Abstract
Understanding salt stress signaling is key to producing salt-tolerant crops. The small ubiquitin-like modifier (SUMO) is a crucial regulator of signaling proteins in eukaryotes. Attachment of SUMO onto substrates is reversible, and SUMO proteases, which specifically cleave the SUMO–substrate linkages, play a vital regulatory role during SUMOylation. We have identified two SUMO proteases, OVERLY TOLERANT TO SALT1 (OTS1) and OTS2, which are localized in the nucleus and act redundantly to regulate salt stress responses in Arabidopsis thaliana. ots1 ots2 double mutants show extreme sensitivity to salt. However, under low-salt conditions, ots1 ots2 double mutants are phenotypically similar to wild-type plants. We demonstrate that salt stress induces a dose-dependent accumulation of SUMO1/2-conjugated proteins in Arabidopsis. ots1 ots2 double mutants constitutively accumulate high levels of SUMO1/2-conjugated proteins even under nonstress conditions and show a further dramatic increase in SUMO1/2-conjugated proteins in response to salt stress. Transgenic lines overexpressing OTS1 have increased salt tolerance and a concomitant reduction in the levels of SUMOylated proteins. Conversely, the ectopic expression of the mutant ots1(C526S) protein lacking SUMO protease activity fails to produce a salt-tolerant phenotype. We show that salt directly affects OTS1-dependent signaling by inducing OTS1 protein degradation. Our results indicate a requirement for OTS1 deSUMOylation activity in plant salt tolerance responses.
INTRODUCTION
Plants tolerate salt stress conditions by a variety of biochemical and physiological mechanisms. In Arabidopsis thaliana, this includes the restoration of salt balance in the cell, increased efficiency of water use, the induction of reactive oxygen species–detoxifying agents, altered gene expression, and a reduction of growth rate (Zhu, 2002). A key goal of plant biology is to identify the molecular signals that allow the plant to mount these responses. Such information will give much insight into how plants perceive varied environmental cues and transduce them into molecular changes leading to a coordinated tolerance response.
Posttranslational modification of proteins plays a critical role in most cellular processes. In eukaryotes, an important form of such modifications is the attachment of a small polypeptide to a target protein. Ubiquitin is the best understood such tag. Apart from ubiquitin, eukaryotes possess other small proteins involved in protein modification that are related in amino acid sequence to ubiquitin. These include RELATED TO UBIQUITIN, which mainly tags Cullin, a component of the SCF (for SKP1/Cullin/F-box) E3 ligase (Larsen and Cancel, 2004). In recent years, another class of ubiquitin-related polypeptide tags called small ubiquitin-like modifiers (SUMO) has emerged as an influential modifier (Melchior, 2000; Johnson, 2004; Hay, 2005; Geiss-Friedlander and Melchior, 2007). SUMO maturation and attachment onto substrates is similar to the ubiquitination process and includes its own set of analogous E1, E2, and E3 enzymes that are involved in activation, conjugation, and ligation. Many important regulatory proteins modified by SUMO have been described in yeast, mammals, and Drosophila. SUMOylation appears to influence the function of substrates in distinct ways, for example, by altering their cellular location, activity, or stability by antagonizing their degradation (e.g., by blocking ubiquitin attachment sites) via the proteasome (Muller et al., 2001; Johnson, 2004).
The role of SUMOylation in plants is just beginning to emerge. SUMOylation has been shown to play a role in various abiotic stresses, abscisic acid signaling, flowering time, and pathogen defense (reviewed in Miura et al., 2007a). However, SUMO conjugation is also essential for normal development, as shown by the embryo-lethal phenotype associated with mutations in either SAE1/2 encoding activating (E1) or SCE1 encoding conjugating (E2) enzymes or in both the SUMO1 and SUMO2 genes (Saracco et al., 2007). Eight different SUMO isoforms are present in Arabidopsis, although only the two nearly identical SUMO1 and SUMO2 isoforms are conjugated onto target proteins upon various types of stress (Kurepa et al., 2003; Lois et al., 2003). Conjugation of SUMO1/2 upon stress is functionally significant. This was shown by analyzing (SUMO E3 ligase) siz1 mutant plants, defective in one of the SUMO E3 ligase proteins, in various stress conditions. siz1 plants have low levels of SUMO conjugation and display a number of phenotypes, including reduced growth, altered response to phosphate starvation, increased salicylic acid levels upon avirulent pathogen infection, reduced basal thermotolerance, and impaired drought and freezing tolerances (Miura et al., 2005, 2007b; Yoo et al., 2006; Catala et al., 2007; Lee et al., 2007).
SIZ1 function underlines the importance of attachment of SUMO onto substrates as a key signaling regulator of diverse pathways. However, attachment of SUMO is reversible, and the same key enzymes involved in generating mature SUMO, the SUMO proteases, also act as isopeptidases that specifically cleave the SUMO–substrate linkages to recycle free SUMO. A small fraction of protein substrates are normally SUMOylated at a given time, implying that SUMO proteases may play a vital regulatory role during SUMOylation (Johnson, 2004). This is illustrated by the SUMO protease–deficient Arabidopsis mutant early in short days4 (esd4). esd4 plants have increased accumulation of SUMOylated protein and show several phenotypes, including early flowering, a significant reduction of growth, as well as other inflorescence development defects (Reeves et al., 2002; Murtas et al., 2003). Thus, ESD4 deSUMOylation activity is required for multiple regulatory pathways of plant development, highlighting the important role of deSUMOylation in signaling. SUMO proteases have also been implicated in plant–pathogen interactions, where avirulence factors like Xanthomonas campestris pv vesicatoria4 (AvrXV4) from the plant pathogenic bacterium Xanthomonas campestris can act as SUMO proteases, presumably to decrease SUMO conjugation onto target proteins to subvert defense in host cells (Roden et al., 2004).
SUMO proteases consist of a variable N-terminal domain and a conserved C-terminal protease domain. At least eight SUMO proteases were found in Arabidopsis, and these were grouped in different families (Kurepa et al., 2003; Chosed et al., 2006; Colby et al., 2006). This high number of SUMO proteases compared with the conjugation and activation enzymes suggests a complex mechanism of modulation of signaling at the level of deSUMOylation. This may rely on specific protease–substrate interactions or preferences by different SUMO proteases for a specific SUMO isoform. For example, extensive in vitro cleavage assays revealed different SUMO substrate preferences for Arabidopsis SUMO proteases. In some cases, this selectivity was determined by the N-terminal domain (Chosed et al., 2006). Indeed, in yeast (Saccharomyces cerevisiae), the N-terminal domain affects Ubiquitin-Like Protease (Ulp1) activity, localization, and target binding properties (Mossessova and Lima, 2000; Li and Hochstrasser, 2003). Overall, variation at the N-terminal domain of SUMO proteases may underlie the molecular basis of the specificity of certain SUMO proteases for defined signaling pathways in Arabidopsis.
Despite the importance of SUMOylation and the key role ESD4 in flowering time, there is no physiological information for the role of any other SUMO proteases in plants. Furthermore, there is no evidence that SUMO conjugate levels are directly modulated by SUMO proteases as part of the plant response to adverse environmental conditions.
Here, we show that two SUMO proteases, OVERLY TOLERANT TO SALT1 (OTS1) and OTS2, act redundantly to regulate salt stress responses in Arabidopsis. ots1 ots2 double mutants show increased salt sensitivity compared with the wild type or single ots1 and ots2 mutants. SUMO1/2-conjugated proteins accumulate at higher levels in ots1 and ots2 single mutants compared with wild-type plants; in the double ots1 ots2 mutant, an even higher level of SUMO1/2 conjugation onto target proteins occurs. Thus, OTS1 and OTS2 have a role in deSUMOylation of target proteins during nonstress conditions. We also show that in wild-type plants, salt stress induces an increase in SUMO1/2-conjugated proteins in a dose-dependent manner. Furthermore, ots1 ots2 double mutants accumulate SUMOylated proteins at much higher levels in response to salt stress. Transgenic lines overexpressing only wild-type OTS1 but not the active site mutant ots1(C526S) have increased salt tolerance and a concomitant reduction in the levels of SUMOylated proteins. We show that OTS1 and OTS2 are nucleus-localized and that salt stress directly affects OTS1 signaling by inducing its turnover.
RESULTS
Isolation of OTS1 as a Component of the Salt Stress Response in Arabidopsis
An activation-tagging screen for Arabidopsis ecotype Columbia (Col-0) plants with increased tolerance to NaCl resulted in the isolation of several candidate insertion mutants. One mutant plant line that showed increased growth on salt-supplemented medium was analyzed further. DNA gel blot analysis revealed a single insertion located upstream of a putative SUMO protease gene (At1g60220) and dubbed OTS1 (see Supplemental Figure 1 online). Despite the phenotype being confirmed in the following generation, the activation of the putative SUMO protease gene expression was lost after the third generation of this line. However, this preliminary information prompted us to further investigate the role of this putative SUMO protease in salt tolerance signaling.
