Abstract
Cellular migration is a fundamental process linked to diverse pathological states such as diabetes and its complications, atherosclerosis, inflammation, and cancer. The receptor for advanced glycation end products (RAGE) is a multiligand cell surface macromolecule which binds distinct ligands that accumulate in these settings. RAGE-ligand interaction evokes central changes in key biological properties of cells, including proliferation, generation of inflammatory mediators, and migration. Although RAGE-dependent signal transduction is critically dependent on its short cytoplasmic domain, to date the proximate mechanism by which this RAGE domain engages and stimulates cytoplasmic signaling pathways has yet to be identified. Here we show that the RAGE cytoplasmic domain interacts with Diaphanous-1 (Dia-1) both in vitro and in vivo. We employed the human RAGE cytoplasmic domain as “bait” in the yeast two-hybrid assay and identified the formin homology (FH1) domain of Dia-1 as a potential binding partner of this RAGE domain. Immunoprecipitation studies revealed that the RAGE cytoplasmic domain interacts with the FH1 domain of Dia-1. Down-regulation of Dia-1 expression by RNA interference blocks RAGE-mediated activation of Rac-1 and Cdc42 and, in parallel, RAGE ligand-stimulated cellular migration. Taken together, these findings indicate that the interaction of the RAGE cytoplasmic domain with Dia-1 is required to transduce extracellular environmental cues evoked by binding of RAGE ligands to their cell surface receptor, a chief consequence of which is Rac-1 and Cdc42 activation and cellular migration. Because RAGE and Dia-1 are implicated in the regulation of inflammatory, vascular, and transformed cell migration, these findings highlight this interaction as a novel target for therapeutic intervention in inflammation, atherosclerosis, diabetes, and cancer.
The receptor for advanced glycation end products (RAGE)5 is a multiligand cell surface macromolecule of the immunoglobulin superfamily which binds diverse ligands, including advanced glycation end products (1), S100/calgranulins (2), high mobility group Box-1 (HMGB1) (3), amyloid-β peptide (Aβ), and β-sheet fibrils (4). RAGE-ligand interaction evokes central changes in cellular properties including stimulation of cellular migration (2, 5-7). Extensive evidence suggests that pharmacological antagonism or genetic modulation of RAGE exerts protection against disease states characterized by up-regulation and accumulation of RAGE ligands, such as the complications of diabetes, atherosclerosis, inflammation, and tumors (2, 7-9).
Our studies have definitively shown that the ligands of RAGE are not simply tethered to this receptor. Rather, studies in vitro and in vivo indicate that RAGE is a signal transduction receptor for these ligand families (6, 9, 10). Both in vitro and in vivo experiments reveal that deletion of the short cytoplasmic domain of RAGE exerts a “dominant negative” (DN) effect in which the signal transduction response to RAGE ligand is blunted (5-7). Studies have shown that activation of RAGE mediates key effects in cellular and in vivo systems. First, ligand-RAGE interaction stimulates cellular motility (7, 11). Second, a number of signal transduction cascades are activated upon ligand-RAGE interaction, including mitogen-activated protein kinases, phosphatidylinositol 3-kinase, Jak/STAT (signal transducers and activators of transcription), and importantly, the Rho GTPases Rac-1 and Cdc42 (2, 5, 7, 10, 12-14). To date, however, the precise molecular mechanism(s) underlying the diverse repertoire of RAGE signaling has yet to be identified. Intriguingly, the cytosolic domain of RAGE lacks endogenous tyrosine kinase activity or any known motifs involved in receptor signaling. Here, we hypothesized that the binding partner(s) of the RAGE cytoplasmic domain may represent a novel mechanism to stimulate signal transduction and thereby mediate pivotal changes in cellular properties linked to disease states.
To establish the precise mechanisms by which the RAGE cytoplasmic domain stimulates cell signaling, we employed the yeast two-hybrid system and report that this domain of RAGE interacts with Diaphanous-1, which we confirmed by multiple in vitro and in vivo methodology. In vivo, RAGE-mediated activation of Rac-1 and Cdc42 and cellular migration is dependent on Dia-1. These studies elucidate a novel signaling paradigm by which extracellular cues stimulated by RAGE ligand binding are transduced through the cytoplasmic domain of the receptor via Dia-1 to stimulate fundamental signaling networks and cell migration.
EXPERIMENTAL PROCEDURES
Cell Lines and Materials—Rat C6 glioma cells were obtained from ATCC (CCL-107) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). SK-BR-3 cells were obtained from ATCC (HTB-30) and maintained in McCoy's 5A media supplemented with 10% fetal bovine serum (Invitrogen). RAGE ligand carboxyl methyl lysine (CML)-human serum albumin (HSA) was prepared by the incubation of HSA (25 mg/ml), sodium cyanoborohydride (10 mm) in sodium phosphate buffer (0.2 m; pH 7.8) and glyoxylic acid at 37 °C for 24 h followed by dialysis. CML modification was characterized using 2,4,6-trinitrobenzenesulfonic acid and gas chromatography mass spectrometry. Endotoxin levels evaluated by chromogenic Limulus assay (Associates of Cape Cod, Inc.) amounted to <0.002 enzyme units/mg of protein in the experimental preparations.
Yeast Two-hybrid Screen—The Matchmaker Two-Hybrid System 2 was used to screen a human lung Matchmaker cDNA library (BD Clontech) with the cytoplasmic domain of RAGE used as bait (corresponding to amino acids 361-404 following the transmembrane domain (GenBank™ accession number AY755619)). To construct the bait plasmid, DNA encoding the RAGE cytoplasmic domain was cloned in-frame with the GAL4 DNA-BD of the pAS2-1 vector (BD Clontech). The yeast strain Y190 was co-transformed with the lung cDNA library (cloned into the GAL4 activation domain of the vector pACT2 (BD Clontech)) and the pAS2-1-RAGE-cytoplasmic domain clone. Colonies that grew successfully on the selective media SD/-His/-Leu/-Trp were screened further for reporter gene β-galactosidase expression by the colony-lift filter assay. Plasmid DNA was isolated from positive clones and transformed into Escherichia coli-competent cells to enable isolation and sequencing. Plasmid DNA was sequenced, and clones were identified by BLAST searches (www.ncbi.nlm.nih.gov/BLAST/). Multiple clones were identified encoding a fragment of the Dia-1 gene corresponding to GenBank™ accession number AF051782. The yeast two-hybrid clone sequence was submitted to GenBank™ (accession number DQ067452).
hDia-1 Full-length cDNA Cloning—To clone the full-length cDNA of Dia-1, the start (5′ end/transcription start site) and finish (3′ end/polyadenylation site) of the human diaphanous mRNA was determined by RNA ligase-mediated rapid amplification of cDNA ends using the GeneRacer kit (Invitrogen).
