Abstract
The clinical severity of Osteogenesis Imperfecta (OI), also known as the brittle bone disease, relates to the extent of conformational changes in the collagen triple helix induced by Gly substitution mutations. The lingering question is why Gly substitutions at different locations of collagen cause different disruptions of the triple helix. Here, we describe markedly different conformational changes of the triple helix induced by two Gly substitution mutations placed only 12 residues apart. The effects of the Gly substitutions were characterized using a recombinant collagen fragment modeling the 63-residue segment of the α1 chain of type I collagen containing no Hyp (residues 877-939) obtained from Escherichia coli. Two Gly → Ser substitutions at Gly-901 and Gly-913 associated with, respectively, mild and severe OI variants were introduced by site-directed mutagenesis. Biophysical characterization and limited protease digestion experiments revealed that while the substitution at Gly-901 causes relatively minor destabilization of the triple helix, the substitution at Gly-913 induces large scale unfolding of an unstable region C-terminal to the mutation site. This extensive unfolding is caused by the intrinsic low stability of the C-terminal region of the helix and the mutation induced disruption of a set of salt bridges, which functions to lock this unstable region into the triple helical conformation. The extensive conformational changes associated with the loss of the salt bridges highlight the long range impact of the local interactions of triple helix and suggest a new mechanism by which OI mutations cause severe conformational damages in collagen.
Considerable effort has been made to elucidate the mechanisms by which Gly substitution mutations of the collagen triple helix cause Osteogenesis Imperfecta (OI),2 also known as brittle bone disease. The collagen triple helix consists of three polypeptide chains each in extended polyproline II conformation and with the characteristic (Gly-X-Y)n repeating amino acid sequence (1-3). The Gly at every third position is necessitated by the close packing of the helix; while the X and Y residues (where X and Y can be any amino acids) contribute directlytothestabilityofthetriplehelixandconferthesequence-dependent properties of collagen (4). Missense mutations that replace the obligatory Gly by another amino acid residue in type I collagen, the major component of bones, are the most common cause of OI (5, 6). The triple helix domain of type I collagen is a heterotrimer composed of two α1 chains and one α2 chain each with more than 1000 amino acids in an uninterrupted (Gly-X-Y)n sequence (7). Nearly 800 Gly replacing mutations from both α1 and α2 chains have been linked to OI, yet, depending on the location and the identity of the Gly substitution, the clinical severity of OI varies from mild increase of bone fragility to the most severe type characterized by death at the prenatal stage (the Type II OI) (6). It remains unclear what molecular properties are related to the sequence locations of the Gly substitutions, which in turn affect the properties of bones.
The prevailing domain model of OI directly links the OI phenotype to the local helix stability surrounding a mutation. The triple helix is not uniform in structure and stability, but organized into thermally stable and labile domains along the axis of the helix (4, 8-11). Mutations disrupting highly stabilizing regions of the helix are considered to be more damaging to the conformation than those in the more flexible, thermally labile regions and thus, predicted to be more severe (12). While the conformational heterogeneity was clearly demonstrated in biophysical characterizations of the full-chain collagens (8, 9, 11), the relations between the conformational heterogeneity and the severity of the OI has not yet been established. In fact, a recent study systematically mapping out the effects of 41 OI mutations on the melting temperature (ΔTm) of type I collagen reported no direct correlation between the ΔTm of the mutants and the severity of OI, and no straight forward relationships between the ΔTm and the local helix stability (11).
Experimentally examining such correlations is difficult because of the large size, and the rope-like conformation of the collagen. Studies using full-chain collagens are often limited to relying on macroscopic parameters such as the ΔTm, which does not imply any specific changes of the conformation (11). Given the long, rope-like structure of the triple helix, a mutation could critically alter the surrounding conformations of the triple helix without causing significant changes in Tm of the whole molecule. On the other hand, the local stability of a segment of the triple helix is often derived from the data obtained from short, synthetic peptides. Despite extensive peptide studies on the stabilizing/destabilizing effects of isolated X or Y residues and of the intrachain and interchain molecular interactions involving both X and Y residues in one Gly-X-Y tripeptide or in two adjacent tripeptides (Gly-X-Y-Gly-X′-Y′), how these interactions modulate each other in the context of the long helix of collagen remain largely unknown (4, 11, 13).
We have developed an E. coli expression system for recombinant collagen fragments to overcome the limitations of the studies using full-chain collagens or short synthetic peptides. The recombinant collagen fragments were used to model the 63-residue segment of the α1 chain of type I collagen corresponding to residues 877-939 (a region containing no hydroxyproline). The optimal size of the fragments allow us to reveal markedly different effects on the triple helix conformation by two OI mutations placed only 12 residues apart, and to correlate the severe OI to a mutation causing more extensive unfolding of a segment of the triple helix known to be involved in the molecular interactions of type I collagen. Furthermore, we were able to demonstrate that the extensive unfolding is caused by the interruption of a set of interchain salt-bridges formed between the Lys and Glu residues of a KGE sequence. Such interchain salt bridges (from a KGE or KGD sequence) are known to stabilize the short peptides (4, 14) and provide essential stability in bacteria collagen (15). In our model system, the stabilizing interactions of the salt bridges extended through a region containing more than 20 amino acid residues C-terminal to the KGE sequence. Thus, by revealing the long range impacts of the salt bridges, our data highlight the critical roles of the salt bridges in modulating the stabilities of the neighboring regions of the triple helix. Based on these findings, a new mechanism by which OI mutations cause severe conformational changes of collagen is proposed.
