Abstract
Leaf senescence in Arabidopsis thaliana is a strict, genetically controlled nutrient recovery program, which typically progresses in an age-dependent manner. Leaves of the Arabidopsis onset of leaf death5 (old5) mutant exhibit early developmental senescence. Here, we show that OLD5 encodes quinolinate synthase (QS), a key enzyme in the de novo synthesis of NAD. The Arabidopsis QS was previously shown to carry a Cys desulfurase domain that stimulates reconstitution of the oxygen-sensitive Fe-S cluster that is required for QS activity. The old5 lesion in this enzyme does not affect QS activity but it decreases its Cys desulfurase activity and thereby the long-term catalytic competence of the enzyme. The old5 mutation causes increased NAD steady state levels that coincide with increased activity of enzymes in the NAD salvage pathway. NAD plays a key role in cellular redox reactions, including those of the tricarboxylic acid cycle. Broad-range metabolite profiling of the old5 mutant revealed that it contains higher levels of tricarboxylic acid cycle intermediates and nitrogen-containing amino acids. The mutant displays a higher respiration rate concomitant with increased expression of oxidative stress markers. We postulate that the alteration in the oxidative state is integrated into the plant developmental program, causing early ageing of the mutant.
INTRODUCTION
In Arabidopsis thaliana, the onset of leaf senescence proceeds within a predictable time window (Jing et al., 2002). The leaf senescence program can be prematurely initiated when a plant is under attack or grows in unfavorable conditions (Gan and Amasino, 1997; Quirino et al., 2000; Schippers et al., 2007). Senescence contributes to relocation of valuable resources from old, dying tissue to young developing parts of the plant. For example, transfer of nutrients supports the filling of grains in order to afford the plant reproductive success. Recent expression profiling studies have identified thousands of transcripts involved in the onset and progression of developmental senescence, termed senescence-associated genes (SAGs) (Buchanan-Wollaston et al., 2003, 2005; Guo et al., 2004). Aside from the fundamental importance of understanding senescence, potential improvements for crop yield or the longevity of ornamental plants might be realized. In this light, a recently identified senescence-associated transcription factor in wheat (Triticum aestivum), which was shown to affect protein content by 30% (Uauy et al., 2006), underlines the impact of senescence research on future agriculture. Despite advances in the understanding of the process of senescence, little is known regarding the mechanism that determines its onset.
During leaf growth, age-related changes (ARCs) occur as a result of the differential regulation of developmental processes. The onset of developmental senescence occurs after certain ARCs have taken place in the leaf (Jing et al., 2005). Ageing occurs throughout leaf development, from leaf initiation to death; however, senescence only refers to the process that leads to the death of the leaf (Lim et al., 2003). By contrast, in ageing research on nonplant species, the terms senescence and ageing are interchangeable. Longevity in these other model organisms may not be regulated by a genetically controlled ageing program, since most organisms reproduce early and, therefore, the ageing process escapes from the force of natural selection (Rose, 1991; Kirkwood, 2002). Nevertheless, many genes have been identified that alter longevity in organisms ranging from yeast to worms, fruit flies, and mammals (Jing et al., 2003; Kirkwood, 2005). On the contrary, for plants, nutrient salvage during leaf senescence is a vital part of development and hence under evolutional selection (Bleecker, 1998). Therefore, the longevity of plants may be fundamentally differently regulated than in other organisms. Although ageing of Arabidopsis is influenced by its reproductive strategy, there is only a weak correlation between the appearance of reproductive structures and the onset of leaf senescence. Male- or female-sterile mutant plants live 20 d longer (Nooden and Penney, 2001); conversely, the lifespan of individual leaves of these mutants is not affected. Thus, individual leaves are useful models for studying the regulation of the ageing program in plants (Jing et al., 2003; Lim et al., 2003; Schippers et al., 2007).
In general, ageing is controlled by programs involved in life maintenance, stress responses, and development (Schippers et al., 2007). In the mid-1950s, it was postulated that reactive oxygen species (ROS) are the main cause of animal ageing, beginning with cumulative damage that results in loss of viability (Harman, 1956). Interestingly, the Arabidopsis longevity mutants oresara1, oresara3, and oresara9 (Woo et al., 2004) and gigantea (Kurepa et al., 1998) all show higher tolerance to oxidative stress. In nonplant species, ROS are mainly produced by mitochondria; however, during leaf senescence in plants, the main ROS source is the chloroplast (Quirino et al., 2000). That said, plant mitochondria produce considerable ROS during the hypersensitive response invoked by plant–pathogen interactions (Finkel and Holbrook, 2000) and under a variety of other cellular circumstances (Sweetlove et al., 2006, 2007). Increased tissue contents of ROS (Navabpour et al., 2003) and exogenously applied causal agents of oxidative stress such as UV-B light and ozone (Miller et al., 1999; John et al., 2001) lead to premature expression of SAGs. These combined data support the notion that ROS influence both the ageing process and the lifespan of Arabidopsis leaves.
The process of senescence correlates with a loss of antioxidant capacity and consequently with an increase in ROS (Zimmermann and Zentgraf, 2005). An important source of ROS production in animals and plants are the membrane-associated NAD(P)H oxidases that generate ROS via lipid oxidation (Finkel and Holbrook, 2000; Mittler, 2002). NAD(P)Hs are well-studied redox molecules that mediate hundreds of reactions and are the basis of almost every metabolic pathway in the cell (Noctor et al., 2006). Pyridine nucleotides are the main redox regulators, since they react slowly with oxygen species in an enzyme-dependent way. ROS production in the chloroplast has been clearly demonstrated to be under the influence of NADP/NADPH ratios in this compartment (Asada, 2006). Production of mitochondrial ROS by the electron transport chain is largely dependent on NAD(P) status as well (Dutilleul et al., 2005; Shen et al., 2006; Sweetlove et al., 2006). The turnover of antioxidant pools such as those of glutathione and ascorbate are maintained by NAD(P)H (Noctor et al., 2006). The ascorbate-deficient mutant vtc1 shows premature senescence when grown under a long photoperiod or when leaves are placed in the dark (Barth et al., 2004). Moreover, a proteomics study of the early senescence mutant old1 revealed that most of the highly upregulated proteins in the mutant belong to the family of glutathione S-transferases (Hebeler et al., 2008). This suggests that glutathione S-transferases in the context of leaf senescence are needed to protect cells against ROS during the process. Taken together, these studies demonstrate that the redox status might be an important determinant for the onset of senescence.
The availability of senescence mutants has given us the opportunity to discover which processes play a role in plant ageing and determine the onset of leaf senescence (Jing et al., 2002, 2005). Here, we report that a mutation in the gene encoding quinolinate synthase (QS/OLD5) results in an early onset of developmental senescence in Arabidopsis. Despite the fact that the genetic lesion of this mutant is in the de novo pathway of NAD synthesis, the mutant exhibits increased content of pyridine nucleotides. Our data indicate that the increased NAD level is due to an increased activity of enzymes involved in NAD salvage. Further characterization of the mutant revealed changes in the levels of transcripts that are suggestive of oxidative stress, while the metabolic profile obtained for the mutant shows increased levels of metabolites of central metabolism. The results are discussed in the context of current theories of ageing and senescence that have been documented previously in the plant and mammalian literature.
RESULTS
Characterization of the old5 Mutant during Developmental Senescence
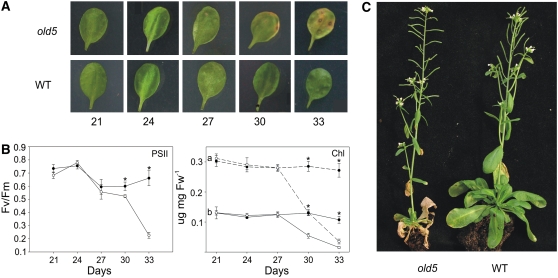
The old5 mutant was identified as a monogenic recessive trait during a screen of an ethyl methanesulfonate–treated population of Arabidopsis plants (Jing et al., 2002) and was shown to display an early onset of senescence and an enhanced senescence response after ethylene treatment (Jing et al., 2005). The mutant shows yellowing of the first leaf pair after 30 d (Figure 1A), whereas the wild type shows senescence symptoms after 40 d. The visual yellowing parallels a decline in chlorophyll content and photochemical efficiency (Figure 1B). Thus, the old5 mutant exhibits the physiological hallmarks of leaf senescence. The early senescence of the mutant results in a decrease in lifespan of ∼2 weeks (Figure 1C).
Figure 1.
The Onset of Leaf Senescence in old5 Mutant Arabidopsis Plants.
(A) Visible leaf yellowing in the old5 mutant line. The first rosette leaves of 21- to 33-d-old wild-type and old5 plants are shown.
(B) Photochemical efficiency (PSII) and chlorophyll (Chl) levels of the old5 mutant line decrease at the onset of senescence (old5, open circles; wild type, closed circles). The photochemical efficiency was determined with a PAM-2000 fluorometer. The values shown are means of three repeats ± sd (indicated by error bars). Asterisks indicate statistical significance versus the wild-type control in each case (* P < 0.05, Student's t test). Fw, fresh weight.
(C) Six-week-old adult wild-type and old5 plants.
To further characterize senescence in this mutant, the expression of two established senescence marker genes was followed by quantitative real-time PCR. First, we examined SAG13, whose expression is associated with oxidative stress and senescence (Weaver et al., 1998). SAG13 expression was detected from day 24 in both the mutant and the wild type. However, the expression in the mutant was ∼10-fold higher at day 24, and this progressed to an ∼10,000-fold higher expression at day 33 than in the wild type (Figure 2). Second, we examined the hallmark of age-induced senescence, SAG12 (Weaver et al., 1998). SAG12 was detected after 30 d only in old5 but not in the wild type. The SAG12 expression correlates with the first symptoms of yellowing at 30 d, implying, together with the expression of the SAG13 marker, that senescence in old5 mutants begins prematurely but still occurs in an age-dependent way.
Figure 2.
Transcript Accumulation of the Senescence-Associated Genes SAG12 and SAG13 in Wild-Type and old5 Mutant Plants.
