Abstract
Okazaki fragment maturation to produce continuous lagging strands in eukaryotic cells requires precise coordination of strand displacement synthesis by DNA polymerase δ (Pol δ) with 5·-flap cutting by FEN1RAD27 endonuclease. Excessive strand displacement is normally prevented by the 3·-exonuclease activity of Pol δ. This core maturation machinery can be assisted by Dna2 nuclease/helicase that processes long flaps. Our genetic studies show that deletion of the POL32 (third subunit of Pol δ) or PIF1 helicase genes can suppress lethality or growth defects of rad27Δ pol3-D520V mutants (defective for FEN1RAD27 and the 3·-exonuclease of Pol δ) that produce long flaps and of dna2Δ mutants that are defective in cutting long flaps. On the contrary, pol32Δ or pif1Δ caused lethality of rad27Δ exo1Δ double mutants, suggesting that Pol32 and Pif1 are required to generate longer flaps that can be processed by Dna2 in the absence of the short flap processing activities of FEN1RAD27 and Exo1. The genetic analysis reveals a remarkable flexibility of the Okazaki maturation machinery and is in accord with our biochemical analysis. In vitro, the generation of short flaps by Pol δ is not affected by the presence of Pol32; however, longer flaps only accumulate when Pol32 is present. The presence of FEN1RAD27 during strand displacement synthesis curtails displacement in favor of flap cutting, thus suggesting an active hand-off mechanism from Pol δ to FEN1RAD27. Finally, RNA-DNA hybrids are more readily displaced by Pol δ than DNA hybrids, thereby favoring degradation of initiator RNA during Okazaki maturation.
The process of DNA replication in eukaryotic cells leads to the generation of a vast number of Okazaki fragments on the lagging strand of the replication fork. Approximately 50,000,000 Okazaki fragments are synthesized when a human cell replicates, and all of these need to be efficiently and accurately matured into continuous lagging strands to ensure genome integrity. Various DNA structures are generated during the synthesis and maturation of Okazaki fragments. These structures constitute the largest pool for potential DNA damage in the cell. Incomplete or poorly processed Okazaki fragments can lead to repeat expansion mutations, small duplication mutations, and to the generation of double-stranded DNA breaks (1).
A large number of activities have been implicated in lagging strand DNA maturation. In the budding yeast Saccharomyces cerevisiae, the RAD27 gene product is the 5′-flap endonuclease FEN1. FEN1RAD27 has been assigned a dominant role in creating ligatable nicks during Okazaki maturation (reviewed in Refs. 2 and 3). Strong support for the importance of RAD27 in Okazaki maturation was initially provided by the dramatic increase of small duplications up to ∼100 nt4 in length flanked by short repeats in rad27-null mutants (4). This unusual class of duplication mutations was proposed to result through ligation of an unremoved flap with the 3′-end of the downstream Okazaki fragment. This type of duplications is caused not only by the lack of RAD27 but also by a subtle rad27-p defect that severely decreases FEN1RAD27 binding to the replication clamp proliferating cell nuclear antigen (PCNA) in vitro (5). Consistent with these genetic observations, biochemical studies showed that FEN1RAD27 is ideally suited to create ligatable nicks from 5′-flaps generated by the lagging strand DNA polymerase δ (6, 7). Based on these biochemical and genetic insights, we and others previously proposed that FEN1RAD27 is part of a minimal core Okazaki maturation machinery that efficiently processes the vast majority of the Okazaki fragments in the cell and can be assisted by a number of auxiliary factors that function under specific circumstances when the core machinery falters (Fig. 1) (reviewed in Ref. 8).
FIGURE 1.
Current model for Okazaki fragment maturation. The main pathway (thick arrows) involves very limited strand displacement by Pol δ, followed by FEN1 cutting of the single nucleotide flap. This process is iterated until all initiator RNA (iRNA; red) is degraded (Nick Translation). Excessive strand displacement is postulated to be rare (dotted arrows). It is reduced by the Exo activity of Pol δ (Idling), and enhanced by Pol32 or Pif1 (Long Flap Formation). The processing of long flaps requires Dna2. Regression of the resulting small flaps to the nick position is accomplished by FEN1 or the Exo activity of Pol δ.
The core maturation machinery consists of three factors: Pol δ, FEN1RAD27 and DNA ligase I. These three factors are held in a complex through interactions with PCNA. FEN1RAD27 and DNA ligase I are both single-subunit enzymes with well characterized biochemical activities (reviewed in Refs. 2 and 9). S. cerevisiae Pol δ consists of three subunits: Pol3 (125 kDa), Pol31 (55 kDa), and Pol32 (40 kDa). The POL3 and POL31 genes are essential for yeast viability. POL32 deletion mutants are viable, but show defects in DNA replication, DNA recombination, and repair and in mutagenesis (10–14). These three subunits of Pol δ are conserved in all organisms; however, the enzyme from Schizosaccharomyces pombe and from higher eukaryotic organisms contains an additional small fourth subunit that increases the stability of the enzyme (15). Biochemical studies of the S. cerevisiae three-subunit Pol δ compared with the two-subunit Pol3-Pol31 enzyme lacking the Pol32 subunit, designated as Pol δ*, indicate that the latter enzyme has a decreased processivity of DNA synthesis and a decreased interaction with PCNA (16). Biochemical and genetic studies indicate that Pol32 is also important for physical interactions of Pol δ with Pol α-primase, suggesting that Pol32 may coordinate enzymatic activities during lagging strand synthesis and Okazaki maturation (12, 17).
We previously noted that defects in the 3′-5′-exonuclease activity of Pol δ can be lethal when combined with rad27Δ or even with minor defects in RAD27 (5). The same combined lethality was observed for POL3 mutants that, although fully proficient for exonuclease activity, show impaired switching from the polymerase to the exonuclease domain (18). Importantly, double mutant combinations that are not lethal show an increased rate of small duplication mutations, a hallmark signature of defects in lagging strand DNA replication. This led us to propose that strand displacement synthesis by the polymerase (Pol) activity of Pol δ is countered by the competing 3′-5′-exonuclease (Exo) activity of the enzyme, also contained within the catalytic Pol3 subunit of polymerase δ. In agreement with this proposal, our in vitro studies showed that the functional interaction between the three enzymatic activities (i.e. polymerase and 3′-exonuclease activities of Pol δ and the 5′-flap endonuclease activity of FEN1) are important for efficient maturation of Okazaki fragments (19). When Pol δ reaches the 5′-end of the downstream Okazaki fragment, it continues an additional 1–2-nt synthesis in a strand displacement mode (20). FEN1 removes the small 5′-flap and creates a ligatable nick for DNA ligase on the border between adjacent fragments, provided that no RNA is left on the downstream Okazaki fragment. Otherwise, the RNA primer is removed by iterative processing through alternate Pol δ and FEN1 action, called nick translation, where a single ribonucleotide is typically removed with each cycle (Fig. 1). This forward movement by Pol δ can be counteracted by its Exo activity. The nuclease activity of Pol δ is generally considered to be a proofreading function to assure high fidelity DNA replication (21). However, the nuclease activity also plays a crucial role in Okazaki fragment maturation (18). Exo-mediated 3′-degradation increases opportunities for generating and maintaining ligatable nicks. In fact, Pol δ has the remarkable ability to idle at a nick. Idling is the iterative process of limited (∼1–2-nt) strand displacement synthesis at the nick, followed by switching to the Exo domain and degradation of the displacing strand until the nick position has been reached again. In this fashion, Pol δ not only has the ability to resolve small 5′-flaps by degradation of the displacing 3′-strand (Fig. 1) but also maintains a dynamic relationship with the nick position. This idling capacity at a nick is unique to the lagging strand Pol δ, and it is not displayed by leading strand Pol ε (20).
This efficient minimal core machinery consisting of Pol δ and FEN1 can be augmented by several auxiliary proteins in order to mature Okazaki fragments that might be difficult to process because of DNA structural problems or if there is an accumulation of long flaps because of lack of flap cutting by FEN1. Among these is Dna2, which has both 3′-5′-helicase and nuclease activity. Its nuclease activity is essential for viability (22, 23). Pif1 is a 5′-3′-helicase that functions in mitochondrial DNA maintenance and in telomere homeostasis (24). It was also suggested to counteract formation of ligatable nicks by creating larger flaps. This conclusion was drawn because deletion of PIF1 rescues the lethality of dna2 mutants (25). Recent biochemical studies have shown that the presence of Pif1 during gap filling by Pol δ favors the generation of longer flaps, thereby providing a biochemical rationale for an essential Dna2 requirement in cells carrying Pif1 (26). Other factors that may act during lagging strand replication include the flap exoendonuclease Exo1, RNase H2, and the RecQ-like helicase Sgs1 (27–29). Interestingly, neither exo1Δ nor rnh2Δ (eliminating RNase H2) mutants cause small duplications.