Human lung mRNA (BD Clontech) was prepared for reverse transcription with GeneRacer to allow selection of only full-length mRNAs according to the manufacturer's instructions (Invitrogen). RNA ligase-mediated rapid amplification of cDNA end-treated lung mRNA was reverse-transcribed using Thermoscript cDNA synthesis kit (Invitrogen) using the GeneRacer Oligo-dT primer (Invitrogen). To amplify the 5′/3′ end of Dia-1, primers were designed on the sense/antisense strand to amplify from within the fragment isolated from the yeast two-hybrid to capture the respective 5′/3′ end. The 5′ end/transcription start site was identified by performing PCR using the GeneRacer 5′ primer (Invitrogen) and a Dia-1-specific 3′ Primer (Dia-5RACE1, 5′-GGCAAAGAAGAGGGTGAAGGGAT-3′) followed by a nested PCR using the GeneRacer 5′ Nested Primer (Invitrogen) and a Dia-1-specific 3′-nested primer (Dia1-5RACE2, 5′-CAGGTAAAGAAGGGGGTGAGGAGA-3′). Similarly, the 3′ end/polyadenylation site was determined using a Dia-1-specific 5′ primer (Dia1-3RACE1, 5′-TATGCCTCCACCTCCCCCATTT-3′) and the GeneRacer 3′ primer (Invitrogen) followed by a nested PCR with Dia-1-specific 5′ primer (Dia-3RACE2, 5′-GCAGCCCCAGTTCTGCCATTT-3′) and the GeneRacer 3′-nested primer (Invitrogen). PCRs were performed with Platinum Taq High Fidelity DNA polymerase (Invitrogen) using a PTC-200 thermocycler (MJ Research). Purified RACE-amplified PCR products were cloned using the TOPO TA cloning system (Invitrogen), and the plasmid DNA was purified by QIAprep Spin Mini-prep (Qiagen). Plasmid DNA was sequenced using the plasmid-specific primers M13F(-20) and M13R and sequenced on a ABI 3100 DNA sequencer (Applied Biosystems).
Using the lung cDNA used in RNA ligase-mediated rapid amplification of cDNA ends experiments, PCR was performed using Dia1-5′ (GTGAACCGGGACATGGAGCC) and Dia1-3′ (GTCAGCGAGCAGGGAGGAGGT), and the resultant PCR product was cloned using the TA TOPO Cloning System (Invitrogen). Cloned Dia-1 cDNA was sequenced using multiple overlapping primers in 500-bp steps to verify the correct sequence of the cloned cDNAs. The resulting full-length Dia-1 cDNA sequence was deposited in GenBank™ as accession number DQ067453.
RAGE/Dia-1 Interaction Studies and in Vitro Binding Assay—The RAGE cytoplasmic domain cDNA (as used for yeast two-hybrid) was cloned in-frame with the GST tag (N terminus tag) in the vector pGEX-4T (Amersham Biosciences). hDia-YIIH was cloned in-frame with the His6 tag (N terminus tag) in the vector pRSET (Invitrogen). All constructs were sequenced to ensure that RAGE and Dia-1 were in-frame with the epitope tags. GST-RAGE tail/GST empty vector was transformed into BL21(DE3) E. coli (Invitrogen), and expression was induced using 1 mm isopropyl 1-thio-β-d-galactopyranoside. Because of low levels of expression in bacteria, His-hDia-YIIH/His empty vector was expressed using the TNT® Quick Coupled Transcription/Translation System according to the manufacturer's instructions (Promega).
Pulldown assays were performed by using the Profound GST Pull-Down Interaction kit (Pierce) according to the manufacturer's instructions. Proteins were detected by either Coomassie Blue or by Western blot. Western blotting was performed using 4-12% NuPAGE Bis-Tris gel with MOPS buffer (Invitrogen), nitrocellulose membranes (Invitrogen), incubation with anti-HisG antibodies (Invitrogen), or streptavidin-horseradish peroxidase (Promega). Protein signal was detected using ECL reagent (GE Healthcare) and exposed to x-ray film.
RAGE/Dia-1 Interaction Studies and Coimmunoprecipitation of the Epitope-tagged RAGE Cytoplasmic Domain and hDia-1—The RAGE cytoplasmic domain and hDia-1 were cloned into epitope tag vectors to allow immunoprecipitation studies to be performed with antibodies independent of RAGE and hDia-1. The RAGE cytoplasmic domain cDNA (as used for yeast two-hybrid) was cloned in-frame with the His6 tag (N terminus tag) in the vector pcDNA4-HisMax (Invitrogen). Dia-1 full-length cDNA was cloned in-frame with the Myc6 tag (N terminus tag) in the vector pCS2+MT (provided by Jan Kitajewski, Columbia University). All constructs were sequenced to ensure the RAGE and Dia-1 were in-frame with the epitope tags. SK-BR-3 cells were seeded overnight at 1 × 106 cells/100-mm dish in complete media and transfected with His-RAGE-cytoplasmic domain and Myc-hDia-1 using FuGENE 6 (Roche Applied Science). Negative controls were included to assess for nonspecific interaction by transfecting separate dishes of cells with His-RAGE-cytoplasmic domain/pcS2-MT empty and Myc-Diaphanous/pcDNA4-HisMax empty. After 48 h cells were stimulated with 10 μg/ml CML-HSA for 1 h, rinsed with ice-cold phosphate-buffered saline, and lysed using MPER lysis buffer (Pierce) containing 1 mm phenylmethylsulfonyl fluoride and Complete Protease Inhibitors (Roche Applied Science). After scraping, cellular debris was pelleted, and immunoprecipitation was performed with 500 μg of cell lysate and 1 μg of anti-HisG/Myc (Invitrogen) for 1 h at 4 °C followed by the addition of protein A/G agarose (Pierce) overnight at 4 °C. Protein A/G-agarose was pelleted and washed three times using MPER lysis buffer followed by resuspension in NuPAGE sample buffer and reducing reagent (Invitrogen) for electrophoresis. Western blotting was performed as described above using anti-HisG or anti-Myc monoclonal antibodies (Invitrogen).
RAGE/Dia-1 Interaction Studies: Immunoprecipitation of Full-length and DN-RAGE to Test for RAGE/Dia-1 Interaction—Full-length RAGE cDNA (1) was generated from lung cDNA (Clontech) by PCR as described previously (15). DN-RAGE was generated using PCR by inserting a stop codon into the cDNA sequence of human RAGE at the end of the transmembrane domain (amino acid number 361) to remove the cytoplasmic tail. Subsequently PCR was performed as for full-length RAGE using the RAGE 5′-UTR1 primer and a reverse primer (RAGE-DNr, 5′-CTTGCCACTAGATGACCCC-3′) and cloned into pcDNA3.1. C6 cells were stably transfected with human full-length (RAGE), cytoplasmic domain-deleted RAGE (DN-RAGE), or empty pcDNA3.1 (mock-transfected) constructs using FuGENE 6 (Roche Applied Science) and by selection with neomycin (G418). Stable clones were screened for RAGE/DN-RAGE expression by Western blot using anti-RAGE IgG (1). RAGE and DN-RAGE C6 stable cells were seeded at 1 × 106 cells/100-mm dish in complete media and grown for 24 h before starvation overnight in serum-free media. Cells were stimulated with 10 μg/ml CML-HSA for 30 min, and cell lysis was performed as above. Immunoprecipitation was performed with 1 μg of monoclonal mouse anti-human RAGE antibody (Chemicon) with all conditions the same as above. Western blots were prepared as above and blotted with monoclonal mouse anti-human Dia-1 antibody (BD Pharmingen).