The relative levels of SAG12 and SAG13 were examined by real-time RT-PCR, using ACT2 as an internal control, of first rosette leaves of 21- to 33-d-old plants (old5, gray bars; wild type, black bars). For SAG13, the relative transcript levels detected in the wild-type plants were arbitrarily assigned a value of 1 after normalization to the ACT2 samples. For SAG12, the relative transcript level detected at day 30 in old5 was set at 1 and used for comparison with day 33. The values shown are means of three repeats ± sd (indicated by error bars). Experiments were repeated and showed comparable results. Asterisks indicate statistical significance versus the wild-type control in each case (* P < 0.05, Student's t test).
OLD5 Encodes a QS
The old5 mutant was identified in the Landsberg erecta (Ler) accession and was crossed to Columbia (Col) to facilitate mapping. The mapping population segregated in a 1:3 ratio (mutant:wild type), demonstrating that the mutant phenotype is encoded by a single recessive locus. In a population of 2500 F2 plants, the location of old5 could be narrowed down to a 60-kb region containing 19 genes in the overlap on BACs K6A12 and MXI22 on chromosome V between markers K6A12-23K and MXI22-12K. From these 19 genes, 11 candidate genes were selected for sequence comparison between the mutant and the Ler wild type. In the sequence of At5g50210, a C to T change in the first exon of the old5 mutant was detected. This gene has previously been annotated as QS (Katoh et al., 2006), which is a component of the pathway of the de novo synthesis of NAD from Asp. A homozygous T-DNA knockout of QS is embryo-lethal (Katoh et al., 2006), demonstrating that the old5 mutation does not result in a complete loss of gene function.
The identity of At5g50210 as the QS/OLD5 gene was confirmed by a complementation test. A 5.1-kb Ler genomic fragment containing the coding sequence, a 2.1-kb 5′ promoter sequence according to the Arabidopsis Gene Regulatory Information Server database (Davuluri et al., 2003), and a 0.5-kb 3′ sequence was transformed into old5 mutant plants. Ten T1 lines were selected on kanamycin, and these all showed absence of the old5 phenotype, as expected since the mutation is recessive. By PCR, we showed that the lines contained both the kanamycin-resistance gene and the homozygous parental old5 point mutation (see Methods). The T2 segregated 3:1 (wild type:mutant), confirming the identity of the OLD5 mutant gene.
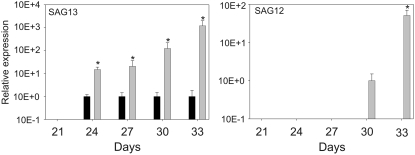
old5 Mutants Have an Amino Acid Change in the SufE/YgdK Domain
The QS/OLD5 protein consists of three domains: a chloroplast-targeting signal, a SufE/YgdK domain, and a QS domain (Figure 3A). The old5 mutation results in a Pro-to Ser amino acid substitution in the SufE/YgdK domain. This domain is highly homologous to the previously identified At SufE/Cp SufE (Xu and Møller, 2006; Ye et al., 2006), the Ygdk and SufE proteins of Escherichia coli (Loiseau et al., 2005), and a putative Arabidopsis gene, At1g67810, which was found by a BLAST search of the OLD5 SufE/YgdK domain (residues 84 to 214) against the Arabidopsis genome (Figure 3B). At SufE/Cp SufE, SufE, and YgdK share functional homology as acceptors of sulfur atoms and stimulators of Cys desulfurase activity. SufE of E. coli (Loiseau et al., 2003) and Cp SufE of Arabidopsis (Ye et al., 2006) stimulate SufS Cys desulfurase activity. The Fe-S cluster, which is inserted by SufS into QS, is absolutely required for enzyme activity in E. coli (Ollagnier-de Choudens et al., 2005). In the mutated Arabidopsis old5 protein, Pro-101 is replaced by a Ser. The Pro-101 residue is present in all Arabidopsis SufE/YgdK-containing proteins, suggesting a conserved role. Utilizing the 3D-Jigsaw protein-modeling server (Bates et al., 2001), the SufE/YgdK domain of OLD5 protein could be modeled with high confidence on the resolved structure of YgdK (Liu et al., 2005). In the model, the mutation from Pro to Ser results in an altered start of the second α-helix (Figure 3C). This possible different folding of the protein might have an influence on its function.
Figure 3.
OLD5 Encodes QS.
(A) Schematic representation of the protein domain organization of OLD5. The first domain is a transit peptide that targets OLD5 to the chloroplast. The other two domains are a SufE/YgdK and a QS domain.
(B) The OLD5 SufE domain shows similarity to SufE proteins from E. coli (SufE and YgdK). This domain is also present in two other Arabidopsis genes, At4g26500, encoding SufE, and At1g67810, a putative pollen-specific SufE protein. The old5 mutation causes a change in a Pro residue that is conserved between all three Arabidopsis proteins (arrow).
(C) Modeling by 3D-Jigsaw server predicts an effect of the mutated Pro on the structure of the SufE/YgdK domain (arrow). The protein is modeled without the chloroplast targeting signal. The three overlapping structures are encoded as follows: yellow, E. coli YgdK structure (Liu et al., 2005); magenta, wild-type Arabidopsis OLD5; cyan, old5 mutant protein.
OLD5 Interacts with a SufS-Like Protein
The YgdK/SufE proteins of E. coli interact with their SufS counterpart (Loiseau et al., 2003, 2005). By analogy, therefore, we expected the SufE/YgdK domain of OLD5 to interact with a SufS-like protein. Recent studies show that the Arabidopsis SufS-like protein Cp NifS interacts with SufE (Xu and Møller, 2006; Ye et al., 2006). Furthermore, it has been suggested that QS might work in an enzyme complex with aspartate oxidase (AO) (Sakuraba et al., 2005), since the product of AO is unstable (Yang et al., 2003).
By a database search, we found that the Arabidopsis genome contains two other putative plastidic SufS-like proteins, Cp NifS2 and Cp NifS3. In order to test whether OLD5 can interact with a Cys desulfurase, a yeast two-hybrid assay was performed. As a positive control, we tested the interaction between Cp NifS and SufE. The results of our screen are summarized in Table 1. On selection medium lacking either His or adenine (Ade), we found that the OLD5 protein interacts with Cp NifS and Cp NifS3. These interactions were found for the mutated old5 protein only on His− selection medium but not on the more stringent Ade− plates. Interestingly, an interaction with AO was also detected, suggesting that OLD5 works as part of an enzyme complex.
Table 1.
Results of Yeast-Two-Hybrid Analysis
| Interaction | 3-Aminotriazole | Ade− |
|---|---|---|
| OLD5–OLD5 | − | − |
| OLD5–Cp NIFS | ++ | ++ |
| OLD5–Cp NIFS2 | − | − |
| OLD5–Cp NIFS3 | ++ | ++ |
| OLD5–AO | ++ | ++ |
| old5–old5 | − | − |
| old5–Cp NIFS | ++ | − |
| old5–Cp NIFS2 | − | − |
| old5–Cp NIFS3 | ++ | − |
| old5–AO | ++ | − |
| SufE–Cp NIFS | ++ | ++ |
| Empty–empty | − | − |
Growth selection on SD medium lacking either His or Ade of yeast strains that were cotransformed with prey and bait vectors that contain the cDNA sequences encoding the different proteins.
Taken together, these results suggest that OLD5 is part of a Cys desulfurase complex, and this was recently confirmed by Murthy et al. (2007).
The old5 Mutation Reduces Cys Desulfurase Activity
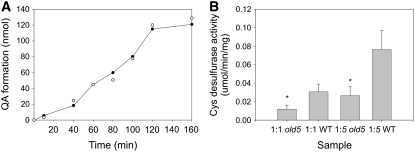
Since the homozygous T-DNA knockout line is embryo-lethal and the old5 allele we isolated is recessive and not lethal, we expected that the old5 mutation must either cause a reduction or a change of enzyme function. In order to assess this, we first performed complementation of an E. coli mutant deficient in QS (ΔNadA) (Murthy et al., 2007). Plasmids containing the full-length open reading frame for wild-type and mutant QS protein lacking the chloroplast localization signal were used (Murthy et al., 2007). Expression of either the wild-type or mutant protein restored growth of the deletion strain on minimal medium lacking nicotinic acid. Subsequently, we wanted to understand if the old5 mutation affects enzyme activity. The QS domain is dependent on a Fe-S cluster, which is provided through its SufE/YgdK domain through stimulation of a Cys desulfurase (Loiseau et al., 2005; Murthy et al., 2007). Previously, the activity of both domains of OLD5 was determined under anaerobic conditions (Murthy et al., 2007). Since the QS domain is not affected by the old5 mutation, we expected that the activity should be similar to that of the wild type. The ability of anaerobically isolated wild-type and old5 protein to catalyze the formation of quinolinic acid was assayed using an established protocol (Murthy et al., 2007). For this purpose, the substrate iminoaspartate, which is a required but highly unstable substrate of QS, was generated from l-Asp by AO. The formation of quinolinate by the mutated old5 protein is shown in Figure 4A and overlaps with that of the wild-type protein. Thus, the enzyme activity of the QS domain is not affected by the mutation. The function of the second domain of the OLD5 protein is to stimulate the Cys desulfurase activity of Cp NifS (Murthy et al., 2007). Testing this activity revealed that the old5 mutation results in an almost threefold lower stimulation than that observed in the wild type (Figure 4B). Taken together, the decreased activity of the SufE domain probably decreases overall activity of the protein, since continuous repairing/reconstituting of the Fe-S cluster is required for the maintenance of QS activity in planta (Murthy et al., 2007).
Figure 4.
OLD5 Enzyme Activity Assay.
(A) Formation of quinolinate by wild-type (closed circles) and mutant (open circles) protein. Experiments were repeated three times and showed comparable results.
(B) Rate of Cys desulfurase activity as induced by either wild type or mutant (old5) QS. The activity was tested at two molar ratios (1:1 and 1:5). Data are means ± se of three experiments. Asterisks indicate statistical significance versus the wild-type control in each case (* P < 0.05, Student's t test).