It is not clear under what conditions these auxiliary Okazaki maturation factors are called into action. One prevailing model is that when flaps have grown to excessive length, they are no longer substrates for FEN1; nor can they be restored to a nick structure by the Exo activity of Pol δ. Long 5′-flaps, perhaps longer than 25 nt, to which DNA-binding proteins are bound (e.g. the single-stranded binding protein RPA) fail to engage FEN1 (reviewed in Ref. 30), whereas backup by the Exo activity of Pol δ is also limited to flaps that are only a few nt long (20). Apparently, efficient and faithful Okazaki fragment maturation requires a proper balance between DNA synthesis, strand displacement synthesis, and the capability for 5′- and 3′-degradation, as indicated in the model in Fig. 1. One prediction of the model is that under conditions where strand displacement synthesis is reduced, there would be less of a need for nuclease degradation of long flaps. Genetic experiments with mutants of the auxiliary Okazaki maturation nuclease DNA2 supported this view in that the temperature sensitivity of conditional dna2 mutants could be suppressed by deletion of the POL32 gene (25). It was suggested that the suppressive effect of the POL32-null mutation was caused by a reduction in strand displacement synthesis by the two-subunit form of Pol δ, resulting in the creation of a smaller number of the infrequent substrates that would require Dna2 for processing. Although consistent with the data, this hypothesis lacked experimental verification.
We have undertaken comprehensive genetic and biochemical studies to understand the role that Pol32 plays in the core machinery required for Okazaki maturation. We found that the distribution of flap sizes is determined by several competing activities in vitro. The length of the flap generated during a processive cycle of strand displacement synthesis by Pol δ is one indicator of the length of the flap to be cut by FEN1. A second indicator is the capacity for 3′-degradation by the Exo activity of Pol δ. Third, the presence of FEN1 curtails processive strand invasion synthesis by Pol δ in favor of flap cutting. Finally, in keeping with its function in Okazaki fragment maturation, Pol δ shows an increased capacity to displace RNA-DNA hybrids. If Pol δ lacks Pol32 (designated Pol δ*), there is decreased processivity of strand displacement as well as reduced 3′-degradation. As a consequence, the size distribution of short flaps (<10 nt) is not affected by Pol32. However, long flaps are only detected when Pol32 is present. These in vitro findings in parallel with our genetic studies provide a compelling explanation of how cells deal with the balance between flap generation and cutting. Importantly, the combination of the exonuclease activity of Pol δ and its third subunit Pol32 provide flexibility to the eukaryotic Okazaki maturation machinery.
EXPERIMENTAL PROCEDURES
Yeast Strains—All strains were derived from ALE100, ALE101, ALE1000, and ALE1001, isogenic to CG379 (MATα ade5-1 his 7-2 leu 2-3, 112trp1-289 ura3Δ). In addition, strains carried alleles of LYS2 in two chromosomal locations (see Ref. 5 and references therein). Completely homozygous isogenic diploid strains were obtained after transformation of the haploid wild type strain with a YEpHO-URA3 plasmid carrying the URA3 selectable marker and HO-endonuclease gene under control of its own promoter. HO-endonuclease causes mating type switching within the population of transformed cells that allows mating and diploid formation. Single colony isolates of diploids that had lost the HO plasmid were taken. Isogenic diploids homozygous for pol3-L523H/pol3-L523H were created by crossing two isogenic isolates of MATa pol3-L523H and MATα pol3-L523H. Diploids with single and multiple heterozygosities for deletion alleles were obtained by transforming homozygous wild type diploids with deletion cassettes carrying selectable markers. The RAD27 gene was replaced with the RAD27: URA3-Blast cassette (31). Other deletions were obtained by inserting antibiotic-resistant cassettes (32). Multiple deletions within the same heterozygous diploid strain were introduced separately by successive transformations. Isogenic diploid strains carrying the triple heterozygous mutations POL3/pol3-5DV RAD27/rad27Δ POL32/pol32Δ were obtained by crossing isogenic isolates of the MATα pol3-5DV (note that pol3-5DV was also referred as pol3-D520V in Ref. 18) with isogenic isolates of the MATa rad27Δ pol32Δ genotype. All crosses and sporulations were carried out at least in duplicate with independent isolates of the relevant strains.
Tetrad Analysis—Effects of eliminating Pol32 or Pif1 on the viability of single and multiple mutants presumably defective in Okazaki maturation were studied in the immediate meiotic progeny of heterozygous diploids. This allowed us to minimize effects due to the accumulation of suppressor mutations that could be selected if multiple mutants were obtained by consecutive transformations of a haploid strain in order to disrupt the genes of interest. Asci of double or triple heterozygous diploids were dissected on YPDA plates and incubated at 23 or 30 °C. In order to allow spores carrying slow growing genotypes to form colonies, plates containing very small colonies were incubated up to 7–10 days. Colonies were then replica-plated onto both YPDA and diagnostic media to identify mating type and gene deletions by the presence of markers. YPDA plates were incubated at 23, 30, and 37 °C to identify possible temperature sensitivities of single and multiple mutants. The presence of the pol3-5DV mutation was determined by diagnostic restriction digestion of a PCR product. We explored genetic interactions throughout this paper in the progeny of multiple heterozygous diploids, where all possible combinations of mutant alleles are represented by colonies that grew from a fresh meiotic progeny.
Replication Proteins—Overexpression and purification of Pol δ and Pol δ-DV (DNA polymerase δ with pol3-5DV mutation) from yeast has been described in detail (33). In order to overproduce Pol δ* and Pol δ-DV*, the overexpression strain contained plasmids pBL335 (GAL-GST-POL3) or pBL335-DV (GAL-GST-pol3-D520V), respectively, together with plasmid pBL338 (GAL-POL31 (16)). After growth, galactose induction, cell lysis, and glutathione affinity chromatography, the preparation was subjected to 3C-protease treatment to remove the glutathione S-transferase purification tag, followed by MonoS chromatography, as described for Pol δ (33). Replication protein A (RPA), PCNA, Replication factor C (RFC), and FEN1 were purified from Escherichia coli overproduction strains (7, 34, 35). All other enzymes were obtained commercially.
DNA Substrates—All oligonucleotides were obtained from Integrated DNA Technologies and purified by polyacrylamide gel electrophoresis or high pressure liquid chromatography before use. They are as follows: Bio-V6, 3′-biotin-T31CCCTTCCCTCTCCCTCCTCTTCTTCCCTCT25CCAAGGTGGTTTGTTTTGGTTGGGTTGA-biotin-5′; C12, 5′-AGGGAAGGGAGAGGGAGGAGAAGAAGGGAG; C6, 5′-GGTTCCACCAAACAAAACCAACCCAAC; RC6, 5′-GGUUCCACCAAACAAAACCAACCCAAC (ribonucleotides underlined).
The 113-nt 5′ and 3′ biotinylated template Bio-V6 was prepared by hybridizing two half-oligonucleotides to a bridging primer followed by ligation with T4 DNA ligase and purification by preparative urea-PAGE (see Fig. 3A). For strand displacement assays, the primer oligonucleotide C12 was 5′-32P-labeled, and 5 pmol of 32P-C12 was hybridized to 7.5 pmol of Bio-V6 and 15 pmol of pC6 or pRC6 (made by phosphorylation of C6 and RC6 with cold ATP). The oligonucleotides were hybridized to the template at 70 °C. For nick translation assays, either primer C6 or RC6 was 5′-32P-labeled, and 5 pmol of the labeled oligonucleotide was hybridized to 7.5 pmol of Bio-V6 together with 15 pmol of C12. For idling assays, 5 pmol of Bio-V6 was hybridized with 7.5 pmol of C12 and 7.5 pmol of pC6. After hybridization, streptavidin was added in a 2-fold molar excess to all template-primer substrates.
FIGURE 3.
