Abstract
The role of the unique plant calpain Defective Kernel 1 (DEK1) in development has remained unclear due to the severity of mutant phenotypes. Here, we used complementation studies of the embryo-lethal mutant to dissect DEK1 protein behavior and to show that DEK1 plays a key role in growth regulation in Arabidopsis thaliana. We show that although full-length DEK1 protein localizes to membranes, it undergoes intramolecular autolytic cleavage events that release the calpain domain into the cytoplasm. The active calpain domain alone is not only necessary for DEK1 function but is sufficient for full complementation of dek1 mutants. A novel set of phenotypes, including leaf ruffling, increased leaf thickness, and abnormalities of epidermal cell interdigitation, was caused by expression of the constitutively active calpain domain. This analysis of the novel phenotypes produced by DEK1 under- and overexpression, as well as DEK1 subcellular localization and protein processing, has revealed a fundamental role for DEK1-mediated signaling in growth regulation.
INTRODUCTION
A highly conserved, unique plant-specific calpain-like molecule (phytocalpain) has been shown to be essential for the correct development of both an embryonic epidermal cell layer and the specialized outer layer of the endosperm (the aleurone) during seed development in maize (Zea mays) and Arabidopsis thaliana (Lid et al., 2002, 2005; Ahn et al., 2004; Johnson et al., 2005). Decreasing expression levels of tobacco (Nicotiana tabacum) phytocalpain has also been shown to disrupt epidermal development during postgerminative growth (Ahn et al., 2004). Phytocalpain genes derive their names from the maize DEFECTIVE KERNEL1 (DEK1) gene, which was the first to be phenotypically characterized and cloned (Becraft and Asuncion-Crabb, 2000; Becraft et al., 2002; Lid et al., 2002). Highly conserved homologs of DEK1 have been described across the plant kingdom, including in more basal plants such as Physcomitrella. Interestingly, these genes occur as singletons in plant genomes, including that of the model plant Arabidopsis.
The epidermal layer is thought to be specified once, during early embryogenesis, in plants (Bruck and Walker, 1985). The subsequent developmental maintenance of the L1/epidermis as a monolayer of closely associated cells requires stringent, but poorly understood, growth control and implies constant signaling both within the epidermal layer and between epidermal and underlying cells. The potential role played by the epidermis in controlling organ growth has also long been a subject of debate. Recent studies suggest that the cells in the epidermis both promote and restrict organ growth by sending signals to underlying layers (most recently reviewed in Savaldi-Goldstein and Chory, 2008). Previous studies have indicated that Arabidopsis DEK1 could be implicated in epidermal signaling processes required for the maintenance of epidermal cell fate and the regulation of cell proliferation. However, how this enigmatic protein functions at the molecular level is still unclear.
Calpains, a class of intracellular Cys proteases, play crucial roles in basic physiological and pathological processes in animals. The best-studied calpain isoforms in humans, μ-calpain and m-calpain, are cytoplasmic and dependent on the presence of calcium for their catalytic activity. Despite extensive study, the sequential/structural determinants of calpain cleavage targets in animals are little understood, making substrate identification difficult (Tompa et al., 2004). In addition, mechanisms of calpain activation remain relatively poorly defined.
Typical mammalian μ-calpain and m-calpains consist of four domains (DI-DIV) and associate with a small calpain subunit (DV and DVI) (Croall and Ersfeld, 2007). Atypical members of the calpain family are often monomeric and lack the calmodulin-like EF-hand containing DIV. Despite this, a dependency on calcium for activity is usually retained, possibly due to calcium-dependent association of the activated protease with internal membranes mediated by DIII (Hood et al., 2004; Shao et al., 2006; Samanta et al., 2007). The calcium binding DIV is not conserved in phytocalpains, although some dependency on Ca2+ for in vitro activity has been reported (Wang et al., 2003). In addition, no homolog of the mammalian small calpain subunit is apparent in the plant genome.
Calpain activation in animal systems has been associated with an autocatalytic removal of the N-Terminal DI, which is thought to be important to significantly lower the Ca2+ requirement for catalytic activity (Cottin et al., 2001; Garcia Diaz et al., 2006). Whether this event is strictly necessary for activation of all calpains remains unclear (Farkas et al., 2004). Unlike mammalian calpains, which have a short N-terminal DI, the phytocalpains have an extended N-terminal region that is predicted to contain 21 transmembrane domains interrupted by an extracellular loop and an extended cytoplasmic juxtamembrane region showing little homology to other proteins (Lid et al., 2002). The C-terminal region is similar to calpain Cys proteinases and consists of the active DIIa, and DIIb, as well as a proposed regulatory domain DIII. The unusual structure of the plant DEK1 calpains raises the question of how they function and why loss of activity leads to such dramatic phenotypes.
Here, we investigated the importance of subcellular localization and Cys proteinase activity for Arabidopsis DEK1 function. Our results show that the activity of the DEK1 calpain domain is necessary for DEK1 function in planta. Like animal calpains, DEK1 undergoes intramolecular proteolysis events, which in the case of DEK1 uncouple the calpain domain from the membrane. We can show that the presence of the cytoplasmic calpain domain alone is sufficient for complementation of the Arabidopsis dek1 mutant phenotype, suggesting that proteolysis of DEK1 may be involved in its activation. Low expression of complementing transgenes revealed additional roles for DEK1 in maintenance of apical meristems and ovule development. In addition, a novel set of phenotypes are associated with overexpression of the calpain domain alone, which suggests that the role of DEK1 in epidermal maintenance may be linked to a fundamental role of DEK1-mediated signaling in growth regulation.
RESULTS
A Full-Length DEK1 cDNA Fused to Green Fluorescent Protein Complements the dek1 Mutant
To generate tools to study the function of DEK1 in more detail, we complemented the dek1 mutant phenotype with a full-length DEK1 cDNA fused at the C terminus to the green fluorescent protein (mGFP) open reading frame (Gifford et al., 2003). DEK1 is expressed strongly in early embryo development and at low levels throughout development; however, attempts to identify the native DEK1 promoter were not successful. We had previously shown that the 35S promoter is not expressed until the late heart stage, making it unsuitable for our complementation experiments (Johnson et al., 2005). The RIBOSOMAL PROTEIN5A (RPS5A) promoter has previously been shown to drive early embryo expression (Weijers et al., 2001). To investigate the expression pattern of pRPS5A and to determine if it was suitable for complementation studies, it was used to drive expression of a nuclear localized reporter (Arabidopsis thaliana meristem L1 [ATML1]-GFP gene fusion; Gifford et al., 2003). The fusion protein from this construct showed a very similar expression pattern to DEK1 in all tissues investigated (Johnson et al., 2005; see Supplemental Figure 1 online), indicating that the RPS5A promoter is suitable for use in complementation studies.
