Abstract
Deletion of any of the core enzymes of the photorespiratory cycle, one of the major pathways of plant primary metabolism, results in severe air-sensitivity of the respective mutants. The peroxisomal enzyme hydroxypyruvate reductase (HPR1) represents the only exception to this rule. This indicates the presence of extraperoxisomal reactions of photorespiratory hydroxypyruvate metabolism. We have identified a second hydroxypyruvate reductase, HPR2, and present genetic and biochemical evidence that the enzyme provides a cytosolic bypass to the photorespiratory core cycle in Arabidopsis thaliana. Deletion of HPR2 results in elevated levels of hydroxypyruvate and other metabolites in leaves. Photosynthetic gas exchange is slightly altered, especially under long-day conditions. Otherwise, the mutant closely resembles wild-type plants. The combined deletion of both HPR1 and HPR2, however, results in distinct air-sensitivity and a dramatic reduction in photosynthetic performance. These results suggest that photorespiratory metabolism is not confined to chloroplasts, peroxisomes, and mitochondria but also extends to the cytosol. The extent to which cytosolic reactions contribute to the operation of the photorespiratory cycle in varying natural environments is not yet known, but it might be dynamically regulated by the availability of NADH in the context of peroxisomal redox homeostasis.
INTRODUCTION
Photorespiration represents one of the major pathways of plant primary metabolism. In terms of mass flow, it actually constitutes the second most important process in the land-based biosphere, exceeded only by photosynthesis. Carbon dioxide losses related to this process can be very high and are further elevated by warmer temperatures and drought, hence reducing the yields of important crops (Tolbert, 1997; Wingler et al., 2000).
The core of the photorespiratory cycle, as revealed by the extensive biochemical analysis of wild-type and mutant plants (Lorimer and Andrews, 1981; Tolbert, 1997; Somerville, 2001), comprises at least eight individual enzymatic reactions distributed over three different types of organelles, namely chloroplasts, peroxisomes, and mitochondria (shown in Supplemental Figure 1 online). The cycle starts in the chloroplast with the synthesis of 2-phosphoglycolate from the Calvin cycle intermediate ribulose 1,5-bisphosphate and oxygen by ribulose-1,5-bisphosphate carboxylase/oxygenase. In the course of the last two reactions of the cycle, catalyzed by the peroxisomal enzyme hydroxypyruvate reductase (HPR1; EC 1.1.1.29) and the plastidial glycerate 3-kinase, the intermediate compound hydroxypyruvate becomes converted to glycerate and eventually to another Calvin cycle intermediate, 3-phosphoglycerate. Although much of this appears as well-established textbook science, considerable gaps still remain concerning our knowledge of the cellular biology and biochemistry of photorespiration. One problem that is as yet only poorly resolved, and which is the focus of this article, concerns the possibility of multiple pathways for the conversion of hydroxypyruvate to glycerate.
From earlier biochemical studies, there is much evidence that this reaction is exclusively catalyzed by HPR1 (Tolbert et al., 1970; Yu and Huang, 1986; Heupel et al., 1991). This enzyme is assumed to operate as part of a peroxisomal multienzyme complex, preventing equilibration of hydroxypyruvate with the cytosol (Reumann, 2000). However, HPR1 might not be the only enzyme that reduces photorespiratory hydroxypyruvate. This can be presumed from specific properties of a barley (Hordeum vulgare) mutant with severely reduced activities of HPR1 (Murray et al., 1989). In contrast with the typical lack of vitality exhibited when other photorespiratory mutants are grown in air, the photosynthetic performance of this mutant was not greatly affected. Therefore, it was hypothesized that a cytosolic hydroxypyruvate reductase (HPR2; EC 1.1.1.81) (Kleczkowski and Randall, 1988) could provide a bypass in this mutant and also serve as an overflow mechanism for the utilization of hydroxypyruvate leaked from peroxisomes under conditions of maximum photorespiration in wild-type plants. The precise genetic defect in this mutant, unfortunately, has not yet been identified, and current EST databases do not exclude the presence of multiple HPR1 genes in barley (http://pgrc.ipk-gatersleben.de/cr-est/). Therefore, it is not known whether this mutant is indeed totally devoid of HPR1 activity. Moreover, the HPR2-encoding gene is not known for any plant species, which precludes studies with genetically characterized mutants. Consistent with this limited evidence, the hydroxypyruvate overflow hypothesis has not yet found much acceptance in the current literature (Wingler et al., 2000; Siedow and Day, 2001; Reumann and Weber, 2006).