EXPERIMENTAL PROCEDURES
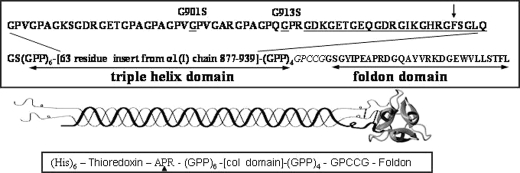
Expression of the Recombinant Fragments—The expression plasmids were built on the (Gly-Pro-Pro)10-foldon construct (generously provided by Dr. Jurgen Engel). The gene coding for the F877 (including the repeating Gly-Pro-Pro sequences and the Gly-Pro-Cys-Cys-Gly-Pro at the C-terminal end) bracketed by BamHI sites were synthesized by GenScript with the codon optimization for bacteria expression, and subsequently cloned into the (Gly-Pro-Pro)10 site (Diagram 1). The fragments were expressed in JM109 and induced by 0.01 mm isopropyl-1-thio-β-d-galactopyranoside. Two Gly → Ser substitutions were introduced by point mutagenesis at position 901 (the G901S) and 913 (the G913S) (Diagram 1) using QuikChange® II site-directed mutagenesis kit (Stratagene).
DIAGRAM 1.
The F877 fragment. The amino acid sequence of the fragment F877 is shown in the upper panel. All the mutations are labeled on top of the sequence, and the two Gly → Ser substitution sites for fragments G901S and G913S are also underlined. The 14-residue Pro-free region (see text) is underlined, and the chymotrypsin digestion site Phe-935 is marked by the arrow. The KGE sequence (position 918-920, see text) is in italics. The residues introduced to form the Cys-knot are shown in italics. The gene expression construct of the fragments is shown in the bottom panel. The His-tagged thioredoxin was removed during the last purification procedure by thrombin digestion at the cleavage site APR marked by the triangle.
The gene product of the expression plasmid is a fusion protein including His-tagged thioredoxin at the N-terminal end (Diagram 1). The purification of the fragments involves two steps. During the first step, the His-tagged fusion protein was purified from the cell extract using TALON® metal affinity resin (Clontech) following the manufacturer's procedure and eluted by 300 mm imidazole. To reduce the contamination of nonspecific binding, the incubation buffer contains 50 mm imidazole. The thrombin cleavage was carried out using Thrombin CleanCleave® kit (Sigma) after incubation overnight at 37 °C. Because of the interaction between the thioredoxin and the collagen fragments, the thioredoxin was removed by gel filtration in the presence of 6 m urea, followed by dialysis against phosphate-buffered saline buffer (10 mm, pH 7, with 127 mm NaCl and 2.7 mm KCl). The final collagen fragments are more than 97% pure, based on the analysis using gel electrophoresis and gel filtration. The molecular weight was confirmed by mass spectrometry.
All three fragments were purified in their trimer form based on gel filtration and analytical ultracentrifugation studies (see “Results”). A set of interchain disulfide bonds between the triple helical domain and the foldon domain is expected to cross-link the three chains and to further increase the stability of the triple helix. However, SDS-PAGE analysis revealed ∼10% trimers dissociated to monomers by boiling in SDS without the addition of reducing agent. Because the cross-linking feature does not affect the current study, the small population of non-cross-linked trimers was not further removed from the preparation.
Circular Dichroism—Circular dichroism (CD) spectra were recorded using an AVIV 200 CD spectrometer equipped with a Peltier thermal controller with 1-mm optical length and 1-nm bandwidth. Baseline correction was done using the solvent and the same cuvette. For structure characterization low concentration samples (0.2 mg/ml) were used to keep the dynode voltage below 500 V in the far UV region. The temperature melt experiments were carried out using 1 mg/ml samples at an average heating rate of ∼0.1 °C/min. SDS-PAGE of samples conducted after the temperature melt experiment are identical with those done before melt with no obvious sign of UV-related fragmentation of the chains. All experiments were repeated at least three times with fresh samples. The concentration of the samples was calibrated using UV absorbance at 280 nm with the extinction coefficient of 0.71 cm-1 (mg/ml)-1.
The fraction of folded triple helix was calculated as F(T) = (θ(T) - θuf)/(θN - θuf), where θ(T) is the observed molar ellipticity at temperature T; θN and θuf are the molar ellipticity of folded and unfolded states, respectively, of the triple helix domain, obtained from linear extrapolation of data from 5 to 10 °C for the folded state and from 50 to 65 °C for the unfolded state. The CD signal at 225 nm in the unfolded state is not zero due to the presence of the foldon domain (see the section below). However, this contribution from foldon will not affect the melting curve of the triple helix domain because the foldon remains folded in the range of temperature used here.
Protease Digestion—Chymotrypsin digestion experiments were carried out using chymotrypsin agarose resin (Sigma, C9134-50UN) in the ratio of 6.5 × 1010 unit/mol protein. Pepsin digestion was carried out using Immobilized Pepsin (Pierce, prod. 20343) with activity > 6000 unit/ml gel. The concentration of protein sample was 1 mg/ml. The digestion experiments for both proteases were carried out following the manufacturer's manual with 0.1 ml of protein at 1 mg/ml for every 100 μl of agarose resin.
