Abstract
Cell-cell and cell-matrix adhesion are crucial during many stages of eukaryotic development. Here, we provide the first example that mucin-type O-linked glycosylation is involved in a developmentally regulated cell adhesion event in Drosophila melanogaster. Mutations in one member of the evolutionarily conserved family of enzymes that initiates O-linked glycosylation alter epithelial cell adhesion in the Drosophila wing blade. A transposon insertion mutation in pgant3 or RNA interference to pgant3 resulted in blistered wings, a phenotype characteristic of genes involved in integrin-mediated cell interactions. Expression of wild type pgant3 in the mutant background rescued the wing blistering phenotype, whereas expression of another family member (pgant35A) did not, revealing a unique requirement for pgant3. pgant3 mutants displayed reduced O-glycosylation along the basal surface of larval wing imaginal discs, which was restored with wild type pgant3 expression, suggesting that reduced glycosylation of basal proteins is responsible for disruption of adhesion in the adult wing blade. Glycosylation reactions demonstrated that PGANT3 glycosylates certain extracellular matrix (ECM) proteins. Immunoprecipitation experiments revealed that PGANT3 glycosylates tiggrin, an ECM protein known to bind integrin. We propose that this glycosyltransferase is uniquely responsible for glycosylating tiggrin in the wing disc, thus modulating proper cell adhesion through integrin-ECM interactions. This study provides the first evidence for the role of O-glycosylation in a developmentally regulated, integrin-mediated, cell adhesion event and reveals a novel player in wing blade formation during Drosophila development.
Cell interactions and adhesion are critical in many diverse processes, from events occurring during embryogenesis and organogenesis to wound healing and the alterations in cell adhesion seen upon tumor formation and metastasis (1). Drosophila wing development has been used as a model system to identify factors responsible for regulating cell adhesion (2–5). Aberrant adhesion of the two epithelial cell layers that comprise the adult wing blade results in separation of the cell layers, creating a localized blister shortly after eclosion. Mutations in the integrin family of cell surface receptors (2, 3, 5–7) as well as proteins that interact with integrins (4, 5, 8–13) have been shown to produce wing blisters, highlighting the central role of integrin-mediated cell adhesion events during wing blade formation. Interestingly, many cell surface and extracellular matrix (ECM)2 proteins that influence wing blistering also interact with integrins in other developmentally regulated cell adhesion events (2, 14, 15).
Cell surface, secreted, and ECM proteins undergo a number of post-translational modifications as they transit the secretory apparatus to their final destinations. Although the roles of classical N-linked glycans and proteoglycans are widely appreciated, recent studies have elucidated crucial roles for other types of glycans during development. The disaccharide GlcNAcβ1–3Fuc on the Notch receptor and its ligands has been shown to regulate receptor/ligand interactions and downstream signaling events (16–19). Protein O-fucosylation and the fucosyltransferase catalyzing this modification have roles in protein folding, trafficking (20, 21), and secretion (22). Most recently, O-linked glucose has been shown to have a modulatory role in Notch signaling events, influencing lateral inhibition and cell-fate specification (23). Taken together, these studies highlight the diversity of glycans found on proteins and the unique functional roles they play during development.
Studies from our laboratory and others have demonstrated the abundant presence of another type of glycosylation, known as mucin-type O-linked glycosylation (referred to as O-linked glycosylation) throughout development in diverse species (24–27). In contrast to the previously mentioned types of glycosylation, mucin-type O-glycosylation is initiated by a large family of enzymes known as the UDP-GalNAc: polypeptide N-acetylgalactosaminyltranferases (PGANTs in Drosophila or ppGalNAcTs in mammals) that transfer N-acetylgalactosamine (GalNAc) to serine or threonine residues of proteins destined to be membrane-bound or secreted (28, 29). Members of this family have unique but overlapping developmental expression patterns, and many show distinct substrate preferences in vitro, suggesting that each enzyme may be responsible for glycosylating a unique subset of proteins in vivo (25, 28, 29). In vitro data further indicate that there exists a hierarchy of activity within the family, with some members initiating the glycosylation of unmodified substrates and others acting only on previously glycosylated substrates, adding GalNAc at positions vicinal to sites previously modified by other family members (30–32). The unique spatial and temporal expression patterns, substrate preferences, and hierarchical action of members of this family suggest a highly regulated process governing the acquisition of this type of glycan.
Mucin-type O-glycans on secreted and membrane-bound proteins are uniquely positioned to mediate many events regulating homeostasis and development. Indeed, mutations in ppGalNAc-T3 are thought to be responsible for familial tumoral calcinosis, a rare human disease characterized by hyperphosphatemia and the development of calcified “tumors” in cutaneous and subcutaneous tissues (33). Mice deficient in ppGalNAc-T1, although viable and fertile, display reduced lymphocyte homing and bleeding disorders (34). Studies from our laboratory have demonstrated a role for another member of this family (pgant35A) in the proper formation of the embryonic tracheal system in Drosophila (35, 36). Finally, mice deficient in the core 1 β3-galactosyltransferase, which adds a galactose to the O-GalNAc of mucin-type O-glycans, die embryonically from defective angiogenesis resulting in fatal brain hemorrhages (37); hypomorphic mutations result in thrombocytopenia and kidney defects later in development (38). Altogether, these studies highlight the diverse consequences of alterations in O-glycosylation that have only recently been discovered due to the inherent complexity of the ppGalNAcT family.
Here, we examine the developmental role of another member of this family, pgant3. Previous work from our laboratory demonstrated that PGANT3 is one of the initiating glycosyltransferases, transferring GalNAc to previously unmodified substrates (25). Additionally, pgant3 gene expression is highly regulated during development (27). In this study we find that a transposon mutation in pgant3 or RNA interference (RNAi) to pgant3 results in wing blistering, implicating O-glycosylation in integrin-mediated cell adhesion occurring during wing blade formation. A reduction in O-glycoproteins was seen along the basal surface of the pgant3 mutant wing imaginal discs along with altered disc morphology. Based on our studies, we propose that O-glycosylation of specific ECM proteins known to bind integrin is required for proper epithelial cell adhesion in the wing disc. This finding provides the first direct evidence for the role of mucin type O-glycans in a developmentally regulated cell adhesion event and identifies a novel protein modification required for proper wing blade formation in the fly.
EXPERIMENTAL PROCEDURES
Fly Strains Used—The stocks used in this study are as follows: Bloomington stocks #5138 (y1, w*; P{w+mC = tubP-GAL4}LL7/TM3, Sb1) (the tubulin-Gal4 driver line); #8860 (w1118, P{w+mW.hs = GawB}BxMS1096) (the wing-specific Gal4 driver line); #1561 (w*; P{w+mW.hs = Gal4-arm}4a, P{w+mW.hs = Gal4-arm}4b/TM3, Sb1, Ser1) (the armadillo-Gal4 driver line); #7748 (w1118; Df(2R)Exel 6283, PXP-UExel6263); #8283 (w1118; CyO, P{w+mc = FRT (w+)Tub-PBac™}2/wgsp-1); #8795 (w*; TigA1/CyO,P{lacZ-un3}276); #8796 (w*; TigX/CyO,P{lacZ-un3}276). Additionally, the following stocks from other sources were used: PBac{PB}pgant3c01318 from the Exelixis Drosophila Stock Collection (39); w; Dr/TM3, Sb1, twi-2XGFP stock (the kind gift of D. Andrew); w; TM6C, cu, Sb, e, ca/Su(Tpl)s1, red, e stock (the kind gift of J. Kennison).
