Abstract
Within their natural habitat plants are subjected to a combination of different abiotic stresses, each with the potential to exacerbate the damage caused by the others. One of the most devastating stress combinations for crop productivity, which frequently occurs in the field, is drought and heat stress. In this study we conducted proteomic and metabolic analysis of Arabidopsis thaliana plants subjected to a combination of drought and heat stress. We identified 45 different proteins that specifically accumulated in Arabidopsis in response to the stress combination. These included enzymes involved in reactive oxygen detoxification, malate metabolism, and the Calvin cycle. The accumulation of malic enzyme during the combined stress corresponded with enhanced malic enzyme activity, a decrease in malic acid, and lower amounts of oxaloacetate, suggesting that malate metabolism plays an important role in the response of Arabidopsis to the stress combination. Cytosolic ascorbate peroxidase 1 (APX1) protein and mRNA accumulated during the stress combination. When exposed to heat stress combined with drought, an APX1-deficient mutant (apx1) accumulated more hydrogen peroxide and was significantly more sensitive to the stress combination than wild type. In contrast, mutants deficient in thylakoid or stromal/mitochondrial APXs were not more sensitive to the stress combination than apx1 or wild type. Our findings suggest that cytosolic APX1 plays a key role in the acclimation of plants to a combination of drought and heat stress.
Unraveling the response of plants to environmental stress conditions is one of the key challenges facing modern biological research (1). Often in the field plants have to endure a combination of different abiotic stresses such as drought and heat, drought and high light, or drought and salinity (2). Several studies showed that the effects of prolonged drought was exacerbated by periods of high temperature, and that the combination of drought and heat had a significant effect on the productivity and growth of different plants and crops (2). A survey of all major U.S. weather disasters between 1980 and 2004 revealed that the damage caused by a combination of a heat wave and prolonged drought was severalfold higher than that caused by drought alone (2).
A genome-wide analysis identified several hundred Arabidopsis transcripts that specifically accumulated in response to a combination of drought and heat (3). Very little overlap was found between transcripts that accumulated in Arabidopsis in response to heat or drought (3), but certain drought- or heat-response-specific transcripts were elevated by the combined stress (3). These results demonstrated that, in addition to transcripts that are specific to the stress combination, components of each independent response are activated during the stress combination (3). A dual role in drought and heat stress was proposed for at least two transcriptional regulators, MBF1 and DREB2A (4–6).
One of the stress-related transcripts that showed enhanced expression in response to the combined drought and heat stress was cytosolic ascorbate peroxidase 1 (Apx1) (3). APX1 plays a key role in regulating H2O2 levels and H2O2 signaling in plant cells (7–9). APX1 is one of several APXs found in Arabidopsis; at least two of which (APX1 and APX2) are found in the cytoplasm (10). Whereas APX1 mRNA can be found in many plant tissues even in the absence of any stress (11), APX2 mRNA is mainly observed in response to high light intensities, heat stress, or wounding (10, 12, 13). Expression of both APX1 and APX2 depends on a signal generated by the redox state of the photosynthetic electron transfer chain in the chloroplasts (14). Despite its cytoplasmic localization, APX1 is required for the protection of chloroplasts against reactive oxygen species (ROS),2 and in its absence the ROS scavenging machinery in the chloroplast is significantly compromised (15).
Ascorbate peroxidases use ascorbate as a reducing agent to catalyze the conversion of H2O2 to water; in this reaction ascorbate is oxidized to monodehydroascorbate. The later is then reduced back to ascorbate by monodehydroascorbate reductase while converting NAD(P)H to NAD(P) (8). Recently it was suggested that NADPH is also required for the activity of NTRC, a plastid localized protein that contains an N-terminal thioredoxin reductase and a C-terminal thioredoxin. NTRC can reduce 2-Cys peroxiredoxins, yet another H2O2-detoxifying system (16). One source of NADPH could be the oxidative decarboxylation of malate to pyruvate by NADP-malic enzyme (NADP-ME, EC 1.1.1.40). The activity of a cytoplasmic ME is enhanced in rice (Oryza sativa L.) in response to NaCl, and transgenic Arabidopsis plants overexpressing the rice ME have a higher NDAPH/NADP ratio than wild type and are more tolerant to salt (17).
In this work we describe a proteomic and metabolic analysis of Arabidopsis plants subjected to a combination of water withholding and high temperature. Several of the proteins that accumulated in response to the combined stress were previously suggested to be involved in the response of plants to oxidative stress, including APX1. Other proteins were involved in malic acid metabolism and CO2 assimilation. A mutant lacking APX1 (9, 11) was significantly more sensitive to the combined stress suggesting that APX1 plays a key role in regulating H2O2 levels in plants under conditions of stress combination.