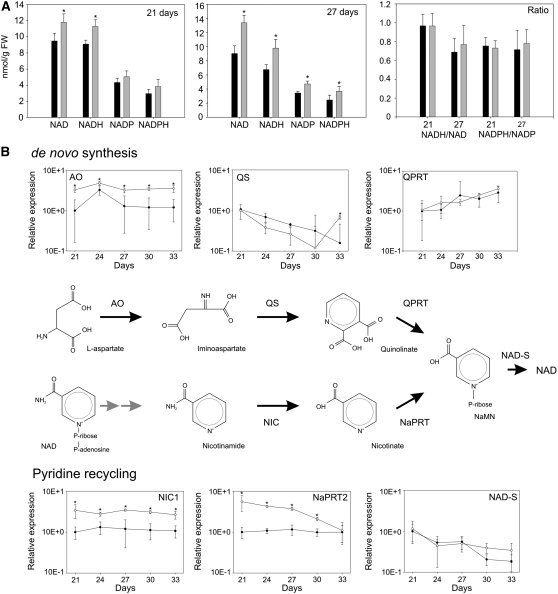
The old5 Mutation Results in Increased Steady State Pyridine Nucleotide Levels
The mutation in the SufE/Ygdk domain might also decrease enzyme activity in planta. Given that a reduced QS activity may be anticipated to result in lower NAD steady state levels, we determined the pyridine nucleotide content in the first leaf pair of 21- and 27-d-old plants. At the 21-d time point, the old5 mutant leaves show no phenotypic difference in comparison with the wild type (Figure 1A), but at 27 d, some changes in gene expression between the wild type and the mutant are detectable (Figure 2). Measurement of pyridine nucleotide content revealed, indeed, that the mutant had significantly increased NAD, NADH, NADP, and NADPH levels after 27 d (Figure 5A). The values we report for NAD, NADP, and NADPH in the wild type are similar to those of previous studies (Chai et al., 2005; Wang and Pichersky, 2007). Interestingly, the NAD/NADH and NADP/NADPH ratios in the mutant do not differ from the wild-type value, suggesting a similar redox balance for the pyridine nucleotides. Complementation of the mutant with the wild-type gene restores the pyridine nucleotide levels (see Supplemental Figure 1 online), demonstrating that the observed changes are a direct result of the old5 mutation.
Figure 5.
Pyridine Nucleotide Biosynthesis Is Increased during Leaf Development and Ageing of old5.
(A) Pyridine nucleotide content in old5 and the wild type at days 21 and 27 (old5, gray bars; wild type, black bars). The values shown are means of six repeats ± sd (indicated by error bars). Asterisks indicate statistical significance versus the wild-type control (* P < 0.05, Student's t test). The ratio of pyridine nucleotides is not affected by the old5 mutation. FW, fresh weight.
(B) Schematic representation of NAD biosynthetic pathways. Transcripts of AO, nicotinamidase (NIC1), and NaPRT2 accumulate in old5 mutants (old5, open circles; wild type, closed circles). The values shown are means of three repeats ± sd (indicated by error bars). Asterisks indicate statistical significance versus the wild-type control (* P < 0.05, Student's t test).
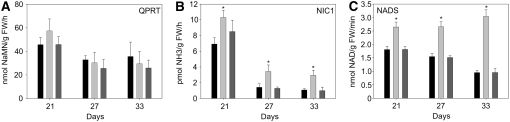
The maintenance of the NAD pool depends on the balance between de novo synthesis, active pyridine salvage, and degradation (Wang and Pichersky, 2007). The de novo synthesis and the pyridine salvage pathway result in the production of nicotinate mononucleotide, which by two enzymatic steps is converted to NAD (Figure 5B). The observed increase in NAD levels is surprising, since the in vitro Cys desulfurase activity is reduced. To understand the regulation of NAD biosynthesis in the mutant, we used expression profiling and enzyme activity measurements (see Methods). First, we determined the expression of genes encoding key enzymes in the pathway of de novo synthesis (Figure 5B). The first step in this pathway is the conversion of l-Asp into iminoaspartate mediated by AO. The transcript level of AO was threefold higher in the old5 mutant than in the wild type at all time points measured. The next step of de novo biosynthesis, the synthesis of the pyridine ring, is controlled by OLD5/QS. The expression of QS decreases with age in both the wild type and the mutant. However, during senescence, the expression of QS is upregulated in the mutant, reflecting a possible increased demand for NAD during the process. The final step, synthesis of nicotinate mononucleotide, requires the quinolinate phosphoribosyltransferase (QPRT), which for both the wild type and the mutant gradually increase in expression with age but do not differ from one another (Figure 5B). In addition, in vitro QPRT activity was tested on whole leaf extracts and no difference was found (Figure 6A). Given that the activity of this enzyme is similar for the wild type and the mutant, it is highly unlikely that the observed increase in pyridine nucleotides can be explained by an increase in de novo synthesis. Taken together, the results show that AO is upregulated at the expression level, while QS likely has a lower activity in vitro and QPRT activity is unchanged in leaf extracts.
Figure 6.
NAD Biosynthesis Enzyme Activities in old5 Mutant, Wild-Type, and Complemented Line Plants Determined in Leaves of 21- to 33-d-Old Plants.
QPRT (A), nicotinamidase (NIC1) (B), and NADS (C). Black bars indicate wild type, light gray bars indicate old5, and dark gray bars indicate the complemented line. Data presented are means ± se (indicated by error bars) of measurements from four independent plants per genotype. Asterisks indicate statistical significance versus the wild-type control in each case (* P < 0.05, Student's t test). FW, fresh weight; NaMN, nicotinate mononucleotide.
Thus, the increased NAD pool suggests that enzymes in the salvage pathway might compensate for the mutation in the de novo pathway. Such a mechanism was previously observed for the enzymes of the salvage pathway of pyrimidine nucleotides following the antisense inhibition of an enzyme of the de novo pathway of pyrimidine biosynthesis (Geigenberger et al., 2005). The salvage of nicotinamide, released from NAD, starts with deamidation by nicotinamidases to nicotinate (Wang and Pichersky, 2007) and subsequent conversion by nicotinate phosphoribosyltransferase (NaPRT). In Arabidopsis, NIC1 is the best characterized nicotinamidase, which, when knocked out, almost completely abolishes the salvage of nicotinamide (Wang and Pichersky, 2007). Overexpression of NIC1 results in an increase in NAD levels; thus, the expression level of NIC1 correlates with NAD content (Wang and Pichersky, 2007). The expression of NIC1 is constitutively higher in the old5 mutant (Figure 5B). The product of NIC1, nicotinate, is converted by NaPRTases, and the Arabidopsis genome encodes two homologs. In old5, At2g23420 (encoding NaPRT2) is upregulated when compared with the wild type, while At4g36940, which encodes the other putative NaPRTase, is expressed at similar levels as the wild type (data not shown). The increase in expression of both NIC1 and At2g23420 suggests an increased activity of the salvage pathway in the old5 mutant. To get a direct measurement of NIC1 activity, we used leaf extracts of 21-, 27-, and 33-d-old plants (Figure 6B). The assay shows constitutively higher activities of NIC in old5, suggesting that the salvage pathway is more active. The final step of NAD biosynthesis occurs through the action of NAD synthetase (NADS), which is similarly expressed in the mutant and the wild type. However, using leaf extracts to determine the activity, we show that NADS activity is 1.5- to 2-fold higher in the old5 mutant (Figure 6C). Thus, the increase in NAD levels is due to a more active salvage pathway in old5, which is supported by an increased activity in NADS.
Taken together, these results demonstrate an increased activity of the salvage pathway in the old5 mutant; this activity causes an increase in the steady state levels of NAD. We show that the alternate biosynthetic routes for NAD production influence each other, either directly or indirectly, in order to maintain sufficient levels of NAD.
old5 Is Characterized by Increased Tricarboxylic Acid Cycle Intermediates
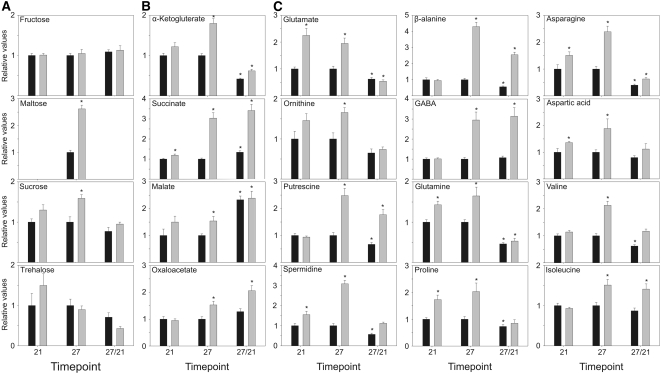
Since previous studies have shown that alterations in NAD/NADH levels can have dramatic consequences on cellular metabolism (Dutilleul et al., 2005; Shen et al., 2006), we next performed a comprehensive metabolite profiling using an established gas chromatography–mass spectrometry protocol capable of quantifying the relative levels of >80 metabolites (Lisec et al., 2006). For this purpose, leaves were harvested, in the middle of the light period, from both the old5 mutant and the wild type at days 21 and 27 and metabolites were measured (see Supplemental Table 1 online). Interestingly, although sugar levels in the old5 mutant were similar to wild-type levels at 21 d, maltose and sucrose levels were significantly higher at 27 d (Figure 7A). Increased sugar levels have been observed previously before the onset of senescence (Diaz et al., 2005; Wingler et al., 2006), suggesting that the old5 lesion invokes an early change in metabolism reminiscent of senescence. Sugars are subsequently processed through glycolysis to form the pyruvate needed to support mitochondrial respiration through the tricarboxylic acid (TCA) cycle (Fernie et al., 2004). After 21 d, we found an increase in the TCA cycle intermediate succinate in the mutant in comparison with the wild type, whereas other intermediates of this cycle were invariant (Figure 7B). However, at this time point, amino acids derived from the TCA cycle intermediate α-ketoglutarate accumulate in old5 to higher levels than in the wild type. These amino acids include Glu, Gln, Pro, and the polyamine spermidine (Figure 7C). Importantly, Asp, which is the precursor for the de novo NAD synthesis, was also significantly upregulated in old5 after 21 d. From Asp, several other compounds are synthesized, of which Asn was observed to accumulate after 21 d in old5. Complementation of the mutant with the wild-type OLD5 gene restored the metabolites to wild-type levels (see Supplemental Table 2 online), demonstrating that the observed metabolite changes are a direct result of the old5 mutation.
Figure 7.
Metabolite Profiling of old5 Leaves.