Mass Spectrometry—For MALDI-TOF MS analysis (matrix-assisted laser desorption ionization time-of flight mass spectrometry), α-cyano-4-hydroxycinnamic acid (CHCA) matrix was prepared as a saturated solution in 50% acetonitrile/0.1% trifluoroacetic acid. The sample and matrix were spotted 1:1 onto a sample plate and allowed to dry. All spectra were acquired using a MALDI time-of-flight mass spectrometer Voyager-DE STR (PE Biosystem, Foster City, CA) in positive ion, linear, delayed extraction mode with a 337-nm nitrogen laser pulsing at 3 Hz. Spectra from 100 individual laser shots were averaged and externally calibrated.
Analytical Ultracentrifugation—The equilibrium sedimentation experiments were performed on a Beckman XLI analytical ultracentrifuge using an An-60 Ti rotor and 12-mm Epon 8-channel short column centerpieces. The data were collected using both interference optics and absorbance at 280 nm at three different temperatures, with loading concentrations of 0.1, 0.3, 0.5, and 1.2 mg/ml and speeds 22K, 30K, and 40K rpm. The data were analyzed using NONLIN software for windows (distributed by the National AUC facility at the University of Connecticut). Results obtained from interference data and from absorbance are in good agreement. The same samples were used for the temperature melt and the AUC experiments.
RESULTS
The Recombinant Collagen Fragments—The complete sequence of the recombinant collagen fragment F877 mimicking the 63-residue region of the α1(I) (residue 877-939) is shown in Diagram 1. To best model this region in the fully folded collagen and eliminate the effects of helix fraying at the ends, repeating sequences of the tripeptide Gly-Pro-Pro with high triple helix propensity were added at both the C and N termini. All together, the recombinant triple helix domain consists of 93 amino acid residues. The C-terminal foldon domain taken from bacteriophage T4 fibritin functions as the nucleation domain to facilitate the folding of the triple helix (16). A short sequence containing two Cys residues (Gly-Pro-Cys-Cys-Gly) was included between the triple helix domain and the foldon domain. When oxidized in folded triple helix conformation, the Cys residues form a set of interchain disulfide bonds, also known as the Cys knot (17), further increasing the stability of the triple helix. Two mutated fragments G901S and G913S containing Gly → Ser substitution at Gly-901 and Gly-913, respectively, were generated to model the effects of OI mutations on the α1 chain of type I collagen. The substitution at Gly-901 causes the milder type of OI while that at Gly-913 is linked to the most severe type OI, type II OI (18).
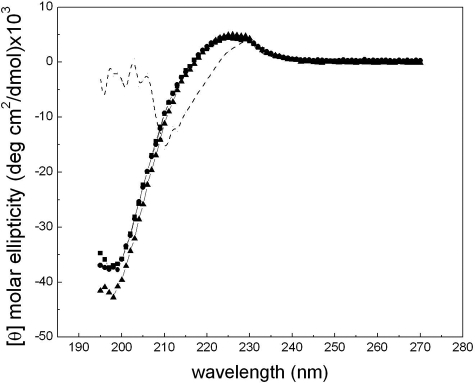
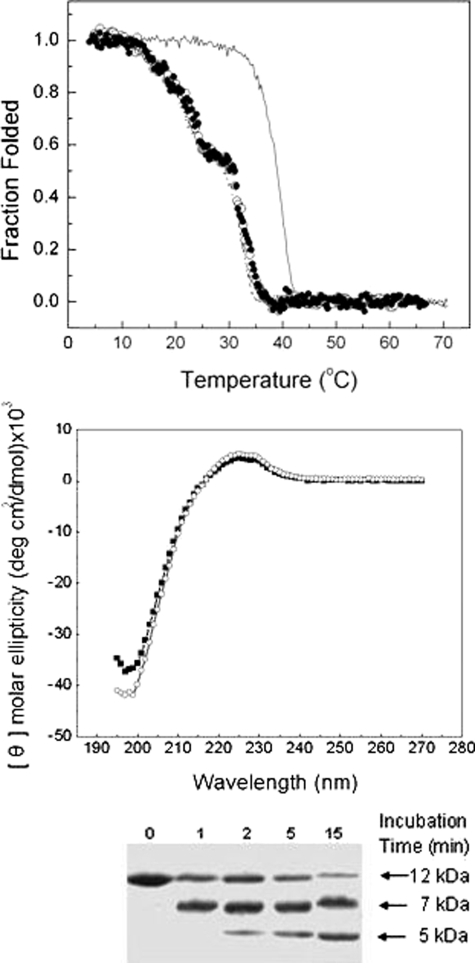
Triple Helix Conformation of All Three Fragments—The CD spectra of the three fragments (F877, G901S, and G913S) at 4 °C are nearly identical despite the presence of Gly substitutions (Fig. 1). The small positive peak at ∼225 nm and the deep negative peak at ∼197 nm are typical of that of a collagen triple helix (3, 19). The foldon domain has contributions to the peak at 225 nm (Fig. 1 and Ref. 16). Thus, the average molar ellipticity of ∼4690 deg cm2/dmol of the three fragments at 225 nm includes ∼20% contribution from the foldon domain. Small differences in the negative peak at 197 nm of the three fragments were observed. While this deep negative peak is often considered a more sensitive characterization of the triple helix conformation, its accurate estimation is often limited by strong absorption of the peptide backbone at the far UV region, especially for large molecules. The close agreement of the molar ellipticity of all three fragments (within experimental error of ± 2%) indicate that the inclusion of the (Gly-Pro-Pro)6 and the segment of triple helix N terminus to the mutation site(s) (residues 877-891) effectively helped the refolding of both G901S and G913S downstream from the mutation sites. The two substitutions appear to cause similarly limited distortions of the triple helix conformation.