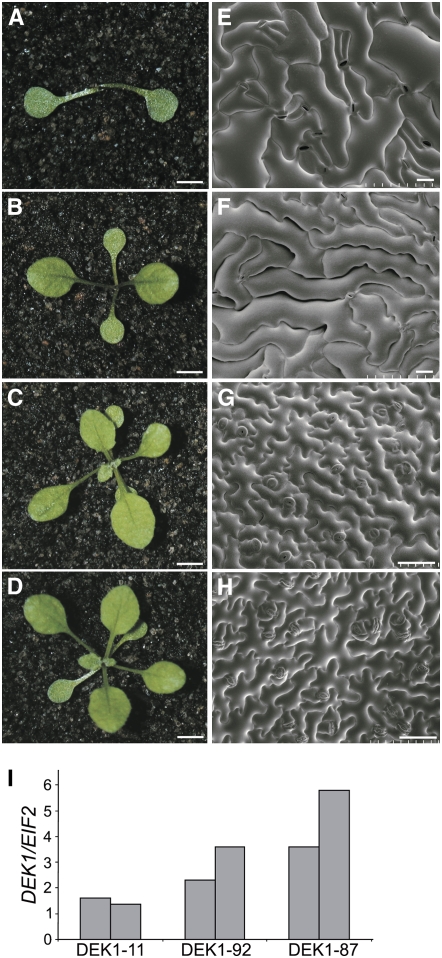
Plants heterozygous for a T-DNA insert in the 22nd intron of the DEK1 gene (dek1-3) were transformed with the pRPS5A:AtDEK1-GFP construct, as homozygous mutants arrest at an early stage of embryogenesis (Johnson et al., 2005). A number of homozygous dek1-3 lines complemented with pRPS5A:AtDEK1-GFP were identified (see Supplemental Figure 2 online) showing a range of phenotypes from similar to DEK1 RNA interference knockdown plants, to wild-type in appearance (Figures 1A to 1D; Johnson et al., 2005). Quantitative real-time PCR (Q-PCR) analysis of DEK1 expression in complementing lines showed that levels of DEK1-GFP transcript and fusion protein expression were variable and correlated with the phenotypes observed (Figure 1I; see Supplemental Figures 3A and 3B online). The dek1-3 plants expressing DEK1-GFP at low levels lacked an observable meristem at early seedling stage or were unable to maintain meristem activity later in development (Figures 1A, 1B, and 1D; see Supplemental Figures 3E to 3H online). In the cotyledons and leaves of these plants, the characteristic epidermal pavement cells of the wild type were replaced by an irregular epidermis of large cells that appear to have gaps between them (Figures 1E and 1F; see Supplemental Figures 3I and 3J online). In addition, epidermal cells in these lines did not interdigitate normally as in the wild type (Figure 1H), suggesting they do not sense and respond correctly to neighboring cells. Plants that recovered meristem activity had perturbed leaf development, with defects resembling loss of adaxial identity frequently noted and abnormal trichome development. These plants also develop callus-like cells on stems and produce sterile inflorescences (see Supplemental Figures 3A, 3C, and 3D online). Sterility was due to the formation of abnormal ovules in which integuments tended to grow out as finger-like projections rather than a continuous sheath surrounding the nucellus (Figure 2I). Several complemented lines did not show any epidermal defects, were fully fertile, and resembled wild-type plants at both seedling and adult stages (Figures 1C and 1G; see Supplemental Figure 3A online). These lines expressed DEK1-GFP at higher levels (Figure 1I; see Supplemental Figure 3B online). The defects seen in lines expressing at low levels are intriguing because they suggest signaling events that maintain epidermal integrity are very sensitive to the levels of DEK1 expression.
Figure 1.
Phenotypes of dek1-3 Plants Complemented with pRPS5A:DEK1-GFP.
(A) to (D) Representative 4-week-old dek1-3 seedlings complemented to varying degrees by pRPS5A:DEK1-GFP. Lines 11 (A), 92 (B), and 87 (C) and a wild-type seedling (D). Bars = 5 mm.
(E) to (H) The adaxial cotyledon epidermis of DEK1-GFP complemented lines 11 (E), 92 (F), 87 (G), and the wild type (H). Bars = 50 μm.
(I) Q-PCR analysis of RNA extracted from influoresences of DEK1-GFP–complemented lines. Lines 11 and 92 (partially complemented) show lower transcript levels of the DEK1-GFP transgene than line 87 (fully complemented). Expression was normalized relative to that of EIF2, and two biological replicates are shown.
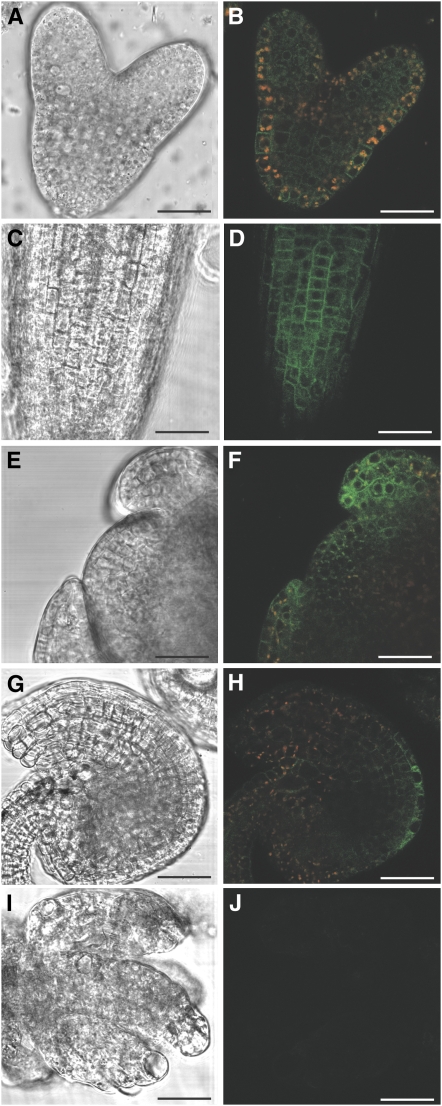
Figure 2.
Expression of pRPS5A:DEK1-GFP.
Differential interference contrast and GFP expression images of DEK1-GFP in dek1-3 complemented line 87 in the heart-stage embryo ([A] and [B]), 7-d-old root ([C] and [D]), floral meristem ([E] and [F]), and ovule ([G] and [H]). Red signal is due to autofluorescence from plastids. Expression of DEK1-GFP is barely detectable in the deformed ovules of complemented line 11 ([I] and [J]). Bars = 25 μm.
In addition to complemented lines, wild-type plants transformed with the pRPS5A:AtDEK1-GFP construct were also obtained. These lines either appeared wild-type or showed phenotypes similar to the partially complemented lines described above. RT-PCR and immunoblot analysis of endogenous and transgene products confirmed that abnormal phenotypes were likely due to cosuppression (see Supplemental Figure 3B online). In cases where transformed wild-type lines expressed high levels of fusion protein, no obvious phenotypes were observed, suggesting DEK1 activity could be regulated posttranscriptionally.
DEK1 Localizes to the Plasma Membrane, Endoplasmic Reticulum, and Cytoplasm
To test whether posttranscriptional regulation and subcellular localization of the DEK1 protein are likely to be important for its function, as is the case in animals, we used confocal microscopy to observe the subcellular localization of DEK1-GFP protein in fully complemented mutant plants. In all tissues studied, some GFP fluorescence was associated with the plasma membrane as expected from the presence of 21 predicted transmembrane domains in DEK1 (Figure 2; see Supplemental Figures 4A and 4D online). A diffuse intracellular pool of DEK1-GFP was also detected, although no signal was present within the nucleus. The intracellular GFP signal was noticeably stronger in a ring around the nucleus, potentially indicating an association with the endoplasmic reticulum (ER), which was subsequently confirmed by colocalization with an ER marker (see Supplemental Figures 4A to 4F online) (Haseloff and Siemering, 2006). Coexpression of the ER marker also identified signal that was not ER localized (see Supplemental Figures 4C and 4F online).
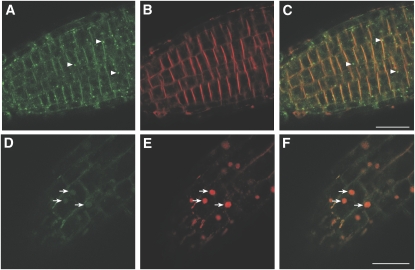
To investigate the cytoplasmic and ER-localized GFP signal further, roots were treated with Brefeldin A (BFA) or Wortmannin (Wm) in the presence of the amphiphilic steryl dye FM4-64. FM4-64 inserts into one side of the plasma membrane bilayer and is thought to enter the cell via internalization of membrane vesicles. Wm, a phosphatidylinositol-3-OH kinase inhibitor, disrupts transport from the trans-Golgi network. Treatment with Wm did not affect the plasma membrane location of DEK1-GFP proteins. Although diffuse cytoplasmic signal was still present, some intracellular GFP fluorescence was observed in small vesicle-like bodies in Wm-treated roots (Figures 3A to 3C). These bodies were not observed in untreated roots and did not accumulate FM4-64 (Figure 3B). BFA disrupts the trafficking of internalized vesicles in Arabidopsis root cells via inhibition of a subset of GDP/GTP guanidine-nucleotide exchange factors (Nebenfuhr et al., 2002). Resulting aggregates of intercellular vesicles, or BFA bodies, can be visualized with FM4-64. DEK1-GFP–expressing root cells treated with BFA and FM4-64 showed only weak GFP fluorescence accumulation in FM4-64–containing BFA bodies, suggesting that the majority of the intracellular signal is not due to endocytosis of DEK1-GFP from the plasma membrane (Figures 3D to 3F).
Figure 3.
Wm and BFA Studies in DEK1-GFP Roots.
FM4-64 is internalized via the endocytic pathway in root cells. Wm disrupts transport from the trans-Golgi network, giving vesicle aggregates that do not contain FM6-64. BFA disrupts the endocytic pathway in roots resulting in BFA bodies, which can be visualized with FM4-64.