RAGE/Dia-1 Interaction Studies: Confocal Microscopy—Mock- and RAGE-transfected C6 cells were seeded at 1 × 104 cells/well of 4-chamber culture slides (BD Biosciences) and allowed to grow for 24 h. Cells were starved overnight and stimulated with 10 μg/ml CML-HSA for 30 min. Cells were washed in phosphate-buffered saline, fixed using 4% paraformaldehyde solution, washed in phosphate-buffered saline, permeabilized, and blocked with 0. 1% Triton X-100 in CAS-block (Zymed Laboratories Inc.). Double antibody confocal microscopy was performed by incubating cells with mouse anti-Dia-1 (BD Pharmingen) overnight at 4 °C followed by incubation at room temperature for 1 h with rabbit anti-RAGE (1). Biotinylated secondary antibodies (anti-mouse and anti-rabbit biotin) were incubated sequentially between primary antibody incubations for 30 min each followed by incubation with streptavidin conjugate Alexa Fluor 488 or Alexa Fluor 555, respectively (Invitrogen). Slides were mounted with Vectorshield mounting media (Vector) and observed with an oil immersion objective using a Nikon E800 microscope. Images were collected using a Bio-Rad Radiance 2000 Confocal System and the Lasersharp 2000 software (Bio-Rad).
Construction of hDia-1 Mutants—hDia-1 truncation mutants including hDia-FH1 and hDia-RhoBD were created from wild-type Myc-hDia-1 using the QuikChange II site-directed mutagenesis kit (Stratagene) by introducing stop codons at amino acid numbers 786 (hDia-FH1) and 253 (hDia-RhoBD). Myc-hDia-YIIH was generated by subcloning hDia-YIIH into the Myc-tag vector pCMV-Myc (BD Clontech).
Coimmunoprecipitation of the Epitope-tagged RAGE Cytoplasmic Domain and hDia-1 Mutants—His-RAGE-cytoplasmic domain and Myc-hDia-1 constructs (hDia-1, hDia-FH1, hDia-YIIH, hDia-RhoBD, empty Myc vector) were transfected into SK-BR-3 cells, and immunoprecipitation and Western blotting were performed as described above with anti-His/Myc antibodies.
Cellular Migration Assays—Chemotaxis assays were performed using the QCM Colorimetric Cell Migration Assay (Chemicon). Cells were seeded per upper chamber fitted with a lower 8-μm porous polycarbonate membrane, and the insert was placed in the lower chamber of a 24-well dish containing Dulbecco's modified Eagle's medium and no-stimulant, CML-HSA RAGE-ligand, or 10% fetal bovine serum (control) and incubated at 37 °C for 5 h according to the manufacturer's instructions (Chemicon). Cells migrated through the porous membrane were incubated with Cell Stain Solution (Chemicon) followed by representative counts of four randomly selected microscope fields. In other studies a distinct RAGE ligand, S100B (Calbiochem), was used to stimulate cells.
GTPase Activation Assays—The activation of GTPases was investigated using EZ-Detect Rac-1, Cdc42, and RhoA activation kits (Pierce). C6 RAGE and RAGE-DN cells were seeded at 1 × 106 cells per 100-mm dish and grown for 24 h. Cells were starved for 24 h by replacing media with Dulbecco's modified Eagle's medium containing no fetal bovine serum. RAGE and RAGE-DN cells were incubated with/without CML-HSA for 1, 5, 15, and 30 min at 37 °C to stimulate GTPase activation. Activated GTPases were purified using the EZ-Detect Rac-1, Cdc42, and RhoA activation kits (Pierce). The amount of GTP-bound Rac-1, Cdc42, and RhoA and the total amount of Rac-1, Cdc42, and RhoA in cell lysates was determined by Western blot with respective antibodies.
GTPases and Migration Assays—Migration assays were performed with dominant negative constructs of Rac1 (N17), Cdc42 (N17), RhoA (N19) (Millipore), and vector alone (Millipore) by cotransfection with the GFP expression vector pMaxGFP (Amaxa) into C6-RAGE cells as described above. Cells were subjected to migration assays as above, and numbers of migrating cells were calculated by counting GFP-positive cells that had passed through the porous membrane.
Dia-1 siRNA—Predesigned Stealth™ Select RNA interference was obtained from Invitrogen corresponding to rat Dia-1 (5′-GAACCAGAAGGAGAGUUCCAGGUCU-3′) and matched RNA interference negative controls (scramble siRNA). To validate Dia-1 knockdown, Dia-1 and control siRNA was transfected into RAGE C6 cells, seeded at 1 × 106 cells/100 mm dish using Lipofectamine 2000 (Invitrogen) and Dia-1 levels assessed by both quantitative PCR and Western blot. To analyze the mRNA levels of Dia-1, RNA was isolated from cells using Trizol (Invitrogen) and reverse-transcribed using the Superscript III reverse transcription-PCR system (Invitrogen). Quantitative-PCR analysis was subsequently performed for Dia-1 levels using Dia-1 Gene Expression Assays (Applied Biosystems), normalized to ribosomal 18 S using the 18 S probe (Applied Biosystems), and analyzed on an Mx3005P real-time PCR system (Stratagene). Protein levels of Dia-1 were detected by Western blot analysis on cell lysates, normalized by correcting to glyceraldehyde-3-phosphate dehydrogenase levels (Chemicon).
Dia-1 siRNA and Migration Assays—To enable quantification of Dia-1/scramble siRNA-transfected cell migration, C6-RAGE cells were cotransfected with siRNA and pMaxGFP expression vector with knockdown of Dia-1 levels assessed using immunofluorescence as described above. Migration assays were performed using siRNA/GFP-cotransfected cells as described above.
Dia-1 Rac-1/Cdc42 Activation Assays—C6-RAGE cells were transfected with Dia-1/scramble siRNA, and Rac-1 activation was assessed using the EZ Detect Rac-1 and Cdc42 activation kit (Pierce) as described above.
Statistical Analyses—In all experiments, unless otherwise indicated, data are reported as the mean ± S.D. in at least three replicates per group. Data were analyzed by post hoc comparisons using 2-tailed t test, and a p value less than or equal to 0.05 was considered significant.
RESULTS
Identification of Dia-1 as a Binding Partner of the RAGE Cytoplasmic Domain Using the Yeast Two-hybrid System—To understand how the RAGE cytoplasmic domain interacts with the intracellular partner(s) to stimulate signal transduction, we screened a yeast two-hybrid human lung cDNA library using the human RAGE cytoplasmic domain as bait. By this approach we identified a potential interaction of the RAGE cytoplasmic domain with human Diaphanous-1 (herein denoted hDia-1), a member of the formin protein family, defined by two key formin homology (FH) domains, FH1 and FH2 (16). The proline-rich FH1 domain of Dia-1 interacts with mediators of the actin cytoskeleton (profilin) and various signal transduction pathway molecules, such as c-src, SH3 domain-containing BAIAP2, and DIP, to mediate cellular movement (17-20). Analysis of potential RAGE-interacting clones revealed the hDia-1 cDNA isolated to span the FH1 domain from amino acid number 607-835 (Fig. 1). This fragment contained a novel 11-amino acid sequence stretch from amino acid number 611, which was not identified in the previously published human Dia-1 sequence (AF051782) (21) (Fig. 1B).
FIGURE 1.
Diaphanous-1 is a binding partner for the RAGE cytoplasmic domain. A, schematic of the hDia-1, as adapted from Krebs et al. (17) and Otomo et al. (36). Domains of hDia-1 in the schematic include the Rho GTPase binding domain (Rho-BD), diaphanous inhibitory domain (DID), dimerization domain (DD), coiled-coil region (CC), formin-homology region 1, 2, and 3 (FH1, -2, -3) and diaphanous autoinhibitory domain (DAD). The arrow from 607 to 835 represents the fragment of hDia-1 isolated from a lung cDNA library using the RAGE cytoplasmic domain as bait. The red box represents the novel FH1 sequence identified. B, protein sequence alignment of the isolated yeast two-hybrid clone of hDia-1 (hDia-YIIH). The alignment includes the GenBank™ accession number AF051782 (21) and the full-length sequence of human Dia-1 generated in this study (hDia-Full). The canonical and novel protein sequence differences of hDia-1 identified in this study are indicated in red.
