Abstract
Seed germination is antagonistically controlled by the phytohormones gibberellic acid (GA) and abscisic acid (ABA). GA promotes seed germination by enhancing the proteasome-mediated destruction of RGL2 (for RGA-LIKE2), a key DELLA factor repressing germination. By contrast, ABA blocks germination by inducing ABI5 (for ABA-INSENSITIVE5), a basic domain/leucine zipper transcription factor repressing germination. Decreased GA synthesis leads to an increase in endogenous ABA levels through a stabilized RGL2, a process that may involve XERICO, a RING-H2 zinc finger factor promoting ABA synthesis. In turn, increased endogenous ABA synthesis is necessary to elevate not only ABI5 RNA and protein levels but also, critically, those of RGL2. Increased ABI5 protein is ultimately responsible for preventing seed germination when GA levels are reduced. However, overexpression of ABI5 was not sufficient to repress germination, as ABI5 activity requires phosphorylation. The endogenous ABI5 phosphorylation and inhibition of germination could be recapitulated by the addition of a SnRK2 protein kinase to the ABI5 overexpression line. In sleepy1 mutant seeds, RGL2 overaccumulates; germination of these seeds can occur under conditions that produce low ABI5 expression. These data support the notion that ABI5 acts as the final common repressor of germination in response to changes in ABA and GA levels.
INTRODUCTION
Mature seeds are the end point of embryogenesis; during seed germination, the plant abandons its embryonic state to initiate the vegetative phase of its life cycle. Embryos within seeds are in a very low metabolic and highly resistant desiccated state. They contain food stores that will fuel seed germination (Kroj et al., 2003). When seeds are nondormant, as in this study, imbibition by water is sufficient to trigger germination.
In Arabidopsis thaliana, the mature seed consists of a protective outer layer of dead tissue, the testa, underneath which the endosperm, a single layer of cells, surrounds the embryo (Debeaujon et al., 2000). Arabidopsis seed germination chronologically involves testa rupture and concomitant endosperm rupture and embryonic axis (i.e., radicle) protrusion (see Supplemental Figure 1 online) (Muller et al., 2006). Rupture events likely involve sugar bond–modifying enzymes such as glucanases and mannanases (Leubner-Metzger, 2003; Kucera et al., 2005), but in Arabidopsis the responsible enzymes remain to be identified. Germination is usually defined as visible embryonic axis protrusion out of the testa (i.e., endosperm rupture) (Kucera et al., 2005). If conditions are optimal, these steps can be completed at 36 h after seed imbibition.
Germination is under tight control by the environment, being affected by light quality, temperature, and water potential (i.e., osmotic stress). Environmental factors eventually determine the relative levels of two phytohormones, gibberellic acid (GA) and abscisic acid (ABA), which exert antagonistic effects on seed germination. GA and ABA levels tend to be negatively correlated: conditions favorable for seed germination are associated with high GA levels and low ABA levels, whereas unfavorable conditions are associated with high ABA levels and low GA levels (Olszewski et al., 2002; Nambara and Marion-Poll, 2005). Prevalent views of how GA and ABA exert their influence to control seed germination emphasize the role of germination repressors, which must be inactivated for germination to occur.
In the mature seed, where germination is repressed, ABA levels are high and GA levels are low. Under normal germination conditions (i.e., moisture and light), GA synthesis starts shortly upon seed imbibition, which is essential for the rupture of both testa and endosperm (Debeaujon and Koornneef, 2000; Lee et al., 2002). At the same time, ABA levels drop rapidly and the role of ABA becomes facultative: after imbibition, a sudden osmotic stress or direct application of ABA (which signals osmotic stresses) efficiently prevents endosperm rupture, delays testa rupture, and confers osmotolerance to the arrested embryo (Lopez-Molina et al., 2001; Muller et al., 2006; Bethke et al., 2007).
ABA has an essential role during the late stages of seed maturation, when it may promote the accumulation of ABA-Insensitive5 (ABI5), a basic Leu-zipper transcription factor (TF). ABI5 activates the transcription of Late Embryonic and Abundant (LEA) genes, whose products confer osmotolerance to the embryo (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). Mature seeds contain high levels of ABA and high amounts of ABI5 and LEA gene products. Under normal germination conditions, ABI5 expression (mRNA and protein) mirrors that of ABA and drops rapidly, becoming undetectable within 12 to 24 h after imbibition. ABA added exogenously or produced in response to osmotic stress prevents germination and confers osmotolerance by stimulating the de novo accumulation of ABI5. However, this ABA-dependent repression of seed germination can occur only within a limited time window of ∼48 h after imbibition (Lopez-Molina et al., 2001, 2002).
Interestingly, the presence of ABI5 protein is not sufficient for its activity, as shown by experiments using constitutive transgenic ABI5 expression (Lopez-Molina et al., 2001). ABA is also necessary to stimulate ABI5 activation, which correlates with ABI5 phosphorylation (Lopez-Molina et al., 2001). Indeed, a recent report showed that two SnRK2-type kinases (SnRK2.2 and SnRK2.3) phosphorylate ABI5 peptides and redundantly mediate ABA-dependent responses during seed germination in Arabidopsis (Fujii et al., 2007).
In contrast with ABA, GA levels are initially very low in dry seeds and rise upon seed imbibition. Increased GA levels lead to the degradation of RGA-LIKE2 (RGL2), a putative GRAS family TF repressing seed germination. RGL2 has a conserved DELLA motif essential for its proteasome-mediated destruction (Lee et al., 2002). The proposed model for RGL2 destruction in response to GA is that GA binds to Arabidopsis GIBBERELLIN-INSENSITIVE DWARF1-like receptors and enhances RGL2 interaction with the F-box protein SLEEPY1 (SLY1), thus facilitating RGL2 ubiquitination and subsequent degradation (Sun and Gubler, 2004; Feng et al., 2008). There are four other DELLA factors present in Arabidopsis: REPRESSOR OF GA1-3 (RGA), GA-INSENSITIVE (GAI), RGL1, and RGL3, and all are expressed during seed germination (Tyler et al., 2004). However, RGL2 is key to repressing seed germination: in the presence of light, only rgl2 mutant seeds can germinate when GA levels are low (e.g., in mutant seeds unable to synthesize GA, such as ga1-3 mutants, or in the presence of inhibitors of GA synthesis) (Lee et al., 2002; Tyler et al., 2004). In the dark, GAI and RGA are also necessary to repress germination in addition to RGL2, since only the triple mutant gai/rga/rgl2 is capable of germination in the absence of GA synthesis (ga1-3 background) (Cao et al., 2005). Thus, although GAI and RGA also play roles in germination, RGL2 is considered to be the main DELLA factor repressing germination (Lee et al., 2002; Tyler et al., 2004). Nevertheless, a recent report has shown that the presence of high RGL2 protein amounts in after-ripened (i.e., a several-month period of dry storage) sly1 mutant seeds did not prevent their germination, indicating that RGL2-repressive activity can be lost over time (Ariizumi and Steber, 2007).
The antagonistic influence of GA and ABA on seed germination tends to be associated with inversely correlated hormone levels. Recent work in Arabidopsis seedlings has unveiled how RGA overaccumulation may stimulate ABA synthesis: RGA was found to stimulate the expression of XERICO, a gene encoding a RING-H2 zinc finger factor promoting ABA accumulation in an unknown fashion (Ko et al., 2006; Zentella et al., 2007).
Here, we propose a model for the regulation of seed germination that integrates the interaction of GA and ABA at the level of metabolism and signaling. We identify ABI5 as the final and common downstream factor active to repress seed germination and find that ABI5 accumulation and activity are controlled by changes in GA and ABA levels. In particular, we show that low GA levels result in an elevation of endogenous ABA levels and that RGL2 overaccumulation is necessary for this response to low GA levels to take place. We provide evidence that this increase in ABA levels could result from RGL2-dependent stimulation of the expression of XERICO to promote endogenous ABA accumulation. In turn, high endogenous ABA levels not only stimulate high ABI5 protein accumulation and activity but, critically, also induce higher RGL2 transcript levels. Furthermore, we show that ABI5 activation is associated with protein phosphorylation involving SnRK2-type kinases. Finally, consistent with this model, we find that the germination of after-ripened sly1 mutant seeds is indeed associated with ABI5 protein disappearance while high levels of RGL2 protein persist.
RESULTS
RGL2 and ABI5 Expression Are Stimulated by Exogenous ABA and by Inhibition of GA Synthesis
The current model for repression of seed germination is that RGL2 is the main DELLA factor repressing seed germination when GA levels are low, whereas ABI5 represses seed germination in response to ABA (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001; Lee et al., 2002; Tyler et al., 2004; Ariizumi and Steber, 2007). We wished to further define the roles of RGL2 and ABI5 in the repression of seed germination. We first studied RGL2 and ABI5 expression in wild-type Columbia (Col-0) seeds upon seed imbibition in the absence or presence of paclobutrazol (PAC), an inhibitor of GA synthesis, and ABA. PAC inhibits ent-kaurene oxidase, a key enzyme of the GA metabolic pathway (Nambara and Marion-Poll, 2005).
Under normal germination conditions in the absence of treatment (see Methods), ABI5 and RGL2 mRNA and protein levels dropped to undetectable levels prior to seed germination (i.e., 24 to 48 h after seed imbibition), consistent with previous results (Figures 1A, 1B, and 1D) (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001; Lee et al., 2002; Tyler et al., 2004; Ariizumi and Steber, 2007). This is in agreement with the proposed role of RGL2 and ABI5 as repressors of seed germination.