OTS1 Has SUMO Protease Activity in Vitro
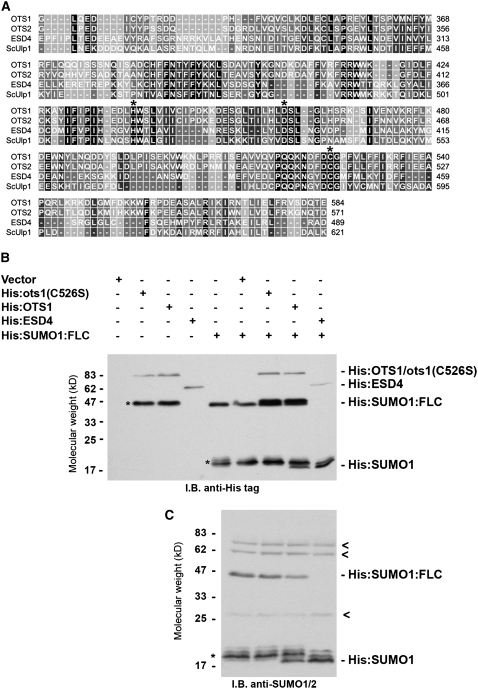
SUMO proteases consist of a conserved catalytic C-terminal domain and a variable N-terminal domain responsible for the regulation of the protein activity (Li and Hochstrasser, 1999, 2003; Mossessova and Lima, 2000). Processing of proSUMO occurs at the C-terminal end, downstream of a conserved di-Gly motif. SUMO protease activity requires the Cys active site, and its substitution into a Ser is sufficient to abolish the protease activity in vitro and in vivo (Li and Hochstrasser, 1999). Alignment of the C-terminal core protease domain of OTS1, OTS2 (a protein with high homology with OTS1), the yeast Ulp1, and Arabidopsis ESD4 showed conservation of three key amino acids previously reported to form the catalytic triad, which includes Cys-526, the putative active site of OTS1 (Figure 1A). Previous reports showed that OTS1 had SUMO protease activity in vitro (Chosed et al., 2006; Colby et al., 2006). Nevertheless, we wanted to determine whether OTS1 activity required Cys-526 by using recombinant His-tagged SUMO1 and SUMO3 from Arabidopsis fused at their C termini to the transcription factor FLC (His:SUMO1:FLC and His:SUMO3:FLC) as proSUMO substrates (Murtas et al., 2003). These were incubated with equal amounts of soluble fractions of Escherichia coli expressing wild-type OTS1 (His:OTS1) or mutant OTS1 [His:ots1(C526S)], in which Cys-526 was converted into a Ser. E. coli expressing His:ESD4 or vector only provided a positive or negative control, respectively, for proSUMO cleavage activity.
Figure 1.
OTS1 Has SUMO Protease Activity.
(A) Alignment of the core protease domain of Arabidopsis OTS1, OTS2, and ESD4 and yeast (Saccharomyces cerevisiae) Ulp1. Asterisks indicate the conserved amino acids forming the catalytic triad. Numbers refer to the amino acid residues of each protein.
(B) and (C) In vitro SUMO protease activity assay. Equal amounts (20 μg) of soluble proteins from E. coli cells transformed with expression vector control (Vector), His:ots1(C526S), His:OTS1, and His:ESD4 were mixed with 200 ng of purified SUMO substrate SUMO1 (His:SUMO1:FLC). Individual components were incubated in the indicated combinations, and the mixture was resolved by 12% SDS-PAGE and probed with anti-His or anti-SUMO1/2 antibodies (C). Individual bands corresponding to different proteins are designated at right. Asterisks indicate additional bands likely derived from nonspecific degradation of His:SUMO1:FLC or His:OTS1/His:ots1(C526S). Arrowheads refer to cross reactions of nonspecific SUMO1 antibodies.
After overnight incubation, the products of the reaction were resolved by SDS-PAGE and analyzed by immunoblotting with anti-His antibodies. Incubation of His:SUMO1:FLC with His:OTS1 or His:ESD4 but not His:ots1(C526S) or vector control resulted in a novel band that migrated at the molecular mass of ∼17 kD (Figure 1B, top). This band could represent the N-terminal, His:SUMO1 portion of His:SUMO1:FLC after cleavage by SUMO proteases. Indeed, immunoblot with anti-SUMO1/2 antibodies revealed a similar ∼17-kD band pattern detected only in correspondence with His:OTS1 or His:ESD4 (Figure 1B, bottom). Despite activity against His:SUMO1:FLC, His:OTS1 and His:ESD4 failed to cleave His:SUMO3:FLC in our assay conditions (see Supplemental Figure 2 online), which is in agreement with previous reports (Chosed et al., 2006; Colby et al., 2006).
These data show that OTS1 has bona fide SUMO1 endopeptidase activity, as it is able to generate mature SUMO1 from the precursor His:SUMO1:FLC, and that this activity is dependent on the catalytic residue Cys-526.
Isolation of ots1 and ots2 Mutants
Phylogenetic analysis revealed that OTS1 (previously known as At Ulp1d) was highly homologous to another SUMO protease encoded by the gene At1g10570 (previously known as At Ulp1c and dubbed OTS2 in this study), which also has been shown to possess SUMO protease activity in vitro (Chosed et al., 2006; Colby et al., 2006). OTS2 was found to be 56% identical to OTS1 at the amino acid level and 73% identical when the comparison was limited to the protease domain (Figure 1). The SUMO protease ESD4, which is involved in flowering time and inflorescence development, is more distantly related to OTS1 and OTS2 (Chosed et al., 2006; Colby et al., 2006). This suggests that OTS1 and OTS2 may have evolved to have similar roles, which are distinct from that played by ESD4 in Arabidopsis.
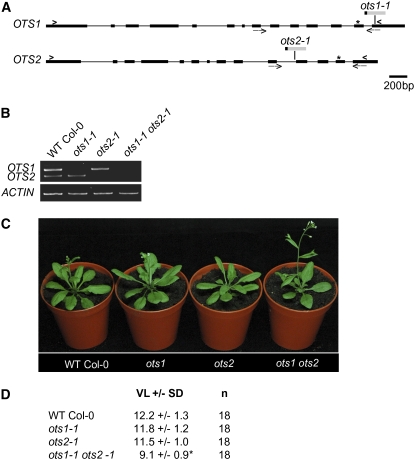
To investigate the role of OTS1 SUMO protease in Arabidopsis, we isolated a T-DNA insertion mutant (Salk_022798) in the OTS1 gene, and this allele was renamed ots1-1 (Figure 2A). Also, to uncover potential redundancy between OTS1 and OTS2, a T-DNA insertion mutant in OTS2 was isolated (Salk_001579; renamed ots2-1) and crossed to ots1-1 to give the double mutant ots1-1 ots2-1. The DNA sequencing of the precise site of T-DNA insertion revealed that in ots1-1 the T-DNA was inserted in the last exon, 49 bp upstream of the stop codon, while in ots2-1 it was located 13 bp upstream of the 11th exon. RT-PCR transcript analysis revealed no full-length OTS1 and OTS2 mRNA from ots1-1 and ots2-1, respectively (see Supplemental Figure 3 online). We then tested whether partial cDNAs could be found. Since all T-DNA insertions were located at different positions at the 3′ end of OTS1 and OTS2, in proximity to the catalytic domain, we performed RT-PCR analysis using oligonucleotides that specifically amplify the 3′ end of the transcripts encoding the catalytic domains of OTS1 and OTS2. No such PCR products could be detected in any of the ots1-1 and ots2-1 T-DNA insertion lines (Figure 2B), indicating that ots1-1 and ots2-1 likely represent loss-of-function alleles of OTS1 and OTS2.
Figure 2.
OTS1 and OTS2 Act Redundantly to Affect Flowering Time.
(A) Schematic representation of the OTS1 and OTS2 genomic DNA regions. Exons are represented as black boxes connected by thin lines (introns), and arrowheads on each sequence represent the positions of the start and stop codons. Asterisks indicate the position of the Cys active site. T-DNA insertion sites are shown as gray boxes with the name of the corresponding allele. Black boxes at the extremities of the T-DNA show the location of the left border. Arrows below the genomic sequence represent the positions of the forward and reverse primers used for RT-PCR analysis.
(B) RT-PCR analysis of OTS1 and OTS2 transcript levels from total RNA derived from young seedlings of the indicated genotypes. OTS1 and OTS2 transcripts were amplified in parallel using the primers (arrows) indicated in (A) for 30 cycles of PCR, and ACTIN was used as a loading control. The experiment was repeated at least four times
(C) Phenotypes of representative 25-d-old ots1 (ots1-1), ots2 (ots2-1), and ots1 ots2 (ots1-1 ots2-1) plants grown in long-day conditions compared with wild-type Col-0.