Construction of Gal4-inducible pgant3 and pgant3IR Vectors and Transgenic Lines—The pUAST plasmid (40) was used to generate a Gal4-inducible construct expressing wild type pgant3 cDNA that was then used to create transgenic flies. The complete coding region from the wild type pgant3 gene was excised from the GH09147 cDNA clone (Invitrogen) using EcoRI and XbaI and cloned into the same sites of pUAST to generate the plasmid pUAS-pgant3. To generate the Gal4-inducible pgant3IR construct, sense (taatacctaggAAGGTGAATGTTACGGAGCGTGTGG) and antisense (taatacctaggCTGCGCCAGCATTACATTCGAAGTG) primers were used to amplify a 500-bp fragment from the catalytic region of pgant3. The PCR product was then cloned stepwise into the AvrII and NheI sites on either side of the white intron in the vector pWIZ (41) to generate a vector containing two inversely oriented pgant3 fragments flanking the white intron. Transformants were produced by Genetic Services Inc. (Cambridge, MA) using methodology based on the procedure described previously (42, 43).
Fly Crosses—Rescue and overexpression experiments were performed using flies from a UAS-pgant35A transgenic line (36) or three independent UAS-pgant3 transgenic lines (w; P{UAS-pgant3#3}, on the third chromosome) (w; P{UAS-pgant3#7}, on the X chromosome) (w; P{UAS-pgant3#10}, on the third chromosome) and the Gal4-driver stocks described herein. All stocks used in the rescue experiments were first crossed into the pgant3c01318 background to generate both Gal4 driver lines and P{UAS-pgant} transgenic lines heterozygous for pgant3c01318. These heterozygous lines were then crossed as follows, and rescue of the wing blister phenotype was assessed by scoring straight winged progeny with or without Sb1: pgant3c01318/CyO; P{UAS-pgant}/P{UAS-pgant} females X pgant3c01318/CyO;P{tubP-GAL4}LL7/TM3, Sb1 males. Crosses to assess the effect of pgant3 overexpression were performed as shown in Table 2.
TABLE 2.
Over-expression of pgant3 results in lethality UAS-pgant3#3 and UAS-pgant3#10 are independent transgenic lines generated as described under “Experimental Procedures.”
| Crosses | Progeny overexpressing pgant3a | Progeny without pgant3 overexpression | Survival of pgant3 overexpressing flies |
|---|---|---|---|
| (%) | |||
| UAS-pgant3#3/UAS-pgant3#3 × Tub-Gal4/TM3, Sb1 | 0 | 312 | 0 |
| UAS-pgant3#10/UAS-pgant3#10 × Tub-Gal4/TM3, Sb1 | 6 | 171 | 4 |
| UAS-pgant3#3/UAS-pgant3#3 × arm-Gal4/TM3, Sb1 | 126 | 213 | 50 |
| UAS-pgant3#3/TM6C, Sb1 × MS1096-Gal4/MS1096-Gal4 | 214 | 211 | 100 |
Assessed by the absence of Sb1
Crosses to generate RNAi to pgant3 were performed by crossing homozygous inverted repeat (IR) transgenic lines (w1118; P{UAS-pgant3IR2#2} or w1118; P{UAS-pgant3IR2#9}) to a tubulin-Gal4-driver line (P{tubP-GAL4}LL7/TM3, Sb1, twi-2XGFP) and comparing progeny with and without Sb1 and GFP. Crosses to the wing-specific driver (MS1096-Gal4) were performed using homozygous w1118, P{w+mW.hs = GawB}BxMS1096 females crossed to homozygous transgene-containing males. All Drosophila crosses were kept on MM media (KD Medical, Inc.) at 25 °C unless specified otherwise.
Mutant Sequencing and Quantitative Reverse Transcription-PCR—The genomic region flanking each transposon insertion was amplified and sequenced according to the previously described procedures (39). To examine the effect of the transposon on pgant3 gene expression levels, pgant3c01318/pgant3c01318 homozygotes, wild type, and transposon excision lines were used to isolate RNA and perform real-time PCR. Briefly, RNA was isolated using the FastRNA Pro Green kit (Q-BIOgene). cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad). PCR primers were designed using Beacon Designer software (Bio-Rad). Quantitative reverse transcription-PCR was performed on a MyiQ real time PCR thermocycler (Bio-Rad) using the SYBR-Green PCR Master Mix (Bio-Rad). Quantitative reverse transcription-PCR to determine expression levels of all pgant family members was performed using the PCR primers listed in supplemental Table 1 with cDNA prepared from wild type, Tub>pgant3IR2#2, and Tub>pgant3IR2#9 larval wing discs.
Wing Disc Fixation and Staining—Imaginal wing discs were stained according to standard procedures and analyzed by confocal microscopy. Mouse monoclonal anti-Tn antibody (Ca3638) (44) (dilution, 1:50) was the kind gift of Dr. Richard Cummings who had acquired the stocks of antibodies and hybridomas from the late Dr. Georg F. Springer. Immunopositive signals were developed using Cy3-conjugated donkey anti-mouse IgM antibody (dilution, 1:100) (Jackson Immuno-Research Laboratories). Stained wing discs were analyzed using the Zeiss LSM 510 confocal laser scanning microscope. Images were processed using the LSM Imager Browser and Photoshop. Measurements of wing disc thickness and O-glycan staining in x-z cross sections were performed in the center of each disc. Values were averaged, and S.D. were calculated. Statistical significance was determined using a Student's t test.
Glycosylation Assays in Vitro—Assays for glycosyltransferase activity were performed as described previously (25). Briefly, media from COS7 cells expressing recombinant pgant3 or pgant35A (25) was harvested and used in the in vitro reactions with [14C]UDP-GalNAc, and the acceptor substrates denoted in Table 3. Reaction products were purified by anion exchange chromatography, and [14C]GalNAc incorporation was measured. Reactions using media from cells expressing empty vector alone yielded background values that were subtracted from each experimental value. Adjusted experimental values were then averaged, and S.D. were calculated. All assays were performed in duplicate. Glycosyltransferase activity is expressed as dpm/h.
TABLE 3.
Peptide substrates derived from proteins expressed in wing discs The bold residues are sites of potential O-glycosylation based on predictions performed by the NetOGlyc server. NA, no evidence from references.
| Peptide | Gene | Protein | Peptide sequence | Wing blisters in mutants |
|---|---|---|---|---|
| if | if | αPS2 integrin | EPQVNQTSFTTYSTSSSSSG | yes |
| lanA1 | LanA | Laminin A | TLPPTTPTTTTTTTTT | yes |
| mew | mew | αPS1 integrin | VGFFKRIRPTDPTLSGNLE | yes |
| scab | scb | αPS3 integrin | VDPVEVTTTLSGGLERTV | yes |
| tigA | tig | Tiggrin | LEGETARPRPPNPAPIVSTPKP | yes |
| tigB | tig | Tiggrin | QQATKVEVEATSEPSFWEKLK | yes |
| tenM1 | ten-m | Tenascin-major | FLLEGVTPTAPPDVPPRNPT | NA |
| tenM2 | ten-m | Tenascin-major | TSNSGTAQGLQSTSASAEATSS | NA |
| tsp | tsp | Thrombospondin | IQIKLVNSTEGPGPMMRNS | NA |
Western Blotting—Protein extracts were prepared from 3rd instar larval wing discs of wild type, pgant3c01318 homozygotes, and transposon excision lines (pgant3c01318revertant#7 homozygotes) as described (45). Samples were electrophoresed under reducing conditions in a 4–12% SDS-PAGE gradient gel. Gels were transferred to nitrocellulose; membranes were blocked with 1× blocking buffer (Sigma), incubated with Tn antibody (dilution, 1:500) or the tiggrin antibody (the kind gift of Drs. L. and J. Fessler) (9) (dilution, 1:500), and developed with horse-radish peroxidase-conjugated secondary antibody (dilution, 1:10,000).