Using an Arabidopsis thaliana hpr1 null mutant, we have identified HPR2. The subsequent analysis of hpr2 knockout mutants revealed elevated leaf hydroxypyruvate levels and altered gas-exchange parameters. Whereas hpr1 and, even more so, hpr2 mutants resemble wild-type plants, the combined deletion of both HPR1 and HPR2 caused a distinct air-sensitivity of the double mutant in combination with dramatic reductions in photosynthetic performance. The remarkably high capacity of the HPR2 pathway suggests that photorespiratory metabolism is not confined to the three organelles in which it was previously thought to occur, namely chloroplasts, peroxisomes, and mitochondria, but apparently also spans the cytosol.
RESULTS
HPR1 Knockout Plants Show Only Slight Visually Noticeable Impairments in Air
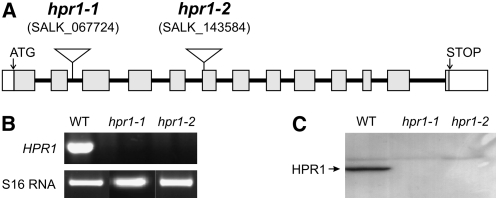
In order to examine the effects of a deletion of HPR1, which is encoded by a single gene in Arabidopsis (Mano et al., 1997), we isolated two allelic knockout mutants. In these lines, further denoted hpr1-1 and hpr1-2, T-DNA insertions reside in the second intron and in the sixth exon of the At1g68010 gene (Arabidopsis Genome Initiative, 2000), respectively (Figure 1A). HPR1-specific transcripts and HPR1 protein were undetectable (Figures 1B and 1C), which confirmed the complete knockout of the gene in each mutant. Moreover, initial measurements indicated very low (∼5% of wild-type levels) NADH-dependent hydroxypyruvate reductase activity in leaf extracts of both lines. While there was evidently no HPR1 present in these mutants, visually noticeable effects of the knockout on plant growth in normal air were small to negligible under a 12-h photoperiod (Figure 2A). Under shorter daylengths of 8 h, we observed somewhat slower growth of the hpr1 mutants than of the wild type, in combination with an approximately 4-week delay in bolting.
Figure 1.
Isolation of hpr1 Knockout Mutants.
(A) Structure of the HPR1 gene and positions of the T-DNA insertions in hpr1-1 and hpr1-2.
(B) HPR1-specific transcripts are undetectable in the two homozygous mutants by RT-PCR analysis of whole leaf RNA using S16 RNA as an internal control and mRNA from Col-0 wild-type leaf for comparison. Identical results were obtained in four independent experiments using different plant individuals for all lines.
(C) HPR1 protein is undetectable in leaf protein extracts from the two homozygous mutants by protein gel blotting with a specific antiserum.
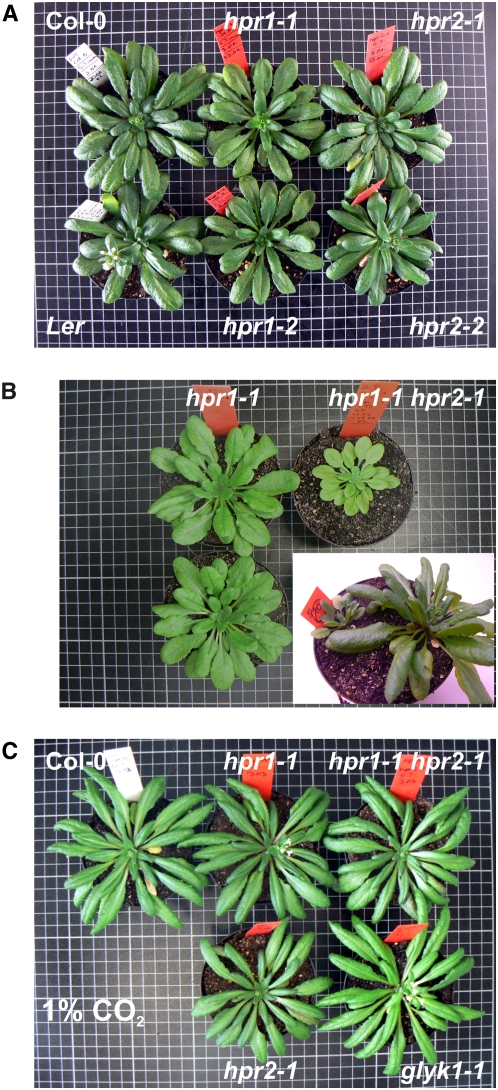
Figure 2.
The Individual Deletion of Either HPR1 or HPR2 Is Well Tolerated in Normal Air, but Combined Deletion Causes Considerable Air Sensitivity.