Further analysis by alignment to the genomic sequence on chromosome 5q31 revealed our sequence to be the correct sequence and the original GenBank™ entry to be due to a deletion during reverse transcription-PCR or cloning (supplemental Fig. 1). We also confirmed this was not due to a splice variant, as in exon 15, where this occurred; we identified no consensus for splicing according to the AG/CT rule (supplemental Fig. 1) or by PCR analysis of cDNA from multiple cell and tissues with primers designed to span this region (data not shown). In addition, the hDia yeast two-hybrid fragment (herein called hDia-YIIH) did not arise by alternative promoter usage or 3′-untranslated region (supplemental Fig. 2). Subsequently, the full-length hDia-1 cDNA sequence was amplified and confirmed that this contained the novel sequence in the FH1 domain. Taken together, these data indicated this to be the correct sequence of hDia-1.
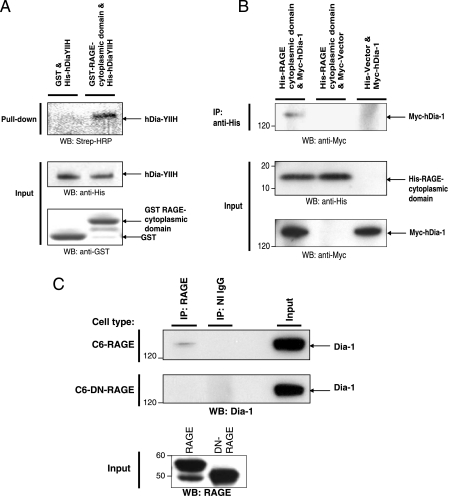
Dia-1 Interacts with the RAGE Cytoplasmic Domain in an in Vitro GST Pulldown Assay—To verify the interaction between the RAGE cytoplasmic domain and hDia-1, we tested the ability of recombinant GST-RAGE-cytoplasmic domain to bind in vitro to hDia-YIIH produced by in vitro transcription/translation. Using GST pulldown assays, hDia-YIIH bound to the GST-RAGE-cytoplasmic domain (Fig. 2A, top panel, second lane). However, hDia-YIIH did not bind to GST alone (Fig. 2A, top panel, first lane). These data confirm the yeast two-hybrid finding that the RAGE cytoplasmic domain interacts in vitro with hDia-1.
FIGURE 2.
Diaphanous-1 interacts with the RAGE cytoplasmic domain: in vitro and in vivo studies. A, the specificity of the interaction of Dia-1 with the RAGE cytoplasmic domain was analyzed in vitro by GST pulldown assays. GST only or GST-RAGE cytoplasmic tail fusion proteins were incubated with in vitro transcribed/translated His-tagged Dia-YIIH or His tag only (empty vector) labeled with biotinylated lysine-tRNA complex (Transcend™ tRNA). GST pulldown assays were performed, and eluted proteins were analyzed by Western blotting (WB) with streptavidin-horseradish peroxidase (Strep-HRP). An equivalent amount of the GST fusion proteins along with a sample from the in vitro transcription/translation reaction were analyzed by Western blotting using anti-His and anti-GST antibodies (lower panel). B, the RAGE cytoplasmic domain and Dia-1 interaction was analyzed by coimmunoprecipitation (IP) experiments with cell lysates from SK-BR-3 cells transiently transfected with His-tagged RAGE cytoplasmic domain or empty vector and Myc-tagged Dia-1 or empty vector. Cell extracts were immunoprecipitated with anti-His or anti-Myc antibodies and analyzed by Western blotting using anti-His and anti-Myc antibodies. C, C6 glioma cells stably expressing RAGE or the cytoplasmic domain-deleted RAGE (DN-RAGE) were analyzed for interaction with endogenous Dia-1 by coimmunoprecipitation experiments. Cells were serum-starved for 24 h followed by stimulation with CML-HSA (10 μg/ml) for 30 min. Cell extracts were then immunoprecipitated with anti-RAGE antibodies and analyzed by Western blotting with anti-Dia-1 antibodies. Total extract used for immunoprecipitation is indicated as input lysate and was analyzed by Western blotting with anti-RAGE and anti-Dia-1 antibodies. All figures (A-C) are representative images of multiple independent experiments.
Dia-1 Interacts with the RAGE Cytoplasmic Domain in Mammalian Cells—Next, we tested whether the RAGE cytoplasmic domain interacts with hDia-1 in vivo and performed coimmunoprecipitation experiments in mammalian cells transfected with the epitope-tagged RAGE cytoplasmic domain and full-length hDia-1. Western blot analysis revealed that the His-tagged RAGE cytoplasmic domain immunoprecipitated Myc-tagged hDia-1 (Fig. 2B, first lane). Importantly, Myc-hDia-1 was not coimmunoprecipitated from cells cotransfected with the empty His-tag vector control (Fig. 2B, third lane). To further confirm that endogenous Dia-1 interacted with the cytoplasmic domain of RAGE in vivo, we performed experiments to investigate if the interaction occurred with the full-length form of RAGE but not to the cytoplasmic domain-deleted DN-RAGE. From C6 RAGE and DN-RAGE-expressing cells, we performed immunoprecipitation studies using antibodies against RAGE, which revealed that from full-length RAGE-expressing cells, Dia-1 was coimmunoprecipitated with RAGE (Fig. 2C, top panel, first lane). However, Dia-1 did not coimmunoprecipitate with RAGE cytoplasmic domain-deleted DN-RAGE-expressing cells (Fig. 2C, second panel from top, first lane). Furthermore, in RAGE-expressing cells isotype-matched IgGs did not immunoprecipitate Dia-1 (Fig. 2C, top panel, second lane). Taken together, these data provided further support in vivo that Dia-1 interacted with RAGE and, specifically with its cytoplasmic domain.
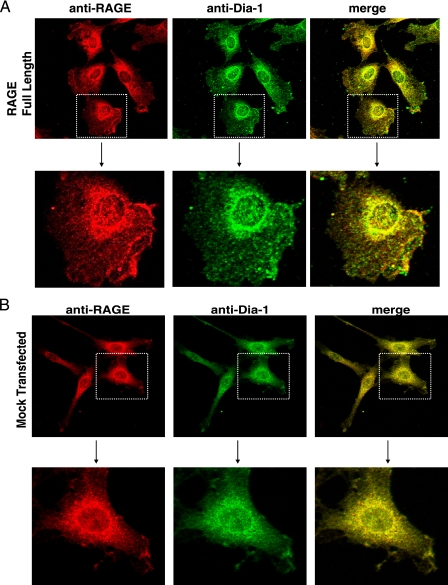
Dia-1 and RAGE Co-localization in the Cell—Next, we investigated the subcellular localization and cellular interaction between RAGE and Dia-1 by confocal microscopy using RAGE and mock C6-transfected cells stimulated with RAGE ligand. In RAGE-expressing cells, RAGE and Dia-1 exhibited a similar punctate cytoplasmic distribution in the perinuclear region which extended to the cell membrane. Extensive co-localization was observed in the merged image (Fig. 3A). Additionally, a significant population of the signal in dot-like structures at the cell membrane was seen (Fig. 3A). Because C6 cells endogenously express rat RAGE, we assessed as a control the colocalization between endogenous rat RAGE and Dia-1 in mock and empty vector-transfected C6 cells. As shown in Fig. 3B, endogenous RAGE and Dia-1 exhibited a pattern similar to the C6-RAGE cells with a punctate cytoplasmic distribution in the perinuclear region, extending to the cell membrane, with extensive co-localization in the merged image.