Strand displacement synthesis by four forms of Pol δ. A, schematic of the substrate. The 113-mer template is Streptavidin-coated Bio-V6. PRI, the 5′-32P-labeled 30-mer primer; BLO, the 5′-phosphorylated 27-mer blocking oligonucleotide. B, in the standard assay (see “Experimental Procedures”), PCNA was preloaded by RFC onto the RPA-coated DNA, the reaction was started by the addition of a 1.4-fold molar excess over DNA of the indicated polymerase, and the assay was allowed to proceed at 30 °C for the indicated times and analyzed by 7 m urea, 12% PAGE, followed by phosphorimaging of the dried gel. Full-length strand displacement products should be 83 nt in length, but 1–2-nt shorter products were mainly observed, because the biotin-streptavidin block inhibited progression of the polymerase. The nick position (55 nt) is indicated (C). Assays were repeated with increasing polymerase for 5 min, and the full-length products (81–83 nt) were quantified.
Strand Displacement and Nick Translation Assays—In all assays in this study, the DNA concentration was 7 nm. Standard 20-μl assays contained 20 mm Tris-HCl, pH 7.8, 1 mm dithiothreitol, 100 μg/ml bovine serum albumin, 8 mm MgAc2, 0.5 mm ATP, 110 mm NaCl, 100 μm each dNTP, 140 fmol of DNA substrate (see above and in the figures), 1 pmol of RPA, 200 fmol of RFC, and 400 fmol of PCNA. The DNA was preincubated with RPA, PCNA, and RFC for 1 min at 30 °C, and the reaction was started by adding the relevant DNA polymerase at the indicated concentrations. Added NaCl was adjusted such that the final concentration, including additions from enzyme storage buffers, was 110 mm. In nick translation assays, FEN1 (200 fmol) was added together with the indicated polymerase. Aliquots were quenched in 20 mm EDTA, 0.4% SDS (final) and heated at 50 °C for 15 min. After the addition of 95% formamide to a 60% final concentration, the samples were heated at 95 °C for 2 min and analyzed on a 12% (strand displacement) or 20% (nick translation) denaturing polyacrylamide gel. The gels were dried and subjected to PhosphorImager analysis. In order to allow accurate quantitation of weak signals, the gels were exposed for several hours and for 2 days. PhosphorImager file intensities were correlated by quantitation of intermediate signals that were in the linear response range for both exposures. Quantification was carried out using ImageQuant software (GE Healthcare). The images in the figures were contrast-enhanced for visualization purposes.
Idling Assays—The above assay was scaled up to 50 μl. It contained unlabeled DNA, 10 μm [α-32P]dGTP, and a 100 μm concentration of the other dNTPs and reaction conditions and enzymes as indicated above and in the legend to Fig. 4. Aliquots of 10 μl were taken after various times, from 1 to 7 min, quenched by the addition of 5 μl of stop buffer (25 mm EDTA, 1% SDS, and a 5 mm concentration each of the relevant nonradioactive dNTP and dNMP). Reaction products were analyzed by thin layer chromatography on polyethyleneimine/cellulose in 0.5 m LiCl/HCOOH, followed by PhosphorImager analysis. Idling rates were calculated from the linear slopes of time courses.
FIGURE 4.
Pol δ* shows inefficient capacity for idling. The inset shows the sequence context of the DNA substrate from Fig. 3A. Idling assays were carried out at a constant DNA concentration of 7 nm and increasing DNA polymerase concentrations, as described under “Experimental Procedures.” PRI, the 5′-32P-labeled 30-mer primer; BLO, the 5′-phosphorylated 27-mer blocking oligonucleotide.
RESULTS
Previous genetic studies have established several networks of interactive factors involved in Okazaki fragment maturation. One network links the nuclease activities of FEN1RAD27 and Dna2, showing that these factors have compensatory activities. Overexpression of DNA2 suppresses the temperature sensitivity of rad27Δ, and overexpression of RAD27 suppresses the temperature sensitivity of a dna2-1 mutant (36). A second network links Dna2 with the helicase Pif1 and the Pol32 subunit of Pol δ. Pif1Δ suppresses the lethality of the dna2Δ mutant, and the additional deletion of POL32 improves growth in the triple mutant (25). A third genetic network includes the core activities for Okazaki maturation. It centers around the nuclease activities of Pol δ and FEN1RAD27, revealing a synthetic lethality or showing a signature hypermutability of small duplications when even mild mutations in POL3 and RAD27 are combined (18). In order to gain additional insights into this core network, we have made a series of multiple mutants that probe the interactions and interdependencies of these networks. We analyzed a large number of spores in the progeny of multiple heterozygous diploids to identify all possible viable single and multiple mutant genotypes based on their associated selection markers or by PCR/restriction digest genotyping. This approach reduces the propagation of potentially unstable mutants to a practically achievable minimum and therefore limits the possibility of generating suppressor mutations of poorly growing mutants. This important technical imposition allowed us to draw conclusions about genetic interactions that are not affected by the generation of suppressor mutants during prolonged growth.
We based our experimental design on the use of two genetic defects (pol32Δ and pif1Δ) that may lead to reduced flap formation via different mechanisms. Pol32 deletion may reduce the capacity of Pol δ for strand displacement on the border between Okazaki fragments, whereas pif1 deletion can eliminate long flaps that may be formed by Pif1 helicase action on Okazaki fragments (26). Asci of double or triple heterozygous diploids were dissected on YPDA plates and incubated at 23 or 30 °C. Different incubation temperatures were used because of previously reported effects of pol32Δ and pif1Δ on the cold or temperature sensitivity of yeast (10, 25).
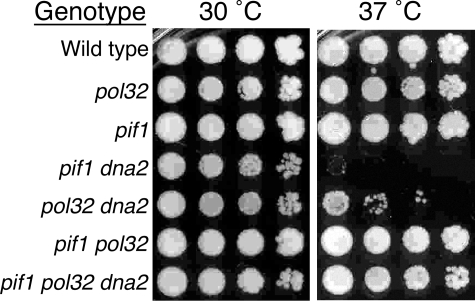
dna2Δ Lethality Is More Efficiently Suppressed by Deletion of PIF1 Than by Deletion of POL32—We dissected the triple heterozygous DNA2/dna2Δ PIF1/pif1Δ POL32/pol32Δ diploids and analyzed a large number of spore colonies for the presence of all possible genotypes and their associated phenotypes (Table 1). As expected, no dna2Δ single mutant could be recovered. Also, as expected from previous studies (25), we isolated viable dna2Δ pif1Δ, pif1Δ pol32Δ, and dna2Δ pif1Δ pol32Δ mutants at approximately equal frequencies. However, dna2Δ pol32Δ double mutants were recovered at a 10-fold lower frequency. Thus, rescue of dna2Δ lethality by pol32Δ occurred much less frequently than that by pif1Δ and possibly relied upon the accumulation of suppressors. Replica plating of all spore colonies listed in Table 1 to three different temperatures, 23, 30, and 37 °C showed that, similar to Ref. 25, the dna2Δ pif1Δ double mutants were temperature-sensitive at 37 °C, whereas dna2Δ pif1Δ pol32Δ triple mutants were capable of growing at 37 °C. This conclusion was verified in a more accurate growth test (Fig. 2) (data not shown). Rare (only three) dna2Δ pol32Δ double mutants showed different degrees of temperature sensitivity (Fig. 2) (data not shown), possibly caused by various suppressor mutations. The temperature-dependent effect of pol32Δ is in agreement with observations that strand displacement synthesis by Pol δ can be greatly reduced at lower temperatures that increase DNA-DNA and RNA-DNA hybrid stability (7). According to our current model, these results indicate that deletion of POL32 may further reduce the generation of long flaps, which are still created in the absence of Pif1-helicase at high temperatures and which require Dna2 for resolution.
TABLE 1.
Effect of POL32 and PIF1 deletions on the viability of dna2Δ
|
DNA2 allele
|
Number of spores with a
genotypea
|
Totalb
|
|||
|---|---|---|---|---|---|
| PIF1+POL32+ | pif1Δ POL32+ | pif1Δ pol32Δ | PIF1+pol32Δ | ||
| DNA2+ | 38 | 56 | 43 | 64 | 201 |
| dna2Δ | 0 | 57 | 43 | 3c | 104 |
Presented are numbers of each genotype within the haploid progeny of two isogenic diploids of the genotype: DNA2+/dna2Δ PIF1+/pif1Δ POL32+/pol32Δ
Plates after dissection were incubated at 23 and 30 °C. The haploid progeny grew equally well at either temperature, so the numbers were combined in this table
The genotype of three rare dna2Δ pol32Δ PIF1+ segregants was confirmed by PCR
FIGURE 2.