(D) Quantification of flowering time as average number of vegetative leaves (VL) of the indicated genotypes with sd. The asterisk indicates a significant difference of flowering time from wild-type Col-0 (P < 0.01).
ots1 ots2 Double Mutants Are Early Flowering
Mutants altered in SUMO conjugation or deconjugation show pleiotropic phenotypes, including small stature and very early onset of flowering (Reeves et al., 2002; Catala et al., 2007). Therefore, we wanted to determine whether OTS1 and OTS2 functions also affected Arabidopsis development. Plants were grown on soil under long-day conditions, and the number of vegetative leaves was scored as a measure of flowering time. Repeated experiments showed that ots1-1 and ots2-2 single mutant plants were similar to wild-type (Col-0) plants in terms of overall growth and flowering time (Figure 2C). However, combining ots1-1 with ots2-1 resulted in a significant (P < 0.01) reduction in flowering time compared with the wild type (Figure 2C). Also in short-day conditions, ots1 ots2 double mutants were earlier flowering compared with the wild type and single mutants. After 10 weeks in short-day conditions, ots1 ots2 bolted after producing 19.8 ± 4.2 (se) leaves (n = 19), while the wild type and single mutants ots1 and ots2 were still vegetative (>28 leaves; n = 20).
Apart from the early-flowering phenotype, double mutant plants did not show any obvious inflorescence defects, and their overall growth was similar to that of the wild type. For instance, primary root elongation rates of young double and single ots1 and ots2 mutant seedlings were not significantly different from those of the wild type (Table 1). Thus, OTS1 and OTS2 have a redundant role with respect to flowering time, but overall, ots1 and ots2 loss of function does not produce any obvious pleiotropic phenotypes under normal growth conditions.
Table 1.
Primary Root Growth Is Not Affected in the ots1 and ots2 Single Mutants or in the ots1 ots2 Double Mutant
| Genotype | Root Length ± sd (cm) |
|---|---|
| Wild type (Col-0) | 5.0 ± 0.5 |
| ots1-1 | 4.9 ± 0.5 |
| Wild type (Col-0) | 4.8 ± 0.3 |
| ots2-1 | 4.8 ± 0.3 |
| Wild type (Col-0) | 4.5 ± 0.7 |
| ots1-1 ots2-1 | 4.5 ± 0.3 |
Primary root growth rate of ots1 and ots2 mutants plants. Wild-type (Col-0), ots1-1 and ots2-1 single mutant, and ots1-1 ots2-1 double mutant seeds were sown on 0.5× MS agar plates and grown for 6 d. Wild-type and mutants seedlings were then transferred to fresh 0.5× MS agar plates so that each plate had both the mutant genotype and the wild-type control (n = 9 each). The root tip position (time point 0) was recorded, and additional root growth was measured 7 d after time point 0.
ots1 ots2 Double Mutants Are Salt-Sensitive
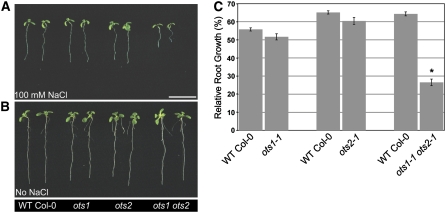
NaCl inhibits root growth in a concentration-dependent manner (Wu et al., 1996). We used this bioassay to test whether ots1 and ots2 single mutants as well as the ots1 ots2 double mutant had altered salt sensitivity. Seeds were sown on regular 0.5× Murashige and Skoog (MS) medium agar plates in the presence or absence of NaCl at a final concentration of 100 mM (a salt concentration that produces a significant induction of protein SUMOylation; see below) and compared with wild-type (Col-0) seeds. Salt-dependent root growth inhibition relative to the nonsalt conditions of 14-d-old seedlings was calculated. In the presence of 100 mM NaCl, neither ots1 nor ots2 single mutants were significantly different from the wild type in terms of root growth inhibition (Figure 3). By contrast, ots1 ots2 double mutants produced a hypersensitive phenotype to salt stress, with much greater reduction of relative root growth compared with the wild type or ots1 and ots2 single mutants. This hypersensitivity phenotype is also seen in the leaf tissues of the ots1 ots2 double mutants after prolonged growth in high salinity (see Supplemental Figure 4 online). These data show that OTS1 and OTS2 have redundant functions with respect to salt stress responses and that abolishing OTS1 and OTS2 functions render Arabidopsis overly salt-sensitive.
Figure 3.
ots1 ots2 Double Mutants Are Salt-Sensitive.
(A) Phenotypes of representative ots1 (ots1-1), ots2 (ots2-1), and ots1 ots2 (ots1-1 ots2-1) 12-d-old seedlings compared with wild-type Col-0. Top, seedlings were grown in the presence of 100 mM NaCl. Bar = 1 cm. Bottom, seedlings were grown in regular MS medium.
(B) Quantification of primary root growth on 100 mM NaCl expressed as a percentage relative to the untreated condition for each of the indicated genotypes. The experiment was performed independently four times with similar results. Error bars indicate se (n = 10 to 20).
OTS1 and OTS2 Are Involved in SUMO Deconjugation in Vivo
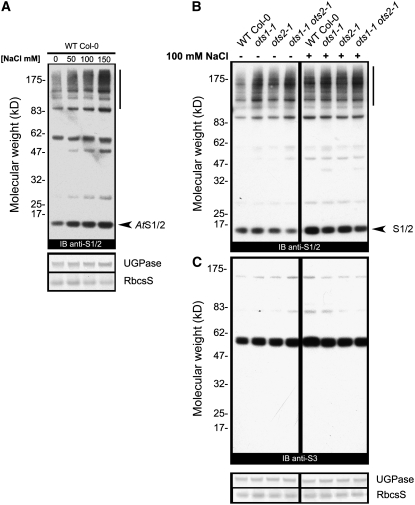
In Arabidopsis, the conjugation of two nearly identical SUMO isoforms, SUMO1 and SUMO2, to target proteins dramatically increases upon a number of abiotic stresses (Kurepa et al., 2003; Catala et al., 2007; Miura et al., 2007b). However, transient exposure of Arabidopsis liquid cultures to salt did not appear to have any effect on the protein SUMOylation pattern (Kurepa et al., 2003). Nevertheless, we wanted to determine if we could observe an effect on SUMO1/2 protein conjugation after prolonged salt stress treatment of Arabidopsis seedlings on agar medium. Total proteins from young wild-type Col-0 seedlings grown in the presence of different salt concentrations were resolved by SDS-PAGE, and any changes in the SUMOylation pattern were monitored by immunoblotting with anti-SUMO1/2 antibodies (Figure 4A). In the absence of supplemented salt, wild-type plants showed a consistent pattern of anti-SUMO1/2 hybridization consisting of a major ∼85-kD band and an ∼15-kD band corresponding to free SUMO1/2. Importantly, very few high molecular mass SUMO1/2 conjugates was detectable, which is consistent with previous reports of the presence of relatively high levels of general deSUMOylation activity within cells (Figure 4A) (Kurepa et al., 2003). Upon salt stress treatment, we observed an accumulation of SUMO1/2-conjugated proteins, and this was directly dependent on the salt concentration in the growth medium (Figure 4A). Interestingly, the steady-state level of free SUMO1/2 increased upon salt treatment, although this was not the result of any obvious increase in SUMO1/2 transcript levels. Overall, we were able to demonstrate that prolonged salt stress leads to SUMOylation of cellular proteins in a dose-dependent manner.
Figure 4.
ots1 ots2 Mutants Are Defective in deSUMOlyation of SUMO1/2 Conjugates.
(A) Immunoblot (IB) analysis of total protein (10 μg loaded for each lane) derived from wild-type Col-0 Arabidopsis seedlings grown for 12 d under long-day conditions in the presence of the indicated NaCl concentrations. Filters were probed with anti-SUMO1/2 (anti At S1/2) antibodies. The vertical bar shows the laddering effect produced by increased accumulation of SUMO1/2 conjugates. The arrowhead indicates free (nonconjugated) SUMO1/2. The bottom panels show immunostaining of UGPase and Ponceau staining of ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) small subunit (RbscS), which served as loading controls. The experiment was repeated three times with similar results.
(B) Immunoblots of total proteins (10 μg loaded for each lane) derived from wild-type Col-0 Arabidopsis seedlings and the indicated genotypes grown in the absence (−) or presence (+) of supplemented 100 mM NaCl for 12 d under long-day conditions. Top, the filter was probed with anti-SUMO1/2 (anti At S1/2) antibodies. The arrowhead and vertical bar indicate free SUMO1/2 and conjugated SUMO1/2, respectively. The experiment was repeated three times with similar results. Bottom, the filter was probed with anti-SUMO3 (anti At S3) antibodies. UGPase immunostaining and Ponceau staining of Rubisco small subunit (RbscS) were used as loading controls.
SUMO proteases process proSUMO and/or remove the SUMO moiety from targets. Thus, OTS1 and OTS2 could be involved in either of these steps as part of their role during salt stress. Immunoblot analysis of extracts derived from ots1 and ots2 single and ots1 ots2 double mutant seedlings grown in the absence of salt revealed an increase in high molecular weight SUMO1/2 conjugate levels when compared with the wild type (Figure 4B). Also, we found a corresponding decrease in the levels of free SUMO1/2 only in ots1 ots2 double mutants.