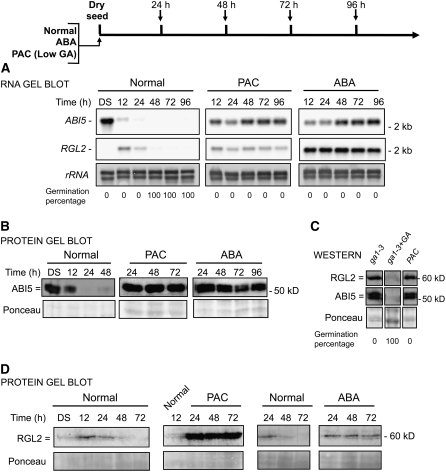
Figure 1.
Low-GA Conditions Stimulate ABI5 Expression, and ABA Stimulates RGL2 Expression.
(A) RNA gel blot analysis of a time course of ABI5 and RGL2 mRNA levels upon wild-type (Col) seed imbibition in the absence (Normal) or presence of 5 μM PAC, an inhibitor of GA synthesis, or 5 μM ABA. Two micrograms of total RNA was used per lane. DS, dry seeds; rRNA, rRNA loading control. Germination percentage at each time point is indicated.
(B) Protein gel blot analysis of ABI5 protein levels from the material in (A). Ten micrograms of total protein was used per lane. Protein extracts were stained with Ponceau S as a loading control prior to detection with antibodies against ABI5.
(C) RGL2 and ABI5 protein levels at 48 h after imbibition in ga1-3 seeds under normal conditions (ga1-3) or with added GA (ga1-3 + GA) and in PAC-treated wild-type seeds (PAC). Ponceau S staining is shown as a loading control. Germination percentage is indicated.
(D) Time course of RGL2 protein levels. Same experiment as in (B) using antibodies against RGL2 (antibodies are shown to be specific in Supplemental Figure 15 online). Ponceau S staining is shown as a loading control.
PAC treatment prevented the germination of wild-type seeds and also prevented the normal drop in ABI5 RNA and protein levels, similar to ABA treatment (Figures 1A and 1B). Also, ga1-3 seeds, which are unable to synthesize GA, accumulated high ABI5 protein levels (Figures 1C and 5C [below]). Exogenous GA was able to reverse the effect of PAC and ga1-3: ABI5 protein could not be detected in wild-type seeds treated with both PAC and GA in the medium or in ga1-3 seeds treated with GA (Figure 1C; see Supplemental Figure 2 online), indicating that the effect on ABI5 levels is specific to GA and not the result of nonspecific effects of the inhibitor or mutation. PAC treatment also led to markedly high and sustained RGL2 protein accumulation (Figure 1D). This was not accompanied by a similar increase in RGL2 mRNA levels (Figure 1A). This is consistent with the proposed model of GA-driven DELLA protein degradation (Lee et al., 2002; Olszewski et al., 2002; Tyler et al., 2004; Zentella et al., 2007).
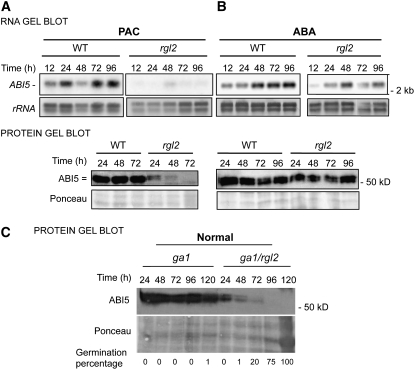
Figure 5.
ABI5 Expression Is Downregulated in rgl2 Seeds under Low-GA Conditions.
(A) RNA and protein gel blot analysis of a time course of ABI5 mRNA and protein levels upon wild-type (Col) and rgl2-13 seed imbibition in the presence of 5 μM PAC, an inhibitor of GA synthesis. Two micrograms of total RNA was used per lane. DS, dry seeds; rRNA, rRNA loading control. Ten micrograms of total protein was used per lane. Protein extracts were stained with Ponceau S as a loading control prior to detection with antibodies against ABI5. Compare with normal conditions in Figure 1.
(B) RNA and protein gel blot analysis of a time course of ABI5 mRNA and protein levels upon wild-type (Col) and rgl2-13 seed imbibition in the presence of 5 μM ABA. Two micrograms of total RNA was used per lane. Ten micrograms of total protein was used per lane. Protein extracts were stained with Ponceau S as a loading control prior to detection with antibodies against ABI5.
(C) Protein gel blot analysis of ABI5 protein levels upon imbibition of ga1-3 and ga1-3/rgl2-13 seeds under normal conditions. Each lane contained protein extracted from 100 seeds, and Ponceau S staining is shown as a loading control. Germination percentage at each time point is indicated.
ABA treatment caused an increase in ABI5 mRNA and protein accumulation, as reported previously (Figures 1A and 1B) (Lopez-Molina et al., 2001). Unexpectedly, ABA also markedly increased and sustained RGL2 mRNA levels (Figure 1A), but only during the 48-h time window previously identified for ABA-dependent ABI5 expression (see Supplemental Figure 3 online) (Lopez-Molina et al., 2001). ABA also stimulated RGL2 protein levels, but it produced a significantly more modest change than that seen in RGL2 mRNA levels (Figures 1A and 1D). This is consistent with previously reported observations that ABA has a minimal, if any, effect on DELLA protein stability; that is, GA persistently drives the ubiquitin-mediated degradation of RGL2 protein, despite the presence of ABA. High ABI5 levels in the absence of GA synthesis and high RGL2 levels in ABA-treated seeds indicated that both ABI5 and RGL2 could participate to repress seed germination under those conditions.
Inhibition of GA Synthesis Activates ABI5
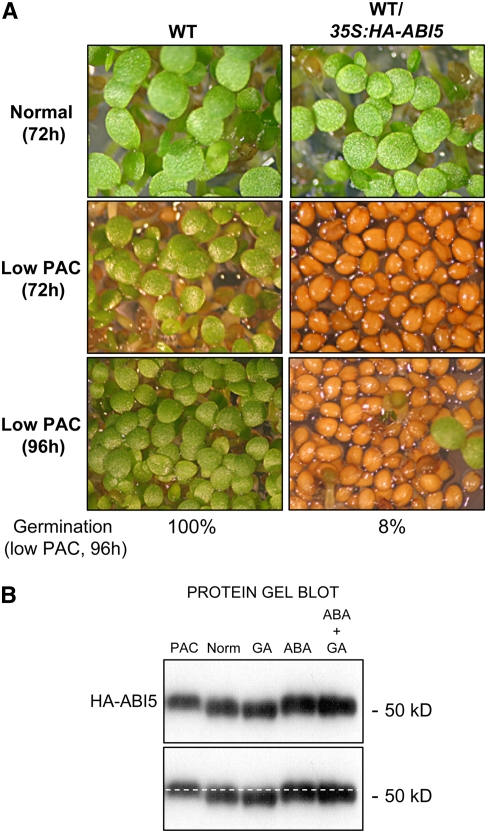
Having shown that ABI5 protein accumulates at low GA levels, we next asked whether ABI5 is active to repress seed germination when GA synthesis is prevented. Previous work has shown that the presence of ABI5 does not necessarily imply that it is actively repressing germination. Indeed, transgenic lines constitutively accumulating ABI5 protein, referred to here as WT/35S:HA-ABI5 plants, germinate normally (Lopez-Molina et al., 2001). However, their germination is hypersensitive to low ABA concentrations, which is also associated with ABI5 phosphorylation (Lopez-Molina et al., 2001). This suggested that ABA stimulates ABI5 activity via phosphorylation events. To examine ABI5 protein activity and phosphorylation, we tested the sensitivity of the ABI5 overexpression line to PAC treatment, examining germination and ABI5 protein mobility. WT/35S:HA-ABI5 seed germination was hypersensitive to low PAC concentrations (0.125 μM), and germination arrest was associated with the presence of a slower migrating form of the ABI5 protein on SDS-PAGE gels, which could be reverted to the faster migrating form by phosphatase treatment (Figures 2A and 2B). However, supplementing the ABA-containing medium with GA did not overcome the repression of seed germination and did not alter ABI5 phosphorylation (Figure 2B). Together, these data suggest that in the presence of ABA or PAC, ABI5 becomes phosphorylated and activated to repress germination.
Figure 2.
Low GA Levels Stimulate ABI5 Activity to Repress Seed Germination and Are Associated with ABI5 Protein Phosphorylation.
(A) Images show wild-type (Ler) transformed plants constitutively expressing HA-ABI5 protein (WT/35S:HA-ABI5 plants) under normal conditions or treated with low PAC concentrations (0.125 μM) that do not prevent wild-type seed germination. Photographs were taken after seed imbibition at the indicated times. Percentage of germination on PAC at 96 h is indicated.
(B) Protein gel blot analysis showing differences in HA-ABI5 protein mobility from protein extracts isolated from WT/35S:HA-ABI5 seeds. Plant material was harvested at 48 h after seed imbibition under the indicated conditions (PAC, 0.125 μM; GA, 50 μM; ABA, 0.5 μM). An antibody to HA was used to reveal HA-ABI5 protein. The gel image is duplicated at bottom with a dashed line to facilitate the comparison of protein mobility in each lane.
SnRK2-Dependent Phosphorylation Controls ABI5 Activity
To further characterize the repression of germination by phosphorylated ABI5, we examined the role of SnRK2-type kinases. Two SnRK2-type kinases (SnRK2.2 and SnRK2.3) have been shown to phosphorylate ABI5 peptides (Fujii et al., 2007); therefore, we attempted to recapitulate this activation with PKABA1, an SnRK2-type Ser/Thr kinase from barley (Hordeum vulgare). We constructed a transgenic system for monitoring ABI5 phosphorylation and activity in a PKABA1-dependent manner in the absence of PAC or ABA treatment. For this purpose, hemagglutinin (HA)-tagged PKABA1 DNA sequences were fused to an estradiol-inducible promoter (ind:HA-PKABA1) (Zuo et al., 2000). The resulting DNA construct was transformed into WT/35S:HA-ABI5 plants. As a negative control, ind:HA-PKABA1 DNA was introduced into wild-type plants. Figure 3A shows HA-PKABA1 induction at 48 h after seed imbibition in wild-type and WT/35S:HA-ABI5 seeds. Control lines transformed with the empty inducible vector displayed no additional bands in the presence of the inducer. As with ABA, induction of HA-PKABA1 protein triggered ABI5 phosphorylation, detected as a slower ABI5 protein mobility, which could be reversed upon phosphatase treatment (Figure 3B). Excess GA in the medium did not prevent HA-PKABA1–dependent phosphorylation of ABI5 (Figure 3B).