Relative metabolite contents of leaves at days 21 and 27 are given together with change over time as indicated by the ratio 27:21 (old5, gray bars; wild type, black bars). Metabolite contents were identified and quantified by gas chromatography–mass spectrometry, and their relative amounts were calculated relative to the wild-type control, as described by Roessner-Tunali et al. (2003). Histograms show the relative amounts of soluble sugars (A), TCA cycle intermediates (B), and amino acids (C). Data are means ± se of six measurements. Asterisks indicate statistical significance versus the wild-type control in each case (* P < 0.05, Student's t test).
At 27 d, all of the measured α-ketoglutarate–derived compounds, including 4-aminobutyric acid (GABA), Orn, putrescine, and β-Ala, were more abundant in old5. Furthermore, the branched chain amino acids Ile and Val, which derive from pyruvate (Binder et al., 2007), were significantly higher after 27 d of growth in the old5 mutant. Ile and Val have previously been documented to accumulate in mutants of the electron-transfer flavoprotein complex, which display accelerated dark-induced senescence (Ishizaki et al., 2005, 2006). Moreover, the Asp precursor oxaloacetate, as well as succinate and malate, are more abundant at 27 d in the mutant. These changes might be indicative of an increased mitochondrial metabolic rate in old5 mutants (Noctor et al., 2007). Previously, it was shown that redox manipulation by DTT treatment of Arabidopsis leaves increases the level of succinate, malate, and oxaloacetate (Kolbe et al., 2006), suggesting that the changes in NAD/NADH levels may be the direct cause of the increase found in TCA cycle intermediates for the old5 mutant.
Comparison of differences in the metabolite levels observed between 21 and 27 d may reveal differential metabolic regulation in the mutant with respect to the wild type. Although the levels of the TCA cycle intermediates are higher in old5 than in the wild type at both time points, the pattern of change (i.e., decrease) between the time points is the same for the wild type and mutants (Figure 7B). However, that is not true for pyridine nucleotide levels, which in the wild type decrease with time, while in old5 they remain stable (Figure 5A).
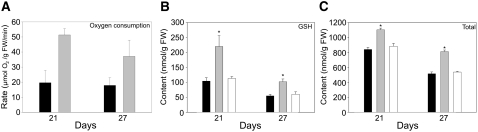
Increased Respiration Rate and Antioxidant Accumulation in old5
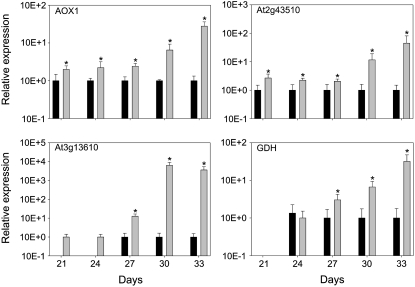
Both increased amounts of organic acids and higher levels of pyridine nucleotide can be attributed to an increased rate of respiration (Priault et al., 2007). The metabolite profile suggests that the changed pyridine nucleotide levels in old5 lead to an altered respiration rate. To analyze that possibility, we determined the oxygen consumption of whole leaves in the dark. Figure 8A shows oxygen consumption at 21 and 27 d. At both time points, the old5 mutant consumes approximately twofold more oxygen than the wild type, demonstrating an altered respiration rate. An overreduction of the mitochondrial electron transport chain gives rise to increased ROS production, which can be alleviated by activation of ALTERNATIVE OXIDASE1 (AOX1) (Noctor et al., 2007). The transcript abundance of AOX1 is also activated directly by TCA cycle intermediates (Vanlerberghe et al., 1997; Gray et al., 2004). Figure 9 shows that AOX1 is constitutively more highly expressed in old5, further supporting the notion that the mutant has a higher respiration rate than the wild type. Higher respiratory rates are generally coupled to an increased production of ROS by complexes I and III of the mitochondrial electron transport chain (Apel and Hirt, 2004; Noctor et al., 2007). The first indication that old5 suffers from ROS stress is SAG13 expression (Figure 2), which is associated with oxidative stress (Miller et al., 1999) and low antioxidant levels (Conklin and Barth, 2004) as well as with senescence. Moreover, the mutant showed a change in expression of two other well-established ROS marker genes: At2g43510 and At3g13610 (Figure 9). The defensin-like gene At2g43510, which has previously been shown to be ubiquitously induced by various ROS (Gadjev et al., 2006), was expressed at higher levels in old5 leaves at all time points and was strongly upregulated after 30 d (Figure 9). The second marker, 2-oxoglutarate–dependent dioxygenase (At3g13610), which is specifically expressed during hydrogen peroxide–related stress (Gadjev et al., 2006), showed a distinct expression after 24 d in old5 that increased rapidly with age. ROS are continuously generated as a by-product of aerobic metabolism and need to be rapidly detoxified by either enzymatic or nonenzymatic pathways, since they are highly toxic (Apel and Hirt, 2004). Protection against oxidative stress is partially provided for by low molecular weight antioxidants such as glutathione and ascorbate. Both the total thiol pool (GSH + GSSX + X) as well as the GSH are significantly increased in the mutant in comparison with the wild type (Figures 8B and 8C). An increase in the reduced pool of GSH provides an efficient protection against the toxic effects of hydrogen peroxide (May and Leaver, 1993).
Figure 8.
Increased Respiration of old5 Coincides with Increased Glutathione Levels.
(A) The oxygen consumption in the dark was measured and quantified with a Clark-type electrode (old5, gray bars; wild type, black bars).
(B) The pool of reduced glutathione shows an approximately twofold increase in the mutant (gray bars) in comparison with the wild type (black bars) and the complemented line (white bars).
(C) Elevated total glutathione pool (GSH + GSSX + X; X = any thiol) is observed in old5 compared with the wild type. Measurement of total glutathione is performed by reducing all disulfides with DTT from leaf tissues of 21- and 27-d-old plants, and the analysis was performed using reverse-phase HPLC.
Asterisks indicate statistical significance versus the wild-type control (* P < 0.05, Student's t test). FW, fresh weight.
Figure 9.
Relative Expression of Marker Genes Indicative of Oxidative Stress.
The relative expression of several genes was determined by real-time PCR and calculated relative to the wild type. The AOX is constitutively higher expressed (old5, gray bars; wild type, black bars). Two oxidative stress marker genes were tested and found to be upregulated in the mutant (At2g23420 and At3g13610). Next to that, GDH is more abundant than in the wild type. The values shown are means of three repeats ± sd (indicated by error bars). Experiments were repeated and showed similar results. Asterisks indicate statistical significance versus the wild-type control in each case (*P < 0.05, Student's t test).
Finally, ROS stress in Arabidopsis has previously been reported to affect nitrate assimilation catalyzed by glutamate dehydrogenase (GDH) (Skopelitis et al., 2006). The altered metabolite profile and increased expression of ROS marker genes coincide with an increased expression of GDH from day 27 in the old5 mutant (Figure 9). Taken together, these results show that the old5 mutation results in an increased respiration rate that is accompanied by an increase in oxidative stress, based on the expression of ROS-inducible genes and an increase in the pool of glutathione.
DISCUSSION
old5 Reveals a Role for NAD in Plant Ageing
Plants differ fundamentally from animals in that they do not have a rigid body plan and that organs can easily be added or removed, making them highly adaptive to the environment. As a result of developmental and adaptive strategies that resist, avoid, and anticipate ageing, it has been suggested that plants do not age at all in any sense recognizable in animals (Thomas, 2002). However, the lifespan of individual leaves clearly resembles animal ageing (Gan, 2003; Jing et al., 2003; Lim et al., 2003; Schippers et al., 2007). That is especially visible in Arabidopsis, in which the onset of leaf senescence occurs in an age-dependent way (Gan and Amasino, 1997; Quirino et al., 2000; Jing et al., 2002). Moreover, the recent identification of genes that affect the onset of leaf senescence (Woo et al., 2001; Jing et al., 2005, 2007) reveals common mechanisms present in the regulation of ageing between plants and animals.
In this study, we show that the old5 mutant displays early age-induced leaf senescence, as indicated by the expression of SAG12. The cloning of old5 identified a mutation in the gene encoding the QS. QS/OLD5 is an essential enzyme in the de novo synthesis of the pyridine nucleotide NAD (Katoh et al., 2006). Pyridine nucleotides have well-characterized roles as redox carriers in processes such as oxidative phosphorylation, the TCA cycle, and as electron acceptors in photosynthesis (Hunt et al., 2004). Next to their role in metabolism, they play a part during several stress conditions, including oxidative stress (Moller, 2001; Hayashi et al., 2005), wound response (Sinclair et al., 2000), and ABA signaling and salt stress (Shen et al., 2006; Wang and Pichersky, 2007). Recently, it was found that the NAD-dependent histone deacetylase SIR2 influences the lifespan for several model organisms, including yeast, Caenorhabditis elegans, mice, and human cells (Lin et al., 2000; Tissenbaum and Guarente, 2001; Lim et al., 2006). Remarkably, SIR2 is involved in both mitotic and postmitotic ageing, as demonstrated by the effect that this protein has on replicative lifespan in yeast and human cells. In Arabidopsis, histone deacetylation was shown to affect both gene regulation and plant development (Tian and Chen, 2001). The current view is that NAD levels serve as a metabolic sensor that is integrated into the developmental program by the effect of SIR2 on chromatin remodeling (Grewal and Moazed, 2003). Here, we show that a mutation that causes an alteration in the level of NAD influences the lifespan of Arabidopsis, suggesting a possible conserved role for NAD on development and ageing in plants.
The old5 Mutation Results in a Metabolite Profile Characteristic of Early Ageing
Redox reactions are the fundamental metabolic processes through which cells convert and distribute the energy that is necessary for growth and maintenance (Noctor, 2006). Changes in the availability of pyridine nucleotide levels have previously been shown to have a dramatic effect on the metabolite profiles of plants (Dutilleul et al., 2005; Shen et al., 2006). In addition, increased NADH levels affect other reactions that produce NADH and disturb the cellular redox balance, resulting in oxidative stress (Moller, 2001; Shen et al., 2006). The increased NAD levels in the old mutant coincide with increases in the level of TCA cycle intermediates and Asp, which is a precursor for NAD biosynthesis. Moreover, nitrogen metabolism is affected, as evidenced by the fact that amino acids deriving from ketoglutarate are more abundant. These data are consistent with those of a previous study on the tobacco (Nicotiana tabacum) nad7 mutant, which also displayed higher NAD/NADH levels concomitant with increased levels of nitrogen-containing amino acids (Dutilleul et al., 2005).