Temperature sensitivity of yeast strains containing the dna2Δ allele. 10-Fold serial dilutions of yeast cell suspensions were spotted onto complete YPDA plates and incubated for 4 days at the indicated temperature. Shown are the representative growth results for one of three independent dna2Δ pol32Δ double mutants tested and for four of the other six viable genotypes tested.
Deletion of POL32 or PIF1 Can Suppress Lethality of rad27Δ pol3-exo Double Mutants—Our hypothesis that the balance between 5′-displacement by Pol δ and 3′- and 5′-degradation by several nucleases determines the efficiency of Okazaki fragment ligation has an important prediction; the lethality caused by the absence of the core pair of Okazaki maturation nucleases activities, Pol δ Exo and FEN1RAD27, may be suppressed by changing the strand displacement parameters. We analyzed the progeny of multiple heterozygous diploids carrying two different pol3-exo mutants (Table 2 and supplemental Tables 1–3). pol3-5DV (D520V) completely lacks the proofreading exonuclease activity of Pol δ, whereas pol3-L523H has a fully active exonuclease activity but is partially defective for switching of the DNA substrate between the Pol and the Exo domain. Each of the pol3 mutations results in lethality when combined with rad27Δ (5, 18). In support of our previous results, sporulation of strains homozygous for pol3-L523H/pol3-L523H and heterozygous for RAD27/rad27Δ yielded only two spurious rad27Δ pol3-L523H colonies of 416 spores examined, which are likely to contain a suppressor. On the other hand, POL32 deletion efficiently suppressed the lethality of the rad27Δ pol3-L523H double mutant at both temperatures of spore germination. Similarly, sporulation of the triple heterozygous mutant POL32/pol32Δ RAD27/rad27Δ POL3/pol3-DV yielded no rad27Δ pol3-DV double mutants but the expected number of pol32Δ rad27Δ pol3-DV triple mutants.
TABLE 2.
Effect of POL32 and PIF1 deletions on the viability of rad27Δ exo1Δ and rad27Δ pol3-exo double mutants
|
Double mutant genotype
|
POL32 or PIF1 allele
|
Number of spores with desired genotype (total viable
spores)a
|
|
|---|---|---|---|
| Germinated at 23 °C | Germinated at 30 °C | ||
| rad27Δ pol3-L523Hb | POL32+ | 1 (112) | 0 (44) |
| pol32Δ | 27 (112) | 13 (44) | |
| PIF1+ | 1 (173) | 0 (87) | |
| pif1Δ | 41 (173) | 0 (87) | |
| rad27Δ pol3-5DV | POL32+ | 0 (36)c | NTd |
| pol32Δ | 15 (37)e | NT | |
| PIF1+ | 0 (18)f | NT | |
| pif1Δ | 0 (18)g | NT | |
| rad27Δ exo1hΔ | POL32+ | 13 (92) | 35 (223) |
| pol32Δ | 0 (92) | 0 (223) | |
| PIF1+ | 38 (212) | 34 (232) | |
| pif1Δ | 3 (212) | 30 (232) | |
Numbers of each genotype observed among meiotic progeny of each analyzed isogenic independent diploids at two different temperatures are presented in supplemental Tables 1–5
Obtained from dissections of two isogenic pol3-L523H/pol3-L523H RAD27/rad27Δ POL32/pol32Δ diploids and three isogenic pol3-L523H/pol3-L523H RAD27/rad27Δ PIF1/pol32Δ diploids, listed in supplemental Tables 1 and 2, respectively
Number of rad27Δ POL32 spore colonies picked from meiotic progeny of four isogenic POL3/pol3-5DV RAD27/rad27Δ POL32/pol32Δ diploids and genotyped for the presence of the pol3-5DV mutation; all analyzed spores are listed in supplemental Table 3
NT, --------------------
Number of rad27Δ pol32Δ spore colonies picked from meiotic progeny of four isogenic POL3/pol3-5DV RAD27/Δrad27 POL32/pol32Δ diploids and genotyped for the presence of the pol3-5DV mutation; all analyzed spores are listed in supplemental Table 3
Number of rad27Δ PIF1 spore colonies picked from meiotic progeny of five isogenic POL3/pol3-5DV RAD27/rad27Δ PIF1/pif1Δ diploids and genotyped for the presence of the pol3-5DV mutation
Number of rad27Δ pif1Δ spore colonies picked from meiotic progeny of five isogenic POL3/pol3-5DV RAD27/Δrad27 PIF1/pif1Δ diploids and genotyped for the presence of the pol3-5DV mutation
Obtained from dissections of five isogenic RAD27/rad27Δ EXO1/exo1Δ POL32/pol32Δ and three isogenic RAD27/rad27Δ EXO1/exo1Δ PIF1/pif1Δ diploids generated by independent crosses, listed in supplemental Tables 4 and 5, respectively
PIF1 deletion was also capable of suppressing the lethality of the rad27Δ pol3-L523H double mutant but only when spores were germinated at 23 °C. Elimination of Pif1 did not rescue the lethality of rad27Δ pol3-5DV even at 23 °C. These data are consistent with a model where reduction of strand displacement synthesis caused by pol32Δ or pif1Δ mutations counteracts the increased strand displacement synthesis in the pol3-DV or pol3-L523H mutant, thereby providing the opportunity for a backup flap exonuclease, such as Exo1, to act in the rad27Δ strain. As discussed above, the temperature dependence of suppression by pif1Δ can be explained by the overall decrease in strand displacement synthesis at lower temperatures.
Deletions of POL32 and PIF1 Cause Lethality of rad27Δ exo1Δ Double Mutants—Next, we investigated the effect of POL32 and PIF1 deletions on the viability of rad27Δ single and rad27Δ exo1Δ double mutants. Exo1 is a 5′-exonuclease acting on double-stranded DNA, and it has an associated 5′-flap endonuclease activity (28). It has been proposed to substitute for FEN1RAD27 during Okazaki fragment maturation in the rad27Δ mutant, because rad27Δ exo1Δ double mutants display either synthetic lethality or slow growth defects (37). We isolated rad27Δ exo1Δ double mutants at the expected frequency, but the spore isolates grew slowly (Table 2) (growth data not shown). Contrary to data from mass genomic synthetic lethality screening (38), we recovered pol32Δ rad27Δ double mutants at the expected frequency (supplemental Tables 4 and 5). Importantly, at each temperature of spore germination, we were unable to recover any pol32Δ rad27Δ exo1Δ triple mutants from the spore progeny of triple heterozygous diploids, indicating that the POL32 deletion eliminated the growth of yeast lacking both FEN1RAD27 and Exo1 (Table 2). Similarly, the pif1Δ rad27Δ exo1Δ triple mutants were mostly nonviable (only three triple mutants were recovered as compared with 38 rad27Δ exo1Δ double mutants) but only when spores were germinated at 23 °C; at 30 °C, the expected number of triple mutants pif1Δ rad27Δ exo1Δ could be recovered. One rational explanation of this result is that in the absence of both FEN1RAD27 and Exo1, maturation of Okazaki fragments proceeds more prominently through the generation of long flaps that are substrates for Dna2 (39). In this model, deletion of POL32 or PIF1 would limit the generation of long flaps, thereby reducing the formation of substrates available for Dna2-mediated processing of the RNA primer, with associated lethality. The cold sensitivity of pif1Δ rad27Δ exo1Δ triple mutant can be explained by an overall reduction in strand displacement synthesis at lower temperature that is further compromised in the PIF1 deletion.
Altogether, our observed genetic interactions indicate that the change in strand displacement by either pol32Δ or pif1Δ can change the requirement in the competing 3′- and 5′-nuclease activities. PIF1 deletion acted as a stronger suppressor compared with POL32 deletion in suppressing dna2 defect. Importantly, the effects of pol32Δ were always stronger than of pif1Δ when the defect in the key Okazaki maturation activity FEN1RAD27 was involved, suggesting that Pol32 participates in the bulk of Okazaki maturation. Therefore, we undertook a biochemical study to test the role of Pol32 in strand displacement synthesis and to characterize the enzymatic cooperation between FEN1 and Pol δ during Okazaki fragment maturation. The biochemical role of Pif1 in the generation of long flaps has recently been addressed elsewhere (26).