FIGURE 3.
RAGE and Dia-1 colocalization in cells. Confocal microscopy was performed using the C6 cells stably transfected with either full-length RAGE (A) or empty vector (Mock) (B) constructs. Cells were treated with RAGE ligand for 30 min, fixed, and immunostained with antibodies to RAGE and Dia-1 followed by respective biotinylated secondary antibodies and streptavidin-linked Alexa-546 (anti-RAGE) and 488 (anti-Dia-1). Higher magnification images of the cells within the stippled boxes in the upper panels are illustrated in the lower panels.
The RAGE Cytoplasmic Domain Interacts with the FH1 Domain of Dia-1—Based on these findings, we prepared truncation mutants of hDia-1 to define the sites within hDia-1 that were critical for interaction with the RAGE cytoplasmic domain (schematically represented in Fig. 4A). To investigate this, cells were cotransfected with His-tagged RAGE cytoplasmic domain and either Myc-tagged hDia-1 wild-type or truncation mutants and immunoprecipitated with His monoclonal antibody. Western blot analysis revealed that both Myc-hDia-1 wild-type and hDia-1 mutants containing the multifunctional proline-rich FH1 domain (hDia-FH1, hDia-YIIH) immunoprecipitated with the His-RAGE cytoplasmic domain (Fig. 4B, left panel, first through third lanes). In contrast, a hDia-1 mutant containing only the N-terminal 252 amino acids (hDia-RhoBD) lacking this FH1 domain (Fig. 4B, left panel, fourth lane) failed to immunoprecipitate the His-RAGE cytoplasmic domain. Note that input controls are illustrated in Fig. 4B, right panel. These data, therefore, support that the interaction between RAGE and Dia-1 occurs between the RAGE cytoplasmic domain and the FH1 domain of hDia-1.
FIGURE 4.
The FH1 domain of hDia-1 is critical for binding to the RAGE cytoplasmic domain. A, schematic diagram of Myc-tagged hDia-1 and truncation mutants used in this study. Numbers indicate amino acid positions in human Dia-1. Rho-BD, Rho GTPase binding domain; DID, diaphanous inhibitory domain; DD, dimerization domain; CC, coiled-coil region; FH1 and -2, formin-homology region 1 and 2; DAD, diaphanous autoinhibitory domain. B, to analyze the domain(s) of Dia-1, which interacts with the RAGE cytoplasmic domain, coimmunoprecipitation (IP) experiments were performed. SK-BR-3 cells were transiently cotransfected with the indicated Myc-tagged hDia-1 constructs and the His-tagged RAGE cytoplasmic domain. Immunoprecipitation was performed with anti-His antibodies followed by Western blotting (WB) with anti-Myc antibodies (left panel). Western blot analysis was performed on input lysate as indicated with anti-Myc and anti-His antibodies for hDia-1 and RAGE cytoplasmic domain constructs, respectively (right panel). Note that the hDia-RhoBD does not bind to the RAGE cytoplasmic domain. All panels (A-B) are representative images of multiple independent experiments.
The RAGE Cytoplasmic Domain Is Required for Ligand-driven Cellular Migration and GTPase Signaling—The key test of these concepts was whether the interaction of the RAGE cytoplasmic domain with hDia-1 was required to effect signal transduction and changes in cellular properties upon RAGE-ligand stimulation. To address this question we first performed studies with RAGE signaling-deficient (DN-RAGE) cells to determine the RAGE-dependent pathways and, second, with RNA interference to silence Dia-1 to investigate its function in RAGE-ligand cellular biology. As studies have elucidated central roles for ligand-RAGE signaling and Dia-1 in modulation of cellular migration in cells such as monocytes/macrophages, T lymphocytes, smooth muscle cells, or cells of tumorigenic origin (6, 7, 22, 23), we focused our experiments on cellular migration and the effector signal transduction pathways (Rho GTPases). In full-length RAGE-expressing cells, migration was triggered by RAGE ligand CML-HSA compared with vehicle treatment (Fig. 5). In contrast, DN-RAGE-expressing cells failed to migrate in response to RAGE-ligand CML-HSA (Fig. 5). A similar effect was seen with the RAGE-ligand, S100B (data not shown), and therefore, we focused our remaining studies on the RAGE-ligand CML-HSA.
FIGURE 5.
The RAGE cytoplasmic domain is required for cellular migration. Migration assays were performed with C6 glioma cells stably expressing RAGE or the cytoplasmic domain deleted RAGE (DN-RAGE) to verify the requirement of the RAGE cytoplasmic domain for RAGE-ligand-driven cellular migration. Cells (1 × 105) were seeded into the top of transwell migration chambers and allowed to migrate for 5 h toward RAGE-ligand or media alone negative control placed in the lower chambers. Cells were fixed and stained using cell stain solution, and three independent fields were counted. Results are expressed as the mean ± S.D. (n = 3).
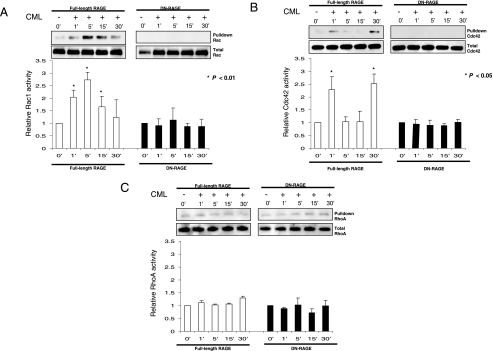
Central to the mechanism of RAGE ligand-directed cellular migration is the Rho family of small GTPases, in particular Rac-1 and Cdc42 (5, 24). Analysis of RAGE ligand activation of Rac-1, Cdc42, and RhoA in RAGE- and DN-RAGE-expressing cells demonstrated a clear and differential effect on each of these GTPases (Fig. 6, A-C).
FIGURE 6.
The RAGE cytoplasmic domain is required for activation of Rac-1 and Cdc42 but not RhoA. GTPase activation assays were performed with C6 glioma cells stably expressing RAGE or the cytoplasmic domain deleted RAGE (DN-RAGE), starved for 24 h, treated with CML-HSA (10 μg/ml), and lysed at the indicated times. GTP activated Rac1 (A), Cdc42 (B), and RhoA (C) were subjected to pulldown assays and examined by Western blotting compared with total GTPase in lysate. Results are expressed as the mean ± S.D. (n = 3). Western blots are representative of three independent experiments.
In RAGE-expressing cells, both Rac1 and Cdc42 were rapidly activated after ligand stimulation, whereas in DN-RAGE-expressing cells no detectable activation or basal signal was seen for either Rac1 or Cdc42. Notably, Rac1, once activated, was sustained for 30 min with a peak at 5 min, whereas Cdc42 demonstrated a biphasic profile of an early transient peak at 1 min and a late elevation of activity at 30 min in response to RAGE-ligand stimulation (Fig. 6, A and B, respectively). In contrast, RhoA activity was not affected by RAGE-ligand stimulation in cells expressing full-length or DN-RAGE (Fig. 6C).