Immunoprecipitation—Equivalent amounts of protein extracts were prepared from 3rd instar larval wing discs from wild type and pgant3c01318 homozygotes. 50 μl of immobilized protein L suspension (Thermo Scientific) was added to 500 μl of protein extract and incubated at 4 °C for 1 h to preclear. The mixtures were centrifuged, and the precleared supernatant was collected. 10 μl of the tiggrin antibody were added to the precleared supernatant and incubated at 4 °C overnight. The following day 50 μl of washed immobilized protein L suspension was added, and incubation was performed at 4 °C overnight. Immunocomplexed proteins were collected by pulse centrifugation. Pellets were washed three times with lysis buffer. The final pellet was resuspended in sample loading buffer, heated to 95 °C for 5 min, and analyzed by reducing SDS-PAGE followed by immunoblotting with the Tn Ab as described.
RESULTS
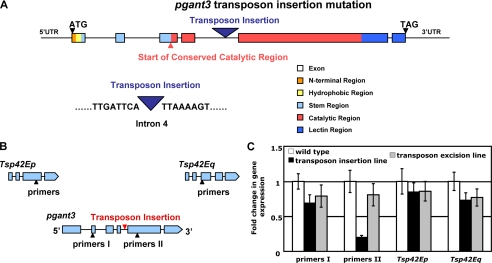
Transposon Insertion Decreases pgant3 Gene Expression and Causes Wing Blistering—pgant3 is one of 12 members of the Drosophila gene family encoding the polypeptide GalNAc glycosyltransferases that are responsible for initiating mucin-type O-linked glycosylation of secreted and membrane-bound proteins (25, 27). pgant3 gene expression is highly regulated both spatially and temporally during development (27). We set out to examine the biological roles of O-glycosylation mediated by this glycosyltransferase using three putative transposon insertions in the pgant3 gene from the Exelixis Drosophila stock collection (39). Genomic sequencing of the insertion sites revealed that only one transposon resided within the pgant3 gene (PBac{PB}pgant3c01318); the line containing this transposon is hereafter designated pgant3c01318. pgant3c01318 contains a piggyBac transposable element in the fourth intron of pgant3, separating exons that encode the conserved catalytic region of the enzyme (Fig. 1A). Quantitative PCR was performed on wild type and homozygous transposon insertion mutants to assess the effect of the transposon on the expression levels of pgant3 as well as the flanking tetraspanin genes (Tsp42Ep andTsp42Eq) (Fig. 1B). As shown in Fig. 1C, pgant3 gene expression 3′ to the transposon insertion site was significantly reduced in the transposon insertion line relative to wild type. Expression of the flanking tetraspanin genes was not affected. This result demonstrates that the transposon insertion specifically affects pgant3 gene expression, thus supporting the use of this line to investigate pgant3 gene function.
FIGURE 1.
pgant3 gene expression is reduced in the pgant3c01318 transposon insertion stock. A, position of the transposon in intron 4 of the pgant3 gene is shown. Exons are represented as boxes, and introns are represented as lines. Functional elements of the pgant3 coding region are shown in color. Nucleotide sequence of the transposon insertion site in the pgant3c01318 stock is shown. B, the genomic region of pgant3 and flanking genes are shown. The positions of the primer pairs used for real-time PCR are shown as triangles. C, real-time PCR analysis of pgant3 transcript levels using the primer pairs shown in B reveals a significant decrease in pgant3 gene expression in pgant3c01318/pgant3c01318 transposon insertion homozygotes relative to pgant3c01318revertant#7/pgant3c01318revertant#7 transposon excision homozygotes or wild type flies. Transcription from the flanking Tsp42Ep and Tsp42Eq genes was unaffected. RNA levels were normalized to 18 S rRNA.
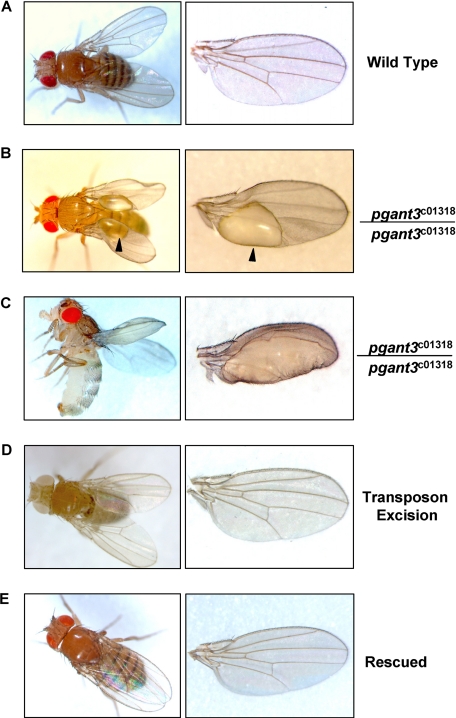
Heterozygous transposon insertion flies were crossed to assess the effect of the transposon on viability. Although a reduction in viability was seen relative to heterozygotes, most homozygous transposon insertion mutants did survive to adulthood. However, a significant number of pgant3c01318 homozygous adults displayed abnormal wings (Fig. 2 and Table 1). Specifically, wings of pgant3c01318 homozygotes developed blisters as they unfolded, which later collapsed into crumpled areas in the wing blade (Fig. 2). Wing blistering was not seen in wild type flies or flies heterozygous for the transposon insertion (Table 1). The extent of blistering varied, with some wings having small blisters and others showing blistering across the entire wing blade (Fig. 2, B and C).
FIGURE 2.
pgant3 transposon insertion results in wing blistering. Wild type (A) pgant3c01318/pgant3c01318 transposon insertion homozygotes (B and C), pgant3c01318revertant#7/pgant3c01318revertant#7 transposon excision homozygotes (D), and rescued homozygous mutants expressing a wild type pgant3 transgene (pgant3c01318/pgant3c01318; UAS-pgant3) (E) are shown. Close up views of wings are shown in the panels on the right.
TABLE 1.