The Generation of Long Flaps by Pol δ Requires Its Pol32 Subunit—The model emerging from the genetic studies predicts that the Pol32 subunit is required for the generation of long flaps during the process of strand displacement synthesis that is initiated when the gap filling polymerase reaches the RNA primer of the previous Okazaki fragment. However, so far, biochemical support for this hypothesis has been lacking. Our previous biochemical studies comparing the three-subunit Pol δ (consisting of Pol3, Pol31, and Pol32) with the two-subunit Pol δ* lacking Pol32 indicate that Pol δ* shows a decreased interaction with PCNA and a decreased processivity of DNA synthesis (12, 16). Based upon these results, it is reasonable to assume that the processivity of strand displacement by Pol δ*is also decreased. However, strand displacement by Pol δ is counteracted by 3′-degradation by the Exo domain of the enzyme, and it is not clear how the latter activity is affected by lack of Pol32. Therefore, we prepared both wild-type Pol δ* and the exonuclease-defective form Pol δ-DV* (containing the pol3-D520V mutation) from yeast overproduction systems. Both enzymes were >98% pure and lacked detectable Pol32 by electrophoretic analysis (data not shown). In order to determine how strand displacement synthesis is affected by Pol32, we first compared Pol δ-DV with Pol δ-DV*. Second, to gain an understanding of how Pol32 affects the enzyme's capacity to switch to its Exo domain and degrade the invading DNA strand, we compared the strand displacement and idling properties of Pol δ with Pol δ*. Idling, the recurring switching between polymerase and exonuclease domains maintains the polymerase at a nick position (Fig. 1) and is inoperative in the exonuclease-deficient Pol δ-DV or Pol δ-DV*.
In our kinetic studies of strand displacement synthesis and nick translation, we have used an oligonucleotide-based system (Fig. 3A). The 113-nt-long oligonucleotide template is primed with a 30-mer primer (PRI) and with a downstream 27-mer blocking oligonucleotide (BLO). In some experiments, the eight 5′-nucleotides of the blocking primer were ribonucleotides, thereby mimicking the structure of an Okazaki fragment (see Fig. 5A). The two oligonucleotides are separated by 25 template dT residues, and an additional 31 dT residues precede the 30-mer primer position. The single-stranded binding protein RPA was bound to both oligo(dT) stretches. Subsequently, PCNA was loaded by RFC in the presence of ATP. Terminal biotin-streptavidin anchors on either side of the template strand prevented PCNA from sliding off of the DNA (7). Upon the addition of one of the four forms of Pol δ that we have investigated, the enzyme incorporated 25 dATPs in a monotonic fashion prior to encountering the blocking oligonucleotide and then initiated strand displacement synthesis. Previous studies have shown that the rate and extent of strand displacement synthesis by Pol δ is dependent on three factors that all relate to the stability of double-stranded DNA. Strand displacement is enhanced when the salt concentration in the assays is reduced, when the temperature is increased, and when the A-T content of the DNA is increased (7). In this paper, our studies were performed at nearly physiological concentrations of 110 mm NaCl, at 30 °C, and with an oligonucleotide templateprimer system where the G-C content of the blocking oligonucleotide (BLO) primer to be displaced is 48% (Fig. 3A).
FIGURE 5.
Processivity of strand displacement synthesis by different forms of Pol δ. Assays were identical to those described under “Experimental Procedures” and in the legend to Fig. 3, except that the DNA polymerase concentration was reduced to 1.4 nm, and aliquots were taken after 15, 30, and 60 s. A, 7 m urea, 12% PAGE analysis of replication with a DNA blocking oligonucleotide (left) or an RNA-DNA blocking oligonucleotide (right). Alternate lanes are marked. B, normalized band intensity of the region between 85 nt and 53 nt, with the total intensity set at 100 for each lane. The migration of various length products is indicated (nick position = 55 nt). Shown are the profiles for the 15, 30, and 60 s time points with Pol δ-DV on the substrate with the DNA blocking oligonucleotide (lanes 7–9). C, normalized band intensity profiles for the 15-s reactions on the substrate with the DNA blocking oligonucleotide with Pol δ (lane 1) and Pol δ-DV (lane 7) and the 30-s reactions with Pol δ*(lane 5) and Pol δ-DV* (lane 11). D, normalized band intensity profiles for the 15-s reactions with Pol δ (lanes 1 and 13) and Pol δ-DV (lanes 7 and 19), on either the DNA or RNA-DNA blocking oligonucleotide. wt, wild type.
At a polymerase/DNA ratio of 1.4, complete strand displacement synthesis, as measured by the generation of extension products ∼82 nt in length, was readily observed with Pol δ-DV, whereas the wild-type enzyme only showed detectable products after 5 min of incubation (Fig. 3B). Very little complete strand displacement was observed with the two-subunit Pol δ-DV*, and none was observed with Pol δ*. We reasoned that poor strand displacement by the enzymes lacking Pol32 could be due to the inherent instability of the DNA·PCNA·Pol δ* complex. If so, increased enzyme concentrations would be predicted to result in increased strand displacement. Strand displacement synthesis by Pol δ and by Pol δ-DV was largely independent of the concentration of each three-subunit polymerase, provided that the enzyme concentration was in excess of the DNA substrate. This indicates that the three-subunit enzyme forms a stable, processive complex with PCNA on the DNA (Fig. 3C). However, when the same experiments were carried out with Pol δ* and with Pol δ-DV*, two important differences were observed. First, saturation of the response required a large excess of enzyme, confirming our hypothesis that the two-subunit enzyme was less stably bound to the DNA. Second, both gap filling synthesis and strand displacement synthesis could be stimulated by including high levels of the enzyme (Fig. 3C). The dissociative behavior of Pol δ* could not be suppressed by the addition of more PCNA, since PCNA remained stably bound to the DNA (data not shown). In addition, even at the highest polymerase concentrations, full strand displacement synthesis did not proceed as efficiently with the two forms of Pol δ* as with the analogous enzymes containing Pol32 (Fig. 3C). Thus, the presence of Pol32 stimulates the formation of long 5′-flaps.
Pol δ* Idles Poorly at a Nick—Successive cycles of 5′-strand invasion by the Pol activity followed by 3′-degradation by the Exo activity results in overall nucleotide turnover (dNTP → dNMP) at the nick, a process called idling. In a previous study, we showed that idling by Pol δ is mainly confined to the initial two nucleotides incorporated during strand invasion of the blocking oligonucleotide (20). In the sequence context of the oligonucleotide system used here, idling would lead to dGTP → dGMP turnover (Fig. 4, inset). Turnover was measured by the inclusion of [α-32P]dGTP in the assay, and the rate of [32P]dGMP formed was measured by thin layer chromatography. The basic idling characteristics of Pol δ* and Pol δ were very similar (supplemental Table 6): (i) dGTP turnover required the presence of both the primer and the blocking oligonucleotide, indicating that idling results from the replicating polymerase synthesizing into downstream double-stranded DNA; (ii) turnover of those dNTPs corresponding to the nucleotides just prior to the nick position was very inefficient, indicating that the switch from the Pol to the Exo domain is mediated only after strand invasion by Pol δ or Pol δ*; (iii) turnover was suppressed in the presence of FEN1, suggesting that nick translation by the coupled action of the polymerase and FEN1 is the dominant process.
Turnover rates by Pol δ increased with increasing enzyme concentration, but this rate saturated when Pol δ was in excess of DNA, indicating that Pol δ remained stably associated with the DNA. (Fig. 4). In contrast, turnover by Pol δ* only saturated at a very high excess of enzyme over DNA, and even at saturating enzyme levels, turnover by Pol δ* was only ∼40% that of wild type. These data indicate that Pol δ* dissociates readily when the enzyme reaches the nick position. However, in addition, Pol δ* may carry out more limited strand invasion than Pol δ and/or dissociate from the invading primer terminus rather than switch to its exonuclease domain so as to commence 3′-degradation. In order to understand how the presence of Pol32 contributes to strand invasion, thereby providing a substrate for FEN1 during Okazaki fragment maturation, it is important to determine how strand invasion is accomplished with or without Pol32 during a single processive cycle of polymerase action. Therefore, we studied this process with the exonuclease-deficient enzymes Pol δ-DV and Pol δ-DV*.