We next wanted to determine whether mutations in OTS1 and OTS2 or both result in an altered SUMOylation pattern upon growth on salt-containing medium. When we compared the levels of SUMO1/2 conjugate accumulation in the wild type, ots1 and ots2 single mutants, and ots1 ots2 double mutants grown in the presence of 100 mM salt, we consistently observed marginally higher levels of SUMOylation only in ots1 ots2 double mutants when compared with the wild type (Figure 4B). This increase was even more evident at 150 mM salt (see Supplemental Figure 5 online). Importantly, higher levels of SUMOlyation in the double mutants were accompanied by a substantial decrease in free SUMO1/2 compared with the wild-type (Figure 4B; see Supplemental Figure 5 online).
We also monitored the pattern of another SUMO isoform, SUMO3, whose role during salt stress is largely unexplored. In our conditions, we could not find any major differences in the SUMO3 pattern between the wild type and the ots1 or ots2 single or the ots1 ots2 double mutant genotypes either in the presence or absence of salt (Figure 4C). Our results suggest that OTS1 and OTS2 SUMO protease activities in vivo are mainly directed at deconjugating SUMO1/2 from its target proteins.
Constitutive OTS1 Activity Results in Salt Tolerance and Reduced SUMOylation upon Salt Stress
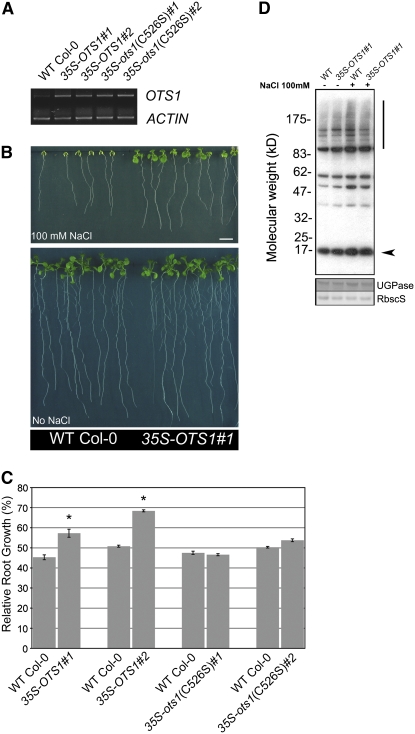
Loss-of-function data indicate that OTS1 and OTS2 deconjugating activities are negatively correlated with salt sensitivity (Figure 5; see Supplemental Figure 4 online). Therefore, it is reasonable to predict that constitutive expression of OTS1 could result in an increased tolerance to salt stress through a reduction of SUMO conjugated proteins. To test this hypothesis, we generated transgenic plants ectopically expressing OTS1 cDNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter. In parallel, we also produced transgenic lines ectopically expressing the SUMO protease–inactive form of OTS1, ots1(C526S). This allowed us to directly test the requirement for SUMO protease activity for salt tolerance in vivo. Two independent, homozygous, single insertion transgenic lines were selected for each construct. We chose these particular lines because RT-PCR analysis confirmed higher levels of transgene-derived expression of OTS1 and ots1(C526S) mutant mRNA compared with the wild type (Figure 5A).
Figure 5.
Overexpression of OTS1 Results in a Salt-Tolerant Phenotype.
(A) RT-PCR transcript analysis (25 cycles) of OTS1 levels in four independent transgenic lines expressing 35S-OTS1 (lines 1 and 2) and 35S-ots1(C526S) (lines 1 and 2) compared with wild-type Col-0. ACTIN was used as a loading control. The OTS1 expression levels in transgenenic lines were verified in more than three independent experiments with similar results.
(B) Phenotype of a representative transgenic line 35S-OTS1#1 compared with the wild type after 15 d of growth on an agar plate in the presence or absence of 100 mM NaCl (Bar = 1 cm).
(C) Quantification of root growth on 100 mM NaCl–supplemented plates expressed as a percentage relative to the untreated condition in four independent transgenic lines, with each one being compared with the wild type. All transgenic lines were tested in at least four independent experiments with similar results. Error bars indicate se (n = 17 to 22). Asterisks indicate significant differences from the wild type (P < 0.01).
(D) Immunoblot with SUMO1/2 antibodies of total protein (10 μg loaded for each lane) derived from 35S-OTS1#1 and wild-type Col-0 seedlings grown for 15 d in the absence (−) or presence (+) of 100 mM NaCl. The experiment was repeated three times with similar results. The arrowhead and vertical bar indicate free SUMO1/2 and conjugated SUMO1/2, respectively. In the bottom panels, UGPase immunostaining and Ponceau staining of Rubisco small subunit (RbscS) were used as loading controls.
When grown in the presence of 100 mM NaCl, 35S-OTS1 transgenic lines fared much better than the wild type grown under the same conditions. Fourteen-day-old 35S-OTS1 lines had significantly longer roots and increased biomass production than wild-type plants (Figures 5A and 5B; Table 2). This phenotype was strictly dependent on OTS1 SUMO protease activity, since lines ectopically expressing the active site mutant version of OTS1 [35S-ots1(C526S)] were not salt-tolerant (Figure 5B; Table 2).
Table 2.
Ectopic Expression of OTS1 but Not ots1(C526S) Resulted in an Increased Biomass Production Relative to the Wild Type When Grown in the Presence of 100 mM NaCl
| Genotype | Relative Fresh Weight ± se (%) |
|---|---|
| Wild type (Col-0) | 68.6 ± 1.2 |
| 35S-OTS1#1 | 82.0 ± 1.6 |
| Wild type (Col-0) | 53.3 ± 2.0 |
| 35S-OTS1#2 | 71.4 ± 0.8 |
| Wild type (Col-0) | 82.2 ± 2.0 |
| 35S-ots1(C526S) line 1 | 87.0 ± 1.5 |
| Wild type (Col-0) | 60.2 ± 1.2 |
| 35S-ots1(C526S) line 2 | 56.1 ± 1.4 |
Biomass production of 35S-ots1 and 35S-ots1(C526S) transgenic plants. Quantification of fresh weight on 100 mM NaCl–supplemented plates expressed as a percentage relative to the untreated condition in four independent transgenic lines, each one compared with the wild type (n = 16 to 24).
To ascertain whether the salt-tolerant phenotype conferred by the overexpression of OTS1 protein is due to an altered SUMO conjugation pattern, we compared the overall SUMO1/2 SUMOylation pattern from protein extracts from 35S-OTS1 and wild-type plants grown in the absence or presence of salt (Figure 5C). In the absence of salt, the SUMOylation pattern of the transgenic plants was similar to that of wild-type plants. However, on salt-supplemented medium, OTS1-overexpressing transgenic lines consistently showed a moderate but significant overall reduction of SUMO conjugates. Our results indicate that increased OTS1 activity induces salt tolerance at least in part by reducing SUMO1/2-dependent SUMOylation levels.
OTS1 mRNA Levels Are Not Affected by Salt Stress, but OTS1 Protein Is Turned Over during the Salt Stress Response
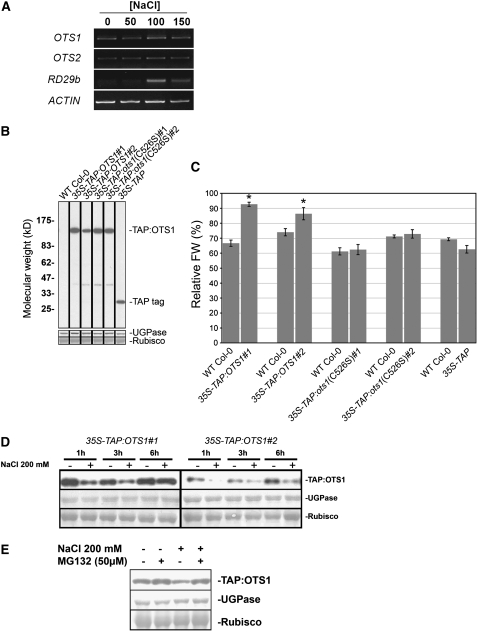
Our data suggest that the control of SUMOylation by OTS1 is an important factor affecting salt tolerance, and it is possible that the levels or the activity of OTS1 itself may be affected by salt stress. However, neither OTS1 nor OTS2 transcripts appeared to be affected by salt in any of the conditions tested (Figure 6A).
Figure 6.
OTS1 Protein Levels Are Downregulated by Salt.
(A) RT-PCR transcript analysis (25 cycles) of OTS1 and OTS2 levels from total cDNA derived from seedlings grown for 12 d on plates supplemented with the indicated salt concentrations. ACTIN served as a loading control, while RD29b, a gene known to be upregulated by salt, was used as a positive control. The RT-PCR analysis was repeated three times with similar results.