EXPERIMENTAL PROCEDURES
Plant Material—Wild-type (Col-0 or WS) and mutant Arabidopsis thaliana plants were grown under a 14/10 light/dark cycle (100 μmol-2.s-1), and drought, heat, and drought and heat combination were applied as previously described (3, 18). For heat treatment plants were transferred to 42 °C for 6, 24, or 48 h (for protein extraction or survival assays). Survival was determined following a recovery period of 72–96 h at 22 °C. For ROS measurements, plants (WS and apx1) were surface-sterilized and grown on ½MS plates, with or without 150 mm sorbitol for 5 days at 21 °C and then exposed to 42 °C for 6 h. ROS levels in seedlings were measured as described (19). Chlorophyll levels were determined using 80% buffered acetone (20). Analysis of microarray data obtained from Rizhsky et al. (3) were performed as described (21). All experiments were performed in triplicates and repeated at least three times. Statistical analysis was performed as described (5). Relative water content was calculated as previously described (4). Water potential of whole plants was measured using a WP4 Dewpoint PotetiaMeter (Decagon, Pullman, WA), as recommended by the manufacturer.
Proteomic Analysis—Protein extraction, two-dimensional gel electrophoresis, imaging and sampling of spots, and protein digestion and mass spectrometry are described in the supplemental materials.
Pathway Analysis—Pathway analysis was performed using “Data base for Annotation, Visualization, and Integrated Discovery” (DAVID) version 2006 (niaid.abcc.ncifcrf.gov/) (22) to discover GO categories and KEGG pathways that are significantly represented in each group. The p value cutoff for this pathway analysis was 0.01.
Enzymatic Assays—Ascorbate peroxidase and malic enzyme activities were determined as described (23, 24). Four-week-old seedlings were collected after 6 h of heat stress, frozen in liquid nitrogen, and ground using a chilled mortar and pestle. All reactions were performed at 25 °C. Enzymes were extracted in 50 mm Hepes-KOH (pH 7.5), 3 mm MgCl2 (for APX) or 50 mm Hepes-KOH (pH 7.4), 10 mm MgCl2, 1 mm EDTA, 5 mm dithiothreitol (for NADP-malic enzyme). Both buffers were supplemented with a protease inhibitor mixture (Sigma) and 10% (v/v) glycerol. Protein concentration was measured using a Bradford reagent (Sigma).
Extraction and Quantification of Metabolites—Ten milligrams of freeze-dried tissue was ground using 3.2-mm chrome-steel beads in a Retsch mixer mill, followed by extraction with 500 μl of 50:50 methanol:water spiked with 2 μg/ml of the internal standards. After the extraction buffer was added the mixture was stirred on the Retsch mill for 1 min at 30 reps/s, sonicated for 1 min in ultrasonic bath, and incubated on dry ice for 5 min. Three cycles of sonication and dry ice incubation were carried out before spinning out the extract 15 min at 18,000 × g. Clear supernatant was transferred into limited volume high-performance liquid chromatography vials and analyzed by LC-MS/MS.
Extracted metabolites were measured using hydrophilic interaction LC-MS/MS as previously described (25). LC-MS/MS was performed on an LC-10ADvp chromatographic system (Shimadzu, Columbia, MD) coupled to a mass spectrometer. LC separation was performed on a Phenomenex 250 × 2 mm Luna 5-μm aminopropyl column (Phenomenex, Torrance, CA). Metabolite quantitation was performed using calibration curves with isotopically labeled internal standards malate-2,3,3-d3 acid and citric-2,2,4-d4 acid (CDN Isotopes, Pointe-Claire, Quebec, Canada), which were added to each sample. Commercially available pure forms of the following compounds were purchased from Sigma-Aldrich and used to prepare calibration curves and estimate the amounts of each one of them in the plant extract: oxaloacetate, malate, phosphoenolpyruvate, NADP, pyruvate, lactate, glycerate 3-phosphate, ribose 5-phosphate, ribulose 5-phosphate, succinate, ADP, glucose 6-phosphate, fructose 6-phosphate, erytrose 4-phosphate, fructose 1,6-biphosphate, and α-ketoglutarate. Starch content was determined as described in a previous study (26).
RESULTS
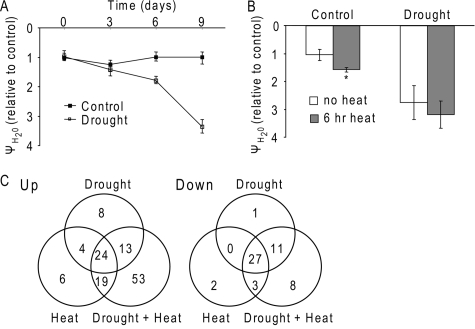
Combination of Drought and Heat Stress in Arabidopsis—To subject Arabidopsis (Col-0) plants to a combination of drought and heat stress we used the same experimental procedure described previously (3, 18). To follow the progression of drought, we monitored the water potential and relative water content of plants. Heat stress was applied 9 days following water withdrawal. At that time the leaf water potential of drought-stressed plants was >3-fold lower than that measured for control plants (Fig. 1A). Relative water content of drought-stressed plants was also lower than that of control, well watered plants, but this difference was smaller (supplemental Fig. S1). After exposing plants to heat stress for 6 h the leaf water potential of control plants was reduced by ∼50% (Fig. 1B), indicating that well watered heat-stressed plants lost water during the 6-h period. In contrast, no significant decrease was observed for the drought-stressed plants subjected to heat stress during this period (Fig. 1B), suggesting that drought-stressed plants exposed to heat stress could not benefit from the cooling effect of evaporation (3, 18). Regardless of the treatment applied, leaf water potential of all plants returned to control levels within 24 h of recovery from stress, and plants were fully recovered (data not shown).