(A) DEK1-GFP localizes to the plasma membrane, cytoplasm, and small intracellular vesicles (arrowheads) in Wm-treated roots.
(B) Root treated in (A) showing localization of FM4-64.
(C) Overlay of DEK1-GFP (A) and FM4-64 (B) in Wm-treated root, showing no colocalization of FM4-64 and GFP.
(D) Localization of DEK1-GFP in BFA-treated root.
(E) FM4-64 labeling of the plasma membrane and BFA bodies (arrows).
(F) Overlay of DEK1-GFP and FM4-64 shows GFP signal is only weakly associated with BFA bodies. Bars = 25 μm for all panels.
In summary, microscopy analysis of DEK1-GFP localization indicates association of fusion proteins with the plasma membrane and endomembrane system. Only a very small proportion of the GFP signal was found to accumulate in BFA bodies, indicating minimal endocytosis of plasma membrane–associated protein. An additional cytoplasmic pool of GFP was also observed.
DEK1 Is Proteolytically Cleaved
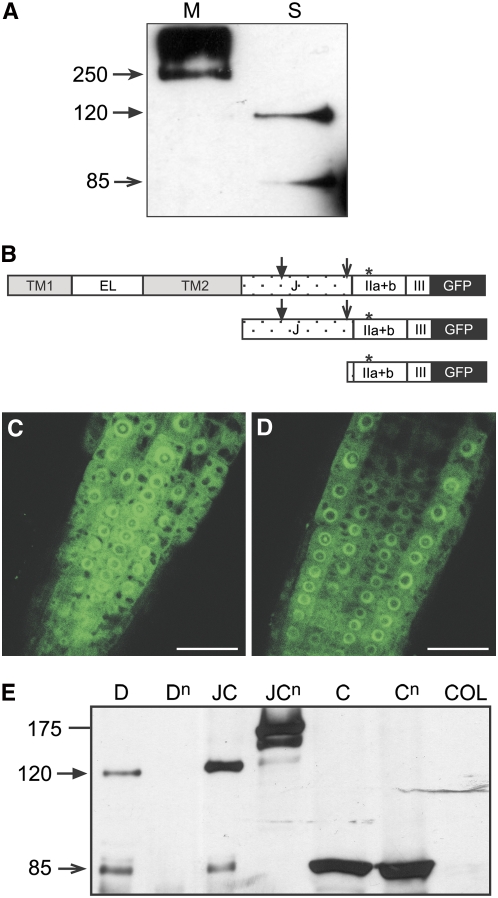
The location of DEK1 in the cytoplasm suggests the DEK1 protein might undergo a regulatory cleavage event, similar to those described in animal calpains. In the case of DEK1, this would release the calpain domain from the membrane into the cytoplasm.To test this possibility, proteins were extracted from plants expressing DEK1-GFP. A probable full-length protein of 265 kD was detected in microsomal-associated protein extracts (Figure 4A). Although no full-length protein was detected in soluble protein extracts, two smaller proteins of sizes ∼85 and 120 kD were visible (Figure 4A). Detection of these products was not affected by treatment with the proteasome inhibitor MG132 in root extracts (data not shown). No free GFP was observed in these fractions, indicating that the cytoplasmic signals observed microscopically were not simply due to cleavage of GFP from the fusion protein. The sizes of the smaller soluble proteins indicate putative cleavage events in the region just before the start of the calpain domain (as observed in animal calpains) and within the juxtamembrane domain (Figure 4B).
Figure 4.
Analysis of DEK1-GFP, JUXTA-CALPAIN-GFP, and CALPAIN-GFP Fusion Proteins.
(A) Immunoblot analysis of DEK1-GFP proteins showing the predicted full-length protein of ∼250 kD in microsomal (M) fractions and cleavage products of 120 and 80 kD in soluble (S) fractions.
(B) Schematic representation of DEK1 derivatives used for complementation studies. Top: DEK1-GFP; middle: JUXTA-CALPAIN-GFP; and bottom: CALPAIN-GFP. Putative cleavage positions of the full-length DEK1-GFP and JUXTA-CALPAIN-GFP protein to give the 120 kD (closed arrow) and 80-kD (open arrow) proteins are indicated. TM1, transmembrane region 1; EL, extracellular loop; TM2, transmembrane region 2; J, juxtamembrane domain; IIa+b, calpain domains IIa and IIb; III, calpain domain III; *, position of point mutation in null versions of calpain domain (see text).
(C) and (D) Localization of JUXTA-CALPAIN-GFP (C) and CALPAIN-GFP (D) in 7-d-old roots of dek1-3 complemented lines. Both fusion proteins localize to the cytoplasm and ER. Bars = 25 μm.
(E) Immunoblot analysis of DEK1-GFP (D), DEK1null-GFP (Dn), JUXTA-CALPAIN-GFP (JC), JUXTA-CALPAIN null-GFP (JCn), CALPAIN-GFP (C), CALPAINnull-GFP (Cn), and Columbia wild-type (COL) soluble protein extracts.
An Active Calpain Domain Is Sufficient to Complement dek1-3 Mutants
To investigate the potential biological significance of the observed cleavage events, we generated two constructs to express synthetic proteins mimicking the cleavage products observed in immunoblots. The first expressed a cytoplasmic protein consisting of the calpain domains IIa, IIb, and III of DEK1 fused to GFP and was designated pRPS5A:CALPAIN-GFP (Figure 4B). The second was designed to express a cytoplasmic protein that encompassed both the juxtamembrane domain and the calpain domains of DEK1 fused to GFP (pRPS5A:JUXTA-CALPAIN-GFP) (Figure 4B). These constructs were transformed into a population segregating dek1-3. A large number of complemented dek1-3 homozygous lines were identified with both the pRPS5A:CALPAIN-GFP (see Supplemental Figure 2C online) and pRPS5A:JUXTA-CALPAIN-GFP transgenes. The majority of complemented lines have a wild-type appearance under long-day conditions, indicating the cytoplasmic calpain domain alone is sufficient for complete complementation of the embryo lethal mutant phenotype (see Supplemental Figures 2 and 5 online). However, the dek1-3 mutation is caused by a T-DNA downstream of the region encoding the transmembrane domains and produces rare transcripts from the region upstream of the T-DNA (see Supplemental Figure 5D online). It was therefore possible that this RNA might encode a truncated, but correctly located, protein containing transmembrane domains. We therefore crossed four independent pRPS5A:CALPAIN-GFP lines that complement the dek1-3 mutation into an dek1-2 mutant background. The dek1-2 allele carries a T-DNA insertion between the 5th and 6th transmembrane domain–encoding region (Johnson et al., 2005). Homozygous complemented dek1-2 mutants were identified in the F2 population for all four lines, confirming that the transmembrane domains are not necessary for complementation by the calpain domain.
The Calpain Domain Is Located in the Cytoplasm and Associates with Membranes
As predicted from their structure, the majority of fluorescent protein expressed from pRPS5A:CALPAIN-GFP and pRPS5A:JUXTA-CALPAIN-GFP appeared to be located intracellularly. However, some signal was still apparent in the cell periphery, and there was strong fluorescence in a ring around the nucleus, suggesting association with both plasma membrane and ER membranes (Figures 4C and 4D). Leaves of CALPAIN-GFP plants were bombarded with an ER-specific marker to further investigate the ER localization. Colocalization studies showed some GFP signal overlapping with the ER marker as well as signal in the cytoplasm (see Supplemental Figures 4G and 4I online).
The potential membrane association of CALPAIN-GFP is intriguing because active calpains in mammals and invertebrates associate both with the plasma membrane and with internal membranes, such as the ER (Shao et al., 2006). To further explore the cellular localization of the plant phytocalpain DEK1, two-phase partitioning of microsome extracts was undertaken using plants expressing either DEK1-GFP or CALPAIN-GFP proteins at high levels. Immunoblot analysis showed full-length DEK1-GFP protein present in both the plasma membrane–enriched (pme) and endomembrane-enriched (eme) fractions. Cleavage products were also detected in both these fractions (see Supplemental Figure 6A online). Although CALPAIN-GFP (85 kD) was detected in soluble protein extracts, partitioning also showed the presence of CALPAIN-GFP associated with the pme and eme fractions (see Supplemental Figure 6A online). Control immunodetection of tubulin (a cytoplamic marker) on the same blot suggested that some of the membrane-enriched fractions contained cytoplasmic contamination (see Supplemental Figure 6B online). This may be because vesicles isolated through two-phase partitioning are mainly in the right side out (i.e, with the cytoplasmic membrane face oriented inwards) orientation, allowing entrapment of small volumes of cytoplasm (Larsson et al., 1984). Calreticulin (an ER marker; Napier et al., 1995) was present both in the pme and eme fractions, suggesting some contamination of the pme fraction with endomembranes (see Supplemental Figure 6C online). However, the relative enrichment of CALPAIN-GFP in the membranous versus soluble fractions compared with tubulin supports the association of CALPAIN-GFP both with plasma membranes and endomembranes. Thus, as suggested by confocal microscopy, two-phase partitioning studies indicate that cytoplasmic CALPAIN-GFP generated either from our synthetic construct or by cleavage of full-length DEK1-GFP is significantly associated with membranes.