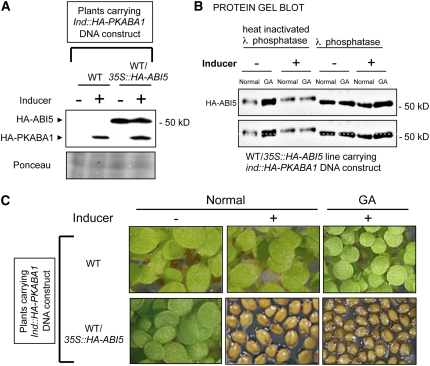
Figure 3.
Coexpression of ABI5 and PKABA1 Is Sufficient to Block Seed Germination, and PKABA1-Dependent Activation of ABI5 Is Associated with ABI5 Phosphorylation.
(A) Protein gel blot analysis of HA-ABI5 and HA-PKABA1 protein levels in wild-type (Wassilewskija) and WT/35S:HA-ABI5 plants transformed with the ind:HA-PKABA1 DNA construct. HA-PKABA1 protein levels could be detected only in the presence of the inducer (50 μM 17β-estradiol). Protein extracts were isolated from plant material at 48 h after seed imbibition under normal conditions with or without inducer. Ponceau S staining is shown as a loading control.
(B) Mobility differences of HA-ABI5 protein isolated from WT/35S:HA-ABI5 lines transformed with a ind:HA-PKABA1 DNA construct. Same protein gel blot analysis as in Figure 2B under the indicated germination conditions. λ phosphatase was inactivated by heat (65°C for 20 min) or left active (30°C for 30 min). The gel image is duplicated at bottom with a dashed line to facilitate the comparison of protein mobility in each lane.
(C) Plant material used in (A) at 120 h after seed imbibition. Excess GA (50 μM) in the medium did not promote germination when PKABA1 protein was present.
When HA-PKABA1 protein was induced in a wild-type background, no effect on seed germination could be observed (Figure 3C). By contrast, induction of HA-PKABA1 protein in WT/35S:HA-ABI5 plants inhibited seed germination, which could not be reversed by additional GA in the medium (Figure 3C; see Supplemental Figure 4A online). Taken together, the data show that ABA responses can be mimicked in vivo by coexpressing PKABA1 and ABI5. They indicate that ABI5-repressive activity may involve an SnRK2-type kinase activity phosphorylating ABI5.
abi5 Germination Is Resistant to PAC, but Unlike That of rgl2, It Occurs without Prior Visible Testa Rupture
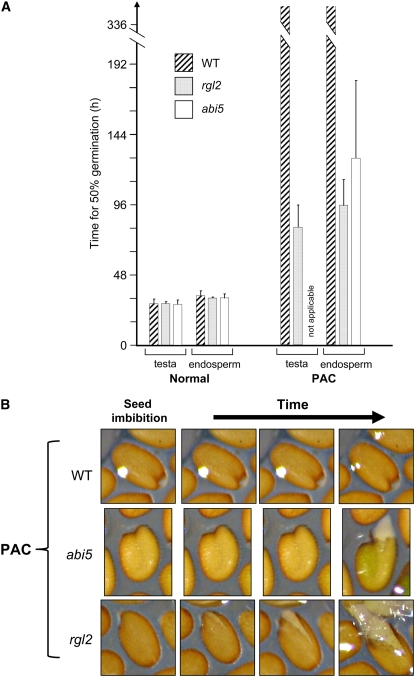
We next asked whether ABI5 is required to arrest seed germination when GA synthesis is inhibited. Normal seed germination involves the rupture of the testa prior to that of the endosperm (see Supplemental Figure 1 online) (Muller et al., 2006). Inhibition of GA synthesis efficiently blocks testa and endosperm rupture in wild-type seeds. If ABI5 is essential for this block, then abi5 mutants should show normal testa and endosperm rupture, even when GA synthesis is inhibited by PAC. To better characterize abi5 germination in the absence of GA synthesis, we monitored germination and testa rupture in PAC-treated wild-type and mutant seeds. For comparison, we included rgl2 mutant seeds, since they also germinate in the presence of PAC (Lee et al., 2002; Tyler et al., 2004). Under normal conditions, wild-type and abi5 seed germination was similar, with seed populations reaching 50% germination at 33 h after imbibition (Figure 4A). PAC-treated wild-type seeds could not germinate but abi5 seeds germinated, with 50% of the population having germinated at 128 h after imbibition (Figure 4A). Under normal conditions, rgl2 and abi5 testa rupture occurred similarly to that in the wild type (Figure 4A). However, in the presence of PAC, abi5 endosperm rupture occurred without prior visible testa rupture (Figure 4). By contrast, testa rupture in rgl2 seeds occurred prior to endosperm rupture (Figure 4). The failure of abi5 seeds to rupture their testa prior to endosperm may hamper radicle emergence and thus delay germination. This may account for the slower abi5 germination relative to rgl2 seeds in the presence of PAC (Figure 4A).
Figure 4.
abi5 Mutant Seed Germination Is Resistant to Low-GA Conditions.
(A) Wild-type, abi5, and rgl2 seeds were sown in a normal germination medium (Normal) or in the presence of PAC. Percentage of seed germination was scored over time, and the resulting germination percentage curves were used to determine the time when half of the seed population had germinated. Histograms show the times obtained for three independent seed batches (P < 0.05).
(B) Differences in early developmental events in abi5 and rgl2 mutant seeds germinating under low-GA conditions. Wild-type, rgl2, and abi5 seeds were photographed at different time points (between 24 and 144 h) upon seed imbibition in the presence of PAC (5 μM). Testa rupture is visible as a small split (see Supplemental Figure 1 online). Note that, unlike rgl2 mutants, abi5 mutants rupture their endosperm without prior visible testa rupture. The white spot on the wild-type seed is a reflection.
rgl2 Seed Germination in the Absence of GA Synthesis Is Associated with Low ABI5 Expression
The above experiments show that when GA synthesis is inhibited, RGL2 overaccumulates and represses both testa and endosperm rupture, whereas ABI5 only represses endosperm rupture. Thus, rgl2 germination in the presence of PAC could be partly due to insufficient ABI5 protein levels. Similarly, absence of testa rupture in PAC-treated abi5 seeds could result from normal RGL2 protein overaccumulation. This prompted the analysis of ABI5 expression in rgl2 mutant seeds and that of RGL2 in abi5 mutant seeds. To test this, we examined RGL2 and ABI5 expression in rgl2 and abi5 mutant seeds in the presence of PAC and ABA (Figure 5; see Supplemental Figure 5 online). In abi5 seeds in the presence of PAC and ABA, RGL2 showed a similar pattern of accumulation to that seen in the wild type under the same conditions, but it accumulated to somewhat higher levels (see Supplemental Figure 5 online). RGL2 protein overaccumulation in PAC-treated abi5 seeds is consistent with their inability to rupture the testa prior to rupturing the endosperm.
ABI5 expression in rgl2 seeds was similar to that in wild-type seeds in the absence or presence of ABA (Figure 5B; see Supplemental Figure 6 online). This is consistent with rgl2 seed germination (i.e., endosperm rupture) being inhibited by ABA in a manner similar to wild-type seeds (see Supplemental Figure 7A online). By contrast, PAC-treated rgl2 seeds had markedly lower ABI5 mRNA and protein levels (Figure 5A). Similarly, ga1/rgl2 double mutants (unable to synthesize GA) failed to accumulate high ABI5 protein levels (Figure 5C). This is consistent with the ability of rgl2 seeds to germinate in the absence of GA synthesis and strongly suggests that ABI5 is the key factor repressing germination.
Transgenic ABI5 Expression in rgl2 Seeds Restores Wild-Type–Like Sensitivity to PAC Treatment
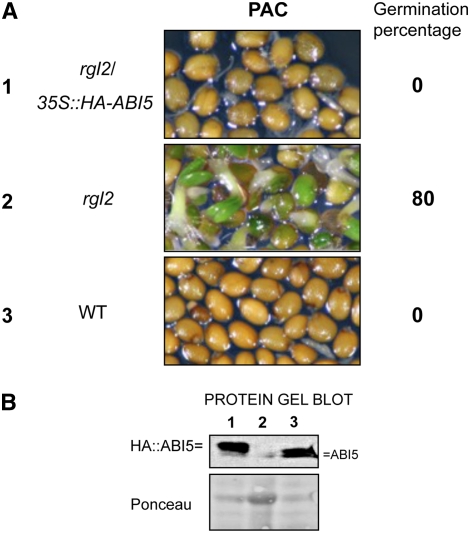
Since our data suggested that ABI5 is key to repressing germination, we asked whether increased ABI5 expression in rgl2 seeds is sufficient to restore wild-type–like germination arrest in the absence of GA synthesis. To that end, the 35S:HA-ABI5 transgene described above was used to transform rgl2 plants. Protein gel blots showed that the transgene produced levels of ABI5 that were similar to wild-type levels (Figure 6B). The resulting rgl2/35S:HA-ABI5 seeds germinated similarly to wild-type seeds under normal conditions. In the presence of PAC, rgl2 seeds could germinate but both rgl2/35S:HA-ABI5 and wild-type seeds failed to germinate (Figure 6A). Thus, transgenic increase of ABI5 protein in PAC-treated rgl2 seeds is sufficient to repress their germination (Figure 6B). This reinforces the notion that ABI5 is a key factor repressing seed germination.