Frequency of flies displaying blistered wings
| Genotype | % Blistereda | nb |
|---|---|---|
| +/+ | 0 | 150 |
| pgant3c01318/+ | 0 | 114 |
| pgant3c01318/+ @ 18 °C | 2 | 279 |
| pgant3c01318/pgant3c01318 | 95 | 129 |
| pgant3c01318/pgant3c01318 @ 18 °C | 99 | 147 |
| pgant3c01318/Df(2R)Exel6283, PXP-UExel6283 | 96 | 190 |
| pgant3c01318revertant#7/pgant3c01318revertant#7 | 0 | 150 |
| pgant3c01318revertant#7/pgant3c013180 | 0 | 213 |
| Tub>pgant3IR2#2 | 97 | 168 |
| Tub>pgant3IR2#9 | 95 | 133 |
| MS1096>pgant3IR2#2 | 23 | 126 |
| MS1096>pgant3IR2#9 | 19 | 145 |
| pgant3c01318/pgant3c01318; UAS-pgant3#3/+ | 0 | 173 |
| pgant3c01318/pgant301318; UAS-pgant3#10/+ | 9 | 163 |
| UAS-pgant3#7/Y;pgant3c01318/pgant3c01318 | 4 | 122 |
| pgant3c01318/pgant3c01318; UAS-pgant35A#5/Tub-Gal4 | 83 | 153 |
| pgant3c01318/pgant3c01318; Tub-Gal4/+ | 90 | 168 |
| pgant3c01318/pgant3c01318; UAS-pgant35A#5/+ | 94 | 121 |
| pgant3c01318/tigX @ 18 °C | 8 | 115 |
| pgant3c01318/tigA1 @ 18 °C | 6 | 91 |
| tigX/+ @ 18 °C | 0 | 119 |
| tigA1/+ @ 18 °C | 0 | 94 |
(number of flies displaying blistered wings/total number of flies of denoted genotype) × 100
n = total number of flies of denoted genotype scored
The pgant3c01318 line was crossed to a deficiency line that uncovered the pgant3 gene (Df(2R)Exel 6283, PXP-UExel6263) to verify that the blistering phenotype is the result of the transposon in pgant3 and not due to mutations elsewhere on the chromosome. Progeny containing the transposon insertion over the deficiency displayed wing blistering (Table 1), lending further support for the role of pgant3 in the wing blistering phenotype.
To conclusively demonstrate that the transposon insertion in pgant3 is responsible for the wing blistering phenotype, the piggyBac transposon was precisely excised from the pgant3 gene using the piggyBac transposase as described previously (39). Transposon excision lines (pgant3c01318revertant#7/ pgant3c01318revertant#7) showed restoration of pgant3 gene expression levels and wing integrity (Fig. 1C and Fig. 2D, and Table 1).
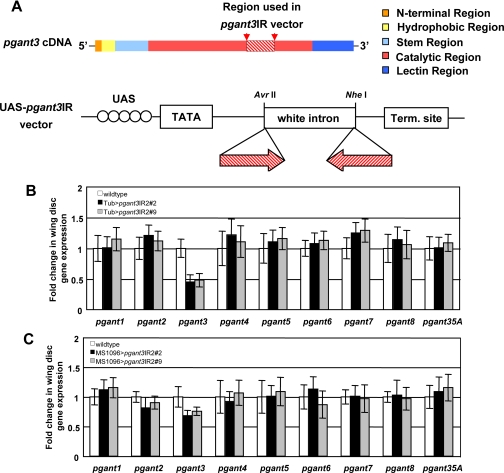
RNAi to pgant3 Causes Wing Blistering—To confirm that decreased levels of pgant3 expression are responsible for the wing blistering observed, we constructed transgenic flies carrying Gal4-inducible pgant3 IR constructs (UAS-pgant3IR) to specifically induce double-stranded RNA to pgant3 and, thus, decrease pgant3 expression via RNAi (Fig. 3). After crossing two independent UAS-pgant3IR transgenic lines to a tubulin-Gal4 driver line (which expresses Gal4 ubiquitously), we found that 95–97% of the progeny expressing pgant3 double-stranded RNA displayed wing blistering (Table 1). Induction of pgant3 RNAi exclusively in the wing disc by crossing UAS-pgant3IR lines to a wing-specific Gal4 driver line (MS1096-Gal4) resulted in 19–23% wing blistering (Table 1). Quantitative PCR verified that pgant3 expression levels were specifically reduced in double-stranded RNA-expressing progeny, whereas expression of other pgant family members was unchanged (Fig. 3, B and C). Additionally, the degree of reduction in pgant3 gene expression with each Gal4-driver correlated with the frequency of wing blistering observed. The tubulin-Gal4 driver resulted in a greater reduction in pgant3 gene expression in wing discs relative to the MS1096-Gal4 driver and also gave a greater incidence of wing blistering (Table 1 and Fig. 3, B and C).
FIGURE 3.
RNAi to pgant3 results in a specific decrease in pgant3 gene expression and induces wing blisters in vivo. A, the segment of the pgant3 coding region used to construct the UAS-pgant3IR vector (below) is shown as a diagonally striped box. Functional elements of the pgant3 coding region are shown in color. Quantitative PCR was employed to assess pgant family member gene expression in wing discs from wild type and two independent UAS-pgant3IR transgenic lines (UAS-pgant3IR2#2 and UAS-pgant3IR2#9) under the control of the tubulin-Gal4 driver (B) or the wing-specific MS1096-Gal4 driver (C). In both instances induction of pgant3 double-stranded RNA resulted in a specific reduction in pgant3 expression without significantly affecting the expression of other family members.
Wild Type pgant3 Expression Restores Wing Integrity but Expression of Another pgant Family Member Does Not—To demonstrate that pgant3 expression is crucial for wing integrity, we constructed Gal4-inducible transgenic lines carrying wild type pgant3 cDNA (UAS-pgant3) to perform rescue experiments. The presence of the UAS-pgant3 expression construct in the homozygous pgant3c01318 transposon background resulted in significant reduction or elimination of wing blistering (Table 1 and Fig. 2E) even in the absence of a Gal4 driver. This was seen with multiple independent transgenic lines (Table 1). However, transgenic lines overexpressing another pgant family member under the control of the tubulin-Gal4 driver (pgant35A) (27, 36) failed to completely rescue wing blistering in the homozygous pgant3c01318 transposon background (Table 1). These results indicate that the restoration of wing integrity is specific to pgant3 gene expression and cannot be achieved by overexpression of another family member. This suggests that PGANT3 is uniquely responsible for glycosylating a subset of proteins required for proper wing blade formation.
Overexpression of pgant3 Results in Lethality—While performing the aforementioned rescue studies, we noticed that progeny containing both UAS-pgant3 and certain Gal4 drivers were not viable. This was seen with multiple independent UAS-pgant3 transgenic lines (Table 2 and data not shown). Overexpression of wild type pgant3 using the tubulin-Gal4 driver was lethal, with most animals dying during embryogenesis and larva stages (Table 2 and data not shown). UAS-pgant3 expression under the control of the armadillo driver (arm-Gal4, whose expression level is less than that of Tub-Gal4) resulted in partial lethality, with 50% of the animals surviving to adulthood. Expression of pgant3 under the control of a wing-specific driver had no effect on viability or wing blade formation (Table 2 and data not shown). These results indicate that although overexpression of pgant3 is tolerated in the wing, specific regulation of pgant3 levels in other tissues and stages is necessary for development to proceed properly. This may be the reason for the highly restricted pattern of expression seen for pgant3 during embryonic development (27).
