PubMed] [Google Scholar]
- 32.Palamakumbura, A. H., and Trackman, P. C. (2002) Anal. Biochem. 300 245-251 [DOI] [PubMed] [Google Scholar]
- 33.Linde, A., Bhown, M., and Butler, W. T. (1980) J. Biol. Chem. 255 5931-5942 [PubMed] [Google Scholar]
- 34.Termine, J. D., Belcourt, A. B., Conn, K. M., and Kleinman, H. K. (1981) J. Biol. Chem. 256 10403-10408 [PubMed] [Google Scholar]
- 35.Cheng, H., Caterson, B., and Yamauchi, M. (1999) Connect. Tissue Res. 40 37-47 [DOI] [PubMed] [Google Scholar]
- 36.Dubois, C. M., Laprise, M. H., Blanchette, F., Gentry, L. E., and Leduc, R. (1995) J. Biol. Chem. 270 10618-10624 [DOI] [PubMed] [Google Scholar]
- 37.Wu, M., Min, C., Wang, X., Yu, Z., Kirsch, K. H., Trackman, P. C., and Sonenshein, G. E. (2007) Cancer Res. 67 6278-6285 [DOI] [PubMed] [Google Scholar]
- 38.Trackman, P. C., Bedell-Hogan, D., Tang, J., and Kagan, H. M. (1992) J. Biol. Chem. 267 8666-8671 [PubMed] [Google Scholar]
- 39.Seyedin, S. M., Thomas, T. C., Thompson, A. Y., Rosen, D. M., and Piez, K. A. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 2267-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balooch, G., Balooch, M., Nalla, R. K., Schilling, S., Filvaroff, E. H., Marshall, G. W., Marshall, S. J., Ritchie, R. O., Derynck, R., and Alliston, T. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18813-18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, Y., and Massague, J. (2003) Cell 113 685-700 [DOI] [PubMed] [Google Scholar]
- 42.Rifkin, D. B. (2005) J. Biol. Chem. 280 7409-7412 [DOI] [PubMed] [Google Scholar]
- 43.Jenkins, G. (2008) Int. J. Biochem. Cell Biol. 40 1068-1078 [DOI] [PubMed] [Google Scholar]
- 44.Bonewald, L. F. (2002) in Principles of Bone Biology (Bilezikian, J., Raisz, L., and Rodan, G., eds) pp. 903-918 Academic Press, London
- 45.Dallas, S. L., Rosser, J. L., Mundy, G. R., and Bonewald, L. F. (2002) J. Biol. Chem. 277 21352-21360 [DOI] [PubMed] [Google Scholar]
- 46.Hildebrand, A., Romaris, M., Rasmussen, L. M., Heinegard, D., Twardzik, D. R., Border, W. A., and Ruoslahti, E. (1994) Biochem. J. 302 527-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breuer, B., Schmidt, G., and Kresse, H. (1990) Biochem. J. 269 551-554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hausser, H., Groning, A., Hasilik, A., Schonherr, E., and Kresse, H. (1994) FEBS Lett. 353 243-245 [DOI] [PubMed] [Google Scholar]
- 49.Kresse, H., and Schonherr, E. (2001) J. Cell. Physiol. 189 266-274 [DOI] [PubMed] [Google Scholar]
- 50.Markmann, A., Hausser, H., Schonherr, E., and Kresse, H. (2000) Matrix Biol. 19 631-636 [DOI] [PubMed] [Google Scholar]
- 51.Riquelme, C., Larrain, J., Schonherr, E., Henriquez, J. P., Kresse, H., and Brandan, E. (2001) J. Biol. Chem. 276 3589-3596 [DOI] [PubMed] [Google Scholar]
- 52.Schaefer, L., Grone, H. J., Raslik, I., Robenek, H., Ugorcakova, J., Budny, S., Schaefer, R. M., and Kresse, H. (2000) Kidney Int. 58 1557-1568 [DOI] [PubMed] [Google Scholar]
- 53.Schaefer, L., Hausser, H., Altenburger, M., Ugorcakova, J., August, C., Fisher, L. W., Schaefer, R. M., and Kresse, H. (1998) Kidney Int. 54 1529-1541 [DOI] [PubMed] [Google Scholar]