DEK1 Likely Undergoes Autocatalytic Cleavage Events
To investigate the possibility that DEK1 undergoes autolytic cleavage to and determine the importance of Cys protease activity for DEK1 function, the active Cys-71 residue was replaced with Ser in the pRPS5A:CALPAIN-GFP, pRPS5A:JUXTA-CALPAIN-GFP, and pRPS5A:AtDEK1-GFP constructs. This mutation has previously been shown to eliminate calpain activity in maize DEK1 (Wang et al., 2003). These constructs were introduced into heterozygous dek1-3 plants. Despite extensive testing, we were unable to identify homozygous dek1-3 plants in the T1 populations, suggesting that CALPAINnull-GFP, JUXTA-CALPAINnull-GFP, and DEK1null-GFP proteins were unable to complement the mutant phenotype and, thus, that Cys protease activity is essential for DEK1 function.
To investigate the effects of loss of protease activity upon DEK1 processing, immunoblots of soluble proteins extracts from all DEK1-GFP variants were analyzed. A soluble product of 85 kD was observed in both pRPS5A:CALPAIN-GFP and pRPS5A:CALPAINnull-GFP expressing plants, corresponding to the size of the cleavage product observed in DEK1-GFP extracts (Figure 4E). Interestingly, in pRPS5A:JUXTA-CALPAIN-GFP expressing plants, products corresponding to cleavage upstream of the calpain domain (85 kD) and within the juxtamamembrane domains (120 kD) was detected, but not the predicted full-length product of 170 kD. This suggests that proteolytic processing of this protein variant also occurs (Figure 4E). In plants expressing JUXTA-CALPAINnull-GFP, the predicted full-length protein of ∼170 kD was observed, but no cleavage product of 85 kD and only a faint band of 120 kD was detected (Figure 4E). Immunoblots of soluble protein extracts of plants expressing pRPS5A:DEK1null-GFP show no soluble products (Figure 4E). These results suggest that DEK1null-GFP and JUXTA-CALPAINnull-GFP are not correctly proteolytically processed. Autolytic cleavage of the N-terminal domain DI is known to occur in animal calpains and is proposed to be involved in regulation of calpain activity. Our results suggest that DEK1 undergoes autolytic cleavage within the juxtamembrane domain and at the start of the calpain domain. These results reinforce complementation studies in suggesting that the cleaved calpain domain is the biologically active form of the DEK1 protein.
The Transmembrane Regions of DEK1 Are Not Required for Calpain Activity but May Play Regulatory Roles
The ability of the calpian domain alone to fully complement the mutant phenotype raises the question of the role of the transmembrane domains of DEK1. One possibility is that these regions play a role in controlling the activation of calpain activity, as is the case in mammalian and invertebrate calpains, where cleavage of a short N-terminal region is often associated with enzyme activation. In the case of DEK1, regulation of cleavage could be in response to either an extracellular or plasma membrane–located signal. To explore the proposed regulatory mechanism and the role of DEK1 in development, the effects of both reduced and increased levels of DEK1 and variants was investigated in transgenic lines. A suite of developmental defects are observed in plants with reduced levels of DEK1, either due to incomplete complementation or cosuppression of the endogenous genes, as previously described (Figure 1; see Supplemental Figure 3 online). These include a tendency to lose meristematic activity in seedlings and female sterility due to ovule defects and were described earlier. Wild-type or complemented mutant plants with levels of pRPS5A:AtDEK1-GFP or pRPS5A: JUXTA-CALPAIN-GFP expression higher than those required for full complementation are phenotypically similar to the wild type. This is consistent with calpain activity being dependent upon a posttranslational activation event as observed in animal calpain systems. One prediction arising from this hypothesis would be that overexpressing the calpain domain alone might bypass the need for this activation event and produce phenotypes not observed upon overexpression of longer protein variants. Our results support this prediction. Plants with high levels of pRPS5A:CALPAIN-GFP exhibit a novel set of phenotypes that may be attributed to an excess of unregulated calpain activity since plants expressing comparable levels of CALPAINnull-GFP show no significant phenotype. When grown in short days, rosette leaves of CALPAIN-GFP overexpressing (OE) plants were much darker green than the wild type and showed severe rumpling of the lamina and serration reminiscent of JAWD mutants (Palatnik et al., 2003) (Figure 5B; see Supplemental Figure 7 online). The phenotypes observed in underexpressing DEK1 variants and overexpressing active calpain lines are effectively opposite and consistent with a deregulated growth throughout developing organs.
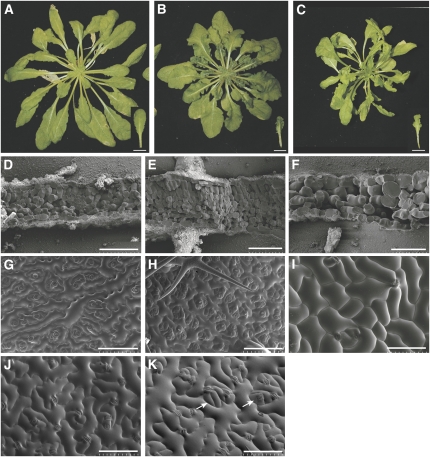
Figure 5.
Phenotypes of CALPAIN-GFP OE Plants and Plants with Low Levels of DEK1 Activity.
(A) to (C) Wild-type plant grown in short-day conditions for 6 weeks (A) has flat and rounded leaves compared with CALPAIN-GFP OE plants that have rumpled, dark-green leaves and a more compact rosette (B). Plants with low levels of DEK1 expression have pale-green leaves and reduced meristematic activity (C). Bars = 1 cm.
(D) to (F) Scanning electron microscopy images of freeze-fractured 10th leaf (inset in [A] to [C]) from the wild type (D), CALPAIN-GFP OE plants (E), and plants with reduced levels of DEK1 (F).
(G) to (I) Adaxial epidermis of the 10th leaf from the apex from the wild type (G), CALPAIN-GFP OE plants (H), and plants with reduced levels of DEK1 (I).
(J) and (K) Adaxial epidermis of mature leaves from wild-type (J) and CALPAIN-GFP OE plants (K). CALPAIN-GFP OE plants show clusters of small cells and stomata (arrows in [K]) interspersed by large pavement cells. Bars = 100 μm.
DEK1 Plays a Role in Regulating Proliferation and Expansion in Developing Leaves
Defects observed in CALPAIN-GFP OE plants are likely due to the uncoupling of DEK1 calpain activity from its normal regulation, since they are not observed upon overexpression of the full-length protein. To understand more about the role of DEK1 in leaf development, the phenotypes of CALPAIN-GFP OE plants were compared in more detail with cosuppressed plants and wild-type plants. Leaves of CALPAIN-GFP OE plants were studied both at a time point during growth (10th dissectable leaf from apex of 8-week-old short-day-grown plants) and when fully expanded. Both classes of leaf showed an increase in both the number of mesophyll cell layers observed and mesophyll cell density (Figures 5D and 5E; see Supplemental Figures 7D and 7E online). In wild-type plants, one palisade mesophyll layer and two to three spongy mesophyll cells layers were apparent in freeze fractures (Figure 5D). However, in CALPAIN-GFP OE plants, up to three tightly packed layers of palisade mesophyll and up to five layers of spongy mesophyll could be distinguished (Figure 5E). The 10th leaves of CALPAIN-GFP OE plants appeared smaller in area and showed an increased epidermal cell density compared with the wild type (Figures 5G and 5H). This difference was particularly marked on the adaxial leaf surface.