Pol32 Stimulates Processive Strand Displacement—Processivity assays were carried out under conditions where the substrate DNA (7 nm) was in 5-fold molar excess over polymerase (1.4 nm). Under those conditions, the polymerase, once dissociated, would be expected to bind a new primer with a higher probability than rebind to the already replicated DNA molecule from which it had dissociated. Product accumulation and the ratio of products formed was followed as a function of time. We also carried out a processivity assay with an even lower concentration of polymerase (0.7 nm) (data not shown). A similar distribution of strand displacement products was observed for Pol δ and for Pol δ-DV when compared with the experiment at 1.4 nm polymerase. However, the extent of DNA synthesis by Pol δ* was too low to allow accurate analysis of displacement products. Therefore, we focus our analysis on the experiments with 1.4 nm polymerase. The analysis will first be discussed for Pol δ-DV, since strand displacement products can be most easily visualized for this enzyme (Fig. 5A).
The distribution of replication products was quantified by dividing the region from just above the 83-nt (full displacement) position to just below the 55-nt (nick) position in 650 equally sized intervals and calculating the normalized intensity in each interval, where the sum of all 650 densities was set to 100 for each lane. In this way, the ratios of the various size products could be compared between different time point samples, although the absolute amounts of products increased with time. This analysis is plotted for Pol δ-DV in Fig. 4B. Interestingly, the normalized product distribution from 55 to ∼65 nt in length, reflecting strand displacement up to 10 nt, was comparable in intensity for the 15, 30, and 60 s time points, indicating that this distribution was the result of a single round of processive strand displacement synthesis accompanied by specific dissociation probabilities at each length. In contrast, the normalized distribution for longer length products, especially those 66–70 nt in length and the full length 80–82 nt displacement products, increased with time, suggesting that they arose, at least in part, from multiple binding and elongation events to the same DNA molecule. Another possible mechanism that may contribute to the observed product distribution is that during the process of strand displacement synthesis, some complexes pause for an extended time and then restart DNA synthesis, thereby enlarging the flap. However, the observation that the total percentage of extended primers increases with incubation times, from 29% after 15 s to 73% after 60 s of incubation, indicates that polymerase molecules recycle. Therefore, polymerase rebinding rather than pausing appears to be the major mechanism by which longer flaps are formed.
We observed no accumulation of ∼72–81-nt-long products consistent with the expected instability of very short DNA hybrids; once strand displacement synthesis had proceeded that far, the instability of the short remaining hybrid presumably led to rapid dissociation of the blocking primer and rapid replication of the remaining template to give full-length products. Our data indicate that processive displacement products up to ∼10 nucleotides in length were synthesized by Pol δ-DV under our experimental conditions.
When this analysis was carried out with the two-subunit Pol δ-DV* under the same conditions, at a 5-fold excess of DNA over polymerase, the accumulation of all replication products was substantially decreased in keeping with the lower stability of DNA·PCNA·Pol δ-DV* complexes. Because of the high noise/signal ratio, the 15 s time point was not further analyzed. However, the normalized distributions of products after 30 and 60 s were very similar, showing a marked decrease in replication products above 57 nt in length when compared with Pol δ-DV (Fig. 5C). These data are consistent with our hypothesis that the Pol32 subunit increases the processivity of strand displacement synthesis.
Surprisingly, however, when we carried out the analogous experiment with Exo-proficient Pol δ and Pol δ*, the normalized strand displacement profiles for these two enzymes were almost superimposable, although less total synthesis was observed with Pol δ* than with Pol δ (Fig. 5C). Very few products over 60 nt in length, indicative of greater than 5 nt of strand displacement, accumulated for either enzyme. Based upon the results observed with the exonuclease-deficient enzymes, one might have expected that the normalized distribution of displacement products made by Pol δ* would be skewed toward the nick position compared with that for Pol δ. The most likely explanation for these observations is based on the reduced idling capacity of Pol δ* (Fig. 4). Although strand displacement synthesis by Pol δ* is probably less efficient than that by Pol δ, as suggested from the comparison between Pol δ-DV* and Pol δ-DV (Fig. 5C), degradation of the invading strand by the exonuclease activity of Pol δ* is also diminished. Therefore, under our experimental conditions, the defects shown by Pol δ*in both forward and reverse movements largely cancel each other out. Significantly, however, the displacement of longer strands accumulate over time with Pol δ but not with Pol δ* (Figs. 3B and 5A).
Pol δ Shows Increased Displacement of RNA—We also carried out this processivity analysis with a substrate that more accurately mimicked that encountered during Okazaki fragment maturation. The 5′-end of the blocking primer now contained eight ribonucleotides, whereas both total length and sequence remained the same (Fig. 5A). The most surprising result from this analysis was that all four forms of Pol δ carried out much more efficient strand displacement synthesis through the RNA-DNA hybrid than through the DNA-DNA hybrid (shown for Pol δ and Pol δ-DV in Fig. 5D). According to a nearest neighbor analysis, the RNA-DNA hybrid used in our study has the same stability as the analogous DNA-DNA hybrid (40). This suggests that Pol δ has an increased capacity to displace RNA primers, perhaps in keeping with its critical role in lagging strand DNA replication. In a quantitative comparison of RNA/DNA versus DNA/DNA substrates, all four enzymes showed increased strand displacement synthesis on the RNA/DNA substrate. Processive products by Pol δ-DV of 65–70 nt in length, resulting from 10–15 nt of strand displacement, were much more abundant on the RNA-DNA than on the DNA-DNA substrate, 23% versus 8% (Fig. 5D). Pol δ-DV* yielded 10–15-nt displacement products to the extent of 6% on the RNA primer and 4% on the DNA primer (Fig. 5A; data not plotted). Wild-type Pol δ and Pol δ* also accumulated significantly more 5–9-nt strand displacement products on the RNA primer (6.6 and 5.4%, respectively) than on the DNA primer (2.8 and 3.2%, respectively). Thus, the 3′-exonuclease activity also restrained invasion of the RNA primer to give a distribution of normalized strand invasion products that is very similar for Pol δ and Pol δ*. Importantly, wild-type Pol δ produced significant full-length extension products on the RNA-DNA substrate (0.2% full-length product after 15 s and 0.6% after 60 s), whereas Pol δ* produced none.
FEN1 Action Restrains Strand Displacement by Pol δ—In one possible model for RNA primer degradation during Okazaki fragment maturation, one could envision that after one cycle of processive strand invasion by the polymerase, the resulting flap is cut by FEN1. The simple prediction from this model is that the quantitative length distribution of products cut by FEN1 would be the same as that generated during processive strand displacement. Alternatively, if the polymerase were allowed to go through several cycles of strand displacement synthesis prior to entry of FEN1, longer products would be made by FEN1 than predicted from the processive strand displacement results. Finally, the possibility exists that the presence of FEN1 in the maturation complex limits processive strand displacement, resulting in shorter products than predicted from strand displacement synthesis. The data presented below suggest that the last model prevails.
In the maturation assay, the blocking oligonucleotide (DNA27 or RNA8DNA19) was 5′-labeled to allow detection of FEN1-dependent degradation products made specifically when the maturation complex reaches the nick position (Fig. 6A). After loading PCNA onto the substrate, the assay was started by the addition of Pol δ together with FEN1. Products were analyzed on a 20% polyacrylamide, 7 m urea gel. Three main general conclusions could be drawn from these studies. First, for each form of Pol δ, the relative distribution of cleavage products, between mononucleotides and oligonucleotides, was independent of the DNA polymerase concentration (Fig. 6, B and C). Second, the relative distribution of cleavage products was also independent of the incubation time, generally between 30 s and 3 min (data not shown). Control assays containing polymerase and no FEN1 showed no cleavage products (Fig. 6B, lanes 2, 3, 9, and 10). Control assays containing FEN1 and no polymerase showed ∼5% cleavage after 30 s and 20% after 3 min. Because this nonspecific background cleavage by FEN1 increased with time and complicated the analysis, incubation times were generally held to 30 s. Background cleavage of the RNA primer by FEN1 was as high as 10% after 30 s (supplemental Fig. 1). Since this background needs to be subtracted from the bona fide signal, S.E. values with the RNA primer were higher than with the DNA primer (Fig. 6D). Nevertheless, it is very evident that maturation with the RNA substrate yielded more long cleavage products than with the DNA substrate; cleavage products in the 6–12 nt range were about 2–3 times as abundant. Third, the product length distribution qualitatively matched the strand displacement capacity of the polymerase; Pol δ-DV, which has the highest strand displacement capacity, also generated the highest proportion of long FEN1 cleavage products (Fig. 6D). However, in quantitative terms, there was no match; long cleavage products were more sparse than might have been suggested from the distribution of products made by processive strand displacement. A comparison is shown in Fig. 7 for the case of 6–12-nt products on the RNA substrate. The analysis for Pol δ shows that 5% of the processive replication products had displaced the RNA primer by 6–12 nt. However, in the presence of FEN1, only 0.9% of the cleavage products were in the 6–12 nt size range. Similarly, Pol δ* also showed 5% processive displacement product in the 6–12 nt range, but in the presence of FEN1, only 0.3% of cleavage products in that range were produced. An about 20-fold disparity was observed for the two exonuclease-deficient enzymes (Fig. 7). For the shorter products, 2–5 nt in length, the disparity was still substantial, RNA strand invasion products being 1.5–5-fold more abundant than the cleavage products actually produced. With all four enzymes, the disparity was just as striking for the substrate with the DNA blocking oligonucleotide. Although the processive strand displacement assays showed that 2% of the products made by Pol δ or by Pol δ* were in the 6–12 nt displacement range, in the presence of FEN1, only 0.2% of products produced with Pol δ and 0.03% of the products produced with Pol δ* were 6–12 nt in size.