Plants were grown in normal air for 6 weeks with the exception of the two plants shown in the inset in (B).
(A) Four individual T-DNA insertion lines isolated during this work in comparison with wild-type plants (12/12-h light/dark cycle; Col-0 for comparison with hpr1-1, hpr1-2, and hpr2-1; Landsberg erecta [Ler] for comparison with hpr2-2).
(B) Two hpr1-1 individuals in comparison with the hpr1-1 hpr2-2 double mutant (10/14-h light/dark cycle). The inset shows a double mutant (left) next to a wild-type plant (right) from a homozygosity screening experiment.
(C) Growth at elevated CO2 normalizes the phenotype of the individual and double mutants. A glycerate 3-kinase–deficient air-hypersensitive photorespiratory mutant, glyk1-1, is shown for comparison (12/12-h light/dark cycle, 1% CO2).
To quantify the possible effects of the hpr1 knockout on gas exchange more precisely, line hpr1-1 was analyzed using a sensitive radiogasometric method (Pärnik and Keerberg, 2007). This method allows the accurate determination of true photosynthetic rates and also distinguishes between (photo)respiratory CO2 fluxes derived from primary photosynthates (metabolite pools saturated with 14C after 10 to 15 min of exposure to 14CO2) and stored photosynthates (pool turnover half-times of >1 h). In agreement with the minor effects of the mutation on growth and development, control and hpr1-1 plants displayed absolute rates of most analyzed processes, including true photosynthesis (6.84 ± 0.26 versus 6.53 ± 0.23 μmol CO2·m−2·s−1, wild type versus hpr1-1; for details, see Supplemental Table 1 online) and photorespiration (0.91 ± 0.08 versus 0.73 ± 0.06 μmol CO2·m−2·s−1), which were not very different from each other and were similar to earlier data obtained with Arabidopsis wild-type plants (Pärnik et al., 2002). Somewhat larger changes were found with respect to the decarboxylation of stored photosynthates. The respective rates were all higher in the hpr1 mutant and were brought about mainly by a higher rate of respiratory decarboxylation of stored photosynthates in hpr1-1 (0.10 ± 0.04 versus 0.23 ± 0.03 μmol CO2·m−2·s−1). In addition, respiration in the dark also occurred at distinctly higher rates in leaves of the hpr1 mutants (0.99 ± 0.05 versus 1.43 ± 0.05 μmol CO2·m−2·s−1), which indicates the accumulation of a significant pool of metabolites with long turnover half-times that become dissimilated during the dark. These alterations to respiratory metabolism could explain the slower growth of the hpr1 mutants relative to the wild type under short-day conditions.
Gene At1g79870 Encodes Arabidopsis HPR2
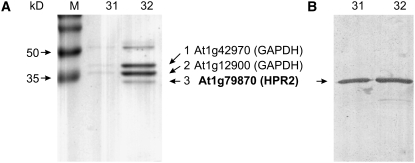
We next set out to identify the enzyme that is capable of replacing the peroxisomal hydroxypyruvate-to-glycerate conversion with such high efficiency. To this end, hpr1 knockout plants were grown and used as a source for enzyme isolation by a combination of ammonium sulfate fractionation, ion-exchange, and affinity chromatography (for details, see Supplemental Table 2 online). The NADH- and NADPH-dependent activities for the reduction of hydroxypyruvate were copurified in all relevant fractions. The final fraction, even if not perfectly pure (Figure 3A), was highly enriched in NADPH-dependent hydroxypyruvate reductase. SDS-PAGE revealed three protein bands in the expected size range, and one of them indeed showed reactivity with an antiserum previously raised against semipurified spinach (Spinacia oleracea) HPR2 (Kleczkowski and Randall, 1988).
Figure 3.
Identification of Arabidopsis HPR2.
(A) SDS-PAGE of the final fraction after purification of HPR2 from hpr1-1 leaves. Protein bands labeled with arrows were excised, trypsin-digested, and analyzed by nanoflow liquid chromatography coupled to mass spectrometry. Numbers 31 and 32 refer to 2′,5′-ADP-Sepharose fractions with the highest specific activity.
(B) Interaction with a semispecific antibody against spinach HPR2 in a protein gel blotting experiment.