When fully expanded leaves were studied, we found that the leaves of CALPAIN-GFP OE plants were ∼45% larger in area than those of wild-type leaves (see Supplemental Figures 7A to 7C online). This difference appears to be attributable to increased cell size in CALPAIN-GFP OE leaves (Figures 5J and 5K; see Supplemental Figures 7F to 7H online). By maturity, CALPAIN-GFP OE and wild-type leaves have equivalent numbers of cells on their adaxial epidermal cell surface (see Supplemental Figure 7I online). However, cell size in mature CALPAIN-GFP OE leaves is much less uniform than in the wild type, with islands of small cells being interspersed by very large pavement cells (Figure 5K; see Supplemental Figure 7J online). Mesophyll cells were also larger in CALPAIN-GFP OE leaves than in the wild type, and mesophyll density and number of mesophyll cell layers were increased compared with the wild type (see Supplemental Figures 7D and 7E online). Thus, total cell number per organ in CALPAIN-GFP OE leaves is likely to be considerably higher than in wild-type leaves. CALPAIN-GFP OE leaves also have a wider lamina than wild-type leaves, suggesting that patterns of growth are disrupted (see Supplemental Figures 7A to 7C online). The ruched appearance of leaves from CALPAIN-GFP OE lines is due to lack of expansion in the midrib area compared with the surrounding lamina. Leaves could be flattened for analysis by bisecting along the midrib and flattening the two resulting halves (see Supplemental Figure 7C online). From these results we concluded that overexpressing CALPAIN-GFP during leaf development caused a severe deregulation in the balance between, and timing of, cell division and expansion during leaf morphogenesis.
The pale-green leaves of cosuppressed plants showed an essentially opposite set of phenotypes to those observed in CALPAIN-GFP OE plants (Figures 5C, 5F, and 5I). Leaves could not be analyzed quantitatively because of the chaotic patterns of leaf initiation in these plants. However, the number of mesophyll layers formed in both young and fully expanded leaves was significantly decreased; in many cases, only two layers of mesophyll cells were present. Palisade mesophyll was very difficult to distinguish. Epidermal cell size was highly variable and often considerably increased compared with wild-type plants at younger stages. However, mature leaves have a mosaic like composition, with areas of both very small and very large cells, making quantitative analysis difficult (see Supplemental Figure 3K online). Similar to the case in cotyledons, the epidermal cells on the adaxial surfaces of these plants were much less interdigitated than in the wild type (Figure 5I).
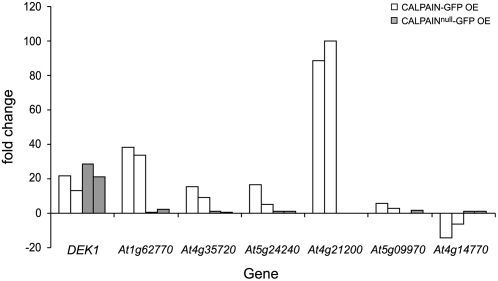
To understand whether DEK1 regulates known growth pathways, RNA was extracted from the four youngest dissectable leaves of CALPAIN-GFP–overexpressing plants, the wild type, and wild-type plants overexpressing CALPAINnull-GFP and used in transcript profiling by microarray analysis. Young leaf material was used to maximize capture of early transcriptional changes attributable to CALPAIN-GFP activity. This analysis confirmed an increase in expression of the DEK1 calpain domain of ∼12-fold in the CALPAIN-GFP–overexpressing material and ∼14-fold in the CALPAINnull-GFP–overexpressing material compared with the wild type (Table 1; see Supplemental Tables 1 and 2 online). Overexpression of CALPAIN-GFP caused a specific change in a number of genes compared with the wild type and CALPAINnull-GFP (see Supplemental Tables 3 and 4 online). A subset of this gene set (DEK1, five overexpressed genes and one underexpressed gene) was validated by Q-PCR (Table 1, Figure 6). An increase in DEK1 transcript in CALPAIN-GFP and CALPAINnull-GFP and the specific change in expression of the six genes in CALPAIN-GFP OE plants was confirmed (Table 1, Figure 6). Despite the similarity of the phenotype of CALPAIN-GFP OE plants to JAWD mutants, no misregulation of the known JAW microRNA (miRNA) targets TCP3 and TCP4 was noted in the microarray data. Q-PCR analysis of TCP3, TCP4, TCP20, and JAW miRNA expression also revealed no significant misregulation in CALPAIN-GFP OE plants (see Supplemental Figure 8 online). Analysis of the data set using the virtual plant, a resource allowing mining of genomic information (http://virtualplant.bio.nyu.edu), showed a highly significant overrepresentation of genes potentially involved in responses to endogenous signals with 14.5% compared with the genome average of 2.4%. Of particular interest was the upregulation of known growth regulators, such as CYP78a7 (Wang et al., 2008) and TINY (Wilson et al., 1996), as well as range of genes encoding enzymes involved in hormone catabolism, such as GA2Ox8 (Zhao et al., 2007). Downregulated genes included a TESMIN/TSO1-like gene and CYP79B2, which has been implicated in auxin production (Mikkelsen et al., 2000). Several genes known to be regulated by gibberellic acid, auxin, ethylene, and brassinosteroids were also present in the data set, suggesting that multiple hormone pathways are likely to be affected by DEK1 activity (see Supplemental Tables 1 to 4 online). Also of interest is a putative phosphoinositol 3/4-kinase family protein containing ubiquitin-like domain, related members of which have been shown to directly interact with components of the 26S proteasome (Galvao et al., 2008). A number of cell wall components, including an invertase/pectin methyltransferase, xyloglucans, expansins, and lipid transfer proteins, were among the most highly upregulated genes. The diversity of genes with changes due to overexpression of CALPAIN-GFP suggest that DEK1 is likely to act through a number of developmental pathways to regulate growth.
Table 1.
Microarray and Q-PCR Analysis of DEK1, Five Upregulated Genes, and One Downregulated Gene in Plants Overexpressing CALPAIN-GFP or CALPAINnull-GFP Proteins
| DEK1 | At1g62770 | At4g35720 | At5g24240 | At4g21200 | At5g09970 | At4g14770 | |
|---|---|---|---|---|---|---|---|
| CALPAIN-GFP | |||||||
| Microarray | 12.68 | 32.19 | 30.81 | 6.75 | 5.21 | 3.24 | −15.20 |
| Q-PCR 1 | 21.79 | 38.42 | 15.40 | 16.37 | 88.66 | 5.97 | −14.00 |
| Q-PCR 2 | 13.23 | 33.55 | 9.27 | 5.10 | 100.100 | 2.70 | −6.20 |
| CALPAINnull-GFP | |||||||
| Microarray | 15.82 | 1.70 | 1.23 | 1.00 | 1.10 | 1.00 | 1.28 |
| Q-PCR 1 | 28.69 | 0.75 | 1.00 | 1.00 | 0.00 | 0.00 | 1.10 |
| Q-PCR 2 | 21.01 | 2.02 | 0.50 | 1.10 | 0.00 | 0.50 | 1.07 |
Values shown represent the fold change in expression compared with Columbia wild type standardized to 1.
Figure 6.
Q-PCR Analysis of RNA Transcribed from Young Leaves of Columbia (Wild-Type), CALPAIN-GFP OE Plants, and Plants Overexpressing CALPAINnull-GFP.
Expression of each gene was normalized relative to that of EIF2, and two biological replicates are shown. The fold change in expression of each gene in CALPAIN-GFP OE plants (white bars) and CALPAINnull-GFP OE plants (gray bars) is compared with expression in Columbia (standardized to 1). CALPAIN-GFP OE plants show specific upregulation of At1g62770, invertase/pectin methylesterase inhibitor; At4g35720, expressed protein; At5g24240, phosphatidylinositol 3- and 4-kinase/ubiquitin; At4g21200, oxidoreductase, 2OG-Fe(II) oxygenase; At5g09970, cytochrome P450; and downregulation of At4g14770, tesmin/TSO-1 like protein. At4g14770, tesmin/TSO-1 like protein. (See also Table 1.)
DISCUSSION
In this study, we have used transgenic approaches to try and further understand the function of an essential gene of plants. Null alleles of DEK1 cause early embryo lethality, a phenotype from which it is almost impossible to draw conclusions about gene function in later plant development. In our previous work, we tried to overcome this problem using an RNA interference knockdown-based approach (Johnson et al., 2005). Although this permitted us to show that DEK1 is required for the maintenance of epidermal cell fate during late embryogenesis, it did not reveal how the DEK1 protein functions in plant development. Here, we used homozygous mutant lines transgenically complemented with GFP fusions to variants of the DEK1 protein to provide in vivo information not only about protein localization but also about regulation of DEK1 activity by protein cleavage.