(B) Immunoblot with PAP (detects the protein A domain of the TAP tag) of 10 μg of total protein derived from independent transgenic lines ectopically expressing TAP:OTS1, TAP:ots1(C526S), and TAP tag alone and compared with wild-type Col-0. The bands corresponding to the TAP:OTS1 and TAP:ots1(C526S) fusion proteins or TAP tag are indicated at right. In the bottom panels, UGPase immunostaining and Ponceau staining of Rubisco large subunit were used as loading controls.
(C) Quantification of fresh weight (FW) on 100 mM NaCl–supplemented plates expressed as a percentage relative to the untreated condition in five independent transgenic lines, with each one being compared with the wild type. Error bars indicate se (n = 16 to 20). Asterisks indicate significant differences from the wild-type control (P < 0.01).
(D) Immunoblot with PAP of 10 μg of total proteins derived from two independent transgenic lines ectopically expressing TAP:OTS1. Seedlings were grown for 8 d in liquid 0.5× MS medium, after which they were treated with 200 mM NaCl (+) or distilled water (−) as a control and harvested at the indicated time points.
(E) Eight-day-old seedlings were incubated for 3 h with MG132 dissolved in DMSO or DMSO alone and subsequently treated with NaCl (200 mM) or distilled water as a control. Ten micrograms of total proteins was fractionated on an 8% SDS-PAGE gel, and TAP:OTS1 was detected with PAP. UGPase immunostaining and Ponceau staining of Rubisco large subunit were used as loading controls.
To ascertain the effect of salt stress on OTS1 protein levels, we generated transgenic lines (in the homozygous ots1-1 background) ectopically expressing a Tandem Affinity Purification (TAP) tag version of OTS1 under the control of the constitutive CaMV 35S promoter (Rigaut et al., 1999; Rohila et al., 2004). Two independent homozygous, single insertion lines of TAP:OTS1 (35S-TAP:OTS1 ots1-1) and TAP:ots1(C526S) [35S-TAP:ots1(C526S) ots1-1] as well as lines ectopically expressing the TAP tag only (35S-TAP ots1-1) were analyzed in detail. Immunoblotting with antibodies raised against the TAP tag revealed the presence of a band migrating at the predicted size of the TAP:OTS1 fusion protein (∼90 kD) in all transgenic lines expressing TAP:OTS1 or TAP:ots1(C526S) but not in the wild type or lines expressing the TAP tag alone (Figure 6B, left). The fusion protein was functional, since upon growth on 100 mM salt, 35S-TAP:OTS1 but not 35S-TAP:ots1(C526S) or 35S-TAP lines showed a substantial increase in biomass production compared with the wild type (Figure 6C).
We hypothesized that any direct changes in protein level caused by salt stress could be evaluated more accurately at early time points after application of salt. To do this, we grew seedlings in liquid culture for 8 d, after which salt was directly added to the culture medium at a final concentration of 200 mM. Plant material was then harvested after 1, 3, and 6 h and the levels of TAP:OTS1 protein were analyzed by immunoblotting. We found that after 1 h of salt treatment, the TAP:OTS1 protein level was significantly reduced compared with the untreated control. Similar results were obtained at 3 and 6 h and for two independent transgenic lines (Figure 6D). Thus, salt has a rapid and direct effect on TAP:OTS1 levels and perhaps mediates TAP:OTS1 protein degradation. We tested the possibility that reduction of TAP:OTS1 levels observed upon salt treatment required proteasome activity. Preincubation of TAP:OTS1-expressing seedling cultures with MG132 (a proteasome inhibitor) for 3 h before salt treatment substantially increased the abundance of TAP:OTS1, overriding the salt-dependent OTS1 degradation (Figure 6E). These results suggest that TAP:OTS1 is proteasomally degraded upon salt stress.
OTS1 and OTS2 Localize to the Nucleus
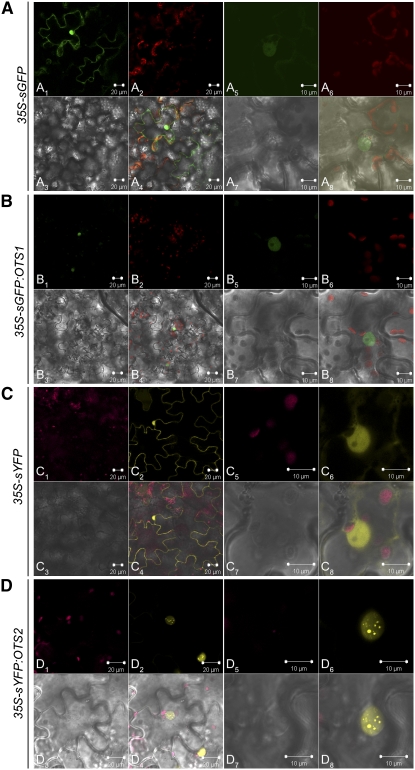
SUMO protease localization is a key determinant for target recognition. Previous reports showed that the SUMO conjugation machinery, the SUMO protease ESD4, and the vast majority of the SUMO conjugated proteins localize mainly to the nucleus (Lois et al., 2003; Murtas et al., 2003; Saracco et al., 2007). To ascertain OTS1 localization, OTS1 was fused to soluble green fluorescent protein (sGFP) and transiently expressed under the control of the CaMV 35S promoter (35S-sGFP:OTS1) in tobacco (Nicotiana tabacum) leaf cells. Confocal analysis revealed that OTS1 was localized exclusively in the nucleoplasm, while the soluble sGFP control was found both in the nucleus and in the cytosol (Figures 7A and 7B). This expression pattern was also observed when we expressed the SUMO protease–inactive mutant ots1(C526S) protein [35S-sGFP:ots1(C526S)] under the same experimental conditions (see Supplemental Figure 6 online).
Figure 7.
OTS1 and OTS2 Are Nuclear Proteins.
Tobacco leaves were transiently transformed with vectors expressing the following fluorescent proteins: sGFP (A), sGFP:OTS1 (B), sYFP (C), and sYFP:OTS2 (D). For each construct, different fluorescent emissions were collected.
(A) sGFP protein localization at low resolution ([A1] to [A4]) and high resolution ([A5] to [A8]). (A1) and (A5), sGFP emission (green); (A2) and (A6), chloroplast autofluorescence (red); (A3) and (A7), bright field; (A4) and (A8), merged images of (A1) to (A3) and (A5) to (A7), respectively.
(B) sGFP:OTS1 protein fusion localization at low resolution ([B1] to [B4]) and high resolution ([B5] to [B8]). (B1) and (B5), sGFP emission (green); (B2) and (B6), chloroplast autofluorescence (red); (B3) and (B7), bright field; (B4) and (B8), merged images of (B1) to (B3) and (B5) to (B7), respectively.
(C) sYFP protein localization at low resolution ([C1] to [C4]) and high resolution ([C5] to [C8]). (C1) and (C5), chloroplast autofluorescence (red); (C2) and (C6), sYFP emission (yellow); (C3) and (C7), bright field; (C4) and (C8), merged images of (C1) to (C3) and (C5) to (C7), respectively.
(D) sYFP:OTS2 protein fusion localization at low resolution ([D1] to [D4]) and high resolution ([D5] to [D8]). (D1) and (D5), chloroplast autofluorescence (red); (D2) and (D6), sYFP emission (yellow); (D3) and (D7), bright field; (D4) and (D8), merged images of (D1) to (D3) and (D5) to (D7), respectively.
Since OTS2 appears to be functionally redundant to OTS1, we expected it to have the same localization. Indeed, soluble yellow fluorescent protein (sYFP):OTS2 fusion protein (35S-sYFP:OTS2) was localized in the nucleus in tobacco cells, indicating that OTS1 and OTS2 proteins might act in the same subcellular compartment (Figures 7C and 7D). However, higher magnification confocal images revealed a punctate pattern of OTS2 accumulation in nuclei, which was not observed with either sGFP:OTS1 or sYFP proteins. Thus, in the tobacco transient assay system, OTS2 may associate with speckle-like bodies in nuclei.
DISCUSSION
SUMO modification of regulatory proteins is emerging as an important mechanism regulating cellular signaling pathways (Muller et al., 2001). A key aspect of SUMO-mediated signaling is the tradeoff between the SUMOylated and the deSUMOylated state for a given protein. SUMO proteases maintain this equilibrium, and they have been implicated as critical players during SUMO-mediated signaling in yeast and mammals (Muller et al., 2001). Less is known about their role in plants. So far, only one bona fide SUMO protease (ESD4 controls floral transition) has been shown to have a role in plant cellular signaling (Reeves et al., 2002; Murtas et al., 2003). Using a combination of genetic, physiological, and biochemical approaches, we were able to show that the SUMO protease enzymes OTS1 and OTS2 are required for salt tolerance in Arabidopsis.