Figure 6.
Transgenic ABI5 Expression in rgl2 Mutant Seeds Restores Wild-Type–Like Germination Responses under Low-GA Conditions.
(A) PAC-treated rgl2-1/35S:HA-ABI5, rgl2-1, and wild-type (Ler) plant material at 96 h after seed imbibition (PAC, 5 μM). Percentage of germination on PAC at 96 h is indicated.
(B) Protein gel blot analysis of HA-ABI5 and endogenous ABI5 protein levels in the plant material shown in (A). HA-ABI5 and endogenous ABI5 protein were detected using antibodies against ABI5. Numbers indicating treatment are as in (A). Ponceau S staining is shown as a loading control.
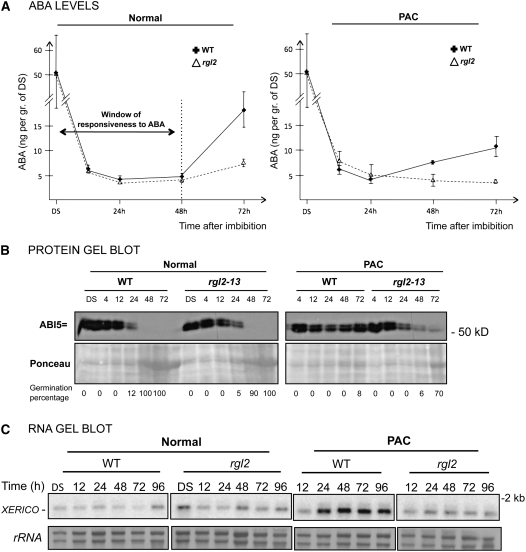
RGL2 Is Necessary to Elevate Endogenous ABA Levels in PAC-Treated Seeds
High ABI5 expression in PAC-treated wild-type seeds could result from increased endogenous ABA levels, whereas low ABI5 expression in PAC-treated rgl2 seeds could be due to low endogenous ABA. Numerous reports have shown that endogenous GA and ABA levels tend to be negatively correlated during seed germination (Olszewski et al., 2002; Nambara and Marion-Poll, 2005). Therefore, we compared endogenous ABA levels in wild-type and rgl2 seeds. ABA levels were measured at different time points upon imbibition in the absence and presence of PAC (Figure 7A). Wild-type and rgl2 dry seeds contained similar ABA amounts. In both genotypes, ABA levels dropped rapidly and massively (∼10-fold) within the first 24 h after seed imbibition in the absence or presence of PAC (Figure 7A). During this time period, ABI5 protein levels mirrored ABA levels in wild-type and rgl2 seeds in each germination condition (Figure 7B).
Figure 7.
rgl2 Mutant Seeds Have Low Endogenous ABA and XERICO mRNA Levels under Low-GA Conditions.
(A) Time course of endogenous ABA levels in wild-type (Col) and rgl2-13 seeds under normal conditions and in the presence of PAC (5 μM). Units are nanograms of ABA per gram of dry seed weighed prior to imbibition. Error bars indicate sd (n = 3).
(B) Protein gel blot analysis of the time course of ABI5 protein levels in wild-type (Col) and rgl2-13 mutant seeds under normal conditions and in the presence of PAC (5 μM). Each lane contained protein extract from 100 seeds from the material used in (A). Percentage germination at each time point is indicated. Ponceau S staining is shown as a loading control.
(C) RNA gel blot analysis of the time course of XERICO mRNA accumulation in wild-type (Col) and rgl2-13 mutant seeds under normal and low-GA conditions. Plant material was as in (A) and (B). rRNA is shown as a loading control.
Under normal conditions, ABA levels in wild-type and rgl2 seeds remained low between 24 and 48 h (Figure 7A, left). At 72 h, ABA levels increased, but this was not associated with an increase in ABI5 protein levels, most likely because ABI5 expression becomes unresponsive to ABA at this stage (Figure 7B; see Supplemental Figure 8A online) (Lopez-Molina et al., 2001, 2002; Belin and Lopez-Molina, 2008). We noticed that ABA levels increased more in wild-type plants than in rgl2 plants (Figure 7A, left). This is likely due to the particular seed batch used, in which rgl2 plants grew slightly slower than wild-type plants (see germination percentages; Figure 7B, left).
In the presence of PAC, ABA levels rose at 48 h in wild-type seeds, and this was associated with an increase in ABI5 protein levels and an absence of seed germination (Figures 7A and 7B, right). This is crucially different from what was observed with ABA levels in wild-type seeds in the absence of PAC, which remained low at 48 h as described above (Figure 7A, left). By contrast, in rgl2 seeds in the presence of PAC, ABA levels remained low at 48 h; thereafter; this was associated with ABI5 protein disappearance and the onset of seed germination (Figures 7A and 7B, right).
These observations support the notion that, in the absence of GA synthesis, rgl2 seeds fail to increase endogenous ABA levels and, as a result, fail to increase ABI5 expression, thus allowing their germination. Recently, it was reported that XERICO, encoding a RING-H2 factor, promotes ABA synthesis (Ko et al., 2006; Zentella et al., 2007). Therefore, we examined XERICO mRNA levels in wild-type and rgl2 seeds during seed germination. XERICO mRNA accumulation was similar in wild-type and rgl2 seeds in the absence or presence of ABA (Figure 7C; see Supplemental Figure 8B online). By contrast, XERICO mRNA levels increased sharply in PAC-treated wild-type seeds at 24 h, remaining elevated thereafter, but did not increase in PAC-treated rgl2 mutant seeds (Figure 7C). This observation suggests that RGL2 stimulates XERICO expression to elevate endogenous ABA levels under low-GA conditions (see Discussion).
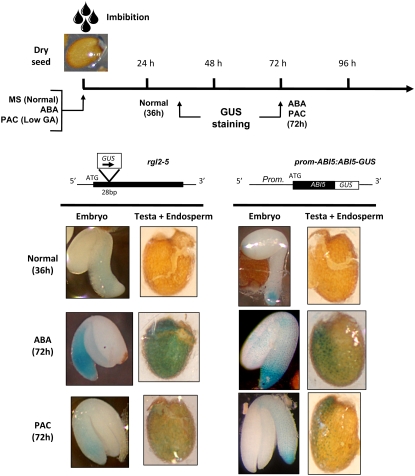
PAC Treatment Induces the Expression of RGL2 and ABI5 in Embryonic Axis and Endosperm
To better document the role of RGL2 and ABI5 in the repression of seed germination, we localized the sites of their expression. We used two previously described transgenic lines to locate RGL2 and ABI5 expression (Figure 8) (Lee et al., 2002; Lopez-Molina et al., 2002). The rgl2-5 line contains a Ds/GUS insertion located 28 bp downstream of the translation start codon, and the prom-ABI5:ABI5-GUS line carries a reporter transgene bearing ABI5 promoter sequences driving the expression of ABI5-coding sequences fused to the blue marker gene β-glucuronidase (GUS) (Lee et al., 2002; Lopez-Molina et al., 2002). Figure 8 shows GUS staining patterns in rgl2-5 and prom-ABI5:ABI5-GUS at 36 h (normal conditions) and 72 h (ABA and PAC conditions) after imbibition. Under normal conditions, the rgl2-5 GUS fusion construct gave blue staining that was only apparent in the embryonic axis, consistent with previous reports (Lee et al., 2002). Even though peak RGL2 mRNA expression was observed at 12 h after seed imbibition, very weak GUS staining was apparent at that time but became slightly stronger at 36 h (Figure 8) (Lee et al., 2002). This difference may reflect the delay to reach peak GUS enzyme accumulation. Stronger and persistent rgl2-5 staining was found both in the embryonic axis and throughout the whole endosperm under both ABA and PAC conditions but was consistently higher on ABA than on PAC, in agreement with RNA gel blot results (Figure 1A).
Figure 8.
RGL2 and ABI5 Expression Is Localized to the Embryonic Axis and Endosperm.
Top, schematic diagram describing the experimental procedure and the times used for GUS staining. Bottom, diagrams above the photographs describe each transgenic line used for GUS staining. Photographs show the blue GUS staining in rgl2-5 and prom-ABI5:ABI5-GUS lines at 36 h after seed imbibition under normal conditions or 72 h after imbibition in presence of ABA (10 μM) and PAC (10 μM). For the ABA and PAC conditions, the observed GUS staining at 72 h is typical of the staining observed between 36 and 96 h.
The prom-ABI5:ABI5-GUS line also gave a weak blue staining at 36 h after seed imbibition under normal conditions, mainly at the tip of the embryonic axis, consistent with previous reports (Figure 8) (Lopez-Molina et al., 2002). In presence of ABA, a stronger blue staining appeared throughout the entire embryo, including cotyledons and the embryonic axis, consistent with previous reports (Penfield et al., 2006). In the nonembryonic tissues, only the micropylar endosperm was significantly stained. By contrast, PAC treatment induced blue staining only in the embryonic axis, while in the nonembryonic tissues, staining remained localized to the micropylar endosperm (Figure 8).
Taken together, these observations are consistent with the notion that, in the absence of GA synthesis, RGL2 stimulates ABI5 expression in the embryonic axis and micropylar endosperm by locally increasing ABA synthesis.