Given that the onset of leaf senescence in old5 is age-induced, it was anticipated that metabolic profiling of the mutant could identify metabolites that are characteristic biomarkers for an ARC (Diaz et al., 2005; Jing et al., 2005). Indeed, several biomarkers specific for the onset of leaf senescence were found in old5. Notably, the accumulation of the branched chain amino acids Val and Ile was previously shown to occur before the onset of senescence (Masclaux et al., 2000; Diaz et al., 2005; Ishizaki et al., 2005). One of the most intriguing metabolites that show an age-related accumulation in both the wild type and old5 is GABA. Studies on Osl2, a mitochondrial GABA transaminase in rice (Oryza sativa) (Ansari et al., 2005), suggested a switch in metabolism at the onset of senescence. Next to that, in plants as well as in animals, the GABA shunt is upregulated in times of high respiratory demand (Sweetlove et al., 2007; Fait et al., 2008). Furthermore, GABA induces senescence in sunflower (Helianthus annuus) by enhancing the synthesis of ethylene (Kathiresan et al., 1997). This is in line with a previous study in which we suggested that the ethylene-induced leaf senescence of old5 is dependent on ARCs (Jing et al., 2005). Thus, using a metabolic profiling approach, useful biomarkers that determine the age and viability of leaves were identified.
Interestingly, we also observed an accumulation of polyamines in the old5 mutant with age. Spermidine has the same precursor as aminocyclopropane, an important source of ethylene that can induce senescence prematurely (Imai et al., 2004). However, the accumulation of the polyamines might not be an ARC per se but may rather reflect a protection mechanism against ROS generation and programmed cell death (Papadakis and Roubelakis-Angelakis, 2005). Moreover, polyamines such as cytokinins are well known for their senescence-retarding effect.
Pyridine Biosynthesis, Crosstalk, and Stress Programming in old5 Mutant
The QS is dependent on a 4Fe-4S cluster for its activity (Loiseau et al., 2005). In E. coli, three Fe-S assembly systems exists, of which one appears to be more efficient in maintaining QS activity. The Arabidopsis OLD5 protein consists of a Fe-S assembly domain fused with the QS domain. This fusion is likely of physiological significance, since the OLD5 protein is highly oxygen-sensitive and localized to the oxygen-rich environment of the chloroplast (Murthy et al., 2007). Therefore, the continuous repairing of the Fe-S cluster in the QS domain by its SufE/Ygdk domain determines the activity of the protein. The analysis of the mutated old5 protein revealed that it weakly interacts with components of the Fe-S biogenesis complex, Cp NifS, and Cp NifS3 in a yeast two-hybrid experiment. Moreover, the activity measurements reveal that the stimulation of Cys desulfurase activity is reduced in the old5 protein. Although the QS activity of the old5 protein in oxygen-free experimental conditions is not altered, the decreased Cys desulfurase activity likely affects the long-term catalytic competence of the enzyme in vivo.
Despite the reduced activity of the mutated old5 protein, plants showed increased levels of NAD/NADH at 21 d. The maintenance of the NAD pool depends on both the de novo synthesis and pyridine salvage. Whereas de novo synthesis is absolutely required for plant survival (Katoh et al., 2006), the recycling pathway is mainly important during stress, when it recycles utilized NAD in an energy-efficient manner (Wang and Pichersky, 2007). In apparent response to the compromised QS, old5 plants exhibit increased expression of AO, the first enzyme in the de novo synthesis of NAD (Figure 5). Thus, these data suggest that the de novo synthesis is transcriptionally regulated at the first step, formation of iminoaspartate from Asp by AO. Iminoaspartate has to be generated in the presence of active QS, since it is a highly unstable intermediate (Murthy et al., 2007). To increase the rate of de novo synthesis, both an increase in substrate and active OLD5 protein are needed. Interestingly, the reduced overall activity of QS in the mutant might mimic increased oxygen stress (Gardner and Fridovich, 1991). Next to that, AO expression was recently found to be highly upregulated during application of oxidative stress (Gadjev et al., 2006), suggesting that AO and QS might play a role in the detection or signaling of ROS.
As a result of the mutation in QS, the salvage pathway is upregulated in old5 mutants. Both NIC1 and a putative NaPRTase show increased expression. Enhanced expression of the salvage pathway enzyme NIC1 has previously been shown to increase the NAD levels in Arabidopsis (Hunt et al., 2007; Wang and Pichersky, 2007). Increased NIC1 expression in the mutant correlates with the measured increased enzyme activity in leaf extracts, suggesting that its activity is regulated at the transcriptional level. By contrast, the increased NAD synthetase activity was associated with a reduced gene expression over time, suggesting that NAD synthetase activity is posttranscriptionally regulated. Despite the old5 mutation, pyridine nucleotide levels in the mutant were increased compared with the wild type. This might be due to the activation of a stress response program, since increased expression of the NAD salvage pathway is reminiscent of the need to replenish the NAD pool during stress conditions (Berglund et al., 1996; Hunt et al., 2007; Wang and Pichersky, 2007). One example of a NAD-consuming enzyme induced by oxidative stress is poly(ADP-ribose) polymerase (De Block et al., 2005), linking increased salvage metabolism to stress. Next to that, the salvage pathway was shown to be activated and necessary for response to osmotic stress in Arabidopsis (Wang and Pichersky, 2007). The compensation for reduced de novo synthesis of NAD by up-regulation of the salvage pathway of pyridine nucleotides is a mechanism that has also been observed for pyrimidine biosynthesis (Geigenberger et al., 2005).
Oxidative Stress and Early Ageing of old5
Longevity is clearly genetically controlled in many species, including yeast, C. elegans, mouse, and human (Jing et al., 2003). Almost 100 years ago it was postulated that there is an inverse relationship between metabolic rate and longevity (Rubner, 1908; Pearl, 1928). Today, it is clear that caloric restriction can greatly extend the maximum lifespan of many species (Sohal and Weindruch, 1996). The discovery of the first gene (PHA-4) absolutely required for lifespan extension by caloric restriction in C. elegans supports a role for ROS generated by mitochondrial oxidative energy metabolism in lifespan determination. This is especially evident by the fact that reduced PHA-4 expression does not suppress the long lifespan of animals with defective electron transport chains (Panowski et al., 2007). Moreover, generation of radical oxygen species by the mitochondria is dependent upon dietary intake (Lee et al., 2008). These results fit within the free radical theory of ageing, which proposes that ROS produced by respiration contribute to the ageing of all organisms (Harman, 1956).
In our study, we demonstrate that the mutant old5 has an increased respiration rate together with increased oxidative stress. The reducing equivalents that are generated from TCA cycle activity are used by the mitochondrial electron transport chain to power the synthesis of ATP (Fernie et al., 2004). The increased NADH levels in the mutant might directly influence the rate of mitochondrial electron transport (Hunt et al., 2004). This results in expression of the AOX, which bypasses phosphorylation and does not contribute to ATP production but alleviates overreduction of the enzymes in the transport chain (Moller, 2001). Transgenic tobacco cells that fail to induce AOX go rapidly into programmed cell death during oxidative stress (Vanlerberghe et al., 2002). It has to be noted that the induction of cell death in plants is highly dependent on genetic factors (Wagner et al., 2004) and may not merely be a direct consequence of ROS-induced damage. The increased glutathione levels and increased expression of hydrogen peroxide–induced stress markers in our mutant fit well with previous studies showing that during oxidative stress mitochondria produce increased amounts of hydrogen peroxide (Sweetlove et al., 2002; Tiwari et al., 2002; Gadjev et al., 2006). Developmental senescence of pea (Pisum sativum) leaves coincides with increases in the levels of superoxide and hydrogen peroxide (Pastori and del Rio, 1997). Furthermore, treatment of Arabidopsis leaves with the herbicide 3-aminotriazole inhibits catalase activity and causes hydrogen peroxide stress and increased expression of SAG genes (Navabpour et al., 2003). Next to that, the senescence-specific transcription factor WRKY53 is induced by hydrogen peroxide (Miao et al., 2004; Miao and Zentgraf, 2007). In general, it is believed that oxidative stress results in damage to the cell, which loses its viability, resulting in early ageing, as denoted by the free radical theory of ageing (Harman, 1956). However, it is more likely that altered ROS levels are implemented in the developmental program of the leaf, causing an early onset of senescence (Wagner et al., 2004; Queval et al., 2007).
By utilizing Arabidopsis leaves as a model system for plants, we have exploited the opportunity to characterize the mechanisms and genetics of ageing. Our study supports a role for ROS signaling in leaf development and ageing. Furthermore, we demonstrate that NAD might have a conserved role in ageing across kingdoms. Since ageing is manifested in many ways, cloning and physiological characterization of other old mutants will allow a fuller elucidation of the subtle differences between the various mechanisms of ageing and senescence.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Ler was used throughout this study. The old5 mutant was obtained from an ethyl methanesulfonate–mutagenized collection (Jing et al., 2005). The T-DNA insertion mutant QS-1 (SALK_079205) was obtained from the Nottingham Arabidopsis Stock Centre. The old5 mutant was complemented by transforming a 5.1-kb Ler genomic fragment containing the wild-type OLD5 gene to the mutant. The 5.1-kb DNA fragment was amplified by PCR using two oligonucleotides, 5′-GAATATCCCAATATAGTACACCACAATCAAATTAAATAC-3′ and 5′-GTTGTTACTAAGTTGCCAAACATTATATGTCTACTATTATG-3′, and cloned into the pGEM-T Easy vector (Promega). The insert was sequenced and transferred as a NotI–NotI fragment to the binary vector pGreen029 (Hellens et al., 2000). Plants were grown on either soil or half-strength Murashige and Skoog medium at 23°C and 65% RH with a daylength of 16 h. The light intensity was set at 120 μmol·m−2·s−1. An organic-rich γ-ray–radiated soil was used (Hortimea Groep).