FIGURE 1.
CD spectra of collagen fragments. The CD spectra of F877 (square), G901S (circle), and G913S (triangle) were taken at 4 °C with a concentration of 0.2 mg/ml. The dashed line is the spectrum of G913S (1 mg/ml) taken at 70 °C to show the typical spectrum of the foldon domain. All data are normalized to molar ellipticity.
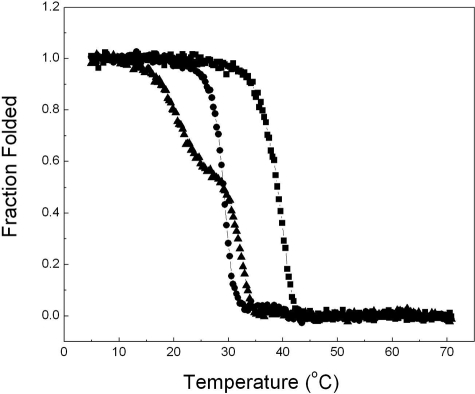
The Effects of OI Mutations on the Thermal Stability—While both form triple helixes at low temperature, the thermal unfolding profiles of the two mutated fragments are, however, considerably different (Fig. 2). The thermal stability was monitored by the change of the CD signal at 225 nm with temperature from 4 to 70 °C. The temperature of the thermal transition (Tm) of F877 is about 40 °C, significantly higher than that of native type I collagen (slightly lower than 37 °C (20)) due to the stabilizing effects of foldon, the Cys-knot and the repeating Gly-Pro-Pro sequences. The Gly → Ser substitution at 901 decreases the thermal stability of F877 and reduces the Tm by ∼10 °C, while the overall unfolding profile remains similar. In contrast, the Gly substitution in G913S affects both the value of Tm and the shape of the unfolding profile. The melting curve reveals two steps in unfolding. The first step occurs at a much lower temperature, between 15 and 25 °C, and involves the decrease of about 40% of the ellipticity at 225 nm. The second melting step takes place between 29 and 35 °C causing the complete unfolding of the triple helix domain.
FIGURE 2.
Temperature melt curves of the collagen fragments. The thermal unfolding of F877 (square), G901S (circle), and G913S (triangle) was monitored by the temperature-induced decrease of the CD signal at 225 nm, and normalized to the fraction folded of the triple helix. The heating rate is ∼0.1 °C/min, and the concentrations of the three fragments are 1 mg/ml.
The loss of the signal at 225 nm in Fig. 2 demonstrates the unfolding of the triple helix domain while the foldon domain remains folded in this temperature range. The Tm of foldon at the comparable concentration is ∼70 °C (16). The presence of the Cys knot will further increase its stability. The CD spectrum of G913S taken at 70 °C (Fig. 1), after the triple helix completely unfolds, shows the typical features of a foldon domain. The analytical ultracentrifugation data further support the trimeric state of the fragments between 4 and 40 °C (Table 1, 40 °C is the highest temperature allowed by the instrument). The average molecular weight of all three samples does not change with temperature. Even when the entire triple helix conformation unfolds for G913S and G901S at 40 °C, they remain a trimer held together by the foldon and/or the Cys-knot. We have observed a low degree of nonspecific aggregation of the sample preparations, which worsens upon storage at 4 °C. This nonspecific aggregation is reflected in the slightly higher molecular weight averages of the samples compared with the expected value of a pure trimer.
TABLE 1.
Molecular weight estimations at different temperatures
|
Monomer mol. wt
|
Molecular weight averages
|
|||
|---|---|---|---|---|
| 4 °C | 15 °C | 40 °C | ||
| kDa | kDa | kDa | kDa | |
| F877 | 12.19 | 43.57 (41.37, 46.57)a | 44.81 (41.00, 46.05) | 35.33 (33.16, 39.72) |
| G901S | 12.22 | 44.70 (43.00, 46.12) | 44.54 (42.75, 46.33) | 37.54 (36.16, 38.02) |
| G913S | 12.22 | 42.74 (39.43, 45.05) | 43.35 (39.04, 45.86) | 36.37 (34.90, 38.43) |
Numbers in parentheses are the error statistics (±95% confidence level) of the estimation. The range is not symmetric with respect to the estimate, reflecting the nature of a nonlinear estimation algorithm.
The close agreement of the molecular weight averages of G913S and F877 at both 4 °C and 15 °C (Table 1) indicates the first melting transition is not caused by the nonspecific aggregation of the sample. Had the 40% signal loss during the first melting step been caused by the dissociation of the aggregates in G913S sample, the molecular weight averages of G913S at 15 °C would be significantly lower than that of the F877, which is clearly not the case. Rather, the similar small decrease of the molecular weight averages with temperature in all three peptides indicates the nonspecific aggregates behave more or less in the same manner in all three samples. The temperature-dependent behavior of the aggregates would contribute similarly, if at all, to the temperature-induced CD signal changes for all three fragments.