FIGURE 1.
Proteomic analysis of Arabidopsis plants subjected to drought, heat stress, or a combination of drought and heat stress. A, leaf water potential in wild-type (Col-0) Arabidopsis plants subjected to drought stress. B, leaf water potential, relative to control, for well watered, or drought-stressed plants before and after 6 h at 42 °C. *, indicates statistical significance (p < 0.05). C, Venn diagrams of protein spots that increased (Up) or decreased (Down) in their intensity in response to drought, heat stress, or a combination of drought and heat stress.
Proteomic Analysis of Plants Subjected to Drought, Heat, or a Combination of Drought and Heat—Soluble proteins from plants subjected to drought (water withholding for 9 days), heat (42 °C for 6 h), or a combination of the two, in three biological replications (for a total of 12 two-dimensional gels), were subjected to proteomics analysis as described in the supplemental materials. As indicated above, the stress treatments were not lethal to plants. In accordance, no evidence was found in our two-dimensional gel for extensive protein degradation in response to either treatment (supplemental Fig. S2).
Out of 123 protein spots that accumulated in response to at least one stress, 53 spots (for a total of 45 different proteins) were found to be unique to the combined drought and heat (Fig. 1C, supplemental Tables S1 and S2). When the proteomics results obtained in this study were compared with our previous microarray study, corresponding changes in steady-state transcript levels were identified for ∼25% of the proteins (supplemental Table S2) (2, 3). The differences between the microarray and proteomic data could be a result of detection threshold or differences in RNA stability and post-transcriptional regulation. It should also be noted that RNA for the microarray and proteins for the proteomics experiments were obtained from plants exposed to the different stresses in two independent experiments. As could be expected (27), low abundance proteins involved in processes such as transcriptional regulation and signaling were underrepresented (data not shown). The average size of proteins in our study was 39.7 kDa and was lower than that reported in a large scale Arabidopsis proteomics analysis (27). Chloroplast proteins are over-represented in our study (16 of the 45 proteins listed in supplemental Table S2 are predicted to be chloroplast-localized proteins), suggesting that the chloroplast might be involved in many of the plants' responses to the combined stress.
Accumulation of Oxidative Stress-related Proteins—Several proteins that accumulated in response to the combined stress were previously implicated in the response of plants to oxidative stress (supplemental Tables S1 and S2). These included mitochondrial glyceraldehyde-3-phosphate dehydrogenase (28), chloroplastic chaperonin 60β (LEN1) (29), cytosolic isoflavone reductase (30), allene oxide (31), and cystine lyase (32). Interestingly, a germin-like protein was also induced by the combined stress. This extracellular protein has an enzymatic activity of oxalate oxidase and was suggested to be involved in generating H2O2 as part of the plants' defense pathways (33).
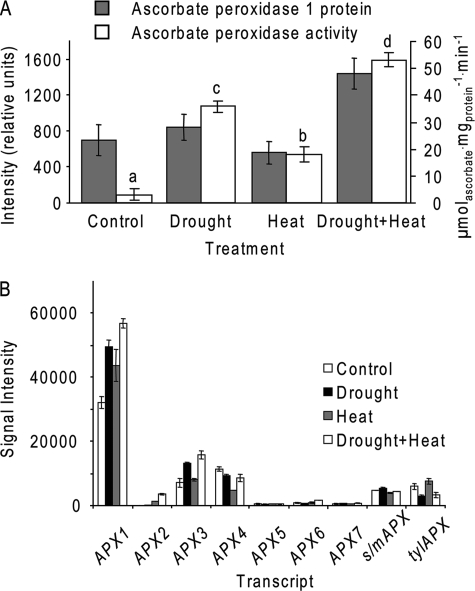
Ascorbate peroxidase 1 (APX1) was yet another protein that accumulated in response to the combined stress. Although no significant accumulation of APX1 was observed under each of the independent stresses, the combined stress resulted in accumulation of APX1 (Fig. 2A). APX1 belongs to a small gene family that includes cytosolic, mitochondrial, peroxisomal, and chloroplastic isozymes (34). We examined therefore the accumulation of nine APX transcripts in response to the different stresses using microarray data (3). Of the nine transcripts examined, four showed a significant response to drought (APX1, -2, and -3 and stromal/mitochondrial APX; s/mAPX). In contrast, only APX1 and APX2 responded to heat stress (both accumulated). The combination of drought and heat resulted in an increased level of four transcripts (APX1, -2, -3, and -6) and reduced amounts of thylakoid APX (tylAPX; Fig. 2B). Although APX2 showed a large -fold increase in response to the different stresses (Fig. 2B), the total amounts of its transcript were low (data not shown). Interestingly, although APX4 levels decreased in response to the combined heat and drought stress (Fig. 2B) protein levels of APX4 actually increased in response to both heat and the combined stress (Table S1). As shown in Fig. 2A, total APX activity increased in response to all stresses. In general the majority of APX activity in protein extracts obtained without ascorbic acid is attributed to APX1 (35, 36). The effect each treatment had on APX activity correlated with the effect it had on transcript levels and protein spot intensity for APX1 with the smallest effect being in plants exposed to heat only and the largest in plants exposed to the combined stress (Fig. 2A). In the absence of stress APX activity was low, probably because the majority of APX protein found in unstressed cells is inactive (37).