Many calpain isoforms in mammals and invertebrates are thought to be localized in the cytoplasm. However, active calpains associate both with the plasma membrane and with internal membranes, such as the ER (Shao et al., 2006). These associations have been shown to be mediated by calpain DIII, which is reported to contain a calcium-regulated phospholipid binding domain (Tompa et al., 2001). Our results show that, as predicted from its structure, full-length DEK1 localizes to the plasma membrane and endomembranes. We have also shown that the functional calpain domain of DEK1 can be released from the membrane through autocatalytic cleavage events. Interestingly, cleaved calpain appears to associate with membranes, suggesting that the phospholipid affinity of animal calpain DIII may be conserved in plants. Our results differ from those of Tian et al. (2007), who were unable to complement dek1 mutants using the endogenous DEK1 or 35S promoters to drive expression of the DEK1 calpain domain (Tian et al., 2007). One possible explanation for this discrepancy is that our GFP fusion proteins behave differently to endogenous proteins. However, our ability to fully complement the dek1 phenotype with these proteins suggests that this is unlikely to be the case. Q-PCR analyses also indicate that our gene fusions are not noticeably overexpressed in fully complemented plants. Whereas Tian et al. detected maize DEK1 largely in FM4-64–labeled endosomal compartments, we did not find this to be the case for DEK1-GFP fusion proteins in Arabidopsis, which are only weakly associated with BFA bodies in roots. Our Wm experiments, two-phase partitioning, and immunoblot and confocal microscopy studies of DEK1-GFP proteins show that the full-length protein is instead likely to be present in the endomembrane system and the plasma membrane. In addition, we show that a cytoplasmic GFP signal is present in pRPS5A:AtDEK1-GFP–expressing plants. We present evidence that this signal represents the cleaved cytoplasmic domain of DEK1 and that this domain associates with the plasma membrane and endomembrane system. Discrepancies in reported DEK1 localization might arise if the anti-DEK1 antibodies used by Tian et al. do not detect the cleaved calpain moiety or the protein in its active conformation. Alternatively, studies in animals have shown that inactive calpains can be very tightly folded, possibly preventing antibody access (Moldoveanu et al., 2001). Additionally, DEK1 in maize could be localized differently to DEK1 in Arabidopsis.
The fact that DEK1 undergoes autolytic cleavage raises obvious comparisons with mammalian and invertebrate calpains, in which DI cleavage is often associated with enzyme activation. Whether these events are necessary for activation is a point of some controversy in animal systems. Current evidence suggests that Drosophila calpains, which have a relatively long DI, require its removal for full activity, whereas canonical mammalian calpains with a short DI may not (Farkas et al., 2004). We have shown that DEK1 is cleaved in at least two positions, one of which is likely to be adjacent to the calpain domain, as in animal calpains, and one of which is near the start of the cytoplasmic domain. We have also shown that expressing synthetic proteins mimicking both cleavage products in planta is sufficient to complement null mutants, although the longer JUXTA-CALPAIN-GFP protein is cleaved in a similar fashion to the full-length DEK1 protein, to release a product equivalent to the CALPAIN-GFP protein and a product encompassing the calpain domain and part of the juxtamembrane region. DEK1 variants in which the active Cys residue has been mutated do not complement dek1-3 mutants. In addition, no cleavage products corresponding to the calpain domain are detected. This indicates that the active Cys is not only essential for calpain function, it is also required for autolytic cleavage of the full-length protein.
Our results, although not fully conclusive, are strongly indicative of a role for autolytic cleavage in DEK1 activation, equivalent to that observed in some animal systems. We hypothesize that the transmembrane and juxtamembrane regions of the DEK1 protein control the cleavage of the calpain domain, which, when released into the cytoplasm, is catalytically active. Regulation, in the case of DEK1, could be in response to either an extracellular or plasma membrane–located signal. Consistent with this theory, overexpression of the full-length DEK1-GFP or the JUXTA-CALPAIN-GFP in mutant backgrounds results in growth phenotypes very similar to the wild type. This suggests the processing of the active calpain domain may be limited by these regions. Overexpressing the CALPAIN-GFP protein revealed the effects of bypassing this regulated activation and leads to severe abnormalities in growth. These defects are effectively the inverse of abnormalities observed in wild-type plants in which expression of DEK1 has been suppressed.
Our results indicate that DEK1 plays a key role in regulating plant growth. To test whether DEK1 acted through any known growth regulatory pathway, we analyzed gene expression during the early leaf development. Although the growth control pathway regulated by the JAW miRNA does not appear to involve DEK1, misregulation of DEK1 activity results in changes in the expression of a number of other known growth regulatory pathways. Interestingly, the CYP78a7 gene is upregulated in CALPAIN-GFP overexpressors and downregulated in plants with suppressed DEK1 expression. Recent work has shown that CYP78a7 functions redundantly with KLUH (CYP78a5) in regulating organ growth, and it is possible that DEK1 acts via this signaling pathway (Anastasiou et al., 2007; Wang et al., 2008). Another known growth regulator, GA2Ox8 (Zhao et al., 2007), is also a potential downstream component of the DEK1 signaling pathway. Ga2Ox8 was found to be dramatically upregulated in Q-PCR analysis and potentially results in less active GAs in young leaves of CALPAIN-GFP OE plants. How much the change in expression of genes such as Ga2Ox8 and CYP787a7 contributes to the CALPAIN-GFP OE phenotype presents an interesting avenue for further investigation. Although misregulated genes identified in our study may not represent direct targets of DEK1, the RPS5A promoter spatially mimics the expression of DEK1 in all cell layers minimizing the risk of off-target effects.
How the role of DEK1 in growth control marries with its previously published role in epidermal development remains an open question. We propose that DEK1 is ideally placed to play a key role in coordinating growth among the cell layers of developing leaves. Classical studies have suggested that de novo epidermal specification occurs only once, during early embryo development in plants, with subsequent maintenance of epidermal fate dependent on signaling between neighboring cells (Bruck and Walker, 1985). This fact might make the epidermis uniquely sensitive to changes in the levels of DEK1 activity. It is possible that when levels of DEK1 activity are limiting, epidermal growth can no longer keep up with that of underlying tissues, leading to a loss of epidermal continuity, and thus the ability to maintain epidermal identity. This would explain the outgrowths of mesophyll-like tissue that occur on the organs and stems of plants showing low levels of DEK1 activity. The observed role of DEK1 in maintaining epidermal cell fate would therefore be essentially an indirect consequence of a fundamental role in coordinating growth.
Our data do not support a direct role for DEK1 in regulation of epidermal specification during postgermination development. In microarray studies, no marked increase in known epidermal markers (for example, ATML1 and ACR4; Sessions et al., 1999; Gifford et al., 2003) is associated with overexpression of CALPAIN-GFP. Moreover, plants expressing low levels of DEK1 (due to cosuppression) still express epidermal markers such as ATML1 and ACR4 both at the RNA and protein level (K.L. Johnson and G.C. Ingram, unpublished data).
Studies into the cell-specific requirements for DEK1 activity, as well as further elucidation of potential DEK1 targets, will further clarify our understanding of how this novel regulator of plant growth functions.
METHODS
Construction of pRPS5A:AtDEK1-GFP Variants and Complementation Studies
Full-length DEK1 cDNA was constructed using the RAFL08-16-B06 (Riken; Carninci et al., 2001; Seki et al., 2002) and APZL41e10R (Kazusa; Asamizu et al., 2000) cDNA clones and amplification of the intervening region from cDNA synthesized by reverse transcription of inflorescence RNA. The mGFP6 open reading frame was amplified from pBSmGFP6 (Gifford et al., 2003) with GFP-F and GFP-R and cloned at the 3′ end of the DEK1 cDNA in pBluescript II (Stratagene) (GI-371). The RPS5A promoter (pSDM7038; Weijers et al., 2001) was inserted into pBIBHyg (Becker, 1990; Becker et al., 1992) and the DEK1-GFP fragment cloned downstream. The JUXTA-CALPAIN-GFP (KJ-33) and CALPAIN-GFP (KJ-32) variants were PCR amplified from GI-371 using JC-F or Calp-F, respectively, and GFP-R and cloned into pGEMT-easy (Promega). Site-directed mutagenesis of Cys-71 residue to Ser to generate the CALPAINnull-GFP construct (KJ-32null) was performed using Calmut-F and Calmut-R. The JUXTA-CALPAIN-GFP, CALPAIN-GFP, and CALPAINnull-GFP fragments were cloned downstream of the RPS5A promoter in pBIBHyg. Plant transformations were performed using Agrobacterium tumefaciens GC3101 (Koncz et al., 1992) and floral dipping (Clough and Bent, 1998) into plants heterozygous for the dek1-3 mutation (Johnson et al., 2005). PCR was used for identification of plants homozygous for the dek1-3 mutation using primers 1-3-F with 1-3-R (wild-type) or 1-3-T (T-DNA) and confirmed by DNA gel blot analysis (see Supplemental Methods online). Genotyping of the dek1-2 allele was as described by Johnson et al. (2005).