Pigment Determination and Measurement of Photochemical Efficiency
For extraction of chlorophyll and carotenoids, samples were incubated overnight with N,N-dimethylformamide at 4°C in darkness. The chlorophyll content was quantified spectrophotometrically according to Wellburn (1994) at 647 and 664 nm. Chlorophyll fluorescence emission was measured from the upper surface of the first leaf at room temperature (23°C) with a pulse-amplitude modulation portable fluorometer (PAM-2000; H. Walz) according to Maxwell and Johnson (2000). Plants were dark-adapted for 1 to 2 h before measurements to ensure complete relaxation of the thylakoid pH gradient. An attached, fully expanded rosette leaf was placed in the leaf clip, allowing air to circulate freely on both sides of the leaf. At the start of each experiment, the leaf was exposed to 2 min of far-red illumination (2 to 4 μmol photons·m−2·s−1) for determination of Fo (minimum fluorescence in the dark-adapted state). Saturating pulses of white light (8000 μmol photons·m−2·s−1) were applied to determine Fm or Fm′ values. PSII efficiency was calculated as (Fm − Fo)/Fm.
RNA Isolation and Real-Time PCR
Total RNA was isolated using TRIZOL reagent (Sigma-Aldrich) according to the manufacturer's protocol. Five hundred nanograms of total RNA was used as template for first-strand cDNA synthesis using 200 units of RevertAid H-minus Moloney murine leukemia virus reverse transcriptase (Fermentas) and an oligo(dT21) primer. Quantitative RT-PCR was performed with the iCycler real-time PCR system (Bio-Rad). Primer pairs for real-time PCR were designed with the open-source PCR primer design program PerlPrimer version 1.1.10 (Marshall, 2004) around an intron to obtain a PCR product of 100 to 300 bp. The primer sequences are available in Supplemental Table 3 online. The presence of a single PCR product was verified by melt-curve analysis and gel electrophoresis. Relative quantification of gene expression was performed using the iCycler iQ software and the gene expression macro of Bio-Rad. All reactions were performed in quadruplicate on separate plates. Expression levels were normalized using Ct values obtained for the housekeeping gene ACT2 (At3g18780). Briefly, real-time PCR amplification was performed with 50 μL of reaction solution containing 2 μL of 10-fold diluted cDNA, 0.5 μL of a 10 mM stock of each primer, 1 μL of 25 mM stock MgCl2 (Fermentas), 5 μL of PCR buffer + Mg (Roche), 1 μL of a 1000× diluted SYBR Green stock (Sigma-Aldrich), 0.5 μL of 100× BSA (New England Biolabs), and 1 unit of Roche Taq polymerase. The amplification program was 94°C for 2 min, 40 cycles at 94°C for 10 s, followed by 60°C for 10 s, and ending with 72°C for 25 s, and amplification was followed by a melt curve analysis.
Bacterial Complementation, Enzyme Purification, and Activity Measurements
Bacterial strain Escherichia coli ΔNadA deletion mutant and plasmid pBAD/Myc-HisB containing the mature coding sequence of wild-type OLD5 were obtained from Murthy et al. (2007). The SufE/Ygdk domain coding sequence of the wild type was replaced with that of the old5 mutant to obtain a pBAD vector that expresses the mutated protein. Cys desulfurase activity was measured as described by Ye et al. (2006). QS enzymatic activity was assayed under anaerobic conditions at 27°C as described by Murthy et al. (2007). The AO protein was kindly provided by Sandrine-Ollagnier.
Determination of Metabolite Levels
Leaf samples were taken at the time points indicated, immediately frozen in liquid nitrogen, and stored at –80°C until further analysis. Extraction was performed by rapid grinding of tissue in liquid nitrogen and immediate addition of the appropriate extraction buffer. The relative levels of metabolites were determined using an established gas chromatography coupled to a time-of-flight mass analyzer protocol as described by Lisec et al. (2006). Data are presented normalized as detailed by Roessner et al. (2001). In short, peaks were assigned and quantified, and all data were normalized to the mean response calculated for the wild-type control of each measured batch; to allow comparison between the samples, individual wild-type values were normalized in the same way. The procedure of extraction and assay of NADs was performed according to the method described by Gibon and Larher (1997). The determination of glutathione contents was performed as described by Kreft et al. (2003).
NAD Biosynthesis Enzyme Activity Assay
Enzyme extracts were prepared from frozen ground leaf tissue. All extraction procedures were performed at 4°C. Twenty-five milligrams of tissue was homogenized in 0.25 mL of an appropriate extraction buffer. The homogenate was centrifuged for 5 min at 2700g, and the supernatant was used in the assays. The extraction buffer for QPRT and NAD synthase consisted of 100 mM MOPS-NaOH, pH 7.4, 5 mM MgCl2, and 0.1 mM EDTA. For nicotinamidase, the buffer was modified as follows: 200 mM MOPS-KOH, pH 7.5, 5 mM MgCl2, 10 mM DTT, and 0.1 mM EDTA. Enzyme assays were performed at 30°C. Nicotinamidase activity was determined as described by Wang and Pichersky (2007). QPRT was assayed as described by Wang et al. (2006). NAD synthase was assayed as described by Wagner and Wagner (1985), with the exception that the NAD produced was determined enzymatically as described by Gibon and Larher (1997).
Yeast Two-Hybrid Experiments
The coding sequences of the selected genes were amplified from cDNA with gene-specific primers (see Supplemental Table 3 online) and cloned in-frame with the GAL4 binding domain of the pGBKT7 vector and the GAL4 activation domain of the pGAD424 vector (Clontech) by restriction with SmaI and SalI. For protein–protein interaction screening, PJ69-4A (James et al., 1996) was transformed according to the Clontech yeast protocols handbook (PR13103) and selected on synthetic dropout (SD) medium lacking Leu and Trp for transformants. Subsequent colonies were dissolved in SD medium and spotted on selection medium for interaction by testing growth on SD medium lacking either Ade or His with the addition of 2 mM 3-amino-1,2,4-triazole. Plates were incubated at 28°C for 2 to 5 nights.
Respiratory Measurements
Assays of oxygen consumption by whole leaves were performed using a Clark-type oxygen electrode. Leaves were placed on a filter containing CO2 buffer, and subsequently the measuring room was filled after calibration with a 2% oxygen-containing gas before measurement. Measurements were performed in six independent samples, and fresh weight and chlorophyll content of the leaves were determined.
Accession Numbers
The Arabidopsis Genome Initiative locus numbers for the major genes discussed in this article are At1g08490 for Cp NifS1, At1g18490 for Cp NifS2, At1g55090 for NADS, At2g01350 for QPRT, At2g22570 for At NIC1, At2g23420 for NaPRT2, At2g29350 for SAG13, At2g43510 for defensin-like, At3g13610 for oxidoreductase, At4g26500 for At SufE, At4g36940 for NaPRT1, At5g07440 for GDH, At5g14760 for AO, At5g26600 for Cp NifS3, At5g50210 for QS/OLD5, and At5g45890 for SAG12.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complementation of the old5 Mutation Reverses the Increased Pyridine Nucleotide Content.
Supplemental Table 1. Metabolite Levels in the First Leaf Pair of 21- and 27-d-Old Plants of old5 Relative to the Wild Type.
Supplemental Table 2. Metabolite Levels in the First Leaf Pair of 21-, 27-, and 33-d-Old Plants of Complemented old5 Relative to the Wild Type.
Supplemental Table 3. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Bert Venema, Margriet Ferwerda, and Anne de Jong for their excellent technical support. Furthermore, we thank Ying Miao and Marcel Proveniers for their support with the yeast two-hybrid experiments, Marinus Pilon and Sandrine Ollagnier-de Choudens for their help with and for providing the materials for the QS and SufE assays, and Joost van Dongen and Silke Karojet for their help with the glove box. We are also grateful to Mariusz Bromke for assistance with glutathione measurements and Alexandra Florian for help in the enzymatic assays.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Paul P. Dijkwel (p.dijkwel@massey.ac.nz).
Online version contains Web-only data.