FIGURE 2.
Response of APX1 to a combination of drought and heat stress. A, APX1 protein and activity in samples obtained from control plants and plants subjected drought, heat, or a combination of drought and heat. Letters and the asterisk indicate statistical significance (p < 0.01 and p < 0.05, respectively). B, abundance of nine different APX transcripts in samples obtained from control plants and plants subjected to drought, heat, or a combination of drought and heat for 6 h. Data were obtained from the microarray experiments described in Ref. 3.
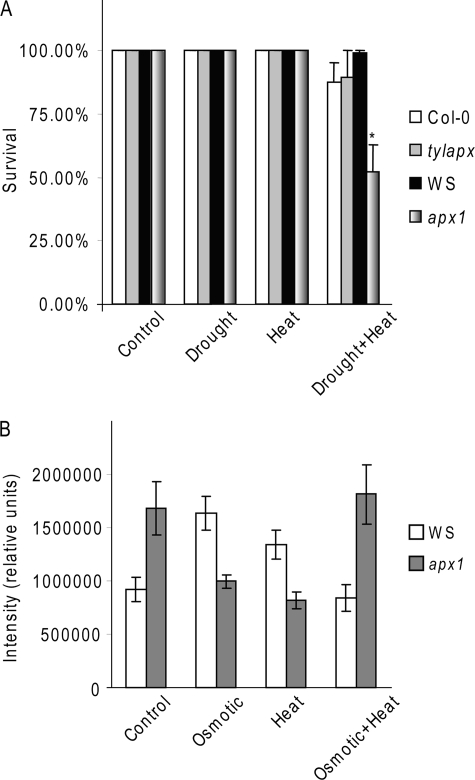
Knock-out Apx1 Plants (apx1) Are More Sensitive to a Combination of Drought and Heat Stress—We used an Apx1-deficient Arabidopsis mutant (9, 11), to further characterize the role of APX1 in the response of plants to a combination of drought and heat stress. We subjected wild-type and apx1 plants grown side-by-side in the same pots to drought, heat stress, and a combination of drought and heat stress and scored plants for survival. When wild-type and apx1 plants were exposed to a short heat stress (6 or 24 h), with or without prior water withholding, and allowed to recover, no differences were observed between wild-type and apx1 (data not shown). When we extended the heat stress period to 48 h we observed a significant reduction in the recovery of apx1 plants subjected to a combination of drought and heat stress (Fig. 3). In contrast, we did not observe a significant decrease in the recovery of wild-type plants or mutants deficient in tylAPX (tylapx; At1g77490 21), subjected to the same stress conditions (Fig. 3A). Similar to tylapx, a mutant deficient in s/mAPX (15) did not show enhanced sensitivity to the combined stress (data not shown). Compared with wild-type plants, apx1 plants that recovered from the combined stress had lower chlorophyll content (supplemental Fig. S3), suggesting that, although they survived the combined stress, their photosynthetic apparatus was affected.
FIGURE 3.
Enhanced sensitivity of knockout Apx1 plants to a combination of drought and heat stress. A, survival of 3-week-old wild-type (Col-0 and WS), tylapx (in Col-0), and apx1 (in WS), exposed to heat stress (42 °C; 48 h) with or without prior water withholding. *, indicates statistical significance (p < 0.05). B, accumulation of ROS in 5-day-old wild-type (WS) and apx1 seedlings grown on plates with or without 150 mm sorbitol and exposed to heat stress (42 °C; 6 h). Differences between WS and apx1 were statistically significant (p < 0.05) under all conditions.
To test whether the enhanced sensitivity of the apx1 mutant to stress combination corresponded with accumulation of ROS we measured hydrogen peroxide in wild-type and apx1 plants subjected to heat, osmotic stress (caused by 150 mm sorbitol in the growth media, which reduced the water potential by 0.53 MPa), and a combination of heat and osmotic stress (Fig. 3B). In the absence of stress, apx1 seedlings accumulated significantly higher amounts of ROS; this is in agreement with the role of APX1 in maintaining ROS levels in the cell (15). When exposed to a single stress (osmotic or heat) wild-type seedlings accumulated higher amounts of ROS than non-treated seedlings, whereas apx1 seedlings had lower ROS amounts than non-treated apx1 seedlings (Fig. 3B). This result suggests that the lack of APX1 could trigger the expression of other stress-response mechanism as was previously demonstrated for apx1 during salinity stress (38). In contrast, when exposed to a combined osmotic and heat stress apx1 seedlings accumulated higher amounts of ROS compared with wild-type suggesting that the alternative mechanisms enhanced in apx1 by the lack of APX1 were not sufficient to prevent ROS accumulation during the stress combination. The reduced ROS amounts in wild-type seedlings exposed to the combined stress compared with either individual stresses is in agreement with the accumulation of APX1 and the increase in APX activity in response to the combined stress.