All three bands were excised, and the proteins were trypsin-hydrolyzed and analyzed using nanoflow liquid chromatography coupled to mass spectrometry. The two larger proteins matched database entries for glyceraldehyde 3-phosphate dehydrogenase isoforms. The smallest protein, which was also shown to interact with the spinach anti-HPR2 antibodies (Figure 3B), was identified as Arabidopsis protein At1g79870. This protein is currently annotated as a putative oxidoreductase family protein containing a d-isomer–specific 2-hydroxyacid dehydrogenase domain (www.Arabidopsis.org). It consists of 313 amino acids with 51% sequence similarity to HPR1. The calculated molecular mass of 34.2 kD is close to the reported mass of spinach HPR2 (∼38 kD) and corresponds with the estimates from SDS-PAGE (Figure 3A). WoLF PSORT prediction (http://wolfpsort.seq.cbrc.jp) indicates confinement to the cytosol, which is in agreement with the reported cellular localization of pea (Pisum sativum) HPR2 (Kleczkowski et al., 1988). Initial analyses with green fluorescent protein–HPR2 fusion proteins in transgenic Arabidopsis also gave no indication for organelle targeting, but the clear detection of green fluorescent protein fluorescence in the cytosol was hampered by the noise produced by the autofluorescence of plant cell walls.
Arabidopsis HPR2 Is an NAD(P)H-Dependent Hydroxypyruvate Reductase
In order to provide additional biochemical evidence that At1g79870 encodes HPR2 and to compare the enzymatic properties of HPR2 with the enzymatic properties of HPR1, we expressed both enzymes in Escherichia coli and tested different substrate cofactor combinations (Table 1). Tag-purified recombinant At1g79870 was most active in the presence of NADPH and hydroxypyruvate, whereas NADH-dependent rates were about four times lower. The enzyme also accepts glyoxylate as a substrate, but at a considerably lower efficiency in the NADH-driven reaction. In accordance with published data on the spinach enzyme (Kleczkowski et al., 1991), catalysis by At1g79870 with all four substrate–coenzyme combinations was strongly inhibited by oxalate. By contrast, inhibitory effects of this compound on HPR1 were much smaller. These combined data from protein identification, interaction with antibodies raised against the spinach enzyme, and catalytic parameters establish At1g70870 as the cytosolic NAD(P)H-dependent hydroxypyruvate reductase, HPR2, in Arabidopsis.
Table 1.
HPR2 Is an NADPH-Dependent Hydroxypyruvate Reductase but Shows Relaxed Substrate and Cofactor Specificity and Is Strongly Inhibited by Oxalate
| Hydroxypyruvate Reductase | Without Oxalate Mean ± sd | With Oxalate Mean ± sd | % |
|---|---|---|---|
| HPR2 | |||
| Hydroxypyruvate:NADH | 1.40 ± 0.12 | 0.300 ± 0.080 | 21 |
| Hydroxypyruvate:NADPH | 5.50 ± 0.43 | 0.700 ± 0.090 | 13 |
| Glyoxylate:NADH | 0.36 ± 0.02 | 0.010 ± 0.001 | 3 |
| Glyoxylate:NADPH | 4.60 ± 0.36 | 0.100 ± 0.026 | 2 |
| HPR1 | |||
| Hydroxypyruvate:NADH | 222.00 ± 15.70 | 147.000 ± 22.000 | 66 |
| Hydroxypyruvate:NADPH | 12.00 ± 1.40 | 11.600 ± 1.100 | 97 |
| Glyoxylate:NADH | 6.40 ± 0.60 | 5.800 ± 0.4000 | 90 |
| Glyoxylate:NADPH | 0.26 ± 0.03 | 0.200 ± 0.020 | 77 |
Mean activities ± sd from three measurements with tag-purified recombinant HPR2 and HPR1 (μmol·min−1·mg−1 protein) or percentage values after inhibition are shown.
HPR2 Knockout Plants Do Not Show Visually Noticeable Impairments in Air
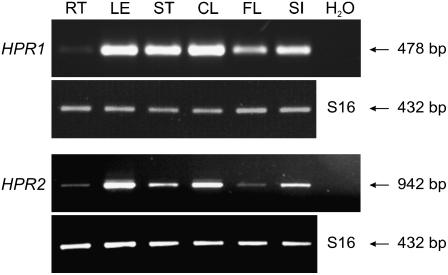
The RT-PCR analysis shown in Figure 4 demonstrates that the expression of HPR2, in comparison with HPR1, is very similarly distributed over the individual organs of Arabidopsis, with highest levels in photosynthesizing organs.
Figure 4.
RT-PCR Analysis of HPR1 and HPR2 Transcripts in Different Organs of Wild-Type Arabidopsis.
Identical results were obtained in three independent experiments using RNA isolated from different plants. RT, roots; LE, rosette leaves; ST, stems; CL, cauline leaves; FL, flowers, SI, siliques. The S16 RNA amplicon was used for internal calibration.