Based on these data, we tested the hypothesis that RAGE ligand-signaling through Rac-1 and/or Cdc42, but not RhoA, mediates cellular motility. Dominant negative mutants of Rac1 (N17Rac1), Cdc42 (N17Cdc42), and RhoA (N19RhoA) were transiently cotransfected with GFP into RAGE-expressing cells, and RAGE ligand-driven migration assays were performed. Both DN-Rac and DN-Cdc42 blocked RAGE ligand-mediated cellular migration, whereas DN-RhoA did not affect RAGE ligand-mediated cellular migration (Fig. 7). These data demonstrate that RAGE-mediated cellular migration occurs through signaling by the GTPases Rac-1 and Cdc42 but not RhoA.
FIGURE 7.
The RAGE cytoplasmic domain is required for Rac-1- and Cdc42-but not RhoA-mediated cellular migration. Migration assays were performed with C6 glioma cells stably expressing RAGE, cotransfected with GFP and DN-Rac1, DN-Cdc42, DN-RhoA, or empty vector. Migration assays were performed as described above, and results are the means ± S.D. (n = 3).
Dia-1 Is Critical for RAGE Ligand-driven Cell Signaling and Migration—Next, we directly probed the role of Dia-1 in RAGE-dependent signaling and cellular migration. In RAGE-expressing cells, Dia-1 siRNA transfection reduced expression of endogenous Dia-1 at both the transcript (Fig. 8A) and protein level (Fig. 8B) by ∼70% compared with the control (scrambled) siRNA-transfected cells.
FIGURE 8.
Dia-1 siRNA blocks RAGE ligand-stimulated cellular migration. C6 glioma cells stably expressing RAGE were transfected with siRNAs against Dia-1 or siRNA control. 48 h after transfection cells were lysed, and Dia-1 levels were assessed by quantitative-PCR (A) or Western blotting (WB) using anti-Dia-1 antibodies (B). C, cells were transfected with either Dia-1 siRNA/GFP and scramble siRNA/GFP and, after 48 h, fixed and immunostained with antibodies to Dia-1 followed by respective biotinylated secondary antibodies and streptavidin-linked Alexa-546 (Dia-1). D, cells were cotransfected with GFP and Dia-1/control siRNA, and migration stimulated by CML-HAS or 10% fetal bovine serum control (FBS) was performed and quantified as described above. The results are the means ± S.D. (n = 3).
To perform migration assays, Dia-1 siRNA was cotransfected with GFP, and individual cells were assessed for knockdown of Dia-1. By assessing GFP levels and Dia-1 by immunofluorescence, we determined that more than 95% of cells positive for GFP showed markedly reduced signal for Dia-1 (Fig. 8C). However, with control scramble siRNA, Dia-1 levels were not affected (Fig. 8C). Therefore, using GFP as a marker for siRNA efficiency, we found that RAGE ligand-stimulated migration was significantly reduced in Dia-1 siRNA-transfected cells compared with scramble control siRNA-transfected cells (Fig. 8D). Importantly, to demonstrate the specificity of RAGE ligand under these experimental conditions, Dia-1 siRNA did not affect migration of cells in response to 10% serum (Fig. 8D). These data illustrated essential roles for Dia-1 in mediating the effects of RAGE ligand stimulation. Furthermore, these data using serum ruled out a nonspecific or toxic effect of Dia-1 siRNA on the ability of these cells to migrate.
Finally, we probed to ascertain if Dia-1 was required for RAGE ligand-stimulated signal transduction pathways mechanistically linked to cellular migration. Silencing of Dia-1, but not scramble control, markedly blocked RAGE ligand-stimulated Rac-1 (Fig. 9A) and Cdc42 (Fig. 9B) activation. Importantly, in Dia-1 siRNA-transfected cells, RAGE protein levels were not affected (Fig. 9C).
FIGURE 9.
Dia-1 siRNA blocks RAGE ligand-mediated activation of Rac1 and Cdc42. C6 glioma cells stably expressing RAGE were transfected with siRNAs against Dia-1 or siRNA control. Cells were starved for 24 h, treated with CML-HSA (10 μg/ml), and lysed at the indicated times as established in Fig. 7. GTP-activated Rac1 (A) or Cdc42 (B) was subjected to pulldown assays and examined by Western blotting compared with total GTPase in lysate. Results are expressed as the mean ± S.D. (n = 3). C, Dia-1 knockdown was confirmed in each experiment by Western blot. Importantly, Dia-1 knockdown did not affect RAGE expression. In all cases equal protein loading was demonstrated by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Western blots (WB). Western blots are representative of three independent experiments.
DISCUSSION
Multiple studies in vitro and in vivo have demonstrated that the RAGE cytoplasmic domain is the critical element required for RAGE ligand-stimulated cellular effects (5-7, 25). However, earlier studies did not define the molecular interactions between the cytoplasmic domain and specific intracellular effector molecules that mediate RAGE-dependent signal transduction. The diverse nature of RAGE signaling and the finding that the RAGE cytoplasmic domain lacked endogenous tyrosine kinase activity strongly suggested that RAGE interacted with cytoplasmic binding partner(s) to trigger the recruitment of downstream effector pathways. Homology modeling with known receptors of similar families or searching for consensus sequences including tyrosine kinase (26), Src homology domains (SH2/3) (27), WW domains (28), or PDZ domains (29) revealed that RAGE has a unique cytosolic tail sequence. Therefore, how RAGE interacts with cytoplasmic binding partner(s) to trigger the recruitment of these downstream effector pathways is a central question in RAGE biology. In this study, using the yeast two-hybrid system, we screened for binding partners with the RAGE cytoplasmic domain and identified a clone of hDia-1 as a potential binding protein. Dia-1 is a member of the formin protein family which functions to re-organize the actin cytoskeleton, regulating cell motility and migration and the transmission of cellular signaling (18, 30), all similar features to the biology of RAGE.
We employed multiple in vitro and in vivo experimental strategies to identify that the RAGE cytoplasmic domain interacts directly with the FH1 domain of hDia-1. The veracity of this interaction was supported by multiple experimental control studies. Specifically, upon deletion of the RAGE cytoplasmic domain (DN RAGE), RAGE antibodies no longer coimmunoprecipitated endogenous Dia-1 in vivo. Furthermore, studies using isotype control IgG and vector-alone controls excluded nonspecific interactions between the RAGE cytoplasmic domain or Dia-1 with these experimental materials. In addition, epitope-tagged RAGE cytoplasmic domain selectively coimmunoprecipitated Dia-1 mutants containing the FH1 domain in vivo but not the Rho binding domain, as would be strongly suggested by the results of the yeast two-hybrid assay.
The FH domains are the defining motifs of the formin family of proteins, which are conserved across a range of species from yeast, drosophila, mice, and humans (30). The FH1 domain of Dia-1 consists of polyproline stretches that have been shown to be targets for SH3- or WW domain-containing proteins involved in receptor signaling (30). Indeed, Dia-1 with more polyproline repeats than any formin homology protein interacts through its FH1 domain with a diverse range of signaling molecules (17, 19, 20). Here, we add the cytoplasmic domain of RAGE to this group of Dia-1 binding partners and illustrate for the first time a precise intracellular mechanism by which RAGE ligand pathway mediates cellular migration and Rac-1 and Cdc42 signaling in a manner requiring Dia-1.