OTS1 and OTS2 Belong to the ULP1 Family of SUMO Proteases in Arabidopsis
In Arabidopsis, eight genes have been identified that encode proteins with high similarity to yeast Ulp1 (Kurepa et al., 2003; Murtas et al., 2003; Chosed et al., 2006; Colby et al., 2006). So far, At ULP1a, At ULP1c (OTS2), At ULP1d (OTS1), and ESD4 have been shown to have At SUMO1/2 endopeptidase activity in vitro (Chosed et al., 2006; Colby et al., 2006) (Figure 1). Despite OTS1 having SUMO1 endopeptidase activity in vitro, its role in vivo is largely directed toward the deconjugation of SUMOylated proteins. This SUMO deconjugating role also applies to OTS2, as shown by the increased levels of SUMOylated protein in the corresponding mutant backgrounds (Figure 4), and to ESD4 (Murtas et al., 2003). Since the major roles of OTS1, OTS2, and ESD4 seem to be mainly deconjugative, the question arises of which of these is the major SUMO maturation protease in Arabidopsis. One possibility is that the SUMO endopeptidase activity could be highly redundant and that most (if not all) Arabidopsis SUMO proteases contribute to some extent to proSUMO maturation. An alternative model could be that one or more Arabidopsis SUMO proteases is involved in proSUMO to SUMO maturation. This model applies in yeast, where only two SUMO proteases, Ulp1 and Ulp2, exist. Although Ulp2 has endopeptidase activity in vitro, Ulp1 is largely responsible for proSUMO maturation in vivo (Li and Hochstrasser, 2000).
In Arabidopsis, the activities of OTS1 and OTS2 largely affect the pool of SUMO1/2 conjugated targets but not of SUMO3 conjugated targets (Figure 4). This is in contrast with ESD4, which was reported to process SUMO3 conjugates in addition to SUMO1/2 conjugates, at least in transgenic plants ectopically expressing At SUMO3 (Murtas et al., 2003). Moreover, OTS1, OTS2, and ESD4 may also have different specificities for SUMOylated target proteins, as inferred by the comparison of the highly pleiotropic phenotype of esd4 mutant plants with the largely wild-type ots1 ots2 double mutants in nonstress conditions (Figure 2). This observation suggests that different proteases may have different targets in Arabidopsis. Alternatively, SUMO proteases might be expressed in different organs, domains, or subcellular compartments; therefore, the SUMO protease-to-target specificity may be dictated mainly by the protease expression pattern. Systematic analysis of the expression pattern of various SUMO proteases in Arabidopsis will clarify this point.
Role of OTS1 and OTS2 during Salt Stress
In Arabidopsis, SUMO1/2 SUMOylation increases upon different types of stress, including heat, cold, and drought (Kurepa et al., 2003; Catala et al., 2007; Miura et al., 2007b). Our results demonstrate a role for SUMOylation also in salt stress. Abundance of SUMO1/2 conjugates increased in a salt concentration–dependent manner and correlated with inhibition of Arabidopsis seedling growth (Figure 5; see Supplemental Figures 4 and 5 online). Moreover, upon salt treatment, the levels of free SUMO1/2 increased strongly in wild-type plants but to a much lesser extent in ots1 ots2 double mutants. This could be partly due to an overall reduced cellular deconjugation activity to recycle free SUMO1/2 from their target proteins in ots1 ots2 mutants. In fact, hyperaccumulation of SUMO1/2 targets due to impaired deconjugation in the ots1 ots2 double mutant results in increased salt sensitivity (Figures 3 and 4), further confirming the prominent role of SUMO1/2 protein conjugates in abiotic stress responses.
Although salt induced SUMO1/2 conjugation and this is directly linked to reduced growth, ots1 ots2 double mutants accumulated SUMO1/2 conjugates even under nonstress conditions (Figure 4). Even though the pattern and the level of SUMOylation in the ots1 ots2 double mutant in the absence of salt were similar to those in wild-type seedlings grown in the presence of 100 mM salt (Figure 4C), ots1 ots2 double mutants showed no obvious signs of growth inhibition under nonstressed conditions. These data suggest that in the ots1 ots2 double mutant, the accumulation of these particular sets of SUMO1/2 conjugates is largely dispensable for growth and survival under nonstress conditions. However, the ots1 ots2 double mutant was hypersensitive to salt, and only catalytically active OTS1-overexpressing plants showed increased salt tolerance with a concomitant decrease in the levels of SUMO1/2 conjugate accumulation, indicating that OTS1- and OTS2-dependent deSUMOylation is a crucial part of the plant response to high salinity. Our data establish a functional link between levels of SUMO1/2 SUMOylation and salt tolerance through OTS1 deSUMOylation activity.
OTS1 protein levels are rapidly downregulated during salt stress, despite OTS1 being constitutively expressed. Downregulation of OTS1 protein is mediated by the proteasome, a process possibly involving ubiquitination of OTS1. High concentrations of salt induce oxidative stress in Arabidopsis (Borsani et al., 2001; Achard et al., 2008), and previous reports showed that, in Arabidopsis cell cultures, the addition of oxidative stress–mimicking agents leads to a dramatic increase in SUMOylation (Kurepa et al., 2003). It is possible, therefore, that the increase in SUMOylation could be in part due to downregulation of specific SUMO proteases and, in the case of salt stress, OTS1 and OTS2. This demonstration that salt stress affects SUMO proteases in plants may shed light on a mechanism for increased SUMO conjugation of target proteins seen during oxidative stress responses. Our data allow us to speculate that the natural salt sensitivity seen in Arabidopsis (Zhu, 2002) could be partly due to an overaccumulation of SUMO1/2 conjugates resulting from the degradation of OTS1 and OTS2.
OTS1 and OTS2 Cellular Localization and Their Respective Substrates
In yeast and higher eukaryotes, SUMO proteases show different domains of subcellular localization, and this is a key determinant of their target binding (Hay, 2007). SUMO proteases in plants exhibit a complex pattern of subcellular localization. ESD4 is mainly distributed to the nuclear pore and indeed interacts with NUCLEAR PORE ANCHOR to regulate flowering time (Murtas et al., 2003; Xu et al., 2007). OTS1 and OTS2 were distributed throughout the nucleus (Figure 7), which is similar to the pattern observed for SUMO1/2 and its stress-related conjugates (Saracco et al., 2007), SIZ1(Miura et al., 2005), and the SUMO conjugating enzyme SCE1 (Lois et al., 2003). This indicates that OTS1 and OTS2 could work in tandem with the SUMO machinery to regulate salt tolerance responses in plant nuclei.
We propose a model whereby Arabidopsis OTS1 and OTS2 SUMO proteases are degraded upon salt stress, resulting in increased SUMOylation. Since OTS1 targets are required in the deSUMOylated state to mount salt-tolerant responses, plants then become susceptible to salt. Modulating the activity of OTS1, therefore, is a key process for withstanding elevated salt conditions, and OTS1 activity may represent an attractive target to engineer salt tolerance in crop species.
Genetic analysis indicates that, like ESD4, OTS1/2 regulates the abundance of many SUMO conjugates (Murtas et al., 2003). Isolation and identification of OTS1/2-specific substrates and reverse genetics of the corresponding genes will uncover how OTS1/2-dependent deSUMOylation allows plants to endure high salinity.
METHODS
Plant Materials and Growing Conditions
For Petri experiments, surface-sterilized Arabidopsis thaliana seeds were resuspended in sterile distilled water and stratified at 4°C for 4 to 6 d in the dark before sowing on agar plates (0.5× MS basal salt mixture [Sigma-Aldrich], 0.75% [w/v] sucrose, and 0.8% [w/v] agar), which contained the indicated concentration of NaCl. Plates were exposed to light, and seeds were germinated and grown vertically for 12 to 16 d under long-day conditions (16 h of light/8 h of dark) at 22°C with a light intensity of 100 μmol·m−2·s−1. For salt experiments, 9 to 12 seeds of the mutant genotype were grown in parallel with wild-type Col-0 seeds on three independent plates.
For soil experiments, seeds were imbibed, stratified in water, and sown on soil (Levington F2). Plants were grown in a 16-h-light/8-h-dark photoperiod in controlled chambers at 23°C, 60% humidity, and 130 μmol·m−2·s−1 light fluence. For short-day experiments, plants were grown in a 9-h-light/15-h-dark photoperiod at 21°C and 100 μmol·m−2·s−1 light fluence.
Isolation of ots1 and ots2 Mutants and Construction of Double Mutants
Independent T-DNA insertion lines (in the Col-0 background) for OTS1 and OTS2 were obtained from the Nottingham Arabidopsis Stock Centre: ots1-1 (N522798) and ots2-1 (N501579), respectively. Insertion-carrying plants were identified by genomic DNA PCR using the T-DNA left border–specific primer LBa1 (SALK T-DNA primer, 5′-TGGTTCACGTAGTGGGCCATCG-3′) in combination with specific primers flanking the insertion site: for ots1-1, LC16 (5′-CGACAAGAAGTGGTTTAGACC-3′); for ots2-1, LC18 (5′-GACAGGGATGCATATTTTGTGAAG-3′). The precise insertion site was verified by sequencing the LBa1-flanking region PCR product with LBb1 (SALK T-DNA primer, 5′-GCGTGGACCGCTTGCTGCAACT-3′). Homozygous T-DNA insertion mutants were identified by genomic DNA PCR, using primer pairs flanking the insertion site: for ots1-1, LC16 and LC17 (5′-GTAACGTAACACTTATTAGATGCC-3′); for ots2-1, LC15 (5′-TTAATCTGTTTGGTTACCCTTGCGG-3′) and LC18.