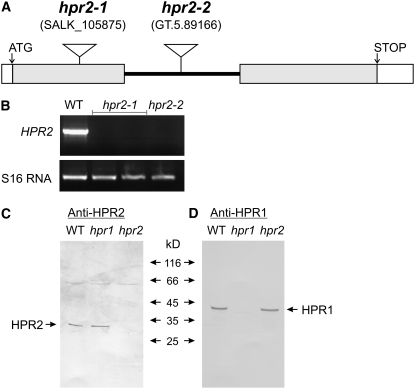
In order to next examine the metabolic function of HPR2, we isolated two allelic knockout mutants, harboring T-DNA insertions in exon 1 (hpr2-1) and intron 1 (hpr2-2) of gene At1g70870 (Figure 5A). HPR2-specific transcripts were undetectable in whole leaf mRNA (Figure 5B) of the respective homozygous lines, which were indistinguishable from wild-type plants in appearance (Figure 2A). Accordingly, HPR2 protein could not be detected in a protein gel blotting experiment (Figures 5C and 5D). These data confirm complete deletion of HPR2 at the mRNA and protein levels. We also observed changes in leaf enzyme activity levels (Figure 6), but these data are of limited diagnostic value. This is because HPR1, which is highly active in the hpr2 knockout plants, shows some reactivity with NADPH (Table 1) (Tolbert et al., 1970).
Figure 5.
Isolation of hpr2 Knockout Mutants.
(A) The structure of the HPR2 gene and positions of the T-DNA insertions in hpr2-1 and hpr2-2.
(B) HPR2-specific transcripts are undetectable in both homozygous mutants by RT-PCR analysis of whole-leaf RNA extracts using the S16 RNA amplicon as an internal control and Col-0 wild-type leaf mRNA for comparison.
(C) and (D) HPR2 (C) and HPR1 (D) proteins are undetectable by protein gel blotting in their respective mutants. This experiment shows the results for hpr1-1 and hpr2-1, which are identical to those obtained with hpr1-2 and hpr2-2.
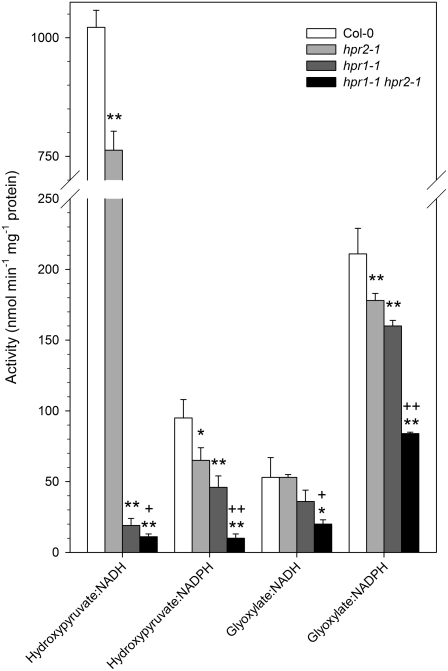
Figure 6.
Activities of Hydroxypyruvate- and Glyoxylate-Reducing Enzymes in Leaves of Wild-Type and Mutant Plants in the Presence of Various Substrate–Cofactor Combinations.
Plants were grown in normal air, and rosette leaves were analyzed at developmental stage 5.1 (Boyes et al., 2001). Mean values ± sd from three measurements each with four different plants per line (n = 12) are shown. Activity significantly different from the wild type (* P < 0.05, ** P < 0.01) and activity significantly different between hpr1-1 and the double mutant (+ P < 0.05, ++ P < 0.01) are indicated. Wild type, white bars; hpr2-1, light gray bars; hpr1-1, dark gray bars; hpr1-1 hpr2-1, black bars.
The Combined Deletion of HPR1 and HPR2 Is Detrimental to Air-Grown Mutants
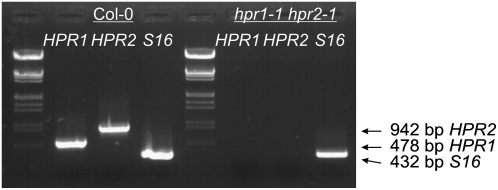
To examine whether the combined knockout of both genes impairs photorespiratory metabolism more strongly than the individual deletion of either HPR1 or HPR2, we next crossed the hpr1-1 mutant with hpr2-1 plants. Individual F2 plants were tested for homozygousity with respect to both gene defects (Figure 7), and the absence of both proteins was verified by protein gel blotting. While these double mutants were still able to grow in air, their performance was considerably reduced in comparison with the hpr1 knockout lines (Figure 2B). Growth under the nonphotorespiratory conditions of air enriched with 1% CO2 restored the phenotype to normal (Figure 2C). These observations show the significance of HPR2 for photorespiratory metabolism in hpr1 knockout plants and suggest a contribution, in addition to HPR1, to photorespiratory hydroxypyruvate turnover in wild-type plants as well.