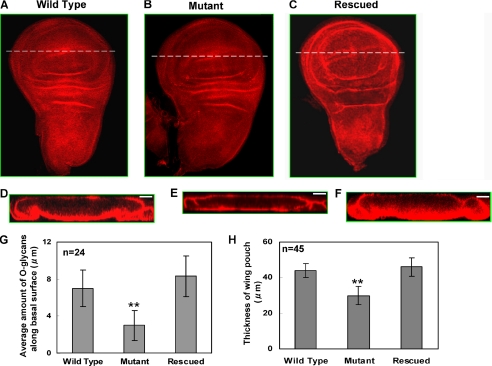
Altered O-Glycan Staining and Wing Disc Morphology is Seen in pgant3c01318/pgant3c01318 Wing Discs—Wing discs from pgant3c01318 homozygotes were stained with an antibody (Tn Ab) that detects the Tn antigen, GalNAcα-S/T (44), to visualize alterations in protein O-glycosylation patterns in the mutant discs. Wing discs from pgant3c01318 homozygotes did not show global changes in O-glycosylation relative to wild type discs (Fig. 4, A and B). However, examination of optical cross-sections through the mutant wing discs revealed a decrease in O-glycoproteins specifically along the basal surface of the columnar epithelial cells (Fig. 4, D and E). The average height of O-glycan staining along the basal surface of pgant3c01318 homozygous mutant wing discs was 3-μm versus 7-μm for wild type discs (Fig. 4G); no significant decrease in O-glycans along the apical surface was observed. Additionally, mutant discs were decreased in thickness relative to wild type (Fig. 4H), with mutants having an average thickness of 30 μm, whereas wild type had an average of 44 μm. Furthermore, basal O-glycan staining and disc thickness were restored in mutant discs expressing wild type pgant3 (Fig. 4, F–H), concomitant with restoration of adult wing blade integrity (Fig. 2E). These results demonstrate that decreased pgant3 gene expression results in localized reductions in O-glycans along the basal region of wing discs followed by separation of epithelial cell layers in the adult wing, both of which can be rescued with wild type pgant3 expression. Because the basal surface of larval wing discs will mediate contacts between epithelial cell layers in the adult wing, this implicates basal O-glycoproteins modified by PGANT3 in these cell adhesion events. Additionally, these studies provide evidence that reduced O-glycosylation results in altered columnar epithelial cell morphology, as mutant discs were significantly thinner than wild type discs.
FIGURE 4.
O-Glycan-specific antibody staining in wing imaginal discs reveals decreased basal O-glycans and altered morphology in pgant3c01318 mutants. Wing imaginal discs from wild type (A and D), pgant3c01318/pgant3c01318 transposon insertion homozygotes (Mutant) (B and E), and transposon homozygotes expressing wild type pgant3 (pgant3c01318/pgant3c01318; UAS-pgant3) (Rescued) (C and F) were stained with the Tn antibody and visualized by confocal microscopy. D–F represent optical X-Z cross-sections of each disc in the region of the white line shown in A–C. Anterior is to the left, and posterior is to the right in A–C. Apical is at the top, and basal is at the bottom in D–F. G, average amount of O-glycan staining seen in wild type, mutant, and rescued discs along the basal surface of the wing pouch (measured as described under “Experimental Procedures”). H, average thickness of the wing pouch in wild type, mutant, and rescued discs. n = total number of wing discs of each genotype analyzed. S.D. are shown. Scale bar = 20 μm. **, p < 0.01.
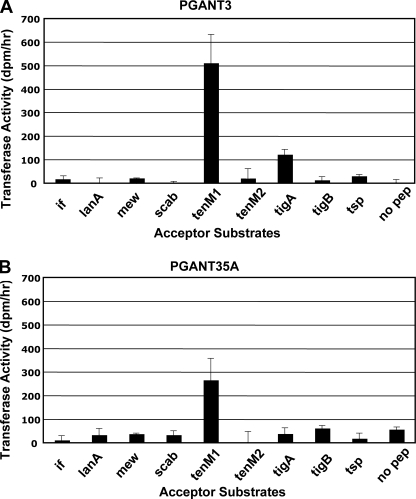
PGANT3 Glycosylates ECM Proteins in Vitro—To identify potential substrates for PGANT3, we compiled a list of secreted and membrane-bound proteins expressed in wing discs that are predicted to have potential sites of O-glycosylation (Table 3) using the NetOGlyc server. This software employs a neural network prediction program based on confirmed sites of O-glycosylation in many diverse proteins. We then performed in vitro glycosylation reactions using recombinant PGANT3 and peptides derived from these proteins to determine which substrates PGANT3 is capable of glycosylating. As shown in Fig. 5A, PGANT3 transfers a substantial amount of GalNAc to tenM1, a specific peptide derived from the tenascin-major protein, and tigA, a peptide derived from the tiggrin protein. No glycosylation was seen for tenM2 or tigB, additional peptides derived from tenascin-major and tiggrin, respectively, suggesting that PGANT3 activity is specific to certain subregions within these proteins. A modest amount of glycosylation was also seen with thrombospondin. No significant transfer was seen to the other peptides tested in this assay, indicating that PGANT3 glycosylates specific regions of certain extracellular matrix proteins in vitro. To demonstrate specificity of the PGANT3 peptide glycosylation observed, similar assays were performed using recombinant PGANT35A. Although PGANT35A was able to transfer GalNAc to tenM1, no glycosylation above background was seen for the other substrates tested (Fig. 5B). This in vitro specificity is in accord with the pgant3-specific in vivo rescue of wing blistering, supporting a unique role for PGANT3 in the wing disc that cannot be compensated for by the activity of another glycosyltransferase family member. The in vitro data presented here suggests that tiggrin is one of the unique candidate targets for PGANT3.
FIGURE 5.
Glycosylation of ECM peptides by PGANT3. Peptides derived from secreted and membrane-bound proteins expressed in wing discs were incubated with recombinant PGANT3 (A) or PGANT35A (B) enzymes in in vitro glycosylation reactions, and transfer of labeled GalNAc was measured. Both PGANT3 and PGANT35A glycosylate the tenM1 peptide, but glycosylation of the tigA peptide was only observed with PGANT3. Rates of GalNAc incorporation (dpm/h) are shown on the vertical axes, and peptide substrates tested are shown on the horizontal axes. Error bars indicate S.D. Background values (no pep = no peptide acceptor present in the reaction) are shown in each graph.
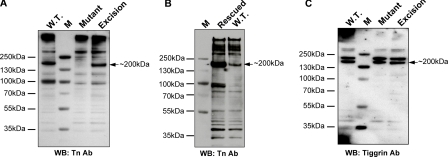
Loss of Specific O-Glycoproteins Is Seen in pgant3c01318 Mutant Wing Discs—Protein extracts from wing discs of wild type, pgant3c01318 homozygotes, and transposon excision lines were run on SDS-PAGE gels, Western-blotted, and probed with the Tn Ab to detect changes in the presence of O-glycosylated proteins. No differences were seen between the majority of bands present in wild type and mutant lanes. However, glycoproteins of ∼68 and ∼200 kDa that were present in wild type and transposon excision samples were absent or severely reduced in staining intensity in pgant3c01318 mutant samples; additionally, there was a slight reduction in intensity within the high molecular weight region at the top of the gel in the pgant3c01318 mutant sample (Fig. 6A). Western blots further demonstrated that glycosylation of the ∼200-kDa band is restored in rescued pgant3c01318 mutant wing discs (expressing wild type pgant3) (Fig. 6B).
FIGURE 6.
Altered patterns of O-glycosylation of wing imaginal disc proteins from pgant3 transposon insertion mutants. A, protein extracts from the wing imaginal discs of wild type (W.T.), pgant3c01318/pgant3c01318 transposon insertion homozygotes (Mutant), and pgant3c01318revertant#7/pgant3c01318revertant#7 transposon excision homozygotes (Excision) were subject to SDS-PAGE electrophoresis, blotted, and probed with the Tn antigen antibody (Tn Ab) to detect O-linked glycoproteins carrying the Tn antigen structure (Gal-NAcα-S/T). Loss or severe reduction in ∼68- and ∼200-kDa bands in the mutant sample relative to the wild type or transposon excision sample is seen. WB, Western blot. B, Western blot of protein extracts from wild type and transposon homozygotes expressing wild type pgant3 (pgant3c01318/pgant3c01318; UAS-pgant3) (Rescued) wing discs probed with the Tn Ab. C, Western blot from panel A was probed with the tiggrin antibody. The position of the ∼200-kDa band is denoted on the right side of each blot. Size markers (lane M) are shown to the left of each panel.