In PAC-Treated Seeds, RGL2 Protein Accumulation Dominates That of GAI and RGA
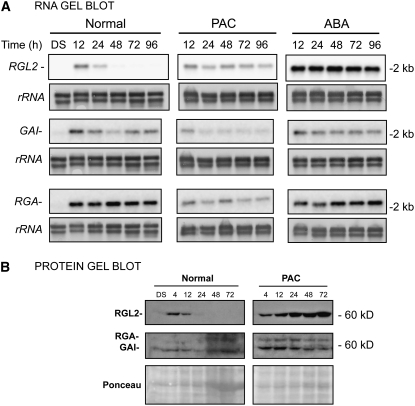
To understand why RGL2 has a prominent role during seed germination, we examined the pattern of GAI and RGA mRNA and protein expression. These factors are of particular interest, since they have been proposed to positively regulate XERICO expression in seedlings (Zentella et al., 2007). In the absence and presence of ABA, the mRNA levels of GAI and RGA increased upon seed imbibition from low levels in dry seeds to constant levels thereafter (Figure 9A). In PAC-treated seeds, GAI and RGA mRNA accumulation decreased significantly relative to normal conditions, while GAI and RGA protein remained constant or decreased over time, in contrast with the very high accumulation of RGL2 protein (Figure 9). Taken together, these results strongly suggest that RGL2 is the predominant DELLA factor accumulating during seed germination in the absence of GA synthesis. RGL1 and RGL3, not studied here, are expressed at lower transcript levels (Tyler et al., 2004).
Figure 9.
RGL2 mRNA and Protein Levels Increase More Relative to Those of RGA and GAI under Low-GA Conditions.
(A) RNA gel blot analysis of the time course of RGL2, GAI, and RGA mRNA levels in wild-type (Col) seeds under normal, PAC, and ABA conditions. Two micrograms of total RNA was used per lane. DS, dry seeds. The RNA gel blot images for RGL2 are repeated from Figure 1 for ease of comparison. rRNA is shown as a loading control.
(B) Protein gel blot analysis of the time course of RGL2, GAI, and RGA protein levels in wild-type seeds under normal and low-GA conditions (PAC). Each lane contained protein extract from 100 seeds from the material used in (A) (see Supplemental Figure 15B online for GAI and RGA antibody specificity). Ponceau S staining is shown as a loading control.
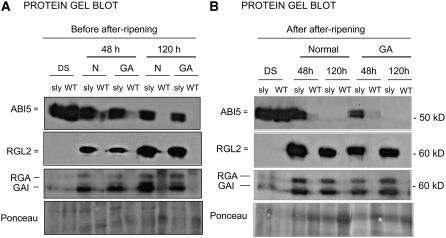
Germination of After-Ripened sly1 Seeds Is Associated with Low ABI5 Accumulation
When freshly harvested, sly1 seeds are highly dormant (i.e., they do not germinate upon seed imbibition), but a long period of after-ripening eventually breaks their dormancy (i.e., they can germinate) (Ariizumi and Steber, 2007). It has been observed that sly1 mutant seed germination takes place despite very high RGL2 and RGA protein levels (Ariizumi and Steber, 2007). This was not readily consistent with the proposed role for RGL2 as the main DELLA factor repressing seed germination. It led the authors to postulate that RGL2 lost its repressive activity during after-ripening. To examine the mechanism by which after-ripening breaks dormancy in sly1 seeds, we compared ABI5 expression between freshly harvested and after-ripened sly1 mutant seeds. As reported previously, freshly harvested sly1 seeds did not germinate and maintained high amounts of RGL2 protein (Figure 10A) (Ariizumi and Steber, 2007). Expectedly, they also accumulated high amounts of ABI5 protein (Figure 10A). After 1 year of after-ripening, sly1 mutant seeds could germinate under normal conditions while accumulating high RGL2, RGA, and GAI protein levels (Figure 10B). However, we observed a drastic downregulation of ABI5 protein levels upon imbibition (Figure 10B). These results are consistent with ABI5 being the final downstream factor repressing germination, even in situations of multiple and massive DELLA protein accumulation.
Figure 10.
ABI5 Is Downregulated in Germinating After-Ripened sly1 Seeds.
(A) Protein gel blot analysis of ABI5, RGL2, GAI, and RGA protein levels in sly1-10 seeds freshly harvested (before after-ripening) in the absence (normal [N]) or presence of 50 μM GA. Plant material was harvested at 48 and 120 h after seed imbibition. DS, dry seeds. Ponceau S staining is shown as a loading control.
(B) Same as in (A) using sly1-10 seeds after-ripened at room temperature for 1 year.
DISCUSSION
In nondormant seeds, moisture is sufficient to trigger germination. However, the environmental conditions encountered by the seed will determine the endogenous levels of GA and ABA, which in turn will define the pace of germination. The possible outcomes for the seed take place between two extreme states: germination is prevented (low GA, high ABA) or unhampered (high GA, low ABA).
The salient finding of this work concerns the roles of ABI5 and RGL2 in germination. It shows that ABI5, whose expression and product activity respond to changes in ABA and GA levels, is the main downstream player repressing the germination of nondormant seeds. ABI5 expression is persistently high in wild-type seeds that are unable to germinate (i.e., in conditions of high ABA or low GA). Conversely, wild-type seed germination is always associated with the disappearance of ABI5 protein.
Concerning RGL2, our findings bring a new perspective on its role during germination by showing that (1) RGL2 mRNA accumulation is strongly stimulated by ABA, (2) RGL2 protein is necessary to elevate endogenous ABA and ABI5 expression levels, specifically when GA levels are low, and (3) RGL2 is necessary to repress testa rupture.
Dry seeds contain endogenous ABA, which played an essential role during embryogenesis to establish seed osmotolerance and dormancy. Dry seeds also contain high levels of ABI5 mRNA and protein, essential to promote the expression of osmotolerance genes, such as Arabidopsis Em1 and Em6 (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). Normal germination conditions trigger a decrease in endogenous ABA levels upon seed imbibition. This leads to a rapid decay in ABI5 mRNA and protein amounts to undetectable levels within 30 h after imbibition (Figures 1A and 1B). Simultaneously, RGL2 expression, very low but detectable in dry seeds, is induced while bioactive GA levels start to rise (Lee et al., 2002; Ogawa et al., 2003). RGL2 mRNA and protein reach peak levels ∼12 h after imbibition but rapidly diminish thereafter. Since exogenous ABA can stimulate the expression of both RGL2 and ABI5 during seed germination (Figure 1), the transient early induction of RGL2 expression could result from the remaining (but fast-disappearing) pool of endogenous ABA. At the protein level, RGL2 accumulation is severely limited, given that its degradation increases as a result of newly synthesized GA. By contrast, no transient stimulation of ABI5 expression is detected, most likely because the pools of mRNA and protein coming from the dry seed mask it. It should be stressed that changes in GA levels do not significantly affect ABI5 protein degradation (Figure 1B).
In summary, the dominant trend under normal conditions (i.e., moisture and light) is the downregulation of RGL2 and ABI5 expression caused by a combination of diminishing ABA levels and increasing GA levels. Low ABA levels also ensure that the disappearing pool of ABI5 protein remains nonphosphorylated and therefore inactive (discussed further below). As a result, normal conditions trigger seed germination, as they lead to a complete shutoff of ABI5 expression and product activity.
Germination under Low-GA Conditions
We altered normal seed germination by repressing GA synthesis upon imbibition using PAC, an inhibitor of GA synthesis. This led to increased endogenous ABA levels between 24 and 48 h (i.e., within the time window when ABA can still stimulate ABI5 expression) (Figure 7A). This was expected, since ABA and GA levels tend to be negatively correlated: low GA levels are associated with higher ABA levels, whereas high GA levels are associated with lower ABA levels (Olszewski et al., 2002; Nambara and Marion-Poll, 2005). For example, ga1-3 mutant seeds, unable to synthesize GA, have higher endogenous ABA levels (Oh et al., 2007). Similarly, aba2-2 mutant seeds, unable to synthesize ABA, have higher endogenous GA levels (Seo et al., 2006). Thus, each hormone appears to somehow regulate the expression of the biosynthetic genes of the other.
A remarkable observation is that inhibition of GA synthesis prevents seed germination due to high ABI5 expression and protein activity, much as in ABA-treated seeds. Thus, higher endogenous ABA levels associated with low-GA conditions may stimulate ABI5 expression. This notion is clearly apparent when studying the germination of rgl2 mutant seeds in the absence of GA synthesis: they fail to increase endogenous ABA levels, and ABI5 expression remains low (Figures 5A and 7; see Supplemental Figure 8A online). High ABI5 expression was also found in ga1 mutant seeds, which are genetically unable to synthesize GA, but not in ga1/rgl2 seeds (Figures 1C and 5C). Further compatible with this view, we also found that inhibition of endogenous ABA synthesis (using norflurazon) in ga1-3 seeds or in PAC-treated wild-type seeds promotes their germination and is associated with a drop in ABI5 protein levels (see Supplemental Figures 9A and 9B online). This is consistent with the observation from Leon-Kloosterziel et al. (1996) that ga1/aba1 mutant seeds are able to germinate.
It remains to be explained why rgl2 seeds fail to elevate endogenous ABA levels. This may involve the expression of XERICO, which encodes a RING-H2 factor that promotes an increase of ABA levels in an unknown manner (Ko et al., 2006). In seedlings, XERICO promotes higher ABA levels when GA levels are low and is positively regulated by RGA and GAI, two essential DELLA factors mediating GA responses in vegetative tissues (Zentella et al., 2007). Chromatin immunoprecipitation assays showed that RGA is associated with XERICO's promoter sequences (Zentella et al., 2007). Since DELLA factors have no known DNA binding domain or activity, RGA and GAI could stimulate XERICO transcription by modulating the activity of DNA binding transcription factors (Zentella et al., 2007). In this work, we observed that PAC-treated rgl2 seeds also failed to express high XERICO mRNA levels as early as 24 h after seed imbibition (i.e., 24 h prior to the rise in ABA levels observed in wild-type seeds). Thus, RGL2 may stimulate XERICO expression to promote ABA synthesis during seed germination in a manner comparable to the effect of RGA and GAI in seedlings. Consistent with this view, we found in preliminary experiments that xerico mutant seeds are insensitive to PAC, although less so than rgl2 mutant seeds (45% of germination at 144 h after imbibition versus 0% for the wild type; see Supplemental Figure 10 online). Moreover, PAC-treated xerico seeds failed to maintain ABI5 protein over time, with levels starting to decay at 96 h after imbibition (see Supplemental Figure 10 online).