Microarray Analysis
For microarrays, total RNA was extracted using the RNeasy kit (Qiagen) from the four youngest dissectable leaves of 8-week-old plants expressing pRPS5A:CALPAIN-GFP or pRPS5A:CALPAINnull-GFP and Columbia plants grown in short-day conditions. Three independent biological samples were taken for each genotype. Samples were assayed on the Affymetrix GeneChip oligonucleotide ATH1 array (Nottingham Arabidopsis Stock Centre [NASC]). Results were analyzed in Genespring GX version 9 (Agilent) with thresholding of the signal values to 5 and then normalization of the data to the 75th percentile, performing baseline transformation to the median of all samples. The normalized values were then used for fold change analysis to identify genes with threefold change in expression ratios between Columbia and CALPAIN-GFP OE plants and not changed more than twofold between Columbia and CALPAINnull-GFP plants. GO terms were analyzed using Virtual Plant (http://www.virtualplant.org/).
Q-PCR
Q-PCR was used to verify the microarray results for seven genes. RNA was extracted from the four youngest dissectable leaves of plants and extracted using the RNeasy kit (Qiagen). cDNA was synthesized from 1 μg of RNA using the Reverse-iT first-strand synthesis kit (ABgene). Transcript abundance of DEK1, EIF2, At1g62770, At4g35720, At5g24240, At4g21200, At5g09970, and At4g14770 were assessed by quantitative RT-PCR in a Bio-Rad iCycler IQ using ABI SYBR Green PCR Master Mix (Applied Biosystems) in 15-μL reactions. Transcript levels were normalized against EIF2 using a cDNA dilution series for each primer set in each experiment. Two independent RNA samples for each genotype were assayed in triplicate. Primer sequences used are described in Supplemental Table 5 online.
Q-PCR analysis of complemented dek1-3 mutants with pRPS5A:AtDEK1-GFP was performed to assess transcript abundance of the complementing transgene. RNA was extracted from five inflorescence tips using the RNeasy Kit (Qiagen) and 1 μg RNA used for cDNA synthesis as above. DEK1 and EIF2 transcript levels were assessed as above.
Analysis of Protein Extracts from Plants
Proteins were extracted from inflorescence tips comprising all unopened flowers and the first two open flowers or leaf material. For total microsome fractions, ∼250 such tips were used for extraction using a modified protocol for small-scale, nonmechanized preparation of microsomal tissue (Terry and Williams, 2002). Changes include harvesting tissue directly into a cooled mortar and pestle rather than ice-cold water and resuspending microsomal pellet in Cytobuster reagent (Merck) containing 0.5% (w/v) Triton X-100 and EDTA-free protease inhibitor cocktail (Roche). For soluble fractions of DEK1-GFP, DEK1null-GFP, and JUXTA-CALPAIN-GFP, 500 mg of leaf material from 6-week-old short-day-grown plants was ground in ice-cold Cytobuster reagent, centrifuged for 5 min at 10,000g and the supernatant used for immunoprecipitation. Microsomal and soluble fractions were immunoprecipitated using the GrabIt ProteinG Plus/ProteinA kit (Merck) and anti-GFP, IgG (Invitrogen) according to the manufacturer's instructions. Soluble fractions of JUXTA-CALPAINnull-GFP, CALPAIN-GFP, and CALPAINnull-GFP were extracted from a single leaf ground in Cytobuster reagent and mixed directly with loading dye without immunoprecipitation. Samples were heated at 65°C for 5 min, and 15-μL aliquots were loaded onto 10% acrylamide gels. Gels were subjected to protein gel blotting onto nitrocellulose membranes (Protran; Schleicher and Schuell) using standard procedures, and GFP-labeled proteins detected using an anti-sheep anti-GFP primary antibody and a horseradish peroxidase–linked donkey secondary antibody (Amersham). Peroxidase activity was detected using the SuperSignal West Pico chemiluminescent substrate (Pierce).
BFA Experiments
Seedlings grown on 0.5% Murashige and Skoog media (brand), 1% agar for 7 d were incubated for 30 min in 50 μm BFA (Sigma-Aldrich) and 4 μm FM4-64 or for 2 h with 33 μm Wm and 4 μm FM4-64. Controls were incubation in 4 μm FM4-64 alone or a water control.
Microscopy Analysis
Fluorescence studies were performed using a Bio-Rad Radiance 2100 confocal microscope or a Leica SP2 confocal laser scanning microscope (Leica Microsystems) with a ×63 water-dipping lens (Leica Microsystems). Cryoscanning electron microscopy studies were performed using a Hitachi S-4700 scanning electron microscope (Hitachi High Technologies).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DEK1, 2035675; EIF2, 2020259; At1g62770, 2026236; At4g35720, 2127972; At5g24240, 3436120; At4g21200, 2127402; At5g09970, 2178212; At4g14770, 2130219; JAW, 1510594065; TCP3, 2009594; TCP4, 2086349; and TCP20, 2092019.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression of pRPS5A:ATML1-GFP.
Supplemental Figure 2. DNA Gel Blot of dek1-3 Mutant Plants Complemented with a pRPS5A:AtDEK1-GFP or pRPS5A:CALPAIN-GFP Transgene.
Supplemental Figure 3. Phenotype and Immunoblot Analysis of dek1-3 Lines Complemented with pRPS5A:AtDEK1-GFP or pRPS5A:CALPAIN-GFP Grown in Long-Day Conditions.
Supplemental Figure 4. Colocalization Studies of DEK1-GFP and CALPAIN-GFP with an ER Marker.
Supplemental Figure 5. Phenotype and Immunoblot Analysis of dek1-3 Lines Complemented with pRPS5A:CALPAIN-GFP Grown in Long-Day Conditions.
Supplemental Figure 6. Immunoblot Analysis of Two-Phase Partitioned Microsomes from DEK-GFP and CALPAIN-GFP Expressing Plants.
Supplemental Figure 7. Phenotype of Mature Leaves of dek1-3 Lines Complemented with pRPS5A:CALPAIN-GFP Grown in Short-Day Conditions.
Supplemental Figure 8. Q-PCR Analysis of JAW, TCP3, TCP4, and TCP20.
Supplemental Table 1. Genes Changed More Than Threefold in CALPAIN-GFP Plants.
Supplemental Table 2. Genes Changed More Than Threefold in CALPAINnull-GFP Plants.
Supplemental Table 3. CALPAIN-GFP Upregulated Genes.
Supplemental Table 4. CALPAIN-GFP Downregulated Genes.
Supplemental Table 5. Primer Sequences.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Ross Walker and Yvonne Stewart for expert technical support, Julia Foreman for advice on microarray analysis, Kevin Hardwick for providing anti-GFP antibody, and Andrew Hudson, Anne Moore, and Peter Doerner for critical comments and discussions. We acknowledge the work of the European Arabidopsis Stock Centre (NASC) Affymetrix service for transcriptome analysis. K.L.J. was supported by a Marie Curie International Incoming Fellowship, and G.C.I. was supported by a Royal Society of London University Research fellowship and a Research Councils UK fellowship.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gwyneth Christina Ingram (gwyneth.ingram@ed.ac.uk).
Online version contains Web-only data.