Figure 7.
Crossing of hpr1-1 and hpr2-1 Leads to Functional Inactivation of Both Genes.
RT-PCR analysis of whole-leaf RNA extracts demonstrates the absence of HPR1 and HPR2 transcripts in the homozygous double mutant. Col-0 wild-type RNA and the S16 mRNA amplicon were used as controls. Identical results were obtained in two independent experiments using different plant individuals for both lines.
The subsequent determination of leaf enzyme activities, alterations to the metabolome, and gas-exchange parameters in the individual and double mutant lines fully supported this notion. Following up from initial analyses with hpr1 and hpr2 plants, all of these experiments (Figures 6, 8, and 9) were performed with the same set of plants grown under identical conditions with a 10-h photoperiod to developmental stage 5.1 (first flower buds visible) (Boyes et al., 2001) to ensure maximum comparability between the wild type and the three mutant lines.
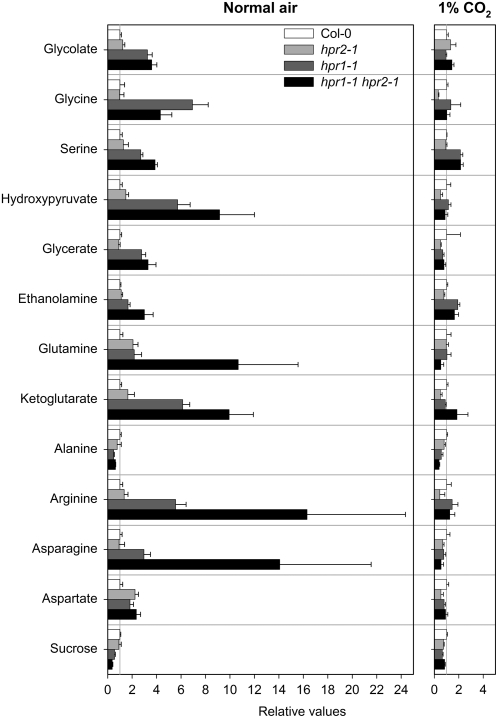
Figure 8.
Alteration of the Leaf Content of Selected Metabolites in the Individual hpr1-1 and hpr2-1 Knockout Plants and in the hpr1-1 hpr2-1 Double Mutant.
Plants were grown in normal air and in air with 1% CO2, and rosette leaves of six individual plants per line were analyzed at developmental stage 5.1 (Boyes et al., 2001). Mutant/wild-type ratios of mean metabolite contents ± sd (n = 6) are shown, where the mean wild-type values are arbitrarily set to 1. Further details are shown in Supplemental Table 3 online. Wild type, white bars; hpr2-1, light gray bars; hpr1-1, dark gray bars; hpr1-1 hpr2-1, black bars.
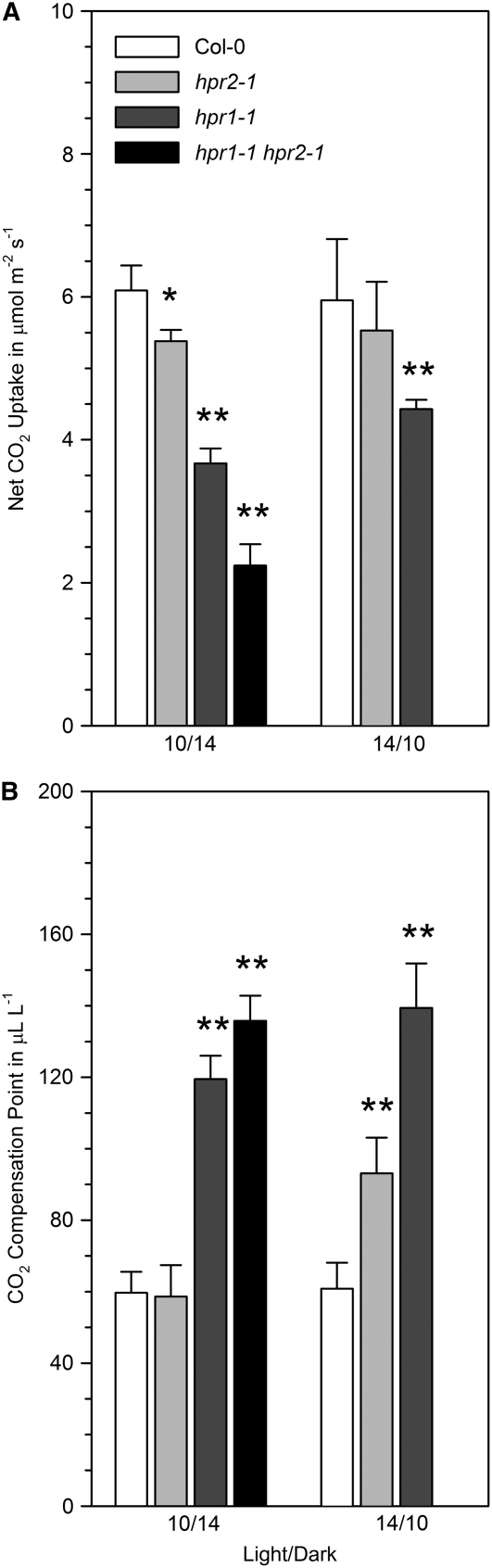
Figure 9.