Double mutant plants were obtained by crossing ots1-1 (pollen donor) with emasculated ots2-1 flowers. F1 plants were checked for the presence of the ots1-1 allele, and double homozygous insertion mutants ots1-1 ots2-1 were identified from 30 to 40 F2 individuals by genomic DNA PCR.
Protein Extraction and Immunoblotting
Arabidopsis seedlings were frozen in liquid nitrogen and homogenized in E buffer (125 mM Tris-HCl, pH 8.8, 1% [w/v] SDS, 10% [v/v] glycerol, and 50 mM sodium metabisulfite) (Martinez-Garcia et al., 1999) with freshly added 5 mM N-ethylmorpholine and EDTA-free protease inhibitor cocktail (Roche). The homogenate was microcentrifuged at 16,000g for 5 min at room temperature, and the supernatant was quantified with Bradford reagent before mixing with 4× SDS-PAGE loading buffer. Equal amounts of proteins for each sample were loaded onto a 4 to 12% NuPAGE Novex Bis-Tris gel run in MES-SDS buffer (Invitrogen) or a standard SDS-PAGE gel. Proteins were then transferred to a polyvinyl difluoride membrane (Bio-Rad) for immunoblot analysis. Filters were blocked in TTBS-milk (5% [w/v] dry nonfat milk, 10 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.1% [v/v] Tween 20) before incubation with primary antibody anti-SUMO1/2 or anti-SUMO3 (Abcam) diluted 1:2000. Filters were washed in TTBS and incubated with secondary antibody (anti-rabbit horseradish peroxidase conjugate [Sigma-Aldrich]) diluted 1:20,000 in TTBS-milk. To detect TAP tag proteins, filters were incubated with peroxidase anti-peroxidase (PAP; Sigma-Aldrich) diluted 1:4000. Filters were washed and incubated with the horseradish peroxidase substrate (Immobilon Western; Millipore) before exposure to film (Kodak).
His tag proteins were detected by incubating with monoclonal anti-His antibodies (Sigma-Aldrich) diluted 1:3000 in the presence of 1× casein blocking solution (Novagen). UDP-glucose pyrophosphorylase (UGPase), a cytosolic protein (Martz et al., 2002), was detected with anti-UGPase rabbit polyclonal antibodies (Agrisera) diluted 1:3000 followed by anti-rabbit alkaline phosphatase–conjugated antibodies diluted 1:5000. Membranes were then submerged in 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium Purple Liquid Substrate (Sigma-Aldrich) to visualize immunocomplexes.
Plasmid Construction
OTS1 and OTS2 open reading frames were amplified by PCR from whole cDNAs from seedlings with oligonucleotides LC1 (5′-CACCATGACGAAGAGGAAGAAGGAAG-3′) and OS12 (5′-TTACTCTGTCTGGTCACTGAC-3′) for OTS1 and LC12 (5′-CACCATGAAGAGACAAAGAGCAATCGAG-3′) and LC15 (5′-TTAATCTGTTTGGTTACCCTTGCGG-3′) for OTS2. PCR fragments were cloned into pENTR/D-TOPO (Invitrogen), sequenced, and renamed pLCG1 for OTS1 and pLCG14 for OTS2. To produce the ots1 active site mutant [ots1(C526S)], pLCG1 was amplified with mutagenic oligonucleotides LC9 (5′-CAGCAGAAAAACGATTTTGATTCTGGTCCGTTTGTGCTCTTCTTC-3′) and LC10 (5′-GAAGAAGAGAAACGGACCAGAATCAAAATCGTTTTTCTGCTG-3′), which carried a single base pair change (underlined), according to the QuikChange site-directed mutagenesis kit directions (Stratagene), and the resulting plasmid (pLCG7) was sequenced. Subsequent subcloning was done using the Gateway system, through LR-mediated recombination (Invitrogen). 35S-OTS1, 35S-ots1(C526S), 35S-GFP:OTS1, and 35S-GFP:ots1(C526S) were produced by recombining pLCG1 and pLCG7 with the binary vectors pGWB2 and pGWB6 (Nakagawa et al., 2007) to give pLCG2 (35S-OTS1), pLCG8 [35S-ots1(C526S)], pLCG5 (35S-sGFP:OTS1), and pLCG17 [35S-sGFP:ots1(C526S)]. TAP:OTS1 fusions were produced by recombining pLCG1 and pLCG7 with ntapi.289 (Rohila et al., 2004) to give pLCG35 (35S-TAP:OTS1) and pLCG36 [35S-TAP:ots1(C526S)]. To produce 35S-sYFP:OTS2, pLCG14 was recombined with the binary vector pEarleyGate104 (Earley et al., 2006) to give pLCG43.
To produce Poly-His:OTS1 and ots1(C526S) expression vectors, pLCG1 and pLCG7 were amplified with oligonucleotides LC46 (5′-AGAATTCATGACGAAGAGGAAGAAGGAAG-3′) and LC38 (5′-TGTCGACTTACTCTGTCTGGTCACTGACACG-3′), which incorporate an EcoRI and a SalI (underlined) site, respectively. The PCR products were subcloned in pGEM-T Easy vectors (Promega) and renamed pLCG55 (OTS1) and pLCG50 [ots1(C526S)]. The resulting EcoRI-SalI fragments were then transferred to pET30a to produce pLCG57 (Poly-His:OTS1) and pLCG54 [Poly-His:ots1(C526S)].
RNA Extraction and RT-PCR
Total RNA was extracted with the Trizol reagent as detailed by the manufacturer's protocol (Invitrogen) starting from 20 to 50 seedlings of the indicated genotype. First-strand cDNA synthesis was performed from 2 μg of total RNA using an oligo(dT) primer with the SuperScript II reverse transcriptase kit (Invitrogen). OTS1 and OTS2 transcripts were amplified by PCR with oligonucleotides OS11 (5′-GATCCTCACTTTGTTCAAGTT-3′)/OS12 and LC15/LC18, respectively. ACTIN2 was amplified with forward primer 5′-CTTACAATTTCCCGCTCTGC-3′ and reverse primer 5′-GTTGGGATGAACCAGAAGGA-3′. RESPONSIVE TO DESICCATION29B (RD29B) was amplified with forward primer 5′-CGAAAACCCCATAGTCCCAAC-3′ and reverse primer 5′-GATACCTTCCGACCAGATAGC-3′. PCR fragments were separated on a 1% (w/v) agarose gel containing a 1:10,000 dilution of Sybr safe (Invitrogen) dye. DNA was visualized with a UV light transilluminator.
Recombinant Proteins and in Vitro SUMO Protease Assay
His:ESD4, pLCG54, pLCG57, and control pET30a vectors were transferred into Eschericha coli BL21 (D3E) cells. E. coli cultures (50 mL) were grown at 37°C to an OD600 of 0.8 before adding isopropylthio-β-galactoside at a final concentration of 1 mM to induce protein expression, which was performed at 28°C for 2 h. Cells were pelleted and resuspended in 1 mL of protein extraction buffer (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.1% [v/v] Triton X-100, 2 mM DTT, 5 mM EDTA, and 100 μg/mL lysozyme) (Li et al., 2005). Cells were incubated at room temperature with agitation for 20 min. Twenty-five units of Benzonase nuclease (Novagen) and MgCl2 (1 mM) were added to the mixture to reduce viscosity, and the extracts were clarified by microcentrifugation. The soluble protein fraction (supernatant) was recovered, and its concentration was determined with the Bradford reagent using BSA as a standard. Poly-His:SUMO1:FLC and Poly-His:SUMO3:FLC (Murtas et al., 2003) were produced in E. coli, affinity-purified using His-select spin columns (Sigma-Aldrich) according to the manufacturer's instructions, and eluted in 200 μL of elution buffer. The eluate was dialyzed overnight at 4°C against reaction buffer I (50 mM Tris-HCl, pH 7.9, 150 mM NaCl, 0.1% [v/v] Triton X-100, and 2 mM DTT).
The SUMO protease assay was performed by mixing 20 μg of the E. coli soluble fractions with 500 ng of substrate Poly-His:SUMO1:FLC or Poly-His:SUMO3:FLC. The reaction mixture was incubated in reaction buffer I overnight at 28°C. The reactions were stopped by adding 4× SDS-PAGE loading buffer, and proteins were resolved on a 12% SDS-PAGE gel. Proteins were transferred to polyvinyl difluoride membranes for immunoblot analysis.