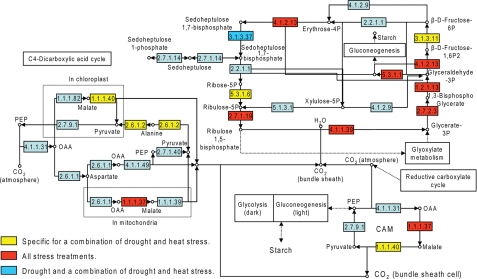
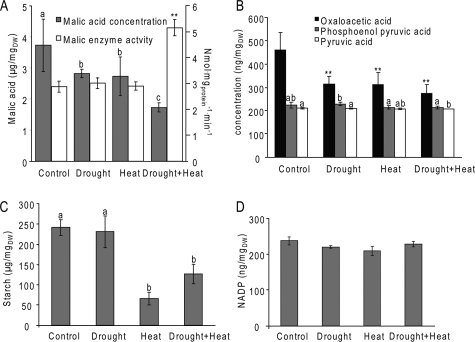
Accumulation of Carbon Fixation-related and Malate Metabolism Proteins—When the proteins that accumulated in response to the combined stress were superimposed on maps of different metabolic pathways we observed an accumulation of proteins involved in carbon fixation (Fig. 4). This was further emphasized when we added to the analysis proteins that were not unique to the combined drought and heat stress, but were also induced by all the stress conditions tested (supplemental Table S1 and Fig. 4). Among these were enzymes involved in the reductive pentose phosphate (Calvin-Benson) cycle, including fructose-1,6-bisphosphatase, a putative ribose-5-phosphate isomerase, and Rubisco (all located in the chloroplast). Interestingly, many of the enzymes identified were related to either C4 or CAM carbon fixation (39). These included a chloroplast NAD-dependent malate dehydrogenase, the cytoplasmic NADP-dependent malic enzyme 2, putative alanine aminotransferase, and a cytoplasmic/mitochondrial glyceraldehyde-3-phosphate dehydrogenase. The accumulation of enzymes involved in carbon fixation in plants with significantly reduced stomatal conductance and photosynthesis rate (3, 18) is intriguing. Because the amount of glycerate 3-phosphate and ribulose 5-phosphate, two key metabolites in the Calvin cycle, did not change significantly during the different stresses (supplemental Fig. S5), CO2 fixation could still be active. This suggests that the source of CO2 for fixation in plants might be carbon cycling from either respiration or the activity of CAM and C4 like enzymes using sugars that are the product of starch degradation (3). This possibility is supported by the fact that the combined stress resulted in a significant increase in the activity of NADP-dependent ME and a concomitant decrease in the amount of malate (Fig. 5A). Amounts of oxaloacetate, which is converted into malic acid by malate dehydrogenase, were reduced in response to all stresses (Fig. 5B). The activity of malate dehydrogenase did not change in response to any of the stresses and was significantly higher than that of ME (data not shown) suggesting that this enzyme is not a rate-limiting step in malate metabolism. Oxaloacetate concentrations were at least 10 times lower than malate concentrations (Fig. 5A and 5B) lending further support to this possibility. Amounts of phosphoenolpyruvate and pyruvate (an immediate product of ME activity) did not change significantly and were also significantly lower than malate amounts (Fig. 5B), suggesting that pyruvate formed by the ME activity was rapidly metabolized. Plants exposed to heat, with or without prior water withholding, had significantly lower amounts of starch (Fig. 5C) suggesting that malate can be formed by the breakdown of starch in response to increased temperatures. Interestingly, when plants were subjected to heat without prior water withholding the amounts of starch were significantly lower than the amounts measured in plants exposed to heat after drought (Fig. 5C). None of the treatments resulted in an increase in total fatty acid content (supplemental Fig. S4). NADP amounts did not change significantly in response to any of the stresses (Fig. 5B), suggesting that, under the conditions tested, plants could still balance the use of NADPH and the reduction of NADP to NADPH, possibly by the activity of ME (17).
FIGURE 4.
Carbon fixation pathways in plants subjected to drought, heat, or a combination of drought and heat. Proteins were assigned to metabolic pathways based on GO annotation. Proteins that accumulated only in response to the combined stress are indicated in yellow. Red and blue indicate proteins that accumulated in response to all three conditions, or drought and the combined stress, respectively. The scheme of carbon fixation pathways was adapted from KEGG (www.genome.jp/dbget-bin/www_bget?path:ot00710). EC 1.1.37 is NAD-dependent malate dehydrogenase, EC 1.1.1.40 is NADP-ME, EC 2.6.1.2 is alanine aminotransferase, EC 5.3.1.6 is ribose-5-phosphate isomerase, and EC 3.1.3.11 is d-fructose-1,6-bisphosphate 1-phosphohydrolase.