Based on our in vitro data and the size of the predominant band absent in the mutant discs (∼200 kDa), we hypothesized that this band may be the tiggrin protein. Western blots probed with the tiggrin antibody revealed that the lower tiggrin band runs in the exact position of the ∼200 kDa band detected with the Tn Ab (Fig. 6C), strongly suggesting that the differentially glycosylated band is tiggrin. Collectively, these results support the notion that pgant3 is responsible for glycosylating specific proteins within the wing discs, one of which may be the ECM protein tiggrin.
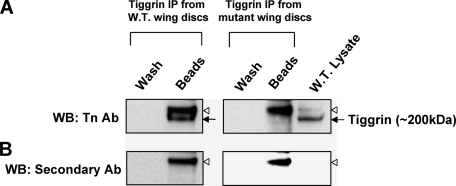
Tiggrin Is an in Vivo Substrate for PGANT3—To conclusively address whether tiggrin represents the ∼200-kDa O-glycosylated band that is lost in pgant3c01318 mutants, we performed immunoprecipitation experiments from wild type wing disc extracts using the tiggrin antibody (Fig. 7). Western blots of immunoprecipitated tiggrin blotted with the Tn Ab revealed that tiggrin is O-glycosylated in wild type wing discs and runs at the same position as the predominant ∼200-kDa band seen on previous Western blots (Fig. 7A). Nonspecific bands detected by the secondary antibody are shown in Fig. 7B. No Tn Ab reactivity was seen in the immunoprecipitated sample from mutant wing discs (Fig. 7A), suggesting that tiggrin is either not O-glycosylated or is unstable/degraded. Although the data from Fig. 6 indicate that tiggrin from mutant discs is present but not glycosylated, we cannot rule out the possibility that the lack of O-glycans may also result in altered protein stability. Nonetheless, these results conclusively demonstrate that tiggrin is a bona fide substrate for PGANT3 in vivo.
FIGURE 7.
Tiggrin is O-glycosylated by PGANT3 in vivo. Immunoprecipitation (IP) of tiggrin from wild type and pgant3c01318/pgant3c01318 mutants was performed as described under “Experimental Procedures.” A, the nonspecific wash (Wash) and immunoprecipitated tiggrin (Beads) from each sample were subject to SDS-PAGE electrophoresis, blotted, and probed with the Tn Ab. Tiggrin from wild type discs is detected with the O-glycan specific antibody, but tiggrin from mutant discs shows no reactivity. Wild type wing disc lysate (W.T. Lysate) was included as a control for detection of O-glycosylated tiggrin. B, Western blots (WB) were reprobed with the secondary antibody alone to identify the nonspecific band present in each sample (denoted with a white triangle to the right of each blot). The position of the specific tiggrin band is denoted with a black arrow.
Genetic Interactions between pgant3 and tiggrin Increase Wing Blistering—To demonstrate an in vivo interaction between tiggrin and pgant3 in mediating the wing blistering phenotype, we performed genetic interaction experiments. Two distinct tiggrin mutants (TigX and TigA1) (14) showed no blistering heterozygously at 18 °C (Table 1); pgant3c01318 heterozygotes displayed 2% wing blistering at 18 °C (Table 1). However, transheterozygotes consisting of pgant3c01318 and TigX or pgant3c01318 and TigA1 both resulted in an increased wing blistering frequency (Table 1). These genetic interactions were only seen at 18 °C. We did not observe genetic interactions between pgant3 and other genes known to be involved in various aspects of wing blade adhesion (if, by, bsk, wb, mys, mew, Ten-m, bs) (data not shown). Thus, pgant3 and tiggrin mutations, when combined, function to exacerbate the same morphological phenotype, further demonstrating that the glycosylation of tiggrin by PGANT3 has a functional consequence in modulating cell adhesive events in the adult wing.
DISCUSSION
Cell adhesion and the factors that regulate it are crucial during many diverse developmental stages. Here we describe for the first time the role of mucin-type protein O-glycosylation in cell adhesion during development. Mutations in the pgant3 gene, which encodes an enzyme responsible for initiating protein O-glycosylation, resulted in aberrant adhesion between the cell layers comprising the wing blade. Epithelial cell adhesion in the Drosophila wing blade is regulated primarily by integrin-ECM interactions (2–5, 8, 14–15). However, a role for O-glycosylation of these proteins in modulating their cell adhesive functions during development has not been previously described. Our data elucidate a novel role for O-glycosylation in cell adhesion during wing development.
Here we show that a known integrin binding ECM protein, tiggrin, is specifically glycosylated by PGANT3 and that the activity of PGANT3 is functionally significant in mediating phenotypic consequences in the developing wing. PGANT3 glycosylates tiggrin both in vitro and in vivo; mutations in pgant3 result in a loss of tiggrin glycosylation (accompanied by wing blistering), whereas expression of wild type pgant3 restores tiggrin glycosylation and wing integrity. Additionally, genetic interaction experiments reveal that the combination of pgant3 and tiggrin mutations resulted in an increased blistering frequency, demonstrating that these genes are acting in the same phenotypic pathway. Prior studies have shown that tiggrin binds integrin (9) and is involved in integrin-mediated cell adhesive events during Drosophila development, including muscle-tendon cell adhesion and wing blade adhesion (14). Indeed, the wing blistering seen in tiggrin mutants (along the posterior portion of the wing blade) (14) is similar to what was often observed in pgant3 mutants. Given this, we propose that O-glycans on tiggrin mediate some aspect of integrin-ECM adhesion in the wing blade. Our Western blots demonstrate that the tiggrin protein is present in pgant3 mutants, suggesting that the loss of O-glycans does not result in significant tiggrin degradation. However, O-glycans on tiggrin may influence more subtle aspects of protein stability as well as transport, secretion, localization, or binding interactions. Indeed, prior work from our laboratory suggests that O-glycosylation by another member of this glycosyltransferase family is involved in protein transport during Drosophila development (36). We are currently investigating the specific mechanistic function of O-glycans on tiggrin.
To address the developmental origin of the wing defect in pgant3 mutants, we examined developing wing imaginal discs. Previous studies have shown that defects in JNK signaling related to integrin function in the developing wing can result in wing blistering (11). However, we did not detect changes in JNK phosphorylation or downstream signaling in pgant3 mutants (data not shown). Detailed morphological examination of mutant wing discs revealed that although global patterns of glycosylation were unchanged, pgant3 mutants displayed a specific reduction in O-glycans along the basal surface of the columnar epithelial cells. Additionally, basal surface glycosylation was restored in mutants expressing wild type pgant3, as was wing integrity and tiggrin glycosylation. These regional alterations in glycosylation are significant, as integrins and ECM proteins present along the basal surface mediate the adhesive contacts that will form as the disc everts and opposing basal regions come in contact with one another (2). Thus, we hypothesize that the wing blisters that form in the adult are most likely the result of an initial reduction in O-glycosylation of proteins normally found along the basal surface of larval wing discs.
Reduced basal glycosylation in larval wing discs was also accompanied by a reduction in the height of the columnar cells comprising the wing disc. Again, expression of wild type pgant3 restored glycosylation and columnar cell height. It has been shown previously that alterations in integrin-ECM interactions can affect cell size and shape in the wing disc (46). We hypothesize that the morphological changes seen in the pgant3 mutants are the result of alterations in integrin-ECM interactions influencing cytoskeletal architecture and cell shape.