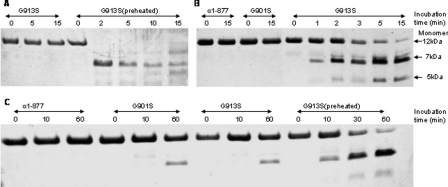
Gly-913 → Ser Induced Unfolding of the Pro-free Region—Proteolytic digestion is often used to assess the conformation of the triple helix (10, 21). When fully folded, the triple helix is known to be resistant to chymotrypsin and pepsin. While there are several pepsin cleavage sites in the construct, chymotrypsin was used to target a unique site, Phe-935, close to the C-terminal end of the triple helix. The protease digestion experiments were carried out at both 4 °C (Fig. 3A) and 15 °C (Fig. 3B). Preheated samples were used as controls in the experiments where the fragments of G913S were preheated to 70 °C for 2 h to induce the complete unfolding of the triple helix, followed by quickly cooling to the specified temperature for the proteolytic digestion. Because the refolding of the triple helix is exceedingly slow (22-24), this preheated sample is expected to contain unfolded chains, which are susceptible to the proteases.
FIGURE 3.
Protease digestion of collagen fragments. A, chymotrypsin digestion at 4 °C. B, chymotrypsin digestion at 15 °C. C, pepsin digestion at 4 °C. The concentrations of the fragments were 1 mg/ml. The SDS-PAGE was performed with the presence of 20 mm dithiothreitol.
All three fragments have the same level of resistance to chymotrypsin at 4 °C (Fig. 3A), indicating similar well folded conformation and consistent with the CD data (Fig. 1). Little digestion by chymotrypsin was observed for all three samples during the 15 min of incubation at 4 °C (gel pictures of F877 and G901S are identical with that of G913S and are not included in Fig. 3A due to space limitation), while the preheated sample was nearly completely digested by the end of the 15 min. Similar results were also seen for pepsin digestion experiments at 4 °C (Fig. 3C). A small degree of digestion was observed in both G913S and G901S after prolonged incubation in pepsin (60 min) reflecting the decreased stability of these two mutated chains.
A significantly higher susceptibility of G913S to chymotrypsin was observed at 15 °C (Fig. 3B) as the G913S starts to get into the first melting step. Clear signs of digestion of G913S were observed after 1 min of incubation with the enzyme, with the digestion nearing completion after 15 min of incubation. On the other hand, both F877 and G901S are resistant to the enzyme after 15 min of incubation. The subsequent MS experiments have confirmed the digested samples of G913S contain a 5K species corresponding to the fragment from the C-terminal to the Phe-935, and a ∼7K species corresponding to the fragment N terminus to the digestion site (data not shown).
We also observed the presence of fragments that were digested by chymotrypsin at the two Tyr residues inside the foldon domain after an extended period of incubation (overnight at 15 °C) in G913S but not in G901S and F877. It appears the removal or the unfolding of the triple helix domain have also destabilized the foldon domain and promoted the digestion at the Tyr residues. Both Tyr residues are not accessible in the fully folded foldon conformation.
A close inspection of the amino acid sequence revealed that the 24 residues between Gly-916 and Gln-939 at the C terminus constitute a very unstable region. This region not only contains the low propensity triplets like GIK and GFS but also is free of imino acid, Pro, which is the most favorable amino acid at the X position for triple helix (4). In fact, the 24-residue Pro-free region represents the longest stretch of sequences without imino acid in fibrillar collagens, and it was identified as the lowest stability region by several studies (4, 10, 12). The significantly increased susceptibility at Phe-935, which is located close to the C-terminal end of the Pro-free region, suggests the nearly complete opening of the Pro-free domain because of the substitution at Gly-913. The unfolding of this 24-residue Pro-free region can readily account for the 40% loss of triple helix signal at 225 nm of the CD spectra (Fig. 2).
The Long Range Stabilizing Effects of the Interchain Salt Bridges—The unfolding of the Pro-free region in G913S must be caused by the interruption of stabilizing factors due to the Gly913 → Ser substitution. The interchain salt bridges between the Lys and the Glu residues at the nearby KGE sequence (position 918-920, Diagram 1) appear to be the most likely candidate. Studies using peptides and molecular modeling revealed that in the homotrimer environment the positively charged Lys residue of a KGE sequence is placed in close contact with the negatively charged Glu residues of a neighboring staggering chain, and form a set of stabilizing interchain salt bridges.
To test the involvement of the KGE sequence, the temperature melt experiment of the wild-type F877 fragment without Gly substitution was carried out in pH 3 buffer. The carboxyl group of Glu-920 (pK ∼ 4.2) is expected to be protonated at pH 3, and the charge-charge interactions involving Glu eliminated. Indeed, at pH 3 the F877 fragment follows the same two-step melting profile as that of G913S at pH 7 (Fig. 4, upper panel). The titration of the Glu-920 in F877 functions the same way as the Gly-913 → Ser mutation to cause the unfolding of the Pro-free domain at 15 °C.
FIGURE 4.
Temperature melt curves of collagen fragment without the interchain salt bridges. Upper panel, the thermal unfolding of F877 in pH 3 buffer (open circle) and of E920A fragment at pH 7 (filled circle) under similar conditions as in Fig. 2. The melt of F877 (solid line) and G913S (dotted line) at pH 7 are included for comparison. Middle panel, the CD scan of E920A at 4 °C (open circle) plotted together with that of F877 (filled square). Lower panel, chymotrypsin digestion experiment of E920A at 15 °C under similar conditions as in Fig. 3.