RGL2 stands out among other DELLA factors during germination because low ABI5 expression is a unique property of rgl2 mutant seeds. Indeed, gai, rga, rgl1, and rgl3 seeds have elevated, wild-type–like ABI5 expression and do not germinate when treated with PAC, in contrast with rgl2 seeds (data not shown) (Tyler et al., 2004). We think this is because RGL2 is achieving the highest relative protein overaccumulation, dominating that of the other DELLAs (Figure 9). This in turn may reflect the way RGL2 mRNA levels are regulated; they are (1) strongly stimulated by ABA and (2) only so during the same time window previously identified for ABA-dependent ABI5 expression (Lopez-Molina et al., 2001) (see Supplemental Figure 3 online). We propose that the elevation of endogenous ABA levels associated with low GA levels triggers a positive autofeedback loop sustaining the RGL2 mRNA levels, as observed in Figure 1 (see model in Figure 11). This is much unlike RGA and GAI: not only are their mRNA levels unresponsive to ABA, but they also decrease when GA levels are low (Figure 9) (the role of RGL1 and RGL3 during germination is minor; U. Piskurewicz and L. Lopez-Molina, unpublished data). Consistent with this hypothesis, we found that aba1 mutant seeds, unable to synthesize ABA, have markedly lower RGL2 mRNA and protein levels when treated with PAC (see Supplemental Figures 11A and 11B online). They also have markedly lower ABI5 mRNA and protein accumulation (see Supplemental Figures 11A and 11B online), consistent with their ability to germinate (Leon-Kloosterziel et al., 1996).
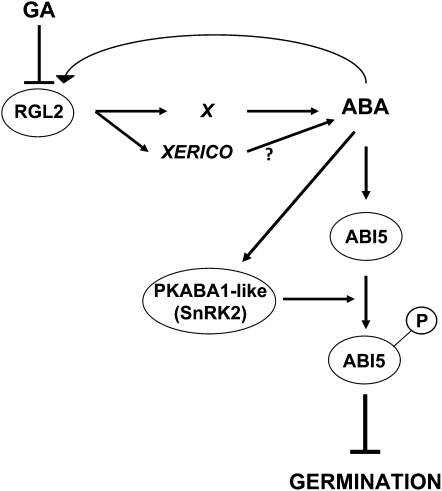
Figure 11.
A Model for the Control of Seed Germination under Low-GA Conditions.
Environmental conditions influence the levels of bioactive GA accumulating upon seed imbibition. When GA levels are low, RGL2 protein overaccumulates, which stimulates endogenous ABA synthesis. This probably involves XERICO and as yet unknown factors (X). In turn, an increase in ABA synthesis sustains ABI5 mRNA and protein expression and activity (through phosphorylation by SnRK2-type kinases) but also RGL2 mRNA levels.
RGL2's importance relative to other DELLAs is only observed when light is present in germination assays. In the dark, germination in the absence of GA synthesis (ga1 background) is observed only with rgl2/gai/rga triple mutants and not with single rgl2 mutants (Cao et al., 2005). Light is perceived by the phytochrome photoreceptors, which in turn stimulate seed germination by promoting the increase in GA levels and decrease in ABA levels (Oh et al., 2006, 2007). This involves in part the destruction of PHYTOCHROME-INTERACTING FACTOR3-LIKE5 (PIL5), a phytochrome-interacting basic helix-loop-helix TF repressing seed germination. In darkness, PIL5 accumulates and associates with GAI and RGA promoter sequences, which stimulate the accumulation of their respective mRNAs. PIL5 promotes higher ABA and lower GA levels in seeds (Oh et al., 2006, 2007) in part through its activation of SOMNUS, a recently discovered factor encoding a CCCH-type ZINC finger protein (Kim et al., 2008) and acting through yet unspecified mechanisms.
In darkness, RGA and GAI may act in parallel with ABI5 as bona fide repressors of seed germination. Alternatively, GAI and RGA could directly stimulate ABI5 gene transcription in the dark or indirectly stimulate ABI5 accumulation and activity by promoting ABA synthesis (e.g., by promoting XERICO expression as in seedlings). Clearly, other unknown dark-specific repressors of seed germination may also be involved.
In conclusion, two features may explain RGL2's important role among other DELLA genes during seed germination: (1) the positive control of its mRNA levels by ABA during the time window of ABA responsiveness; and (2) the localization of its expression. Indeed, GUS staining shows that RGL2 is predominantly expressed in the embryonic axis and endosperm cells. In those cells, the elevation of endogenous ABA levels ensures high ABI5 expression and protein activity under low-GA conditions, thus preventing endosperm rupture and embryonic axis elongation.
Seed Germination in the Presence of ABA
Exposing seeds to ABA has a mild stimulatory effect on RGL2 protein levels despite a marked increase in RGL2 mRNA levels. This is likely due to excess endogenous GA levels that limit RGL2 protein accumulation. Indeed, additional GA in the medium does not lower RGL2 protein levels relative to normal conditions (see Supplemental Figure 12 online). Unsurprisingly, XERICO mRNA levels are similar in the absence or presence of ABA, likely because of low RGL2 protein accumulation. This ensures that ABI5 expression and activity are mainly driven by exogenously provided ABA. It is likely that the same holds true under osmotic stress conditions.
It remains an open question how ABA stimulates RGL2 transcription. Among the known TFs whose expression is stimulated by ABA, none seemed to be required to induce RGL2 expression under ABA conditions (tested on abi5, abi3, and abi4 seeds; data not shown). Similar questions remain open for other essential genes controlling seed germination, such as ABI3 (Lopez-Molina et al., 2002).
Previous work showed that ABA-dependent stimulation of ABI5 activity to repress seed germination is associated with protein phosphorylation (Lopez-Molina et al., 2001). Therefore, it is not surprising that low GA levels stimulated ABI5 phosphorylation and activity, since they elevate endogenous ABA levels (Figures 2 and 7A).
The SnRK2 family of plant-specific kinases can mediate ABA-dependent responses, as originally demonstrated with PKABA1 in barley aleurone cells (Anderberg and Walker-Simmons, 1992; Gomez-Cadenas et al., 1999). ABA induces PKABA1 expression to block the GA-dependent induction of α-amylase expression (Gomez-Cadenas et al., 2001; Shen et al., 2001; Zentella et al., 2002). Together with other SnRK2 kinases, PKABA1 was later shown to phosphorylate basic domain/leucine zipper (bZIP) factors homologous to ABI5 in vitro (Johnson et al., 2002; Kobayashi et al., 2005; Chae et al., 2007). The importance of SnRK2 kinases was recently confirmed by genetic experiments in Arabidopsis (Fujii et al., 2007). Those authors also showed SnRK2-dependent phosphorylation events in ABI5 peptides.
The role of SnRK2-type kinases in ABI5 protein phosphorylation and activation was investigated. We showed that we could mimic all of the germination responses observed in response to ABA by inducing the PKABA1 kinase in plants constitutively accumulating ABI5 in the absence of ABA treatment (Figure 3). Thus, a model accounting for ABA-dependent activation of ABI5 is that ABA induces the expression of SnRK2-type kinases, such as SnRK2.2 and SnRK2.3 (Fujii et al., 2007), which phosphorylate and activate ABI5 (see model in Figure 11). Consistent with the hypothesis that ABI5 activation is essential to arrest germination in the absence of GA synthesis, we found that snrk2-2/snrk2-3 double mutant seeds are insensitive to PAC (see Supplemental Figure 13 online).
The potential mechanisms of ABI5 inactivation could be limited to its protein destruction. We previously described an ABA-dependent protein, ABI Five Binding Protein, facilitating ABI5 degradation (Lopez-Molina et al., 2003). However, other mechanisms can be envisaged, such as ABI5 dephosphorylation. Recessive mutations in abi1 and abi2 were shown to be hypersensitive to ABA, suggesting that they are negative regulators of ABA signaling (Gosti et al., 1999; Merlot et al., 2001). It remains to be investigated whether they could directly dephosphorylate ABI5. Phosphatases acting on ABI5 may be recruited by other signaling pathways, such as that of ethylene, which inhibits ABA signaling in seeds (Beaudoin et al., 2000). Mutants defective in ethylene signaling were recovered in genetic screens suppressing the ABA-resistant phenotype of a dominant allele of ABI1, encoding a PP2C phosphatase (Beaudoin et al., 2000).
ABI5 and similar bZIP factors likely activate gene transcription by binding to ABA-responsive elements (ABREs) (Carles et al., 2002; Kang et al., 2002). Comparison of the ga1-3 seed transcriptome in the absence or presence of GA has shown that genes downregulated by GA have a statistically significant enrichment in ABRE motifs in their promoter sequences. This is consistent with ABI5 (and other bZIP factors) stimulating, when GA levels are low, the transcription of ABRE-containing GA-sensitive genes. In these experiments, endogenous ABA levels were not affected by the presence or absence of GA, perhaps because they were only measured at early time points (Ogawa et al., 2003).
The molecular mechanisms underlying testa and endosperm rupture are not fully understood. Several reports on various plant species have suggested that enzymes modifying sugar bonds (e.g., mannanases and cellulases) and other digestive enzymes (e.g., chitinases) orchestrate rupture processes (Leubner-Metzger, 2003; Kucera et al., 2005). However, in Arabidopsis, neither the identity of the enzymes nor their spatial and temporal activities have been firmly identified.