Although tiggrin appears to be the major O-glycosylated protein affected in pgant3 mutants, our in vitro and Western blot data suggest that PGANT3 may glycosylate other proteins in addition to tiggrin. In vitro biochemical assays revealed that other wing disc ECM proteins, such as tenascin-major (∼300 kDa) and thrombospondin (∼120 kDa), are glycosylated by PGANT3. No significant glycosylation was seen for the integrin peptides tested. Tenascin-major was also glycosylated by another PGANT family member (PGANT35A), suggesting that its glycosylation is not PGANT3-specific. Western blots further revealed an additional glycosylated band (∼68 kDa) that is decreased in pgant3 mutants. These results suggest that PGANT3 may glycosylate a subset of basal ECM proteins (including tiggrin) in the developing wing disc. The aforementioned ECM proteins are also known to specifically interact with αPS2 integrin to mediate cell adhesion in a number of developmental contexts (9, 10, 13, 14). We postulate that glycosylation of ECM proteins in addition to tiggrin could also play a role in the wing blade adhesion. We are currently trying to identify the ∼68-kDa band.
Our studies indicate that pgant3 is uniquely responsible for glycosylating certain proteins such as tiggrin, which are required for proper wing development and that the activity of another family member is not compensatory. This unique requirement for pgant3 in the developing wing disc is demonstrated by in vivo rescue experiments where wild type pgant3 can restore wing integrity, but overexpression of another related family member (pgant35A, which is normally expressed in the wing disc and is also an initiating peptide transferase) cannot. This result is likely due to isoform-specific substrate preferences that were also observed in vitro. These data highlight the significance of substrate preferences seen for PGANT family members in vitro and illustrate the functional consequences of this specificity in vivo.
The notion of unique roles for pgant3 during development is further supported by the observation that overexpression of pgant3 is lethal, whereas pgant35A overexpression has no obvious effect. Although we currently do not know the specific nature of this lethality (most pgant3 overexpressing animals died during embryonic and larval stages), these results suggest that control of PGANT3 enzymatic levels during development is critical. Indeed, we know from previous in situ experiments that pgant3 gene expression is specifically regulated during embryonic development, being found in a very restricted subset of tissues, including the wing disc (27). The unique substrate preferences of PGANT3 may necessitate very strict spatial and temporal regulation of enzymatic activity during development. The role of pgant3 in other tissues and stages is currently under investigation.
In summary, we have identified a novel role for mucin-type O-glycosylation during development. Here, we present evidence for the requirement of O-glycans in cell adhesion processes during development. Previous work has demonstrated the role of O- and N-linked carbohydrates and sugar-binding proteins in cell adhesion mediating lymphocyte homing and extravasation within the mammalian immune system (34, 47–49). The presence of mucin type O-glycans on secreted and membrane-bound proteins across many species suggests a more widespread role for O-glycans in cell adhesive events in many developing organ systems. Building upon the studies presented here, it will be interesting to investigate the role of O-glycans in other developmental contexts.
Supplementary Material
Acknowledgments
We thank Drs. L. Tabak, J. Kennison, J. Kassis, D. Hursh, L. Angerer, and M. Mortin for many helpful discussions. We thank Drs. R. Cummings and T. Ju for the kind gift of the Tn antibody and Drs. L. Fessler and J. Fessler for the kind gift of the tiggrin antibody. Finally, we also thank Drs. J. Kennison, D. Andrew, F. Shoeck, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for fly stocks and other reagents.
This work was supported, in whole or in part, by the National Institutes of Health (the Intramural Research Program of the NIDCR). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.

Author's Choice—Final version full access.
Footnotes
The abbreviations used are: ECM, extracellular matrix; ppGalNAcT or PGANT or pgant, UDP GalNAc:polypeptide N-acetylgalactosaminyltransferase; IR, inverted repeat; JNK, c-Jun NH2-terminal kinase; RNAi, RNA interference; Ab, antibody.
References
- 1.Hynes, R. O., and Zhao, Q. (2000) J. Cell Biol. 150 89-96 [DOI] [PubMed] [Google Scholar]
- 2.Brower, D. L. (2003) Curr. Opin. Cell Biol. 15 607-613 [DOI] [PubMed] [Google Scholar]
- 3.Brower, D. L., Bunch, T. A., Mukai, L., Adamson, T. E., Wehrli, M., Lam, S., Friedlander, E., Roote, C. E., and Zusman, S. (1995) Development 121 1311-1320 [DOI] [PubMed] [Google Scholar]
- 4.Prout, M., Damania, Z., Soong, J., Fristrom, D., and Fristrom, J. W. (1997) Genetics 146 275-285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh, E. P., and Brown, N. H. (1998) Genetics 150 791-805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araujo, H., Negreiros, E., and Bier, E. (2003) Development 130 3851-3864 [DOI] [PubMed] [Google Scholar]
- 7.Bloor, J. W., and Brown, N. H. (1998) Genetics 148 1127-1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, N. H., Gregory, S. L., Rickoll, W. L., Fessler, L. I., Prout, M., White, R. A., and Fristrom, J. W. (2002) Dev Cell 3 569-579 [DOI] [PubMed] [Google Scholar]
- 9.Fogerty, F. J., Fessler, L. I., Bunch, T. A., Yaron, Y., Parker, C. G., Nelson, R. E., Brower, D. L., Gullberg, D., and Fessler, J. H. (1994) Development 120 1747-1758 [DOI] [PubMed] [Google Scholar]
- 10.Graner, M. W., Bunch, T. A., Baumgartner, S., Kerschen, A., and Brower, D. L. (1998) J. Biol. Chem. 273 18235-18241 [DOI] [PubMed] [Google Scholar]
- 11.Lee, S. B., Cho, K. S., Kim, E., and Chung, J. (2003) Development 130 4001-4010 [DOI] [PubMed] [Google Scholar]
- 12.Subramanian, A., Wayburn, B., Bunch, T., and Volk, T. (2007) Development 134 1269-1278 [DOI] [PubMed] [Google Scholar]