FIGURE 5.
Enzymatic and metabolic analysis of plants subjected to drought, heat, or a combination of drought and heat. A, malic enzyme activity and malate content; B, oxaloacetate, phosphoenolpyruvate, and pyruvate content; C, starch content; D, NADP concentrations, in control plants and plants subjected to drought, heat, or a combination of drought and heat for 6 h. Average rates and standard errors (n = 3) are shown for all experiments, letters and ** indicate statistical significance (p < 0.05 and p < 0.01, respectively).
DISCUSSION
Many of the proteins that accumulated in plants in response to the combined stress treatment were related to oxidative stress (supplemental Table S2). This result was supported by our findings that in apx1 plants the combination of water deficiency and heat resulted in increased ROS levels (Fig. 3B). Several different Apx transcripts accumulated in response to the stress combination (even those that were not affected by drought or heat alone; Fig. 2B). However, we could only identify two proteins: the cytoplasmic APX1 and the chloroplast-localized APX4 by our proteomics analysis (supplemental Table S2 and Fig. 2A). It should, however, be noted that we only studied soluble proteins, and therefore any effect the various stresses had on the amount of the thylakoid APX protein could not have been detected. Despite its cytosolic localization, APX1 is crucial for the protection of the chloroplast from ROS (15). The enhanced susceptibility of apx1 plants to a combination of drought and heat stress (Fig. 3), combined with our observations that in the absence of APX1 plants accumulate H2O2 (11, 15), suggest that under conditions of stress combination H2O2 accumulation in the cytosol could trigger cell injury and death (Fig. 3A). Our results therefore provide genetic evidence for a link between H2O2 removal in the cytosol and plant survival during a combination of drought and heat stress. In addition, they provide the first demonstration for a gene (Apx1) that is required for tolerance to a combination of two different stresses.
The cellular sources of ROS production during a combination of drought and heat stress are unknown at present. During drought stress ROS are thought to be produced in the chloroplast and peroxisomes (40, 41), whereas during heat ROSs are thought to be produced in chloroplasts and mitochondria (42). The detection of an extracellular germin-like protein that could have an oxalate oxidase activity in plants subjected to drought and heat could suggest that during the stress combination ROS are also produced at the apoplast. Because the path from all of these compartments to the nuclei transverses the cytosol, APX1, which is considered a major hydrogen peroxide-scavenging enzyme in the cytosol (34), could affect the diffusion of ROSs such as hydrogen peroxide from these different compartments to the nuclei. Thus, when APX1 is absent more ROS could reach the nuclei during the stress combination and effect or alter cellular survival.
The accumulation of malic enzyme during the combined drought and heat stress is intriguing. This enzyme is part of the CO2 concentration mechanism of C4 (43) and CAM (39) plants. Nevertheless, in Arabidopsis, which is a C3 plant, it could not function in a classic CAM or C4 concentration mechanism, especially because it does not appear from our proteomic or transcriptomic analysis that phosphoenolpyruvate carboxylase is elevated in response to drought or drought combined with heat (supplemental Table S2) (Ref. 3, and data not shown). However, it is possible that in an attempt to avoid the metabolic strain that occurs during stress malic enzyme protein and activity are enhanced (Fig. 5). Malic enzyme converts malate to pyruvate, generating NADPH and CO2 thus providing NADPH for the reduction of monodehydroascorbate to ascorbate in the ascorbate-glutathione H2O2 detoxification pathway (Asada-Foyer-Halliwell pathway (35)). The CO2 released during the ME reaction could also be utilized by Rubisco and the Calvin cycle pathway. Such recycling of CO2 can reduce the formation of ROS during severe stress that is caused by limited CO2 supply when stomata are closed to reduce water loss (44). In support of this hypothesis the amounts of glycerate 3-phosphate and ribulose 5-phosphate did not change significantly in response to either of the independent stresses or the combined stress (supplemental Fig. S5), and the level of Calvin cycle enzymes increased during the different stresses (supplemental Table S1). Taken together our results suggest that, although photosynthesis is significantly reduced under the stress combination (3), the Calvin cycle is still active possibly using recycled CO2. The source of malate for this reaction could be starch breakdown that occurs in Arabidopsis in response to heat or drought combined with heat (Fig. 5). Transcripts involved in starch breakdown, as well as in sucrose biosynthesis, are elevated in Arabidopsis in response to drought or the combination of drought and heat stress (3). Pyruvate that accumulates during this process could then be used for fatty acid biosynthesis (45). However, we could not detect an increase in the amount of several different fatty acids tested (supplemental Fig. S4). Conversion of oxaloacetate to malate can also help maintain NADP/NADPH ration as part of the “malate valve” during stress (46).