FIGURE 6.
Coupling between Pol δ and FEN1 at a nick. A, DNA substrates. B, nick translation assays, as described under “Experimental Procedures,” were carried out on the substrate with the DNA oligonucleotide, for 30 s at 30 °C, at increasing polymerase concentrations. Assays contained 7 nm DNA substrate; standard levels of RPA, PCNA, and RFC; 10 nm FEN1 when present; and either 14 or 28 nm Pol δ or Pol δ-DV or 14, 45, or 160 nm Pol δ*orPol δ-DV*. C, the region between that of a dinucleotide and a 14-mer from the gel in B was contrast-enhanced. D, an assay like in B under identical reaction conditions was carried out with a 7 nm concentration of either the DNA or RNA-DNA blocking oligonucleotide containing substrate, 14 nm each DNA polymerase, and 10 nm FEN1. Products in the 2–5 nt and the 6–12 nt size range were quantified. Note the ∼12-fold difference in y scale for the 2–5-nt and 6–12-nt products. Polymerase-independent cleavage products by FEN1 alone (lane 12 in B) were subtracted as background. wt, wild type.
FIGURE 7.
FEN1 action restrains strand displacement synthesis by Pol δ. Replication products 61–67 nt in length, corresponding to 6–12 nt of strand displacement synthesis, made during processive synthesis on the substrate with the RNA blocking oligonucleotide from Fig. 5A, were quantified on the left. Polymerization-dependent FEN1 cleavage products 6–12 nt in length made under the exact same assay conditions on the same substrate (from Fig. 6D) are shown on the right. Note the ∼15-fold difference in y scales. wt, wild type.
DISCUSSION
Our previous genetic and biochemical studies of Okazaki fragment maturation (see Introduction) have provided compelling evidence that the critical function of the 3′-Exo activity of Pol δ during this process is to prevent uncontrolled strand displacement synthesis by the polymerase, through idling at the nick (Fig. 1). Pol δ uniquely carries out limited strand invasion synthesis when the downstream RNA-DNA is encountered. Although dissociation of Pol δ when it reaches the nick has also been observed (41), at the rapid time scale of our experiments, dissociation of the enzyme is not a major factor of consideration. When Pol δ carries out strand displacement synthesis, the substrate for FEN1 action is generated on the fly, allowing for rapid and efficient degradation of the RNA portion of an Okazaki fragment. Because DNA ligase I does not catalyze ligation of RNA-DNA nicks, the entire RNA portion of an Okazaki fragment has to be degraded before ligation can occur. This process occurs in multiple steps, each step consisting of strand displacement and hand-off of the flap to FEN1, followed by flap cutting and a hand-off of the nicked substrate back to the polymerase. One cycle generally releases a monoribonucleotide. We consider this recurring hand-off between polymerase and FEN1 (nick translation in Fig. 1) to be the dominant mechanism for Okazaki fragment maturation in wild-type cells. One rationale for taking such tightly controlled minimal steps is that the generation of deleterious long flaps can be prevented. Nevertheless, they do occur, in vitro and in vivo, and this paper has addressed the factors that contribute to the generation of long flaps and their resolution.
In order to understand the formation of long flaps, a closer biochemical examination of the core machinery was required, since several aspects of this mechanism had remained unexplored. The coupling between the polymerase and FEN1 had not been closely investigated. In addition, most previous biochemical studies had focused on model DNA substrates rather than on those containing short RNA segments that are more representative of Okazaki fragments. We determined in this study how each of the kinetic and coupling parameters was affected by the presence of Pol32 and by the presence of the Pol δ Exo activity in vitro and investigated in vivo the genetic interaction of these functions.
Okazaki Maturation Uses an Active Hand-off from Pol δ to FEN1—Despite the comparable stability of RNA-DNA hybrids and DNA-DNA hybrids, Pol δ was more efficient in strand displacement synthesis of an RNA than the analogous DNA hybrid. In particular, displacement over 6 nt in length was substantially enhanced (Fig. 5D). Since RNA flaps are cut by FEN1 with the same efficiency as DNA flaps (42), the 2-fold increase in longer-sized RNA oligonucleotides produced by FEN1 is entirely consistent with an increased displacement of RNA strands (Fig. 6D).
For each DNA polymerase investigated, we noticed a qualitative relationship between the capacity for strand displacement by the polymerase and the sizes of fragments produced when FEN1 is included in the experiment. However, significantly, a quantitative relationship is lacking. Our intent had been to test a passive hand-off model between Pol δ and FEN1 (i.e. products formed during one processive cycle of extension and strand displacement synthesis would become substrates for FEN1 upon polymerase dissociation). According to this model, the distribution of FEN1 cleavage products should quantitatively match the distribution of processive extension products. However, this is not the case, particularly not for long extension products (Fig. 7). FEN1 cleavage products 6–12 nt in size occurred about 20–100-fold less frequently than predicted from the processive extension products formed in the absence of FEN1. Therefore, the presence of FEN1 actively curtailed strand displacement by Pol δ, hence the designation of an active hand-off model. Whether FEN1 does so through direct capture of the DNA substrate from the DNA·Pol δ complex or through allosteric action on Pol δ remains to be established.
Pol32 Is Required for the Production of Long Flaps—Under our experimental conditions, the generation of long flaps (i.e. those that are represented by complete strand displacement synthesis of the 27-mer blocking oligonucleotide) during a cycle of processive strand displacement synthesis is very rare and is only observed to a measurable extent with the exonuclease-deficient Pol δ-DV and with RNA substrates (Fig. 5D). When strand displacement synthesis is allowed to proceed for an extended time, full-length products were observed with wild-type Pol δ but not with Pol δ* lacking Pol32. Interestingly, the distribution of short displacement products by the wild-type enzyme was not affected by the presence of Pol32. As discussed above, we reason that Pol δ* actually does carry out reduced strand displacement synthesis, but it also idles more poorly (Fig. 4). Therefore, the two defects cancel each other out. However, once strand displacement synthesis has proceeded beyond a certain length, an estimated 5–10 nt in length, control by idling appears to be lost. Under those conditions, Pol δ is more efficient than Pol δ* in extending these short flaps into very long flaps.
Flexibility of Okazaki Maturation in Vivo—The results of our synthetic lethality studies are in agreement with the roles proposed for Pol32 and Pif1 in the generation of long 5′-flaps. Based on the current biochemical results, the presence of the Pol32 subunit can lead to the generation of long flaps via excessive strand displacement synthesis by Pol δ. Earlier it was established that Pif1-helicase can lead to long 5′-flap formation through DNA unwinding (26). Elimination of either of these factors restored growth to the pol3-exo- rad27Δ strain (Table 2). Presumably, the elimination of Pol32 or Pif1 reduces the number of long flaps, which would occur at deleterious numbers in the absence of Pol δ Exo and FEN1RAD27, to the amount that can be handled by Dna2 and Exo1.
On the contrary, when both Exo1 and FEN1RAD27 were absent, the functions of Pol32 and Pif1 became essential, indicating the existence of a pathway requiring longer flaps at some stage of Okazaki maturation (e.g. to allow Dna2 cleavage of displaced RNA primers).