Such a critical stabilizing effect of the KGE sequence on the Pro-free region is further supported by the study of a new fragment E920A. In E920A, the salt bridges are permanently removed by a Glu-920 → Ala substitution. As expected, the E920A fragment forms a stable triple helix as that of F877 at low temperature (Fig. 4, middle panel), but follows the two-step temperature melt profile as that of G913S (Fig. 4, upper panel). Furthermore, E920A shows the same susceptibility to chymotrypsin as G913S does at 15 °C (Fig. 4, lower panel), indicating the same degree of unfolding of the Pro-free region.
It is also interesting to note the nearly identical temperature melt profiles of F877 at pH 3 with that of G913S and E920A at pH 7 (Fig. 4, upper panel). The pH-induced conformational changes appear to be dominated by the interactions involving Glu-920, despite the presence of several other titratable residues, including the Glu-890 residue in the N-terminal region. Although the Glu-890 is adjacent to another positively charged residue Arg (Diagram 1), no stabilizing effects were found in RGE sequence in peptide models (13), which is consistent with the lack of effects of protonation of Glu-890 seen here.
From these data we can conclude that the Pro-free region in F877 constitutes a micro-unfolding region, which has intrinsic low thermal stability and prefers an unfolded conformation. However, this Pro-free region maintains the triple helix conformation under physiological conditions because of the salt bridges of the KGE sequence at its N-terminal end. Without the salt bridges, the 24-residue Pro-free region will unfold as an isolated cooperative unit.
DISCUSSION
The distinct effects of the two adjacently placed Gly → Ser mutations on the triple helix conformation underscore the close dependence of the regional properties of the triple helix on both the local amino acid residues and the long range stabilizing interactions. The different conformational changes are caused by the region C-terminal to the substitution sites. Located adjacent to an unstable Pro-free region and to the set of stabilizing salt bridges, the Gly substitution in G913S induces breakage of the salt bridges and an opening of more than 20 amino acids. This extensive opening is not seen in G901S because the set of salt bridges is not affected by the localized conformational changes at Gly-901 and the salt bridges, in turn, act to limit the spreading of the effects of the substitution at Gly-901 to reach the Pro-free zone. The effects of the Gly substitutions are thus modulated by the intrinsic helix propensity of residues in the immediate vicinity of the mutation site and the long range effects of specific stabilizing interactions, such as the interchain salt bridges.
The stabilizing effects of the interchain salt bridges of KGE and KGD sequence are well characterized in synthetic peptides (4) and more recently, in bacteria collagen (15). The inclusion of a KGE or KGD sequence increases the melting temperature of peptides with 24-30 amino acid residues by ∼15 °C. The range of this stabilizing effect of the salt bridges in collagen had never been characterized before. Our data has, for the first time, demonstrated experimentally that the stabilizing effects of the salt bridges can extend at least 15-20 residues C-terminal to the KGE sequence. The chymotrypsin digestion site Phe-935 is placed 15 residues C-terminal to the Glu-920. The effective digestion at Phe-935 may require the unfolding of a few more residues C terminus to Phe-935. Experiments are underway to investigate if the stabilizing effects of the salt bridges can extend to the similar range N-terminal to the sequence.
Knowledge of the long range effects of molecular interactions of the triple helix is fundamental for our ability to infer the structures and molecular dynamics of a segment of the triple helix from its constituent sequence. The Pro-free region has been identified as a thermally labile region based on the low helix propensity of the residues in the region (4, 10, 12), but little is known about its conformation in the context of the long helix of type I collagen. Here, we have demonstrated that this Pro-free region per se has the potential to be in the unfolded state. In fact, the isolated opening of the 24-residue Pro-free region in E920A exemplifies the cooperative unfolding of a micro-unfolding domain of collagen triple helix whose size, location, and functions have been implicated in various functions of collagen (8, 10). According to our data on F877, however, the Pro-free region is kept in triple helix conformation by the nearby salt bridges and unfolds cooperatively only with the rest of the helix.
The structural changes of the Gly substitutions appear to be small and localized when embedded in highly stable triple helical peptides (1, 25). The lowered thermal stability of the mutated chains is consistent with the structural studies indicating local distortions of the helix and interruption of H-bond networks. Such conformational changes at Gly-913 appear to be enough to affect the orientation and the alignment of the charged residues involved in the salt bridges at the nearby KGE sequence. In contrast, these salt bridges likely remain intact in G901S. NMR studies showed nearly identical backbone dynamics of a Gly located 15 residues C-terminal from Gly-901 in peptides with and without the Gly substitution, suggesting little conformational changes in this region (26). The KGE sequence located 18 residues away is thus, expected to maintain its normal conformation in G901S and its stabilizing effects on the Pro-free region. These observations accentuate the pivotal roles of the salt bridges of KGE in maintaining the triple helix conformation and in modulating the effects of the OI mutations. If the opening of the Pro-free region in G913S is the direct consequence of losing the stabilizing salt bridges, the presence of the salt bridges in G901S may act to limit the propagation of the effects of the mutation into the Pro-free region to avoid the large scale unfolding.
The more significant effects on the thermal stability of the Gly → Ser substitution at Gly-913 than at Gly-901 were also characterized in synthetic peptides (27). However, because each of the peptides contained a very short stretch of the helix, this study could not relate the more severe effects of the Gly-913 substitution to the disruption of the salt bridges and to the unfolding of the Pro-free domain.