References
- Ansari, M.I., Lee, R.-H., and Chen, S.-C.G. (2005). A novel senescence-associated gene encoding γ-aminobutyric acid (GABA):pyruvate transaminase is upregulated during rice leaf senescence. Physiol. Plant. 123 1–8. [Google Scholar]
- Apel, K., and Hirt, H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 373–399. [DOI] [PubMed] [Google Scholar]
- Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141 391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, C., Moeder, W., Klessig, D.F., and Conklin, P.L. (2004). The timing of senescence and response to pathogens is altered in the ascorbate-deficient Arabidopsis mutant vitamin c-1. Plant Physiol. 134 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, P.A., Kelley, L.A., MacCallum, R.M., and Sternberg, M.J.E. (2001). Enhancement of protein modelling by human intervention in applying the automatic programs 3D-JIGSAW and 3D-PSSM. Proteins 5 (suppl.): 39–46. [DOI] [PubMed] [Google Scholar]
- Berglund, T.L., Kalbin, G., Strid, A., Rydström, J., and Ohlsson, A.B. (1996). UV-B- and oxidative stress-induced increase in nicotinamide and trigonelline and inhibition of defensive metabolism induction by poly(ADP-ribose) polymerase inhibitor in plant tissue. FEBS Lett. 380 188–193. [DOI] [PubMed] [Google Scholar]
- Binder, S., Knill, T., and Schuster, J. (2007). Branched-chain amino acid metabolism in higher plants. Physiol. Plant. 129 68–78. [Google Scholar]
- Bleecker, A.B. (1998). The evolutionary basis of leaf senescence: Method to the madness? Curr. Opin. Plant Biol. 1 73–78. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Earl, S., Harrison, E., Mathas, E., Navabpour, S., Page, T., and Pink, D. (2003). The molecular analysis of leaf senescence: A genomics approach. Plant Biotechnol. J. 1 3–22. [DOI] [PubMed] [Google Scholar]
- Buchanan-Wollaston, V., Page, T., Harrison, E., Breeze, E., Lim, P.O., Nam, H.G., Lin, J.F., Wu, S.H., Swidzinski, J., Ishizaki, K., and Leaver, C.J. (2005). Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 42 567–585. [DOI] [PubMed] [Google Scholar]
- Chai, M.F., Chen, Q.J., An, R., Chen, Y.M., Chen, J., and Wang, X.C. (2005). NADK2, an Arabidopsis chloroplastic NAD kinase, plays a vital role in both chlorophyll synthesis and chloroplast protection. Plant Mol. Biol. 59 553–564. [DOI] [PubMed] [Google Scholar]
- Conklin, P.L., and Barth, C. (2004). Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ. 27 959–970. [Google Scholar]
- Davuluri, R.V., Sun, H., Palaniswamy, S.K., Matthews, N., Molina, C., Kurtz, M., and Grotewold, E. (2003). AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Block, M., Verduyn, C., De Brouwer, D., and Cornelissen, M. (2005). Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 41 95–106. [DOI] [PubMed] [Google Scholar]
- Diaz, C., Purdy, S., Christ, A., Morot-Gaudry, J.-F., Wingler, A., and Masclaux-Daubresse, C. (2005). Characterization of new markers to determine the extent and variability of leaf senescence in Arabidopsis thaliana: A metabolic profiling approach. Plant Physiol. 138 898–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul, C., Lelarge, C., Prioul, J.L., De Paepe, R., Foyer, C.H., and Noctor, G. (2005). Mitochondria-driven changes in leaf NAD status exert a crucial influence on the control of nitrate assimilation and the integration of carbon and nitrogen metabolism. Plant Physiol. 139 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait, A., Fromm, H., Walter, D., Galili, G., and Fernie, A.R. (2008). Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 13 14–19. [DOI] [PubMed] [Google Scholar]
- Fernie, A.R., Carrari, F., and Sweetlove, L.J. (2004). Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 7 254–261. [DOI] [PubMed] [Google Scholar]
- Finkel, T., and Holbrook, N.J. (2000). Oxidants, oxidative stress and the biology of ageing. Nature 408 239–247. [DOI] [PubMed] [Google Scholar]
- Gadjev, I., Vanderauwera, S., Gechev, T.S., Laloi, C., Minkov, I.N., Shulaev, V., Apel, K., Inzé, D., Mittler, R., and Van Breusegem, F. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, S. (2003). Mitotic and postmitotic senescence in plants. Sci. Aging Knowledge Environ. 28 Re7. [DOI] [PubMed] [Google Scholar]
- Gan, S., and Amasino, R.M. (1997). Making sense of senescence (molecular genetic regulation and manipulation of leaf senescence). Plant Physiol. 113 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, P.R., and Fridovich, I. (1991). Quinolinate synthetase: The oxygen-sensitive site of de novo NAD(P)+ biosynthesis. Arch. Biochem. Biophys. 284 106–111. [DOI] [PubMed] [Google Scholar]
- Geigenberger, P., Regierer, B., Nunes-Nesi, A., Leisse, A., Urbanczyk-Wochniak, E., Springer, F., van Dongen, J.T., Kossmann, J., and Fernie, A.R. (2005). Inhibition of de novo pyrimidine synthesis in growing potato tuber leads to a compensatory stimulation of the pyrimidine salvage pathway and a subsequent increase in biosynthetic performance. Plant Cell 17 2077–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon, Y., and Larher, F. (1997). Cycling assay for nicotinamide adenine dinucleotides: NaCl precipitation and ethanol solubilization of the reduced tetrazolium. Anal. Biochem. 251 153–157. [DOI] [PubMed] [Google Scholar]
- Gray, G.R., Maxwell, D.P., Villarimo, A.R., and McIntosh, L. (2004). Mitochondria/nuclear signaling of alternative oxidase gene expression occurs through distinct pathways involving organic acids and reactive oxygen species. Plant Cell Rep. 23 497–503. [DOI] [PubMed] [Google Scholar]
- Grewal, S.I., and Moazed, D. (2003). Heterochromatin and epigenetic control of gene expression. Science 301 798–802. [DOI] [PubMed] [Google Scholar]
- Guo, Y., Cai, Z., and Gan, S. (2004). Transcriptome of Arabidopsis leaf senescence. Plant Cell Environ. 27 521–549. [Google Scholar]
- Harman, D. (1956). Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 2 298–300. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., Takahashi, H., Tamura, K., Huang, J., Yu, L.H., Kawai-Yamada, M., Tezuka, T., and Uchimiya, H. (2005). Enhanced dihydroflavonol-4-reductase activity and NAD homeostasis leading to cell death tolerance in transgenic rice. Proc. Natl. Acad. Sci. USA 102 7020–7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebeler, R., Oeljeklaus, S., Reidegeld, K.A., Eisenacher, M., Stephan, C., Sitek, B., Stühler, K., Meyer, H.E., Sturre, M.J., Dijkwel, P.P., and Warscheid, B. (2008). Study of early leaf senescence in Arabidopsis thaliana by quantitative proteomics using reciprocal 14N/15N labeling and difference gel electrophoresis. Mol. Cell. Proteomics 7 108–120. [DOI] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42 819–832. [DOI] [PubMed] [Google Scholar]
- Hunt, L., Holdworth, M.J., and Gray, J.E. (2007). Nicotinamidase activity is important for germination. Plant J. 51 341–351. [DOI] [PubMed] [Google Scholar]
- Hunt, L., Lerner, F., and Ziegler, M. (2004). NAD—New roles in signaling and gene regulation in plants. New Phytol. 163 31–44. [DOI] [PubMed] [Google Scholar]
- Imai, A., et al. (2004). Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol. 135 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki, K., Larson, T.R., Schauer, N., Fernie, A.R., Graham, I.A., and Leaver, C.J. (2005). The critical role of Arabidopsis electron-transfer flavoprotein:ubiquinone oxidoreductase during dark-induced starvation. Plant Cell 17 2587–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki, K., Schauer, N., Larson, T.R., Graham, I.A., Fernie, A.R., and Leaver, C.J. (2006). The mitochondrial electron transfer flavoprotein complex is essential for survival of Arabidopsis in extended darkness. Plant J. 47 751–760. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing, H.C., Anderson, L., Sturre, M.J.G., Hille, J., and Dijkwel, P.P. (2007). Arabidopsis CPR5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J. Exp. Bot. 58 3885–3894. [DOI] [PubMed] [Google Scholar]
- Jing, H.C., Hille, J., and Dijkwel, P.P. (2003). Ageing in plants: Conserved strategies and novel pathways. Plant Biol. 5 455–464. [Google Scholar]
- Jing, H.C., Schippers, J.H.M., Hille, J., and Dijkwel, P.P. (2005). Ethylene-induced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J. Exp. Bot. 546 2915–2923. [DOI] [PubMed] [Google Scholar]
- Jing, H.C., Sturre, M.J., Hille, J., and Dijkwel, P.P. (2002). Arabidopsis onset of leaf death mutants identify a regulatory pathway controlling leaf senescence. Plant J. 32 51–63. [DOI] [PubMed] [Google Scholar]
- John, C.F., Morris, K., Jordan, B.R., Thomas, B., and A-H-Mackerness, S. (2001). Ultraviolet-B exposure leads to upregulation of senescence-associated genes in Arabidopsis thaliana. J. Exp. Bot. 52 1367–1373. [PubMed] [Google Scholar]
- Kathiresan, A., Tung, P., Chinnappa, C.C., and Reid, D.M. (1997). γ-Aminobutyric acid stimulates ethylene biosynthesis in sunflower. Plant Physiol. 115 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, A., Uenohara, K., Akita, M., and Hashimoto, T. (2006). Early steps in the biosynthesis of NAD in Arabidopsis start with aspartate and occur in the plastid. Plant Physiol. 141 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood, T.B.L. (2002). Evolution of ageing. Mech. Ageing Dev. 123 737–745. [DOI] [PubMed] [Google Scholar]
- Kirkwood, T.B.L. (2005). Understanding the odd science of aging. Cell 120 437–447. [DOI] [PubMed] [Google Scholar]
- Kolbe, A., Oliver, S.N., Fernie, A.R., Stitt, M., van Dongen, J.T., and Geigenberger, P. (2006). Combined transcript and metabolite profiling of Arabidopsis leaves reveals fundamental effects of the thiol-disulfide status on plant metabolism. Plant Physiol. 141 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft, O., Hoefgen, R., and Hesse, H. (2003). Functional analysis of cystathionine γ-synthase in genetically engineered potato plants. Plant Physiol. 131 1843–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa, J., Smalle, J., Van Montagu, M., and Inze, D. (1998). Oxidative stress tolerance and longevity in Arabidopsis late-flowering mutant gigantea is tolerant to paraquat. Plant J. 14 759–764. [DOI] [PubMed] [Google Scholar]
- Lee, K.P., Simpson, S.J., Clissold, F.J., Brooks, R., Ballard, J.W., Taylor, P.W., Soran, N., and Raubenheimer, D. (2008). Lifespan and reproduction in Drosophila: New insights from nutritional geometry. Proc. Natl. Acad. Sci. USA 105 2498–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, C.-S., Potts, M., and Helm, R.F. (2006). Nicotinamide extends the replicative life span of primary human cells. Mech. Ageing Dev. 127 511–514. [DOI] [PubMed] [Google Scholar]
- Lim, P.O., Woo, H.R., and Nam, H.G. (2003). Molecular genetics of leaf senescence in Arabidopsis. Trends Plant Sci. 8 272–278. [DOI] [PubMed] [Google Scholar]
- Lin, S.J., Defossez, P.A., and Guarente, L. (2000). Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289 2062–2063. [DOI] [PubMed] [Google Scholar]
- Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., and Fernie, A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protocols 1 387–396. [DOI] [PubMed] [Google Scholar]