Intriguingly, and perhaps not surprisingly, the biology of RAGE bears striking parallels to that of Dia-1. For example, in tumor cells including C6 glioma cells as used in the current study, RNA interference strategies to specifically reduce Dia-1 expression identified roles for Dia-1 in the regulation of Rac-1 and Cdc42 and cellular motility (33). Multiple studies have demonstrated Dia-1 to be an effector of RhoA and its activation coupled to binding of Rho-GTP (34-39) and that Dia-1 is linked to Rac-1 activation (33, 40, 41). In this context RAGE action has been clearly linked to Rac-1 as well (5, 24, 42). Huttunen et al. (5) showed that amphoterin (HMGB1)-RAGE interaction-mediated neuroblastoma cell neurite outgrowth in a manner suppressed by DN RAGE as well as DN-Rac or DN-Cdc42. Interestingly, our data revealed that RAGE ligand-RAGE interaction did not affect RhoA activation, although detectable levels of activated RhoA were present in cells, which does not rule out the role of RhoA in the activation of Dia-1 in RAGE-ligand signaling. Future studies will address the precise requirements or not of RhoA in RAGE-Dia-1 interactions and signal transduction.
RAGE ligand-stimulated cellular migration contributes integrally to multiple facets of RAGE biology. For example, in diabetes pathogenesis and complications, blockade of RAGE sharply limited influx of macrophages and lymphocytes into pancreatic islets in NOD/scid mice (43), suppressed smooth muscle cell migration into injured arterial tissue in diabetic Zucker rats (44), and mitigated macrophage and smooth muscle cell infiltration into early atherosclerotic plaques in diabetic mice deficient in apolipoprotein E (8). Even in the absence of diabetes, RAGE ligands mediate cellular migration in inflammation and tumors, as examples (2, 7).
The present work links signal transduction and cellular migration via Dia-1 to a cell surface membrane receptor. Similar links have been previously demonstrated with Dia-1 and other formin family members. Most recently, Dia-1 has been linked to G-coupled receptors via interaction with the RhoGEF LARG and the Gα12/13 proteins (45) and directly to the transient receptor potential Polycystin 2 (TRPP2) channel involved in Ca2+ signaling (32). Together with the finding that the interaction of formin homologue overexpressed in spleen (FHOS) with the short cytoplasmic domain of the CD21 receptor (31), our data support that the family of formin molecules may, in addition to their profound intracellular effects, contribute to signal transduction mechanisms stimulated through cell surface receptors.
Taken together, we have identified a novel facet in the biology of RAGE in which extracellular cues stimulated by RAGE ligand binding are transduced through the cytoplasmic domain of RAGE to stimulate fundamental signaling networks, specifically activation of Rac-1 and Cdc42, and cellular migration in a manner requiring Dia-1. Because cellular migration critically contributes to diabetes and its complications, tumor metastases, and inflammatory disorders, particularly in RAGE ligand-enriched environments, our findings highlight novel approaches to mitigate RAGE-dependent tissue injury.
Supplementary Material
Acknowledgments
We are grateful to Dr. Jan Kitajewski for the pcS2 plasmid and helpful discussions. We thank Jean-Luc Wautier and Marie-Paul Wautier for providing CML-HSA.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) DQ067452 and DQ067453.
This work was supported, in whole or in part, by National Institutes of Health Grants HL60901 and CA 087677 (United States Public Health Service). This work was also funded by the Juvenile Diabetes Research Foundation and by the Surgical Research Fund. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: RAGE, receptor for advanced glycation end products; DN, dominant negative; CML, carboxyl methyl lysine; HSA, human serum albumin; GST, glutathione S-transferase; Bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl) propane-1,3-diol; MOPS, 4-morpholinepropanesulfonic acid; GFP, green fluorescent protein; siRNA, small interfering RNA; hDia-1, human Diaphanous-1.
References
- 1.Neeper, M., Schmidt, A. M., Brett, J., Yan, S. D., Wang, F., Pan, Y. C. P., Elliston, K., Stern, D., and Shaw, A. (1992) J. Biol. Chem. 267 14998-15004 [PubMed] [Google Scholar]
- 2.Hofmann, M. A., Drury, S., Fu, C. F., Qu, W., Taguchi, A., Lu, Y., Avila, C., Kambham, N., Bierhaus, A., Nawroth, P., Neurath, M. F., Slattery, T., Beach, D., McClary, J., Nagashima, M., Morser, J., Stern, D., and Schmidt, A. M. (1999) Cell 97 889-901 [DOI] [PubMed] [Google Scholar]
- 3.Hori, O., Brett, J., Slattery, T., Cao, R., Zhang, J., Chen, J. X., Nagashima, M., Lundh, E. R., Vijay, S., Nitecki, D., Morser, J., Stern, D., and Schmidt, A. M. (1995) J. Biol. Chem. 270 25752-25761 [DOI] [PubMed] [Google Scholar]
- 4.Yan, S. D., Chen, X., Fu, J., Chen, M., Zhu, H., Roher, A., Slattery, T., Zhao, L., Nagashima, M., Morser, J., Migheli, A., Nawroth, P., Stern, D., and Schmidt, A. M. (1996) Nature 382 685-691 [DOI] [PubMed] [Google Scholar]
- 5.Huttunen, H. J., Fages, C., and Rauvala, H. (1999) J. Biol. Chem. 274 19919-19924 [DOI] [PubMed] [Google Scholar]
- 6.Sakaguchi, T., Yan, S. F., Yan, S. D., Belov, D., Rong, L. L., Sousa, M., Andrassy, M., Marso, S. P., Duda, S., Arnold, B., Liliensiek, B., Nawroth, P. P., Stern, D. M., Schmidt, A. M., and Naka, Y. (2003) J. Clin. Investig. 111 959-972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taguchi, A., Blood, D. C., del Toro, G., Canet, A., Lee, D. C., Qu, W., Tanji, N., Lu, Y., Lalla, E., Fu, C., Hofmann, M. A., Kislinger, T., Ingram, M., Lu, A., Tanaka, H., Hori, O., Ogawa, S., Stern, D., and Schmidt, A. M. (2000) Nature 405 354-360 [DOI] [PubMed] [Google Scholar]
- 8.Park, L., Raman, K. G., Lee, K. J., Lu, Y., Ferran, L. J., Chow, W. S., Stern, D., and Schmidt, A. M. (1998) Nat. Med. 4 1025-1031 [DOI] [PubMed] [Google Scholar]