References
- Ahn, J.W., Kim, M., Lim, J.H., Kim, G.T., and Pai, H.S. (2004). Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 38 969–981. [DOI] [PubMed] [Google Scholar]
- Anastasiou, E., Kenz, S., Gerstung, M., MacLean, D., Timmer, J., Fleck, C., and Lenhard, M. (2007). Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13 843–856. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7 175–180. [DOI] [PubMed] [Google Scholar]
- Becker, D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Becraft, P.W., and Asuncion-Crabb, Y. (2000). Positional cues specify and maintain aleurone cell fate in maize endosperm development. Development 127 4039–4048. [DOI] [PubMed] [Google Scholar]
- Becraft, P.W., Li, K., Dey, N., and Asuncion-Crabb, Y. (2002). The maize dek1 gene functions in embryonic pattern formation and cell fate specification. Development 129 5217–5225. [DOI] [PubMed] [Google Scholar]
- Bruck, D.K., and Walker, D.B. (1985). Cell determination during embryogenesis in Citrus jambhiri. II. Epidermal differentiation as a one-time event. Am. J. Bot. 72 1602–1609. [Google Scholar]
- Carninci, P., Shibata, Y., Hayatsu, N., Itoh, M., Shiraki, T., Hirozane, T., Watahiki, A., Shibata, K., Konno, H., Muramatsu, M., and Hayashizaki, Y. (2001). Balanced-size and long-size cloning of full-length, cap-trapped cDNAs into vectors of the novel lambda-FLC family allows enhanced gene discovery rate and functional analysis. Genomics 77 79–90. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cottin, P., Thompson, V.F., Sathe, S.K., Szpacenko, A., and Goll, D.E. (2001). Autolysis of mu- and m-calpain from bovine skeletal muscle. Biol. Chem. 382 767–776. [DOI] [PubMed] [Google Scholar]
- Croall, D.E., and Ersfeld, K. (2007). The calpains: Modular designs and functional diversity. Genome Biol. 8 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas, A., Tompa, P., Schad, E., Sinka, R., Jekely, G., and Friedrich, P. (2004). Autolytic activation and localization in Schneider cells (S2) of calpain B from Drosophila. Biochem. J. 378 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvao, R.M., Kota, U., Soderblom, E.J., Goshe, M.B., and Boss, W.F. (2008). Characterization of a new family of protein kinases from Arabidopsis containing phosphoinositide 3/4-kinase and ubiquitin-like domains. Biochem. J. 409 117–127. [DOI] [PubMed] [Google Scholar]
- Garcia Diaz, B.E., Gauthier, S., and Davies, P.L. (2006). Ca2+ dependency of calpain 3 (p94) activation. Biochemistry 45 3714–3722. [DOI] [PubMed] [Google Scholar]
- Gifford, M.L., Dean, S., and Ingram, G.C. (2003). The Arabidopsis ACR4 gene plays a role in cell layer organisation during ovule integument and sepal margin development. Development 130 4249–4258. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., and Siemering, K.R. (2006). The uses of green fluorescent protein in plants. Methods Biochem. Anal. 47 259–284. [PubMed] [Google Scholar]
- Hood, J.L., Brooks, W.H., and Roszman, T.L. (2004). Differential compartmentalization of the calpain/calpastatin network with the endoplasmic reticulum and Golgi apparatus. J. Biol. Chem. 279 43126–43135. [DOI] [PubMed] [Google Scholar]
- Johnson, K.L., Degnan, K.A., Ross Walker, J., and Ingram, G.C. (2005). AtDEK1 is essential for specification of embryonic epidermal cell fate. Plant J. 44 114–127. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Nemeth, K., Redei, G.P., and Schell, J. (1992). T-DNA insertional mutagenesis in Arabidopsis. Plant Mol. Biol. 20 963–976. [DOI] [PubMed] [Google Scholar]
- Larsson, C., Kjellbom, P., Widell, S., and Lundborg, T. (1984). Sidedness of plant plasma mebrane vesicles purified by partitioning in aqueous two-phase systems. FEBS Lett. 171 271–276. [Google Scholar]
- Lid, S.E., Gruis, D., Jung, R., Lorentzen, J.A., Ananiev, E., Chamberlin, M., Niu, X., Meeley, R., Nichols, S., and Olsen, O.A. (2002). The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 99 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lid, S.E., Olsen, L., Nestestog, R., Aukerman, M., Brown, R.C., Lemmon, B., Mucha, M., Opsahl-Sorteberg, H.G., and Olsen, O.A. (2005). Mutation in the Arabidopisis thaliana DEK1 calpain gene perturbs endosperm and embryo development while over-expression affects organ development globally. Planta 221 339–351. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275 33712–33717. [DOI] [PubMed] [Google Scholar]
- Moldoveanu, T., Hosfield, C.M., Jia, Z., Elce, J.S., and Davies, P.L. (2001). Ca2+-induced structural changes in rat m-calpain revealed by partial proteolysis. Biochim. Biophys. Acta 1545 245–254. [DOI] [PubMed] [Google Scholar]
- Napier, R., Trueman, S., Henderson, J., Boyce, J., Hawes, C., Fricker, M., and Venis, M.A. (1995). Purification, sequencing and functions of calreticulin from maize. J. Exp. Bot. 46 1603–1613. [Google Scholar]
- Nebenfuhr, A., Ritzenthaler, C., and Robinson, D.G. (2002). Brefeldin A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol. 130 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 257–263. [DOI] [PubMed] [Google Scholar]
- Samanta, K., Kar, P., Ghosh, B., Chakraborti, T., and Chakraborti, S. (2007). Localization of m-calpain and calpastatin and studies of their association in pulmonary smooth muscle endoplasmic reticulum. Biochim. Biophys. Acta 1770 1297–1307. [DOI] [PubMed] [Google Scholar]
- Savaldi-Goldstein, S., and Chory, J. (2008). Growth coordination and the shoot epidermis. Curr. Opin. Plant Biol. 11 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296 141–145. [DOI] [PubMed] [Google Scholar]
- Sessions, A., Weigel, D., and Yanofsky, M.F. (1999). The Arabidopsis thaliana MERISTEM LAYER 1 promoter specifies epidermal expression in meristems and young primordia. Plant J. 20 259–263. [DOI] [PubMed] [Google Scholar]
- Shao, H., Chou, J., Baty, C.J., Burke, N.A., Watkins, S.C., Stolz, D.B., and Wells, A. (2006). Spatial localization of m-calpain to the plasma membrane by phosphoinositide biphosphate binding during epidermal growth factor receptor-mediated activation. Mol. Cell. Biol. 26 5481–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry, M., and Williams, L. (2002). Molecular Plant Biology, Vol. 2. (Oxford, UK: Oxford University Press).
- Tian, Q., Olsen, L., Sun, B., Lid, S.E., Brown, R.C., Lemmon, B.E., Fosnes, K., Gruis, D.F., Opsahl-Sorteberg, H.G., Otegui, M.S., and Olsen, O.A. (2007). Subcellular localization and functional domain studies of DEFECTIVE KERNEL1 in maize and Arabidopsis suggest a model for aleurone cell fate specification involving CRINKLY4 and SUPERNUMERARY ALEURONE LAYER1. Plant Cell 19 3127–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, P., Buzder-Lantos, P., Tantos, A., Farkas, A., Szilagyi, A., Banoczi, Z., Hudecz, F., and Friedrich, P. (2004). On the sequential determinants of calpain cleavage. J. Biol. Chem. 279 20775–20785. [DOI] [PubMed] [Google Scholar]
- Tompa, P., Emori, Y., Sorimachi, H., Suzuki, K., and Friedrich, P. (2001). Domain III of calpain is a ca2+-regulated phospholipid-binding domain. Biochem. Biophys. Res. Commun. 280 1333–1339. [DOI] [PubMed] [Google Scholar]
- Wang, C., Barry, J.K., Min, Z., Tordsen, G., Rao, A.G., and Olsen, O.A. (2003). The calpain domain of the maize DEK1 protein contains the conserved catalytic triad and functions as a cysteine proteinase. J. Biol. Chem. 278 34467–34474. [DOI] [PubMed] [Google Scholar]
- Wang, J.W., Schwab, R., Czech, B., Mica, E., and Weigel, D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243. [DOI] [PMC free article] [PubMed]
- Weijers, D., Franke-van Dijk, M., Vencken, R.J., Quint, A., Hooykaas, P., and Offringa, R. (2001). An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128 4289–4299. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Yu, X., Foo, E., Symons, G.M., Lopez, J., Bendehakkalu, K.T., Xiang, J., Weller, J.L., Liu, X., Reid, J.B., and Lin, C. (2007). A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiol. 145 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.