- Liu, G., Li, Z., Chiang, Y., Acton, T., Montelione, G.T., Murray, D., and Szyperski, T. (2005). High-quality homology models derived from NMR and x-ray structures of E. coli proteins YgdK and SufE suggest that all members of the YgdK/SufE protein family are enhancers of cysteine desulfurases. Protein Sci. 14 1597–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau, L., Ollagnier-de Choudens, S., Lascoux, D., Forest, E., Fontecave, M., and Barras, F. (2005). Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli. J. Biol. Chem. 280 26760–26769. [DOI] [PubMed] [Google Scholar]
- Loiseau, L., Ollagnier-de-Choudens, S., Nachin, L., Fontecave, M., and Barras, F. (2003). Biogenesis of Fe-S cluster by the bacterial suf system: SufS and SufE form a new type of cysteine desulfurase. J. Biol. Chem. 278 38352–38359. [DOI] [PubMed] [Google Scholar]
- Marshall, O.J. (2004). PerlPrimer: Cross-platform, graphical primer design for standard, bisulphite and real-time PCR. Bioinformatics 20 2471–2472. [DOI] [PubMed] [Google Scholar]
- Masclaux, C., Valadier, M.H., Brugière, N., Morot-Gaudry, J.F., and Hirel, B. (2000). Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211 510–518. [DOI] [PubMed] [Google Scholar]
- Maxwell, K., and Johnson, G.N. (2000). Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 51 659–668. [DOI] [PubMed] [Google Scholar]
- May, M.J., and Leaver, C.J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103 621–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55 853–867. [DOI] [PubMed] [Google Scholar]
- Miao, Y., and Zentgraf, U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19 819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.D., Arteca, R.N., and Pell, E.J. (1999). Senescence-associated gene expression during ozone-induced leaf senescence in Arabidopsis. Plant Physiol. 120 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7 405–410. [DOI] [PubMed] [Google Scholar]
- Moller, I.M. (2001). Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 561–591. [DOI] [PubMed] [Google Scholar]
- Murthy, N.M., Ollagnier-de-Choudens, S., Sanakis, Y., Abdel-Ghany, S.E., Rousset, C., Ye, H., Fontecave, M., Pilon-Smits, E.A., and Pilon, M. (2007). Characterization of Arabidopsis thaliana SufE2 and SufE3: Functions in chloroplast iron-sulfur cluster assembly and NAD synthesis. J. Biol. Chem. 282 18254–18264. [DOI] [PubMed] [Google Scholar]
- Navabpour, S., Morris, K., Allen, R., Harrison, E., A-H-Mackerness, S., and Buchanan-Wollaston, V. (2003). Expression of senescence-enhanced genes in response to oxidative stress. J. Exp. Bot. 54 2285–2292. [DOI] [PubMed] [Google Scholar]
- Noctor, G. (2006). Metabolic signaling in defense and stress: The central roles of soluble redox couples. Plant Cell Environ. 29 409–425. [DOI] [PubMed] [Google Scholar]
- Noctor, G., De Paepe, R., and Foyer, C.H. (2007). Mitochondrial redox biology and homeostasis in plants. Trends Plant Sci. 12 125–134. [DOI] [PubMed] [Google Scholar]
- Noctor, G., Queval, G., and Gakière, B. (2006). NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 57 1603–1620. [DOI] [PubMed] [Google Scholar]
- Nooden, L.D., and Penney, J.P. (2001). Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae). J. Exp. Bot. 52 2151–2159. [DOI] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens, S., Loiseau, L., Sanakis, Y., Barras, F., and Fontecave, M. (2005). Quinolinate synthetase, an iron-sulfur enzyme in NAD biosynthesis. FEBS Lett. 579 3737–3743. [DOI] [PubMed] [Google Scholar]
- Panowski, S.H., Wolff, S., Aguilaniu, H., Durieux, J., and Dillin, A. (2007). PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447 550–555. [DOI] [PubMed] [Google Scholar]
- Papadakis, A.K., and Roubelakis-Angelakis, K.A. (2005). Polyamines inhibit NADPH oxidase-mediated superoxide generation and putrescine prevents programmed cell death syndrome induced by the polyamine oxidase generated hydrogen peroxide. Planta 220 826–837. [DOI] [PubMed] [Google Scholar]
- Pastori, G.M., and del Rio, L.A. (1997). Natural senescence of pea leaves (an activated oxygen-mediated function for peroxisomes). Plant Physiol. 113 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl, R. (1928). The Rate of Living. (London: University of London Press).
- Priault, P., Vidal, G., De Paepe, R., and Ribas-Carbo, M. (2007). Leaf age-related changes in respiratory pathways are dependent on complex I activity in Nicotiana sylvestris. Physiol. Plant. 129 152–162. [Google Scholar]
- Queval, G., Issakidis-Bourguet, E., Hoeberichts, F.A., Vandorpe, M., Gakière, B., Vanacker, H., Miginiac-Maslow, M., Van Breusegem, F., and Noctor, G. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52 640–657. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F., Noh, Y.S., Himelblau, E., and Amasino, R.M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5 278–282. [DOI] [PubMed] [Google Scholar]
- Roessner, U., Willmitzer, L., and Fernie, A.R. (2001). High-resolution metabolic profiling of genetically and environmentally diverse potato tuber systems: Identification of phenocopies. Plant Physiol. 127 749–764. [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali, U., Hegemann, B., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., and Fernie, A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.R. (1991). Evolutionary Biology of Aging. (New York: Oxford University Press)
- Rubner, M. (1908). Das Problem der Lebensdauer und Seine Beziehungen Zu Wachstum und Ernarhrung. (Muenchen, Germany: R. Oldenburg).
- Sakuraba, H., Tsuge, H., Yoneda, K., Katunuma, N., and Ohshima, T. (2005). Crystal structure of the NAD biosynthetic enzyme quinolinate synthase. Biol. Chem. 280 26645–26648. [DOI] [PubMed] [Google Scholar]
- Schippers, J.H.M., Jing, H.C., Hille, J., and Dijkwel, P.P. (2007). Developmental and hormonal control of leaf senescence. In Senescence Processes in Plants, Vol. 26, S. Gan, ed (Oxford, UK: Blackwell Publishing), pp. 145–170.
- Shen, W., Wei, Y., Dauk, M., Tan, Y., Taylor, D.C., Selvaraj, G., and Zou, J. (2006). Involvement of a glycerol-3-phosphate dehydrogenase in modulating the NADH/NAD+ ratio provides evidence of a mitochondrial glycerol-3-phosphate shuttle in Arabidopsis. Plant Cell 18 422–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair, S.J., Murphy, K.J., Birch, C.D., and Hamill, J.D. (2000). Molecular characterization of quinolinate phosphoribosyltransferase (QPRtase) in Nicotiana. Plant Mol. Biol. 44 603–617. [DOI] [PubMed] [Google Scholar]
- Skopelitis, D.S., Paranychianakism, N.V., Paschalidis, K.A., Pliakonis, E.D., Delis, I.D., Yakoumakis, D.I., Kouvarakis, A., Papadakis, A.K., Stephanou, E.G., and Roubelakis-Angelakis, K.A. (2006). Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18 2767–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal, R.S., and Weindruch, R. (1996). Oxidative stress, caloric restriction, and aging. Science 273 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove, L.J., Fait, A., Nunes-Nesi, A., Williams, T., and Fernie, A.R. (2007). The mitochondrion: An integration point of cellular metabolism and signalling. Crit. Rev. Plant Sci. 26 17–43. [Google Scholar]
- Sweetlove, L.J., Heazlewood, J.L., Herald, V., Holtzapffel, R., Day, D.A., Leaver, C.J., and Millar, A.H. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32 891–904. [DOI] [PubMed] [Google Scholar]
- Sweetlove, L.J., Lytovchenko, A., Morgan, M., Nunes-Nesi, A., Taylor, N.L., Baxter, C.J., Eickmeier, I., and Fernie, A.R. (2006). Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 103 19587–19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, H. (2002). Ageing in plants. Mech. Ageing Dev. 123 747–753. [DOI] [PubMed] [Google Scholar]
- Tian, L., and Chen, Z.J. (2001). Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc. Natl. Acad. Sci. USA 98 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum, H.A., and Guarente, L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410 227–230. [DOI] [PubMed] [Google Scholar]
- Tiwari, B.S., Belenghi, B., and Levine, A. (2002). Oxidative stress increased respiration and generation of reactive oxygen species, resulting in ATP depletion, opening of mitochondrial permeability transition, and programmed cell death. Plant Physiol. 128 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uauy, C., Distelfeld, A., Fahima, T., Blechl A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314 1298–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., Robson, C.A., and Yip, J.Y. (2002). Induction of mitochondrial alternative oxidase in response to a cell signal pathway down-regulating the cytochrome pathway prevents programmed cell death. Plant Physiol. 129 1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe, G.C., Vanlerberghe, A.E., and McIntosh, L. (1997). Molecular genetic evidence of the ability of alternative oxidase to support respiratory carbon metabolism. Plant Physiol. 113 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, D., Przybyla, D., Op den Camp, R., Kim, C., Landgraf, F., Lee, K.P., Würsch, M., Laloi, C., Nater, M., Hideg, E., and Apel, K. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306 1183–1185. [DOI] [PubMed] [Google Scholar]
- Wagner, R., and Wagner, K.G. (1985). The pyridine-nucleotide cycle in tobacco: Enzyme activities for the de novo synthesis of NAD. Planta 165 532–537. [DOI] [PubMed] [Google Scholar]
- Wang, G., and Pichersky, E. (2007). Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 49 1020–1029. [DOI] [PubMed] [Google Scholar]
- Wang, K., Conn, K., and Lazarovits, G. (2006). Involvement of quinolinate phosphoribosyl transferase in promotion of potato growth by a Burkholderia strain. Appl. Environ. Microbiol. 72 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, L.M., Gan, S., Quirino, B., and Amasino, R.M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37 455–469. [DOI] [PubMed] [Google Scholar]
- Wellburn, A.R. (1994). The spectral determination of chlorophyll-a and chlorophyll-b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144 307–313. [Google Scholar]
- Wingler, A., Purdy, S., MacLean, J.A., and Pourtau, N. (2006). The role of sugars in integrating environmental signals during the regulation of leaf senescence. J. Exp. Bot. 57 391–399. [DOI] [PubMed] [Google Scholar]
- Woo, H.R., Chung, K.M., Park, J.-H., Oh, S.A., Ahn, T., Hong, S.H., Jang, S.K., and Nam, H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, H.R., Kim, J.H., Nam, H.G., and Lim, P.O. (2004). The delayed leaf senescence mutants of Arabidopsis, ore1, ore3, and ore9 are tolerant to oxidative stress. Plant Cell Physiol. 45 923–932. [DOI] [PubMed] [Google Scholar]
- Xu, X.M., and Møller, S.G. (2006). AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 25 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Savchenko, A., Yakunin, A., Zhang, R., Edwards, A., Arrowsmith, C., and Tong, L. (2003). Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643. J. Biol. Chem. 278 8804–8808. [DOI] [PubMed] [Google Scholar]
- Ye, H., Abdel-Ghany, S.E., Anderson, T.D., Pilon-Smits, E.A.H., and Pilon, M. (2006). CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J. Biol. Chem. 281 8958–8969. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., and Zentgraf, U. (2005). The correlation between oxidative stress and leaf senescence during plant development. Cell. Mol. Biol. Lett. 10 515–534. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.