Effects on Net Photosynthetic Rates and CO2 Compensation Points.
(A) Net photosynthetic rates of plants grown with a 10-h or a 14-h light period.
(B) CO2 compensation points at 21% oxygen of plants grown with a 10-h or a 14-h light period.
Fully expanded rosette leaves were analyzed at developmental stage 5.1 (Boyes et al., 2001). Bars show mean values ± sd from measurements with five individual plants per line. Asterisks indicate significance levels in comparison with the wild type (* P < 0.05, ** P < 0.01). The double mutant flowers very early at the longer photoperiod, which rendered the acquisition of comparable data impossible for these conditions. Wild type, white bars; hpr2-1, light gray bars; hpr1-1, dark gray bars; hpr1-1 hpr2-1, black bars.
HPR1 and HPR2 Are the Major Hydroxypyruvate-Reducing Enzymes in Leaves
The deletion of either the HPR1 or HPR2 enzyme resulted in reduced activities with any of the tested substrate–cofactor combinations (Figure 6). This broad-range effect was to be expected due to the relaxed substrate and cofactor specificities of HPR1 and, even more so, of HPR2, as already discussed. The comparison of the total NADH- plus NADPH-dependent capacity for hydroxypyruvate reduction shows that ∼6% of the total activity of the wild type was left in the hpr1 mutant. This fraction was further reduced to ∼2% of total activity after additional deletion of HPR2. The enzymatic nature of this residual activity in the double mutant is not clear, but its presence is not surprising: despite the severe metabolic defect, the double mutant still survives and grows, although very slowly, in normal air.
Knockout of Both HPR1 and HPR2 Alters Steady State Metabolite Profiles
In comparison with the effects of other mutations on the photorespiratory pathway, such as an up to 200-fold accumulation of glycerate in a glycerate kinase–deficient mutant (Boldt et al., 2005), alterations to the leaf metabolome of the individual hpr1 and hpr2 knockout lines were relatively small (Figure 8). They are large, however, in comparison with many broad-range metabolite profiles (Roessner et al., 2001; Schauer et al., 2006) and become much more distinct in the double mutant (Figure 8; see Supplemental Table 3 online). Hydroxypyruvate levels are about sixfold higher in hpr1 plants and ninefold higher in the double mutant than in wild-type plants. It is interesting that the hydroxypyruvate level is also somewhat (albeit not statistically significantly) elevated in the HPR2-deficient plants. Growth under elevated CO2 restored hydroxypyruvate levels to normal in all three mutant lines. This supports our hypothesis that HPR1 and HPR2 are involved in photorespiratory carbon metabolism, both in a nonobligatory manner, wherein HPR1 catalyzes most of the hydroxypyruvate-to-glycerate conversion under many environmental conditions.
The leaf glycerate content remains essentially unaltered in the hpr2 mutant and is increased in hpr1 and the double knockout plant (Figure 8; see Supplemental Table 3 online). This increase is an unexpected response and possibly related to the reported irreversibility of the HPR2 reaction (Kleczkowski and Randall, 1988). By contrast, the HPR1 reaction is fully reversible. The elevated leaf Gly levels of the hpr1 and the double knockout mutants suggest that these mutants are also impaired in their conversion of Gly to Ser, with the intriguing effect that the double mutant is less severely affected than the hpr1 mutant. The increased Ser contents, especially in the hpr1 and double mutants, could be responsible for the enhanced level of ethanolamine, most of which is produced by decarboxylation of free Ser (Rontein et al., 2003). It also indicates that Ser:glyoxylate aminotransferase operates close to equilibrium in the photorespiratory cycle.