Plant Transformation and Analysis of the Transgenic Plants
Agrobacterium tumefaciens cells (GV301) were transformed with binary vectors pLCG2, pLCG8, pLCG35, pLCG36, and ntapi.289 and cultured for transformation of Arabidopsis wild-type (Col-0) or ots1-1 mutant plants by the floral dipping method (Clough and Bent, 1998). T1 seeds were selected on agar plates containing kanamycin (50 mg/L) and hygromycin (10 mg/L) or BASTA (20 mg/L glufosinate ammonium), and >50 T1 resistant lines were identified for each construct. T2 seeds were harvested, and 10 independent lines for each construct were examined for the number of T-DNA insertion loci by selection on kanamycin or BASTA and expression levels of the transgene. Two independent lines that showed a 1:3 (sensitive:resistant) ratio and high levels of transgene expression were transferred to soil. T3 seeds derived from 9 to 12 T2 plants were selected to find homozygous individuals. In this study, all transgenic plants are from the T3 generation.
Tobacco Transient Expression and Confocal Microscopy
Binary vectors expressing the fusion proteins sGFP:OTS1, sGFP:ots1(C526S), and sYFP:OTS2 (pLCG5, pLCG17, and pLCG43, respectively) or just sGFP and sYFP (pGWB6 and EarleyGate104, respectively) were used to transform Agrobacterium cells. Luria-Bertani cultures supplemented with antibiotic selection (100 mg/L kanamycin and 100 mg/mL rifampicin) were inoculated with Agrobacterium and grown for 4 to 5 h at 28°C with shaking. Cells were harvested, washed with 10 mM MgCl2, and resuspended in 10 mM MgCl2 to an OD600 of 0.2 before acetosyringone was added to a final concentration of 200 μM. Cell cultures were left at room temperature for 1 to 2 h before injection into wild-type tobacco (Nicotiana tabacum) leaves. Transgene-derived expression was monitored 2 to 3 d after infiltration by confocal microscopy on a Zeiss LSM510-UV microscope using 20× Planapo objectives for low-resolution analysis and 63× water-immersion objectives (Zeiss) for high-resolution image collection. Fluorophores were excited using an argon laser at 488 nm (GFP) or 514 nm (YFP). Fluorescent light was collected after passage through a HFT488 and a NFT545 beam splitter, a 505- to 530-nm band-pass filter for GFP detection, and a 560-nm long-pass filter to detect chloroplast fluorescence. Otherwise, fluorescence was collected after passage through a HFT458/514, NFT635 VIS, or 606- to 724-nm filter to detect chloroplast fluorescence or a NFT515 beam splitter and a 530-nm long-pass filter to detect YFP. Bright-field images were collected using the transmitted light detector. Laser power, detector gain, and offset were kept fixed for all of the different specimens to allow comparison between the different samples.
Measurements and Statistical Analysis
Primary root length was measured with the ImageJ software package. Mutant genotypes were compared with the wild type, and significant differences were calculated with a Tukey's error rate of 0.01 using analysis of variance.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative data library under the following accession numbers: At1g60220 (OTS1), At1g10570 (OTS2), and At4g26840 (SUMO1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Activation Tagging of OTS1.
Supplemental Figure 2. OTS1 Does Not Cleave His:SUMO3:FLC.
Supplemental Figure 3. ots1-1 and ots2-1 Are Knockout Alleles.
Supplemental Figure 4. Shoot Growth Is Reduced in ots1 ots2 Double Mutants in the Presence of Salt.
Supplemental Figure 5. ots1 ots2 Double Mutants Overaccumulate SUMOylated Proteins upon Growth on High Concentration of NaCl.
Supplemental Figure 6. ots1(C526S) Localizes to the Nucleus.
Supplementary Material
Acknowledgments
We thank Mike Blatt for critical reading of the manuscript, Prisca Campanoni for her help with confocal microscopy, and George Coupland for providing the SUMO:FLC constructs and His:ESD4. This work was supported by the Biological and Biotechnological Research Council.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ari Sadanandom (a.sadanandom@bio.gla.ac.uk).
Online version contains Web-only data.
References
- Achard, P., Renou, J.P., Berthome, R., Harberd, N.P., and Genschik, P. (2008). Plant DELLAs restrain growth and promote survival of adversity by reducing the levels of reactive oxygen species. Curr. Biol. 18 656–660. [DOI] [PubMed] [Google Scholar]
- Borsani, O., Valpuesta, V., and Botella, M.A. (2001). Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 126 1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala, R., Ouyang, J., Abreu, I.A., Hu, Y., Seo, H., Zhang, X., and Chua, N.-H. (2007). The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19 2952–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed, R., Mukherjee, S., Lois, L.M., and Orth, K. (2006). Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem. J. 398 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Colby, T., Matthai, A., Boeckelmann, A., and Stuible, H.-P. (2006). SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K.W., Haag, J.R., Pontes, O., Opper, K., Juehne, T., Song, K., and Pikaard, C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45 616–629. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander, R., Melchior, F. (2007). Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell. Biol. 8 947–956. [DOI] [PubMed] [Google Scholar]
- Hay, R.T. (2005). SUMO: A history of modification. Mol. Cell 18 1–12. [DOI] [PubMed] [Google Scholar]
- Hay, R.T. (2007). SUMO-specific proteases: A twist in the tail. Trends Cell Biol. 17 370–376. [DOI] [PubMed] [Google Scholar]
- Johnson, E.S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73 355–382. [DOI] [PubMed] [Google Scholar]
- Kurepa, J., Walker, J.M., Smalle, J., Gosink, M.M., Davis, S.J., Durham, T.L., Sung, D.-Y., and Vierstra, R.D. (2003). The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J. Biol. Chem. 278 6862–6872. [DOI] [PubMed] [Google Scholar]
- Larsen, P.B., and Cancel, J.D. (2004). A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J. 38 626–638. [DOI] [PubMed] [Google Scholar]
- Lee, J., et al. (2007). Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 49 79–90. [DOI] [PubMed] [Google Scholar]
- Li, S.J., Hankey, W., and Hochstrasser, M. (2005). Preparation and characterization of yeast and human desumoylating enzymes. Methods Enzymol. 398 457–467. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (1999). A new protease required for cell-cycle progression in yeast. Nature 398 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (2000). The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20 2367–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.J., and Hochstrasser, M. (2003). The Ulp1 SUMO isopeptidase: Distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois, L.M., Lima, C.D., and Chua, N.-H. (2003). Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20 251–257. [DOI] [PubMed] [Google Scholar]
- Martz, F., Wilczynska, M., and Kleczkowski, L.A. (2002). Oligomerization status, with the monomer as active species, defines catalytic efficiency of UDP-glucose pyrophosphorylase. Biochem. J. 367 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior, F. (2000). SUMO—Nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16 591–626. [DOI] [PubMed] [Google Scholar]
- Miura, K., Jin, J.B., and Hasegawa, P.M. (2007. a). Sumoylation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10 495–502. [DOI] [PubMed] [Google Scholar]
- Miura, K., Jin, J.B., Lee, J., Yoo, C.Y., Stirm, V., Miura, T., Ashworth, E.N., Bressan, R.A., Yun, D.-J., and Hasegawa, P.M. (2007. b). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19 1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, K., Rus, A., Sharkhuu, A., Yokoi, S., Karthikeyan, A.S., Raghothama, K.G., Baek, D., Koo, Y.D., Jin, J.B., Bressan, R.A., Yun, D.-J., and Hasegawa, P.M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102 7760–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova, E., and Lima, C.D. (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5 865–876. [DOI] [PubMed] [Google Scholar]
- Muller, S., Hoege, C., Pyrowolakis, G., and Jentsch, S. (2001). SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2 202–210. [DOI] [PubMed] [Google Scholar]
- Murtas, G., Reeves, P.H., Fu, Y.-F., Bancroft, I., Dean, C., and Coupland, G. (2003). A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., Toyooka, K., Matsuoka, K., Jinbo, T., and Kimura, T. (2007). Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104 34–41. [DOI] [PubMed] [Google Scholar]
- Reeves, P.H., Murtas, G., Dash, S., and Coupland, G. (2002). early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129 5349–5361. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17 1030–1032. [DOI] [PubMed] [Google Scholar]
- Roden, J., Eardley, L., Hotson, A., Cao, Y., and Mudgett, M.B. (2004). Characterization of the Xanthomonas AvrXv4 effector, a SUMO protease translocated into plant cells. Mol. Plant Microbe Interact. 17 633–643. [DOI] [PubMed] [Google Scholar]
- Rohila, J.S., Chen, M., Cerny, R., and Fromm, M.E. (2004). Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38 172–181. [DOI] [PubMed] [Google Scholar]
- Saracco, S.A., Miller, M.J., Kurepa, J., and Vierstra, R.D. (2007). Genetic analysis of SUMOylation in Arabidopsis: Conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 145 119–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.J., Ding, L., and Zhu, J.K. (1996). SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8 617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X.M., Rose, A., Muthuswamy, S., Jeong, S.Y., Venkatakrishnan, S., Zhao, Q., and Meier, I. (2007). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/Megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, C.Y., Miura, K., Jin, J.B., Lee, J., Park, H.C., Salt, D.E., Yun, D.-J., Bressan, R.A., and Hasegawa, P.M. (2006). SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 142 1548–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.