In the C3 plant Digitalis lanata net photosynthesis was reduced by ∼70% during drought stress, whereas the demand for light energy was reduced by only 40% (47). It was suggested that this balance was achieved by breakdown of starch and reassimilation of CO2 (47). The elevated levels of Calvin cycle enzymes observed under all stress condition in our study, in conjugation with our metabolic and enzymatic measurements (supplemental Table S2 and Figs. 2 and 3), could suggest that Arabidopsis plants subjected to heat, drought, or drought combined with heat stress are cycling CO2 through a similar pathway. Further studies are, however, required to address this possibility.
Plants can utilize several different mechanisms to avoid oxidative stress resulting from low water potential that leads to stomatal closure and low CO2 (44). These include photorespiration (48), non-photochemical quenching or thermal dissipation (49, 50), cyclic electron transport (49, 51), and pseudocyclic electron transport (also referred to as the Mehler-ascorbate peroxidase reaction or the water-water cycle (7, 49)). Although the NADPH formed by the activity of ME could be used for detoxifying ROS formed by the excessive stress, it is conceivable that CO2 recycling (47) is yet another mechanism that prevents excess light energy from damaging cells when environmental conditions limit CO2 availability. Malic enzyme therefore could play a dual role during stress by providing NADPH for antioxidant regeneration and CO2 for carbon fixation at the expense of starch breakdown.
Supplementary Material
Acknowledgments
We thank Drs. Kathleen Schegg, David Quilici, and Rebekah Woolsey from the proteomic center at the University of Nevada, Reno for their help in two-dimensional gels, MS, and data base searches. We thank Drs. John C. Cushman and David Shintani (the University of Nevada, Reno) for important comments and useful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants RR-03-008 and P20 RR16464-05 from the IDeA Network of Biomedical Research Excellence. This work was also supported by National Science Foundation Grants IBN-0420033, NSF-0431327, and IOS-0743954 and The Nevada Agricultural Experimental Station (Publication 03055517). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, references, Tables S1 and S3, and Figs. S1–S5.
Footnotes
The abbreviations used are: ROS, reactive oxygen species; APX, ascorbate peroxidase; tylAPX, thylakoid APX; ME, NADP-dependent malic enzyme; DAVID, Data base for Annotation, Visualization, and Integrated Discovery; LC-MS/MS, liquid chromatography-tandem mass spectrometry.
References
- 1.Cushman, J. C., and Bohnert, H. J. (2000) Curr. Opin. Plant Biol. 3 117-124 [DOI] [PubMed] [Google Scholar]
- 2.Mittler, R. (2006) Trends Plant Sci. 11 15-19 [DOI] [PubMed] [Google Scholar]
- 3.Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., Davletova, S., and Mittler, R. (2004) Plant Physiol. 134 1683-1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki, N., Bajad, S., Shuman, J., Shulaev, V., and Mittler, R. (2008) J. Biol. Chem. 283 9269-9275 [DOI] [PubMed] [Google Scholar]
- 5.Suzuki, N., Rizhsky, L., Liang, H., Shuman, J., Shulaev, V., and Mittler, R. (2005) Plant Physiol. 139 1313-1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006) Proc. Natl. Acad. Sci. U. S. A 103 18822-18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asada, K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601-639 [DOI] [PubMed] [Google Scholar]
- 8.Apel, K., and Hirt, H. (2004) Annu. Rev. Plant Biol. 55 373-399 [DOI] [PubMed] [Google Scholar]
- 9.Davletova, S., Schlauch, K., Coutu, J., and Mittler, R. (2005) Plant Physiol. 139 847-856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panchuk, I., Volkov, R. A., and Schoffl, F. (2002) Plant Physiol. 129 838-853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pnueli, L., Liang, H., Rozenberg, M., and Mittler, R. (2003) Plant J. 34 187-203 [DOI] [PubMed] [Google Scholar]
- 12.Mullineaux, P. M., Karpinski, S., and Baker, N. R. (2006) Plant Physiol. 141 346-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, C. C., Ball, L., Fryer, M. J., Baker, N. R., Karpinski, S., and Mullineaux, P. M. (2004) Plant J. 38 499-511 [DOI] [PubMed] [Google Scholar]
- 14.Karpinski, S., Escobar, C., Karpinska, B., Creissen, G., and Mullineaux, P. M. (1997) Plant Cell 9 627-640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davletova, S., Rizhsky, L., Liang, H., Shengqiang, Z., Oliver, D. J., Coutu, J., Shulaev, V., Schlauch, K., and Mittler, R. (2005) Plant Cell 17 268-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spinola, M. C., Perez-Ruiz, J. M., Pulido, P., Kirchsteiger, K., Guinea, M., Gonzalez, M., and Cejudo, F. J. (2008) Physiol. Plant. 133 516-524 [DOI] [PubMed] [Google Scholar]