A study using full chain type I collagen (10) revealed similar long range effects of OI mutation. The Gly substitution at position 25 and 36 of the α1(I) cause an opening of 70+ residues in the relatively stable N-anchor domain (the first 85 amino acid residues). It is intriguing how the relatively small changes of a Gly substitution can induce such a large scale unfolding in a region consisting of residues with relatively high helix propensity. The heterotrimeric environment of the type I collagen may be an important factor. The nonequivalent structure may aggravate the conformational effects of the substitution. Other critical stabilizing factors not yet identified may also play a role. The structural flexibility of a micro-unfolding region, identified by the low helix propensity of constituent residues, next to the N-anchor domain was postulated to mark the boundary of the domain and to help limit the spreading of the mutation induced conformational effects into the neighboring region (10). Interestingly, there is a KGE sequence at position 108 in both α1(I) and α2(I) chains, just 5 residues C-terminal to the postulated micro-unfolding region. It remains interesting to see if the salt bridges of this sequence play any role in limiting the propagation of the conformational effects of the mutations.
The fully folded N-terminal regions of both G901S and G913S appear to follow the renucleation and bi-directional propagation mechanism of the folding of collagen with Gly mutations (25). When modeled in short synthetic peptides, the 15 amino acid residues (residue 886-900) immediately N terminus to G901S fail to refold without the addition of a renucleation domain GPO(GAO)3 (25, 26). The triple helix formation in the N-terminal region of G901S and G913S can start from the repeating GPP sequence (diagram 1), which shares the same two major features with GPO(GAO)3: a high triple helix propensity and high imino acid content (25). NMR studies reveal an ordered structure around the Gly-901 substitution site when attached to the renucleation sequence GPO(GAO)3, although with a degree of helix untwisting and alteration of the H-bond network. Similar refolding of the N terminus of G913 is expected, since the 12 amino acid residues between Gly-901 and Gly-913 are fairly similar to the region N-terminal to Gly-901 in both composition and triple helix propensity.
The crucial position of the Pro-free region suggests the severe effects of the mutation at Gly-913 correlate with its more pronounced clinical consequences compared with the substitution at Gly-901. The Pro-free region overlaps with the proteoglycan binding site (28). The region also contains the lysine hydroxylase and lysyl oxidase binding site; the Lys-930 is known to be hydroxylated and glycosylated for cross-linking in type I collagen. Conformational changes in this region can potentially affect these interactions and result in detrimental effects on the properties and functions of bones. It is therefore, no surprise to find that entire Pro-free region is included in the lethal zone at position 910-964 (6). While the current study is conducted in a homotrimer environment modeling the sequence of the α1(I) chain only, significant destabilization of the Pro-free region by the Gly-913 substitution in the heterotrimeric collagen is expected because of the highly analogous sequences of the α1(I) and α2(I) chains in this region. Besides, the KGE sequence for the salt bridges in α1(I) is conserved in the α2(I) chain at the same position. The nonequivalent sequences of heterotrimers are often considered to be more disruptive than homotrimers with the potential to cause the bend or the kink of the triple helix (1). Two altered α1(I) chains appears to be effective to cause the unfolding of the N-anchor domain even when the α2(I) is normal (10). Thus, the salt bridges are expected to play the same critical stabilizing roles in the heterotrimer environment, and their disruption cause similar effects to the structure as seen in the homotrimer.
CONCLUSION
To summarize, by revealing the stabilizing effects of interchain salt bridges on the Pro-free region, our data demonstrated that the effects of the OI mutations are modulated by both the helix propensity of surrounding amino acid residues and the long range molecular interactions of the triple helix. Together, these findings support a novel explanation for the varied effects of mutations at different locations. The conformation of the collagen triple helix is maintained by certain specific stabilizing interactions, including but not limited to the interchain salt bridges. Mutations placed in the vicinity of these interactions tend to cause more disruptions of the overall conformation and, possibly, lead to more severe phenotypes of OI. Indeed, a survey of the OI mutation data base on Uniprot revealed that 90% of Gly mutations placed within 5 residues of a KGE or KGD sequence in both α1 and α2 chains are linked to severe cases of OI, suggesting the general nature of the critical effects of the salt bridges in maintaining the conformation and in the functions of collagen.
Acknowledgments
We thank Dr. Jurgen Engel (University of Basel, Switzerland) for generously sharing the GPP10-foldon clone with us. We thank Drs. Barbara Brodsky, Adele Boskey and Neville Kallenbach for insightful comments. We especially want to thank Dr. Michael Blumenstein for numerous helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant 3S06-GM060654-06S1. This work was also supported by National Science Foundation (NSF) Grant DBI0521709, PSC-CUNY Grants 60042-34 35 and 67874-0036, and the Gender Equity Program at Hunter (NSF Advanced Transformation Award). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OI, osteogenesis imperfecta; Hyp, hydroxyproline (three-letter code); O, hydroxyproline (single-letter code); Hyl, hydroxyllysine (three-letter code).