- 54.Schaefer, L., Macakova, K., Raslik, I., Micegova, M., Grone, H. J., Schonherr, E., Robenek, H., Echtermeyer, F. G., Grassel, S., Bruckner, P., Schaefer, R. M., Iozzo, R. V., and Kresse, H. (2002) Am. J. Pathol. 160 1181-1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaefer, L., Raslik, I., Grone, H. J., Schonherr, E., Macakova, K., Ugorcakova, J., Budny, S., Schaefer, R. M., and Kresse, H. (2001) FASEB J. 15 559-561 [DOI] [PubMed] [Google Scholar]
- 56.Schonherr, E., Broszat, M., Brandan, E., Bruckner, P., and Kresse, H. (1998) Arch. Biochem. Biophys. 355 241-248 [DOI] [PubMed] [Google Scholar]
- 57.Bi, Y., Stuelten, C. H., Kilt, T. Waldhawa, S., Iozzo, R. V., Robey, P. G., Chen, X. D., and Young, M. F. (2005) J. Biol. Chem. 280 30481-30489 [DOI] [PubMed] [Google Scholar]
- 58.Droguett, R., Cabello-Verrugio, C., Riquelme, C., and Brandan, E. (2006) Matrix Biol. 25 332-341 [DOI] [PubMed] [Google Scholar]
- 59.Kagan, H. M., Williams, M. A., Williamson, P. R., and Anderson, J. M. (1984) J. Biol. Chem. 259 11203-11207 [PubMed] [Google Scholar]
- 60.Young, G. D., and Murphy-Ullrich, J. E. (2004) J. Biol. Chem. 279 38032-38039 [DOI] [PubMed] [Google Scholar]
- 61.Yamauchi, M., and Mechanic, G. L. (1988) Collagen (Nimni, M. E., ed) pp. 157-172, CRC Press, Inc., Boca Raton, FL
- 62.Bonar, L. C., and Glimcher, M. J. (1970) J. Ultrastruct. Res. 32 545-548 [DOI] [PubMed] [Google Scholar]
- 63.Domenicucci, C., Goldberg, H. A., and Sodek, J. (1997) Connect. Tissue Res. 36 151-163 [DOI] [PubMed] [Google Scholar]
- 64.Cronlund, A. L., Smith, B. D., and Kagan, H. M. (1985) Connect. Tissue Res. 14 109-119 [DOI] [PubMed] [Google Scholar]
- 65.Boak, A. M., Roy, R., Berk, J., Taylor, L., Polgar, P., Goldstein, R. H., and Kagan, H. M. (1994) Am. J. Respir. Cell Mol. Biol. 11 751-755 [DOI] [PubMed] [Google Scholar]
- 66.Feres-Filho, E. J., Choi, Y. J., Han, X., Takala, T. E., and Trackman, P. C. (1995) J. Biol. Chem. 270 30797-30803 [DOI] [PubMed] [Google Scholar]
- 67.Gacheru, S. N., Thomas, K. M., Murray, S. A., Csiszar, K., Smith-Mungo, L. I., and Kagan, H. M. (1997) J. Cell. Biochem. 65 395-407 [DOI] [PubMed] [Google Scholar]
- 68.Shanley, C. J., Gharaee-Kermani, M., Sarkar, R., Welling, T. H., Kriegel, A., Ford, J. W., Stanley, J. C., and Phan, S. H. (1997) J. Vasc. Surg. 25 446-452 [DOI] [PubMed] [Google Scholar]
- 69.Hong, H. H., Uzel, M. I., Duan, C., Sheff, M. C., and Trackman, P. C. (1999) Lab. Investig. 79 1655-1667 [PubMed] [Google Scholar]
- 70.Bose, K. K., Chakraborty, J., Khuder, S., Smith-Mensah, W. H., and Robinson, J. (2000) J. Occup. Environ. Med. 42 582-587 [DOI] [PubMed] [Google Scholar]
- 71.Goto, Y., Uchio-Yamada, K., Anan, S., Yamamoto, Y., Ogura, A., and Manabe, N. (2005) Virchows Arch. 447 859-868 [DOI] [PubMed] [Google Scholar]
- 72.Shibanuma, M., Mashimo, J., Mita, A., Kuroki, T., and Nose, K. (1993) Eur. J. Biochem. 217 13-19 [DOI] [PubMed] [Google Scholar]
- 73.Oleggini, R., Gastaldo, N., and Di Donato, A. (2007) Matrix Biol. 26 494-505 [DOI] [PubMed] [Google Scholar]
- 74.Murphy-Ullrich, J. E., Schultz-Cherry, S., and Hook, M. (1992) Mol. Biol. Cell 3 181-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kagan, H. M., and Cai, P. (1995) Methods Enzymol. 258 122-132 [DOI] [PubMed] [Google Scholar]