- 13.Torgler, C. N., Narasimha, M., Knox, A. L., Zervas, C. G., Vernon, M. C., and Brown, N. H. (2004) Dev. Cell 6 357-369 [DOI] [PubMed] [Google Scholar]
- 14.Bunch, T. A., Graner, M. W., Fessler, L. I., Fessler, J. H., Schneider, K. D., Kerschen, A., Choy, L. P., Burgess, B. W., and Brower, D. L. (1998) Development 125 1679-1689 [DOI] [PubMed] [Google Scholar]
- 15.Prokop, A., Martin-Bermudo, M. D., Bate, M., and Brown, N. H. (1998) Dev. Biol. 196 58-76 [DOI] [PubMed] [Google Scholar]
- 16.Bruckner, K., Perez, L., Clausen, H., and Cohen, S. (2000) Nature 406 411-415 [DOI] [PubMed] [Google Scholar]
- 17.Haines, N., and Irvine, K. D. (2003) Nat. Rev. Mol. Cell Biol. 4 786-797 [DOI] [PubMed] [Google Scholar]
- 18.Moloney, D. J., Panin, V. M., Johnston, S. H., Chen, J., Shao, L., Wilson, R., Wang, Y., Stanley, P., Irvine, K. D., Haltiwanger, R. S., and Vogt, T. F. (2000) Nature 406 369-375 [DOI] [PubMed] [Google Scholar]
- 19.Okajima, T., Xu, A., and Irvine, K. D. (2003) J. Biol. Chem. 278 42340-42345 [DOI] [PubMed] [Google Scholar]
- 20.Sasaki, N., Sasamura, T., Ishikawa, H. O., Kanai, M., Ueda, R., Saigo, K., and Matsuno, K. (2007) Genes Cells 12 89-103 [DOI] [PubMed] [Google Scholar]
- 21.Sasamura, T., Ishikawa, H. O., Sasaki, N., Higashi, S., Kanai, M., Nakao, S., Ayukawa, T., Aigaki, T., Noda, K., Miyoshi, E., Taniguchi, N., and Matsuno, K. (2007) Development 134 1347-1356 [DOI] [PubMed] [Google Scholar]
- 22.Ricketts, L. M., Dlugosz, M., Luther, K. B., Haltiwanger, R. S., and Majerus, E. M. (2007) J. Biol. Chem. 282 17014-17023 [DOI] [PubMed] [Google Scholar]
- 23.Acar, M., Jafar-Nejad, H., Takeuchi, H., Rajan, A., Ibrani, D., Rana, N. A., Pan, H., Haltiwanger, R. S., and Bellen, H. J. (2008) Cell 132 247-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingsley, P. D., Ten Hagen, K. G., Maltby, K. M., Zara, J., and Tabak, L. A. (2000) Glycobiology 10 1317-1323 [DOI] [PubMed] [Google Scholar]
- 25.Ten Hagen, K. G., Tran, D. T., Gerken, T. A., Stein, D. S., and Zhang, Z. (2003) J. Biol. Chem. 278 35039-35048 [DOI] [PubMed] [Google Scholar]
- 26.Tian, E., and Ten Hagen, K. G. (2007) Glycobiology 17 820-827 [DOI] [PubMed] [Google Scholar]
- 27.Tian, E., and Ten Hagen, K. G. (2006) Glycobiology 16 83-95 [DOI] [PubMed] [Google Scholar]
- 28.Hang, H. C., and Bertozzi, C. R. (2005) Bioorg. Med. Chem. 13 5021-5034 [DOI] [PubMed] [Google Scholar]
- 29.Ten Hagen, K. G., Fritz, T. A., and Tabak, L. A. (2003) Glycobiology 13 1-16 [DOI] [PubMed] [Google Scholar]
- 30.Pratt, M. R., Hang, H. C., Ten Hagen, K. G., Rarick, J., Gerken, T. A., Tabak, L. A., and Bertozzi, C. R. (2004) Chem. Biol. 11 1009-1016 [DOI] [PubMed] [Google Scholar]
- 31.Ten Hagen, K. G., Bedi, G. S., Tetaert, D., Kingsley, P. D., Hagen, F. K., Balys, M. M., Beres, T. M., Degand, P., and Tabak, L. A. (2001) J. Biol. Chem. 276 17395-17404 [DOI] [PubMed] [Google Scholar]
- 32.Ten Hagen, K. G., Tetaert, D., Hagen, F. K., Richet, C., Beres, T. M., Gagnon, J., Balys, M. M., VanWuyckhuyse, B., Bedi, G. S., Degand, P., and Tabak, L. A. (1999) J. Biol. Chem. 274 27867-27874 [DOI] [PubMed] [Google Scholar]
- 33.Topaz, O., Shurman, D. L., Bergman, R., Indelman, M., Ratajczak, P., Mizrachi, M., Khamaysi, Z., Behar, D., Petronius, D., Friedman, V., Zelikovic, I., Raimer, S., Metzker, A., Richard, G., and Sprecher, E. (2004) Nat. Genet. 36 579-581 [DOI] [PubMed] [Google Scholar]
- 34.Tenno, M., Ohtsubo, K., Hagen, F. K., Ditto, D., Zarbock, A., Schaerli, P., von Andrian, U. H., Ley, K., Le, D., Tabak, L. A., and Marth, J. D. (2007) Mol. Cell. Biol. 27 8783-8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ten Hagen, K. G., and Tran, D. T. (2002) J. Biol. Chem. 277 22616-22622 [DOI] [PubMed] [Google Scholar]
- 36.Tian, E., and Ten Hagen, K. G. (2007) J. Biol. Chem. 282 606-614 [DOI] [PubMed] [Google Scholar]
- 37.Xia, L., Ju, T., Westmuckett, A., An, G., Ivanciu, L., McDaniel, J. M., Lupu, F., Cummings, R. D., and McEver, R. P. (2004) J. Cell Biol. 164 451-459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander, W. S., Viney, E. M., Zhang, J. G., Metcalf, D., Kauppi, M., Hyland, C. D., Carpinelli, M. R., Stevenson, W., Croker, B. A., Hilton, A. A., Ellis, S., Selan, C., Nandurkar, H. H., Goodnow, C. C., Kile, B. T., Nicola, N. A., Roberts, A. W., and Hilton, D. J. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16442-16447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thibault, S. T., Singer, M. A., Miyazaki, W. Y., Milash, B., Dompe, N. A., Singh, C. M., Buchholz, R., Demsky, M., Fawcett, R., Francis-Lang, H. L., Ryner, L., Cheung, L. M., Chong, A., Erickson, C., Fisher, W. W., Greer, K., Hartouni, S. R., Howie, E., Jakkula, L., Joo, D., Killpack, K., Laufer, A., Mazzotta, J., Smith, R. D., Stevens, L. M., Stuber, C., Tan, L. R., Ventura, R., Woo, A., Zakrajsek, I., Zhao, L., Chen, F., Swimmer, C., Kopczynski, C., Duyk, G., Winberg, M. L., and Margolis, J. (2004) Nat. Genet. 36 283-287 [DOI] [PubMed] [Google Scholar]
- 40.Brand, A. H., and Perrimon, N. (1993) Development 118 401-415 [DOI] [PubMed] [Google Scholar]
- 41.Lee, Y. S., and Carthew, R. W. (2003) Methods 30 322-329 [DOI] [PubMed] [Google Scholar]
- 42.Rubin, G. M., and Spradling, A. C. (1982) Science 218 348-353 [DOI] [PubMed] [Google Scholar]
- 43.Spradling, A. C., and Rubin, G. M. (1982) Science 218 341-347 [DOI] [PubMed] [Google Scholar]
- 44.Avichezer, D., Springer, G. F., Schechter, B., and Arnon, R. (1997) Int. J. Cancer 72 119-127 [DOI] [PubMed] [Google Scholar]
- 45.Pickup, A. T., and Banerjee, U. (1999) Dev. Biol. 205 254-259 [DOI] [PubMed] [Google Scholar]
- 46.Dominguez-Gimenez, P., Brown, N. H., and Martin-Bermudo, M. D. (2007) J. Cell Sci. 120 1061-1071 [DOI] [PubMed] [Google Scholar]
- 47.Ellies, L. G., Sperandio, M., Underhill, G. H., Yousif, J., Smith, M., Priatel, J. J., Kansas, G. S., Ley, K., and Marth, J. D. (2002) Blood 100 3618-3625 [DOI] [PubMed] [Google Scholar]
- 48.Ellies, L. G., Tsuboi, S., Petryniak, B., Lowe, J. B., Fukuda, M., and Marth, J. D. (1998) Immunity 9 881-890 [DOI] [PubMed] [Google Scholar]
- 49.Gauguet, J. M., Rosen, S. D., Marth, J. D., and von Andrian, U. H. (2004) Blood 104 4104-4112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.