References
- 1.Bella, J., Eaton, M., Brodsky, B., and Berman, H. M. (1994) Science 266 75-81 [DOI] [PubMed] [Google Scholar]
- 2.Rich, A., and Crick, F. H. C. (1961) J. Mol. Biol. 3 483-506 [DOI] [PubMed] [Google Scholar]
- 3.Ramachandran, G. N., Doyle, B. B., and Boult, E. R. (1968) Biopolymers 6 1771-1775 [DOI] [PubMed] [Google Scholar]
- 4.Persikov, A. V., Ramshaw, J. A. M., and Brodsky, B. (2005) J. Biol. Chem. 280 19343-19349 [DOI] [PubMed] [Google Scholar]
- 5.Byers, P. H. (1993) in Osteogenesis Imperfecta, Connective Tissue and Its Heritable Disorders (Royce, P. M., and Steinmann, B., eds), pp. 317-350, Wiley-Liss, New York
- 6.Marini, J. C., Forlino, A., Cabral, W. A., Barnes, A. M., Antonio, J. D. S., Milgrom, S., Hyland, J. C., Korkko, J., Prockop, D. J., Paepe, A. D., Coucke, P., Symoens, S., Glorieux, F. H., Roughley, P. J., Lund, A. M., Kuurila-Svahn, K., Hartikka, H., Cohn, D. H., Krakow, D., Mottes, M., Schwarze, U., Chen, D., Yang, K., Kuslich, C., Troendle, J., Dalgleish, R., and Byers, P. H. (2007) Hum. Mutat. 28 209-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kielty, C. M., Hopkinson, I., and Grant, M. E. (1993) in Connective Tissue and Its Heritable Disorders (Royce, P. M., and Steinmann, B., eds), pp. 103-147, Wiley-Liss, New York
- 8.Privalov, P. L., Tictopulo, E. I., and Tischenko, V. M. (1979) J. Mol. Biol. 127 203-216 [DOI] [PubMed] [Google Scholar]
- 9.Privalov, P. L. (1982) Adv. Prot. Chem. 35 1-104 [PubMed] [Google Scholar]
- 10.Makareeva, E., Cabral, W. A., Marini, J. C., and Leikin, S. (2006) J. Biol. Chem. 281 6463-6470 [DOI] [PubMed] [Google Scholar]
- 11.Makareeva, E., Mertz, E. L., Kuznetsova, N. V., Sutter, M. B., DeRidder, A. M., Cabral, W. A., Barnes, A. M., McBride, D. J., Marini, J. C., and Leikin, S. (2008) J. Biol. Chem. 283 4787-4798 [DOI] [PubMed] [Google Scholar]
- 12.Bachinger, H. P., Morris, N. P., and Davis, J. M. (1993) Am. J. Med. Genet. 45 152-162 [DOI] [PubMed] [Google Scholar]
- 13.Persikov, A. V., Ramshaw, J. A. M., Kirkpatrick, A., and Brodsky, B. (2002) J. Mol. Biol. 316 385-394 [DOI] [PubMed] [Google Scholar]
- 14.Venugopal, M. G., Ramshaw, J. A., Braswell, E., Zhu, D., and Brodsky, B. (1994) Biochemistry 33 7948-7956 [DOI] [PubMed] [Google Scholar]
- 15.Mohs, A., Silva, T., Yoshida, T., Amin, R., Lukomski, S., Inouye, M., and Brodsky, B. (2007) J. Biol. Chem. 282 29757. [DOI] [PubMed] [Google Scholar]
- 16.Frank, S., Kammerer, R. A., Mechling, D., Schulthess, T., Landwehr, R., Bann, J., Guo, Y., Lusting, A., Bachinger, H. P., and Engel, J. (2001) J. Mol. Biol. 308 1081-1089 [DOI] [PubMed] [Google Scholar]
- 17.Mechling, D. E., and Bachinger, H. P. (2000) J. Biol. Chem. 275 14532-14536 [DOI] [PubMed] [Google Scholar]
- 18.Byers, P. H., Wallis, G. A., and Willing, M. C. (1991) J. Med. Genet. 28 433-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel, J., Chen, H. T., Prockop, D. J., and Klump, H. (1977) Biopolymers 16 601-622 [DOI] [PubMed] [Google Scholar]
- 20.Leikina, E., Mertts, M. V., Kuznetsova, N., and Leikin, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, No 3 1314-1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruckner, P., and Prockop, D. J. (1981) Anal. Biochem. 110 360-368 [DOI] [PubMed] [Google Scholar]
- 22.Engel, J., and Prockop, D. J. (1991) Annu. Rev. Biophys. Biophys. Chem 20 137-152 [DOI] [PubMed] [Google Scholar]
- 23.Engel, J., and Bachinger, H. P. (2000) Matrix Biology 19 235-244 [DOI] [PubMed] [Google Scholar]
- 24.Xu, Y., Bhate, M., and Brodsky, B. (2002) Biochemistry 41 8143-8151 [DOI] [PubMed] [Google Scholar]
- 25.Hyde, T. J., Bryan, M. A., Brodsky, B., and Baum, J. (2006) J. Biol. Chem. 281 36937-36943 [DOI] [PubMed] [Google Scholar]
- 26.Liu, X., Kim, S., Dai, Q.-H., Brodsky, B., and Baum, J. (1998) Biochemistry 37 15528-15533 [DOI] [PubMed] [Google Scholar]
- 27.Yang, W., Battineni, M. L., and Brodsky, B. (1997) Biochemistry 36 6930-6935 [DOI] [PubMed] [Google Scholar]
- 28.DiLullo, G. A., Sweeney, S. M., Körkkö, J., Ala-Kokko, L., and Antonio, J. D. S. (2002) J. Biol. Chem. 277 4223-4231 [DOI] [PubMed] [Google Scholar]