Analysis of rgl2 and abi5 mutants showed that RGL2 is necessary to repress testa rupture in the absence of GA synthesis, while ABI5 repressed endosperm rupture in the absence of GA synthesis as well as on ABA (Figure 4). Therefore, it is not surprising that ABA achieves a poor repression of testa rupture, since it leads to modest RGL2 protein levels (Figure 1D). However, under very high ABA conditions (25 μM ABA), testa rupture occurred faster in rgl2 mutant seeds relative to wild-type seeds (see Supplemental Figure 7B online). This suggests that sufficient RGL2 protein can accumulate in response to ABA to delay testa rupture.
SLY1 is an F-box protein necessary to promote DELLA protein degradation in Arabidopsis (McGinnis et al., 2003; Dill et al., 2004). sly1 mutant seeds are highly dormant and accumulate large amounts of DELLA factors, including RGL2. Surprisingly, sly1 mutant seeds eventually germinate after a long period of after-ripening, despite maintaining markedly high RGL2, RGA, and GAI protein levels (Figure 10) (Ariizumi and Steber, 2007). This observation directly contradicted a recently proposed model describing RGL2 and other DELLA factors as promoters of seed dormancy and repressors of seed germination (Penfield et al., 2006). Therefore, it was proposed that RGL2 loses its repressive activity during the after-ripening of sly1 mutant seeds (Ariizumi and Steber, 2007).
We could correlate strong ABI5 protein accumulation with the absence of germination in freshly harvested sly1 seeds as well as ABI5 protein disappearance in germinating after-ripened sly1 seeds. This observation strongly supports the importance of downregulating ABI5 expression to allow seed germination. Conceivably, the loss of high ABI5 expression in nondormant sly1 seeds could result from the loss of function of any of the signaling components described here (RGL2, XERICO, and ABI5 machinery) or from the failure to sustain high endogenous ABA levels, perhaps as a consequence of damaging oxidation events occurring during seed after-ripening (Oracz et al., 2007). We tested the latter hypothesis by treating dormant sly1 seeds with norflurazon, an inhibitor of ABA synthesis. Indeed, norflurazon triggered the germination of sly1 dormant seeds, and this was associated with a marked drop in ABI5 protein levels (see Supplemental Figure 14 online).
METHODS
Statistics
Average values were obtained from a minimum of three independent seed batches. Within a seed batch, measurements were performed at least twice, giving consistent results. Errors bars in histograms correspond to sd values. We used Student's t test (two-tailed assuming unequal variance) to compare average mean values in order to determine if their differences were statistically significant (P < 0.05).
Plant Material
The Arabidopsis thaliana mutant rgl2-13 in the Col-0 background was isolated by Tyler et al. (2004) and obtained from T.P. Sun. xerico (Col-0) and ga1-3/rgl2-13 seeds were also obtained from T.P. Sun. The abi5-3 (Col-0 ecotype) mutant was obtained from R.R. Finkelstein and first described by Finkelstein and Lynch (2000). The rgl2-1 and rgl2-5 mutants (Landsberg erecta [Ler] ecotype; obtained from J. Peng) were described by Lee et al. (2002), and the 35S:HA-ABI5 line (Ler ecotype) was described by Lopez-Molina et al. (2001). The aba1-1 allele was obtained from M. Koornneef, sly1-10 from C. Steber, and snrk2-2/snrk2-3 from J.K. Zhu. The rgl2-5 line contains a Ds/GUS insertion located 28 bp downstream the translation start codon, and the prom-ABI5:ABI5-GUS line carries ABI5 promoter sequences driving the expression of ABI5 coding sequences fused to GUS (Lee et al., 2002; Lopez-Molina et al., 2002).
Germination Assays
Throughout this study, we used nondormant seeds in the absence of seed stratification. We refer to seeds sown under normal conditions when seeds were imbibed in a standard germination medium (see below) and provided with light. ABA conditions and low-GA conditions, respectively, mean that ABA (5 μM) and PAC (5 μM), an inhibitor of GA synthesis, were added to the medium.
All seed batches compared in this study were harvested on the same day from plants grown side by side (i.e., identical environmental conditions). Dry siliques were obtained ∼8 weeks after planting and left for a further 4 weeks at room temperature prior to seed harvesting. Seeds were then permanently stored at 4°C. Seeds obtained in this manner lacked dormancy. A minimum of three independently grown seed batches were used to measure percentage of testa and endosperm rupture events.
For germination assays, seeds were surface-sterilized as described (Lopez-Molina and Chua, 2000) and sown on plates with Murashige and Skoog medium containing 0.8% (w/v) Bacto-Agar (Applichem). Medium was supplemented with GA3 (G7645; Sigma-Aldrich), ABA (A1049; Sigma-Aldrich), PAC (46046; Riedel-de Haen), or norflurazon (PS1044; Sigma-Aldrich) according to the germination condition examined. Plates were incubated in a climate-controlled room (20 to 25°C, 16 h of light per day, light intensity of 80 μE·m−2·s−1, humidity of 70%). Between 100 and 300 seeds were examined with a Stemi 2000 (Zeiss) stereomicroscope and photographed with a high-resolution digital camera (Canon Power G6; 7.1 megapixels) at different times of seed imbibition. Photographs were enlarged electronically for measurement of testa and endosperm rupture events.
Plasmid Constructs and Plant Transformation
DNA manipulations were performed according to standard methods (Sambrook et al., 1989). The 35S:HA-ABI5 binary vector was described previously (Zuo et al., 2000; Lopez-Molina et al., 2001). A barley (Hordeum vulgare) PKABA1 cDNA was first described by Gomez-Cadenas et al. (1999) and obtained from R. Johnson. The PKABA1 open reading frame (ORF) DNA sequence was amplified with 5′-CGACTCGAGATGTATCCATATGACGTGCCGGACTACGCCTCCCTCATGGATCGGTACGAGGTGGTG-3′ and 5′-CGAACTAGTTCACAACGGGCACACGAAGTC-3′. The first primer contains an XhoI site and the HA sequence (MYPYDVPDYASL), while the second contains a SpeI site. Both restriction sites were used for cloning into pER8 (Zuo et al., 2000). The resulting ind:HA-PKABA1 binary vector was transformed into the previously described WT/35S:HA-ABI5 line (Zuo et al., 2000; Lopez-Molina et al., 2001). Transgenic Arabidopsis lines were generated using the Agrobacterium tumefaciens vacuum-infiltration method (Bechtold and Pelletier, 1998). Seeds (T1) from infiltrated plants were plated in selection medium as described (Zuo et al., 2000; Lopez-Molina et al., 2001).
RNA Extraction and RNA Gel Blots
Total RNA extraction was as described by Vicient and Delseny (1999). RNA concentrations were measured using a spectrophotometer (GeneQuant pro; Biochrom). RNA gel blot hybridizations were by standard procedures; RNA immobilized on membranes was stained with methylene blue and used as a loading control (Sambrook et al., 1989). RGL2 and GAI full-length ORF DNA probes were amplified from cDNA using the primers described below for RGL2 and GAI. For ABI5 and RGA, full-length ORF DNA probes were amplified from cDNA with 5′-ATGGTAACTAGAGAAACGAAGTTG-3′/5′-TTAGAGTGGACAACTCGGGTTCCTC-3′ and 5′-ATGAAGAGAGATCATCACCAATTCC-3′/5′-TCAGTACGCCGCCGTCGAGAGTTTCC-3′. Probes were radiolabeled according to standard procedures, and membranes were exposed to phosphor imager screens for quantification of signal intensity.
Antibody Production and Protein Gel Blot Analysis
DNA fragments containing RGL2 ORF (amplified with 5′-ATCGTCGACGAATGAAGAGAGGATACGGAGAAACATG-3′ and 5′-ATGTCGACTCAGGCGAGTTTCCACGCCGAGGTTG-3′) and GAI ORF (amplified with 5′-GCTGGATCCATGAAGAGAGATCATCATCATCATCATCATCAAG-3′ and 5′-GCTGTCGACCTAATTGGTGGAGAGTTTCCAAGCCGAGG-3′) were cloned into the SalI and BamHI/SalI sites of pET28a (Novagen), respectively, and the recombinant proteins were induced and purified using a commercial kit according to the manufacturer's instructions (His-Trap; Amersham). Polyclonal anti-RGL2 and anti-GAI antisera (also recognizing RGA) were obtained from rabbits immunized with His-tagged full-length RGL2 and GAI. Antibodies were further affinity-purified using His-RGL2 and His-GAI immobilized on nitrocellulose filters (Sambrook et al., 1989). Plant protein extracts were resolved under reducing conditions using 10% SDS/polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride membranes (Immobilon-P; Millipore), which were incubated with primary affinity-purified ABI5 antibody (diluted 1:3000; 60 μg/mL) for 4 h at room temperature, with affinity-purified RGL2 antibody (diluted 1:1500; 60 μg/mL) for 16 h in 4°C, or with affinity-purified GAI antibody (diluted 1:3000; 60 μg/mL) for 1 h at room temperature. After a first wash of membranes (5% milk in Tris-buffered saline [TBS] for anti-RGL2, TBS for anti-GAI, and TBS + 0.05% Tween for anti-ABI5), the secondary antibody, peroxidase-conjugated anti-rabbit antibody (Amersham Pharmacia; diluted 1:10,000), was added and membranes were incubated for 1 h at room temperature in TBS supplemented with 5% nonfat dry milk. After incubation, membranes were washed twice (10 min each) with TBS containing 0.05% Tween 20 for anti-ABI5 and TBS for anti-RGL2 and anti-GAI. After the final wash, immune complexes were detected on x-ray film (Fuji medical x-ray film) in its linear range using the ECL kit according to the manufacturer's specifications (Amersham Pharmacia).