- 17.Cheng, Y., and Long, M. (2007) Biotechnol. Lett. 29 1129-1134 [DOI] [PubMed] [Google Scholar]
- 18.Rizhsky, L., Liang, H., and Mittler, R. (2002) Plant Physiol. 130 1143-1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, L., Ciftci-Yilmaz, S., Harper, J., Cushman, J. C., and Mittler, R. (2008) Plant Physiol. 148 280-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989) Biochim. Biophys. Acta 975 384-394 [Google Scholar]
- 21.Miller, G., and Mittler, R. (2006) Ann. Bot. (Lond.) 98 279-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis, G., Jr., Sherman, B. T., Hosack, D. A., Yang, J., Gao, W., Lane, H. C., and Lempicki, R. A. (2003) Genome Biol. 4 P3. [PubMed] [Google Scholar]
- 23.Mittler, R., and Zilinskas, B. A. (1991) Plant Physiol. 97 962-968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winter, K., Foster, J. G., Edwards, G. E., and Holtum, J. A. (1982) Plant Physiol. 69 300-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajad, S. U., Lu, W., Kimball, E. H., Yuan, J., Peterson, C., and Rabinowitz, J. D. (2006) J. Chromatogr A. 1125 76-88 [DOI] [PubMed] [Google Scholar]
- 26.Smith, A. M., and Zeeman, S. C. (2006) Nat. Protoc. 1 1342-1345 [DOI] [PubMed] [Google Scholar]
- 27.Baerenfaller, K., Grossmann, J., Grobei, M. A., Hull, R., Hirsch-Hoffmann, M., Yalovsky, S., Zimmermann, P., Grossniklaus, U., Gruissem, W., and Baginsky, S. (2008) Science 320 938-941 [DOI] [PubMed] [Google Scholar]
- 28.Sweetlove, L. J., Heazlewood, J. L., Herald, V., Holtzapffel, R., Day, D. A., Leaver, C. J., and Millar, A. H. (2002) Plant J. 32 891-904 [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa, A., Tanaka, H., Nakai, M., and Asahi, T. (2003) Plant Cell Physiol. 44 255-261 [DOI] [PubMed] [Google Scholar]
- 30.Babiychuk, E., Kushnir, S., Belles-Boix, E., Van Montagu, M., and Inze, D. (1995) J. Biol. Chem. 270 26224-26231 [DOI] [PubMed] [Google Scholar]
- 31.Stenzel, I., Hause, B., Miersch, O., Kurz, T., Maucher, H., Weichert, H., Ziegler, J., Feussner, I., and Wasternack, C. (2003) Plant Mol. Biol. 51 895-911 [DOI] [PubMed] [Google Scholar]
- 32.Maeda, H., Song, W., Sage, T. L., and Dellapenna, D. (2006) Plant Cell 18 2710-2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter, C., Graham, R. A., and Thornburg, R. W. (1998) Plant Mol. Biol. 38 929-943 [DOI] [PubMed] [Google Scholar]
- 34.Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004) Trends Plant Sci. 9 490-498 [DOI] [PubMed] [Google Scholar]
- 35.Asada, K. (1992) Physiol. Plant. 85 235-241 [Google Scholar]
- 36.Mittler, R., and Zilinskas, B. A. (1992) J. Biol. Chem. 267 21802-21807 [PubMed] [Google Scholar]
- 37.Mittler, R., and Zilinskas, B. A. (1994) Plant J. 5 397-405 [DOI] [PubMed] [Google Scholar]
- 38.Ciftci-Yilmaz, S., Morsy, M. R., Song, L., Coutu, A., Krizek, B. A., Lewis, M. W., Warren, D., Cushman, J., Connolly, E. L., and Mittler, R. (2007) J. Biol. Chem. 282 9260-9268 [DOI] [PubMed] [Google Scholar]
- 39.Cushman, J. C., and Bohnert, H. J. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 305-332 [DOI] [PubMed] [Google Scholar]
- 40.Asada, K. (2006) Plant Physiol. 141 391-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Breusegem, F., and Dat, J. F. (2006) Plant Physiol. 141 384-390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki, N., and Mittler, R. (2006) Physiol. Plant 126 45-51 [Google Scholar]
- 43.Sheen, J. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 187-217 [DOI] [PubMed] [Google Scholar]
- 44.Mittler, R. (2002) Trends Plant Sci. 7 405-410 [DOI] [PubMed] [Google Scholar]
- 45.Schwender, J., Ohlrogge, J., and Shachar-Hill, Y. (2004) Curr. Opin. Plant Biol. 7 309-317 [DOI] [PubMed] [Google Scholar]
- 46.Scheibe, R. (2004) Physiol Plant 120 21-26 [DOI] [PubMed] [Google Scholar]
- 47.Stuhlfauth, T., Scheuermann, R., and Fock, H. P. (1990) Plant Physiol. 92 1053-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raines, C. A. (2006) Plant Cell Environ. 29 331-339 [DOI] [PubMed] [Google Scholar]
- 49.Niyogi, K. K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 333-359 [DOI] [PubMed] [Google Scholar]
- 50.Szabo, I., Bergantino, E., and Giacometti, G. M. (2005) EMBO Rep. 6 629-634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson, G. N. (2005) J. Exp. Bot. 56 407-416 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.