In conclusion, our results show a remarkable flexibility of Okazaki maturation machinery, where different pathways are capable of taking the responsibility for creating ligatable nicks. Based on our results, the key parameter determining which pathway prevails is the capacity for strand displacement synthesis by Pol δ on the border between adjacent Okazaki fragments. Pol32 can affect the distribution of maturation intermediates by two mechanisms. Pol32 stimulates the production of long flaps during the bulk of Okazaki fragment maturation. In addition, it is important for the efficient resolution of small 5′-flaps through 3′-degradation of the displacing DNA strand, during idling and in the trimming of small flaps left after Dna2 action (Fig. 1). Although the relevance of the 3′-degradation function is clear, the potential benefits of producing rare long flaps require further investigation.
Supplementary Material
Acknowledgments
We thank John Majors for critical discussions during the course of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM32431 (to P. M. B.) and by the Intramural Research Program of the National Institutes of Health, Project 1 Z01 ES065073, of NIEHS (to J. S., M. A. R., and D. A. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–6 and Fig. 1.
Footnotes
The abbreviations used are: nt, nucleotide(s); Pol δ, DNA polymerase δ; Pol δ-DV, Pol δ with Pol3-D520V; Pol δ* and Pol δ-DV*, two-subunit enzymes lacking Pol32; Pol, polymerase function of Pol δ; Exo, 3′-exonuclease function of Pol δ; RFC, replication factor C; RPA, replication protein A; PCNA, proliferating cell nuclear antigen.
References
- 1.Gordenin, D. A., Kunkel, T. A., and Resnick, M. A. (1997) Nat. Genet. 16 116-118 [DOI] [PubMed] [Google Scholar]
- 2.Liu, Y., Kao, H. I., and Bambara, R. A. (2004) Annu. Rev. Biochem. 73 589-615 [DOI] [PubMed] [Google Scholar]
- 3.Garg, P., and Burgers, P. M. (2005) Cell Cycle 4 221-224 [PubMed] [Google Scholar]
- 4.Tishkoff, D. X., Filosi, N., Gaida, G. M., and Kolodner, R. D. (1997) Cell 88 253-263 [DOI] [PubMed] [Google Scholar]
- 5.Jin, Y. H., Obert, R., Burgers, P. M., Kunkel, T. A., Resnick, M. A., and Gordenin, D. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 5122-5127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maga, G., Villani, G., Tillement, V., Stucki, M., Locatelli, G. A., Frouin, I., Spadari, S., and Hubscher, U. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14298-14303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayyagari, R., Gomes, X. V., Gordenin, D. A., and Burgers, P. M. (2003) J. Biol. Chem. 278 1618-1625 [DOI] [PubMed] [Google Scholar]
- 8.Garg, P., and Burgers, P. (2005) Crit. Rev. Biochem. Mol. Biol. 40 115-128 [DOI] [PubMed] [Google Scholar]
- 9.Tomkinson, A. E., Vijayakumar, S., Pascal, J. M., and Ellenberger, T. (2006) Chem. Rev. 106 687-699 [DOI] [PubMed] [Google Scholar]
- 10.Gerik, K. J., Li, X., Pautz, A., and Burgers, P. M. (1998) J. Biol. Chem. 273 19747-19755 [DOI] [PubMed] [Google Scholar]
- 11.Huang, M. E., Rio, A. G., Galibert, M. D., and Galibert, F. (2002) Genetics 160 1409-1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson, E., Garg, P., and Burgers, P. M. (2004) J. Biol. Chem. 279 1907-1915 [DOI] [PubMed] [Google Scholar]
- 13.Gibbs, P. E., McDonald, J., Woodgate, R., and Lawrence, C. W. (2005) Genetics 169 575-582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lydeard, J. R., Jain, S., Yamaguchi, M., and Haber, J. E. (2007) Nature 448 820-823 [DOI] [PubMed] [Google Scholar]
- 15.Podust, V. N., Chang, L. S., Ott, R., Dianov, G. L., and Fanning, E. (2002) J. Biol. Chem. 277 3894-3901 [DOI] [PubMed] [Google Scholar]
- 16.Burgers, P. M., and Gerik, K. J. (1998) J. Biol. Chem. 273 19756-19762 [DOI] [PubMed] [Google Scholar]
- 17.Huang, M. E., Le Douarin, B., Henry, C., and Galibert, F. (1999) Mol. Gen. Genet. 260 541-550 [DOI] [PubMed] [Google Scholar]
- 18.Jin, Y. H., Garg, P., Stith, C. M., Al Refai, H., Sterling, J., Weston, L., Kunkel, T., Resnick, M. A., Burgers, P. M., and Gordenin, D. A. (2005) Mol. Cell. Biol. 25 461-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin, Y. H., Ayyagari, R., Resnick, M. A., Gordenin, D. A., and Burgers, P. M. (2003) J. Biol. Chem. 278 1626-1633 [DOI] [PubMed] [Google Scholar]
- 20.Garg, P., Stith, C. M., Sabouri, N., Johansson, E., and Burgers, P. M. (2004) Genes Dev. 18 2764-2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon, M., Giot, L., and Faye, G. (1991) EMBO J. 10 2165-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, K. H., Kim, D. W., Bae, S. H., Kim, J. A., Ryu, G. H., Kwon, Y. N., Kim, K. A., Koo, H. S., and Seo, Y. S. (2000) Nucleic Acids Res. 28 2873-2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budd, M. E., Choe, W., and Campbell, J. L. (2000) J. Biol. Chem. 275 16518-16529 [DOI] [PubMed] [Google Scholar]
- 24.Boule, J. B., and Zakian, V. A. (2006) Nucleic Acids Res. 34 4147-4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budd, M. E., Reis, C. C., Smith, S., Myung, K., and Campbell, J. L. (2006) Mol. Cell. Biol. 26 2490-2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossi, M. L., Pike, J. E., Wang, W., Burgers, P. M., Campbell, J. L., and Bambara, R. A. (2008) J. Biol. Chem. 283 27483-27493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen, J. Z., Qiu, J., Shen, B., and Holmquist, G. P. (2000) Nucleic Acids Res. 28 3649-3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran, P. T., Erdeniz, N., Dudley, S., and Liskay, R. M. (2002) DNA Repair (Amst.) 1 895-912 [DOI] [PubMed] [Google Scholar]
- 29.Ii, M., and Brill, S. J. (2005) Curr. Genet. 48 213-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao, H. I., Veeraraghavan, J., Polaczek, P., Campbell, J. L., and Bambara, R. A. (2004) J. Biol. Chem. 279 15014-15024 [DOI] [PubMed] [Google Scholar]
- 31.Sommers, C. H., Miller, E. J., Dujon, B., Prakash, S., and Prakash, L. (1995) J. Biol. Chem. 270 4193-4196 [DOI] [PubMed] [Google Scholar]
- 32.Goldstein, A. L., and McCusker, J. H. (1999) Yeast 15 1541-1553 [DOI] [PubMed] [Google Scholar]
- 33.Fortune, J. M., Stith, C. M., Kissling, G. E., Burgers, P. M., and Kunkel, T. A. (2006) Nucleic Acids Res. 34 4335-4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henricksen, L. A., Umbricht, C. B., and Wold, M. S. (1994) J. Biol. Chem. 269 11121-11132 [PubMed] [Google Scholar]
- 35.Gomes, X. V., Gary, S. L., and Burgers, P. M. (2000) J. Biol. Chem. 275 14541-14549 [DOI] [PubMed] [Google Scholar]
- 36.Budd, M. E., and Campbell, J. L. (1997) Mol. Cell. Biol. 17 2136-2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tishkoff, D. X., Boerger, A. L., Bertrand, P., Filosi, N., Gaida, G. M., Kane, M. F., and Kolodner, R. D. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 7487-7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., Andrews, B., Tyers, M., and Boone, C. (2001) Science 294 2364-2368 [DOI] [PubMed] [Google Scholar]
- 39.Bae, S. H., Bae, K. H., Kim, J. A., and Seo, Y. S. (2001) Nature. 412 456-461 [DOI] [PubMed] [Google Scholar]
- 40.Sugimoto, N., Nakano, S., Yoneyama, M., and Honda, K. (1996) Nucleic Acids Res. 24 4501-4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langston, L. D., and O'Donnell, M. (2008) J. Biol. Chem. 283 29522-29531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murante, R. S., Rumbaugh, J. A., Barnes, C. J., Norton, J. R., and Bambara, R. A. (1996) J. Biol. Chem. 271 25888-25897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.