Analysis of Endogenous ABA
ABA was extracted with 80% methanol containing 1% acetic acid, and [2H6]ABA (Icon Isotopes) was added to the extract as an internal standard. ABA-containing fractions were purified as described previously (Priest et al., 2006) and then subjected to liquid chromatography–selected reaction monitoring using a system consisting of a quadrupole/time-of-flight tandem mass spectrometer (Q-Tof Premier; Waters) and an Acquity Ultra Performance liquid chromatograph (Waters) equipped with a reverse-phase column (Acquity UPLC BEH-C18; Waters).
Phosphatase Experiments
The methods used were as described previously (Lopez-Molina et al., 2001).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RGL2 (At3g03450), ABI5 (At2g36270), RGA (At2g01570), GAI (At1g14920), PKABA1 (AB058923), and XERICO (At2g04240).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Early Developmental Steps upon Seed Imbibition.
Supplemental Figure 2. GA Treatment Prevents High ABI5 Protein Accumulation in PAC-Treated Seeds.
Supplemental Figure 3. ABA Regulates RGL2 Expression within the Same Developmental Window Previously Characterized for ABI5.
Supplemental Figure 4. Coexpression of ABI5 and PKABA1 Is Sufficient to Block Seed Germination.
Supplemental Figure 5. RGL2 Expression in abi5 Mutant Is Similar to That in the Wild Type under All Germination Conditions Examined.
Supplemental Figure 6. ABI5 Expression in rgl2 Mutant Is Similar to That in the Wild Type under Normal Germination Conditions.
Supplemental Figure 7. Inhibition of Endosperm Rupture by ABA Is Similar between rgl2 and the Wild Type; Testa Rupture of rgl2 Seed Is Moderately Resistant to High ABA Concentrations.
Supplemental Figure 8. ABI5 and XERICO mRNA Expression Levels in Samples from Figure 7.
Supplemental Figure 9. Inhibition of ABA Synthesis in ga1 or in PAC-Treated Wild-Type Seeds Triggers Seed Germination and Correlates with Decreased ABI5 Protein Levels.
Supplemental Figure 10. Germination of xerico Mutant Seeds under Low-GA Conditions Coincides with ABI5 Protein Disappearance.
Supplemental Figure 11. Seeds of aba1-1 Mutant Germinate under Low-GA Conditions, and This Coincides with RGL2 and ABI5 Protein Disappearance.
Supplemental Figure 12. Exogenous GA Does Not Affect RGL2 mRNA and Protein Levels Relative to Normal Conditions.
Supplemental Figure 13. snrk2.2/snrk2.3 Double Mutant Seed Germination Is Resistant to Low-GA Conditions.
Supplemental Figure 14. Dormant sly1-10 Mutant Germination in the Presence of Norflurazon, an Inhibitor of ABA Synthesis, Coincides with ABI5 Protein Disappearance but Not with That of RGL2, GAI, or RGA.
Supplemental Figure 15. Antibody Specificity.
Supplementary Material
Acknowledgments
We are especially grateful to Pierre Vassalli for help and numerous suggestions in the writing of the manuscript. We are greatly indebted to the following colleagues for their generous gifts: Russell Johnson for pKABA1 cDNA, Tai-Ping Sun for rgl2-13, ga1-3/rgl2-13, and xerico mutants, Ruth Finkelstein for abi5-3 mutant, Jingrong Peng for rgl2-1 and rgl2-5 mutants, Marteen Koornneef for aba1-1 and ga1-3 mutants, Camille Steber for sly1-10 mutant, and Jian-Kang Zhu for the snrk2-2/snrk2-3 double mutant. We thank Christophe Belin and Michel Goldschmidt-Clermont for critical comments on the manuscript. We thank Richard Chappuis and Christian Megies for technical help in generating transgenic lines. N.K. received support from the Naito Foundation. This work was supported by grants from the Swiss National Science Foundation and by the State of Geneva.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Luis Lopez-Molina (luis.lopezmolina@unige.ch).
Online version contains Web-only data.
References
- Anderberg, R.J., and Walker-Simmons, M.K. (1992). Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 89 10183–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi, T., and Steber, C.M. (2007). Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Belin, C., and Lopez-Molina, L. (2008). Arabidopsis seed germination responses to osmotic stress involve the chromatin modifier PICKLE. Plant Signaling and Behavior 3 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, P.C., Libourel, I.G., Aoyama, N., Chung, Y.Y., Still, D.W., and Jones, R.L. (2007). The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 143 1173–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, D., Hussain, A., Cheng, H., and Peng, J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113. [DOI] [PubMed] [Google Scholar]
- Carles, C., Bies-Etheve, N., Aspart, L., Leon-Kloosterziel, K.M., Koornneef, M., Echeverria, M., and Delseny, M. (2002). Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 30 373–383. [DOI] [PubMed] [Google Scholar]
- Chae, M.J., Lee, J.S., Nam, M.H., Cho, K., Hong, J.Y., Yi, S.A., Suh, S.C., and Yoon, I.S. (2007). A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol. Biol. 63 151–169. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I., and Koornneef, M. (2000). Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Leon-Kloosterziel, K.M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., Thomas, S.G., Hu, J., Steber, C.M., and Sun, T.P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H., Verslues, P.E., and Zhu, J.K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Verhey, S.D., Holappa, L.D., Shen, Q., Ho, T.H., and Walker-Simmons, M.K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA 96 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cadenas, A., Zentella, R., Walker-Simmons, M.K., and Ho, T.H. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13 667–679. [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.R., Wagner, R.L., Verhey, S.D., and Walker-Simmons, M.K. (2002). The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 130 837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.Y., Choi, H.I., Im, M.Y., and Kim, S.Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.H., Yamaguchi, S., Lim, S., Oh, E., Park, J., Hanada, A., Kamiya, Y., and Choi, G. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277. [DOI] [PMC free article] [PubMed]
- Ko, J.H., Yang, S.H., and Han, K.H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47 343–355. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y., Murata, M., Minami, H., Yamamoto, S., Kagaya, Y., Hobo, T., Yamamoto, A., and Hattori, T. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44 939–949. [DOI] [PubMed] [Google Scholar]
- Kroj, T., Savino, G., Valon, C., Giraudat, J., and Parcy, F. (2003). Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073. [DOI] [PubMed] [Google Scholar]
- Kucera, B., Cohn, M.A., and Leubner-Metzger, G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15 281–307. [Google Scholar]
- Lee, S., Cheng, H., King, K.E., Wang, W., He, Y., Hussain, A., Lo, J., Harberd, N.P., and Peng, J. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Kloosterziel, K.M., Gil, M.A., Ruijs, G.J., Jacobsen, S.E., Olszewski, N.E., Schwartz, S.H., Zeevaart, J.A., and Koornneef, M. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10 655–661. [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger, G. (2003). Functions and regulation of beta-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci. Res. 13 17–34. [Google Scholar]
- Lopez-Molina, L., and Chua, N.H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., Kinoshita, N., and Chua, N.H. (2003). AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 17 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., McLachlin, D.T., Chait, B.T., and Chua, N.H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32 317–328. [DOI] [PubMed] [Google Scholar]
- McGinnis, K.M., Thomas, S.G., Soule, J.D., Strader, L.C., Zale, J.M., Sun, T.P., and Steber, C.M. (2003). The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot, S., Gosti, F., Guerrier, D., Vavasseur, A., and Giraudat, J. (2001). The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 25 295–303. [DOI] [PubMed] [Google Scholar]
- Muller, K., Tintelnot, S., and Leubner-Metzger, G. (2006). Endosperm-limited Brassicaceae seed germination: Abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 47 864–877. [DOI] [PubMed] [Google Scholar]
- Nambara, E., and Marion-Poll, A. (2005). Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56 165–185. [DOI] [PubMed] [Google Scholar]
- Ogawa, M., Hanada, A., Yamauchi, Y., Kuwahara, A., Kamiya, Y., and Yamaguchi, S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W.I., and Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47 124–139. [DOI] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., Lee, H.S., Sun, T.P., Kamiya, Y., and Choi, G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski, N., Sun, T.P., and Gubler, F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.): S61–S80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oracz, K., El-Maarouf Bouteau, H., Farrant, J.M., Cooper, K., Belghazi, M., Job, C., Job, D., Corbineau, F., and Bailly, C. (2007). ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 50 452–465. [DOI] [PubMed] [Google Scholar]
- Penfield, S., Gilday, A.D., Halliday, K.J., and Graham, I.A. (2006). DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16 2366–2370. [DOI] [PubMed] [Google Scholar]
- Priest, D.M., Ambrose, S.J., Vaistij, F.E., Elias, L., Higgins, G.S., Ross, A.R., Abrams, S.R., and Bowles, D.J. (2006). Use of the glucosyltransferase UGT71B6 to disturb abscisic acid homeostasis in Arabidopsis thaliana. Plant J. 46 492–502. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Seo, M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48 354–366. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Gomez-Cadenas, A., Zhang, P., Walker-Simmons, M.K., Sheen, J., and Ho, T.H. (2001). Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Mol. Biol. 47 437–448. [DOI] [PubMed] [Google Scholar]
- Sun, T.P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55 197–223. [DOI] [PubMed] [Google Scholar]
- Tyler, L., Thomas, S.G., Hu, J., Dill, A., Alonso, J.M., Ecker, J.R., and Sun, T.P. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M., and Delseny, M. (1999). Isolation of total RNA from Arabidopsis thaliana seeds. Anal. Biochem. 268 412–413. [DOI] [PubMed] [Google Scholar]
- Zentella, R., Yamauchi, D., and Ho, T.H. (2002). Molecular dissection of the gibberellin/abscisic acid signaling pathways by transiently expressed RNA interference in barley aleurone cells. Plant Cell 14 2289–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella, R., Zhang, Z.L., Park, M., Thomas, S.G., Endo, A., Murase, K., Fleet, C.M., Jikumaru, Y., Nambara, E., Kamiya, Y., and Sun, T.P. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., and Chua, N.H. (2000). Technical advance. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.