- 9.Kislinger, T., Tanji, N., Wendt, T., Qu, W., Lu, Y., Ferran, L. J., Taguchi, A., Olson, K., Goova, M. T., Hofmann, M. A., Cataldegirmen, G., D'Agati, V., Pischetsrieder, M., Stern, D. M., and Schmidt, A. M. (2001) Arterioscler. Thromb. Vasc. Biol. 21 905-910 [DOI] [PubMed] [Google Scholar]
- 10.Kislinger, T., Fu, C., Huber, C., Qu, W., Taguchi, A., Yan, S. D., Hofmann, M. A., Yan, S. F., Pischetsrieder, M., Stern, D., and Schmidt, A. M. (1999) J. Biol. Chem. 274 31740-31749 [DOI] [PubMed] [Google Scholar]
- 11.Schmidt, A. M., Yan, S. D., Brett, J., Mora, R., Nowygrod, R., and Stern, D. (1993) J. Clin. Investig. 91 2155-2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander, H. M., Tauras, J. M., Ogiste, J. S., Hori, O., Moss, R. A., and Schmidt, A. M. (1997) J. Biol. Chem. 272 17810-17814 [DOI] [PubMed] [Google Scholar]
- 13.Huang, J. S., Guh, J. Y., Chen, H. C., Hung, W. C., Lai, Y. H., and Chuang, L. Y. (2001) J. Cell Biochem. 81 102-113 [DOI] [PubMed] [Google Scholar]
- 14.Yeh, C. H., Sturgis, L., Haidacher, J., Zhang, X. N., Sherwood, S. J., Bjercke, R. J., Juhasz, O., Crow, M. T., Tilton, R. G., and Denner, L. (2001) Diabetes 50 1495-1504 [DOI] [PubMed] [Google Scholar]
- 15.Hudson, B. I., Carter, A. M., Harja, E., Kalea, A. Z., Arriero, M., Yang, H., Grant, P. J., and Schmidt, A. M. (2007) FASEB J. 22 1572-1580 [DOI] [PubMed] [Google Scholar]
- 16.Higgs, H. N. (2005) Trends Biochem. Sci. 30 342-353 [DOI] [PubMed] [Google Scholar]
- 17.Krebs, A., Rothkegel, M., Klar, M., and Jockusch, B. M. (2001) J. Cell Sci. 114 3663-3672 [DOI] [PubMed] [Google Scholar]
- 18.Tominaga, T., Sahai, E., Chardin, P., McCormick, F., Courtneidge, S. A., and Alberts, A. S. (2000) Mol. Cell 5 13-25 [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara, T., Mammoto, A., Kim, Y., and Takai, Y. (2000) Biochem. Biophys. Res. Commun. 271 626-629 [DOI] [PubMed] [Google Scholar]
- 20.Satoh, S., and Tominaga, T. (2001) J. Biol. Chem. 276 39290-39294 [DOI] [PubMed] [Google Scholar]
- 21.Lynch, E. D., Lee, M. K., Morrow, J. E., Welcsh, P. L., Leon, P. E., and King, M. C. (1997) Science 278 1315-1318 [PubMed] [Google Scholar]
- 22.Miyata, T., Hori, O., Zhang, J., Yan, S. D., Ferran, L., Iida, Y., and Schmidt, A. M. (1996) J. Clin. Investig. 98 1088-1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan, S. S., Wu, Z. Y., Zhang, H. P., Furtado, G., Chen, X., Yan, S. F., Schmidt, A. M., Brown, C., Stern, A., LaFaille, J., Chess, L., Stern, D. M., and Jiang, H. (2003) Nat. Med. 9 287-293 [DOI] [PubMed] [Google Scholar]
- 24.Wang, L., Li, S., and Jungalwala, F. B. (2007) J. Neurosci. Res. 86 1254-1266 [DOI] [PubMed] [Google Scholar]
- 25.Harja, E., Bu, D. X., Hudson, B. I., Chang, J. S., Shen, X., Hallam, K., Kalea, A. Z., Lu, Y., Rosario, R. H., Oruganti, S., Nikolla, Z., Belov, D., Lalla, E., Ramasamy, R., Yan, S. F., and Schmidt, A. M. (2008) J. Clin. Investig. 118 183-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbieri, M. A., Ramkumar, T. P., Fernadez-Pol, S., Chen, P. I., and Stahl, P. D. (2004) Curr. Top. Microbiol. Immunol. 286 1-20 [PubMed] [Google Scholar]
- 27.Schlessinger, J., and Lemmon, M. A. (2003) Sci. STKE 2003, RE12. [DOI] [PubMed]
- 28.Komuro, A., Imamura, T., Saitoh, M., Yoshida, Y., Yamori, T., Miyazono, K., and Miyazawa, K. (2004) Oncogene 23 6914-6923 [DOI] [PubMed] [Google Scholar]
- 29.Assanasen, C., Mineo, C., Seetharam, D., Yuhanna, I. S., Marcel, Y. L., Connelly, M. A., Williams, D. L., de, l. L.-M., Shaul, P. W., and Silver, D. L. (2005) J. Clin. Investig. 115 969-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallar, B. J., and Alberts, A. S. (2003) Trends Cell Biol. 13 435-446 [DOI] [PubMed] [Google Scholar]
- 31.Gill, M. B., Roecklein-Canfield, J., Sage, D. R., Zambela-Soediono, M., Longtine, N., Uknis, M., and Fingeroth, J. D. (2004) J. Cell Sci. 117 2709-2720 [DOI] [PubMed] [Google Scholar]
- 32.Bai, C. X., Kim, S., Li, W. P., Streets, A. J., Ong, A. C., and Tsiokas, L. (2008) EMBO J. 27 1345-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamana, N., Arakawa, Y., Nishino, T., Kurokawa, K., Tanji, M., Itoh, R. E., Monypenny, J., Ishizaki, T., Bito, H., Nozaki, K., Hashimoto, N., Matsuda, M., and Narumiya, S. (2006) Mol. Cell. Biol. 26 6844-6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe, N., Madaule, P., Reid, T., Ishizaki, T., Watanabe, G., Kakizuka, A., Saito, Y., Nakao, K., Jockusch, B. M., and Narumiya, S. (1997) EMBO J. 16 3044-3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nezami, A. G., Poy, F., and Eck, M. J. (2006) Structure 14 257-263 [DOI] [PubMed] [Google Scholar]
- 36.Otomo, T., Otomo, C., Tomchick, D. R., Machius, M., and Rosen, M. K. (2005) Mol. Cell 18 273-281 [DOI] [PubMed] [Google Scholar]
- 37.Lammers, M., Rose, R., Scrima, A., and Wittinghofer, A. (2005) EMBO J. 24 4176-4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, F., and Higgs, H. N. (2005) J. Biol. Chem. 280 6986-6992 [DOI] [PubMed] [Google Scholar]
- 39.Li, F., and Higgs, H. N. (2003) Curr. Biol. 13 1335-1340 [DOI] [PubMed] [Google Scholar]
- 40.Meng, W., Numazaki, M., Takeuchi, K., Uchibori, Y., ndo-Akatsuka, Y., Tominaga, M., and Tominaga, T. (2004) EMBO J. 23 760-771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arakawa, Y., Bito, H., Furuyashiki, T., Tsuji, T., Takemoto-Kimura, S., Kimura, K., Nozaki, K., Hashimoto, N., and Narumiya, S. (2003) J. Cell Biol. 161 381-391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bassi, R., Giussani, P., Anelli, V., Colleoni, T., Pedrazzi, M., Patrone, M., Viani, P., Sparatore, B., Melloni, E., and Riboni, L. (2008) J. Neuro-Oncol. 87 23-33 [DOI] [PubMed] [Google Scholar]
- 43.Chen, Y., Yan, S. S., Colgan, J., Zhang, H. P., Luban, J., Schmidt, A. M., Stern, D., and Herold, K. C. (2004) J. Immunol. 173 1399-1405 [DOI] [PubMed] [Google Scholar]
- 44.Zhou, Z., Wang, K., Penn, M. S., Marso, S. P., Larson, M. A., Forudi, F., Zhou, X., Qu, W., Lu, Y., Stern, D. M., Schmidt, A. M., Lincoff, A. M., and Topol, E. J. (2003) Circulation 107 2238-2243 [DOI] [PubMed] [Google Scholar]
- 45.Goulimari, P., Knieling, H., Engel, U., and Grosse, R. (2008) Mol. Biol. Cell 19 30-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.