Furthermore, the levels of the important metabolites of nitrate assimilation, α-ketoglutarate and Gln, were increased in both hpr1 and hpr2 mutants. In contrast with the effect observed for Gly, these effects were exacerbated in the double mutant. They correspond to the altered respiratory metabolism observed during the radiogasometric measurements (see Supplemental Table 1 online), and it appears as if these changes are to some extent due to similar substrate requirements of the respiratory and photorespiratory pathways (Nunes-Nesi et al., 2008). While the changes above are likely directly related to the modifications of photorespiratory and energy metabolism, several other changes were also observed. Most prominent among these was the decrease in Ala content coupled with enhanced levels of Arg, Asn, and Asp in the hpr1 mutant and the double knockout plant (see Supplemental Table 3 online). Looking at the unaltered levels of a broad range of other metabolites, however, the impact of the genetic perturbation in the mutant lines studied here was highly specific.
Knockout of Both HPR1 and HPR2 Alters Photorespiratory 13C Fluxes
Given that isotope analyses are potentially far more sensitive than steady state measurements, we adopted an isotope tracing approach (Roessner-Tunali et al., 2004) to confirm that the hpr2 mutant was impaired in the photorespiratory conversion of hydroxypyruvate to glycerate. To this end, leaf discs were fed with 10 mM [13C]Glc via the transpiration stream, and the accumulation of isotope in the intermediates of the photorespiratory pathway was followed during the time course of the experiment (Table 2). Considering the complexity involved in evaluating such data sets (Baxter et al., 2007), we restricted our analyses to the relevant intermediates of the photorespiratory pathway and our calculations to the total isotope accumulation in each of these pools.
Table 2.
Redistribution of Label following Feeding of Leaf Discs with [U-13C]Glc in the Individual hpr1-1 and hpr2-1 Knockout Plants and the hpr1-1 hpr2-1 Double Mutant
| Redistribution of Label (μmol C1 equivalents·g−1 fresh weight·h−1)
|
||||
|---|---|---|---|---|
| Metabolite | Col-0 | hpr1-1 | hpr2-1 | hpr1-1 hpr2-1 |
| Gly | 0.013465 ± 0.002167 | 0.044554 ± 0.010226 | 0.016896 ± 0.001430 | 0.076195 ± 0.009705 |
| Ser | 0.157030 ± 0.021351 | 0.271353 ± 0.030008 | 0.148851 ± 0.027776 | 0.067525 ± 0.011927 |
| Hydroxypyruvate | 0.000007 ± 0.000003 | 0.000727 ± 0.000198 | 0.000022 ± 0.000005 | 0.000745 ± 0.000200 |
| Glycerate | 0.004239 ± 0.000328 | 0.009824 ± 0.002895 | 0.006039 ± 0.000597 | 0.007983 ± 0.002068 |
Values are presented as means ± se from four determinants per line. Numbers in boldface indicate values that were determined by the t test to be significantly different (P < 0.05) from the wild type.
When the labeling patterns of the mutants are compared with one another, they reveal that both HPR1 and HPR2 play roles in the conversion of hydroxypyruvate to glycerate. Not surprisingly, hpr1 accumulated over 103 times as much label in hydroxypyruvate as the wild type did, whereas hpr2 accumulated 3 times as much label as the wild type, showing that HPR1 plays the dominant role in photorespiration. Interestingly, the double mutant accumulated over 106 times as much label in hydroxypyruvate as the wild type. While this tendency corresponds with the expected additive effects of the two mutations in the wild type, the difference to hpr1 is not significant if one considers the errors of each determination.
The changes in labeling of the other metabolite pools were far less dramatic, with hpr1, hpr2, and the double mutant accumulating 3-, 1.3-, and 6-fold more label in Gly; 1.7-, 0.9-, and 0.4-fold more label in Ser; and 2.4-, 1.7-, and 2.0-fold more label in glycerate, respectively. All of these alterations could be anticipated considering the crossover theorem of Crabtree and Newsholme (1987). From these kinetic data, which support and extend the results from our analysis of steady state metabolite levels, we infer a contribution of HPR2 to photosynthetic–photorespiratory metabolism not only in HPR1-deficient plants but also in wild-type plants.
Knockout of Either HPR1 or HPR2 Alters Photosynthetic Gas Exchange
Considering the possibility that light intensity and daylength could affect peroxisomal redox homeostasis and hence the partitioning of hydroxypyruvate between HPR1 and HPR2, we analyzed photosynthetic gas-exchange parameters with two sets of plants, one grown under a 10-h photoperiod (same set as for Figures 6 and 8) and one grown under a 14-h photoperiod (Figure 9). Both photoperiods are longer than that used for the radiogasometric measurements, and the data from these two different approaches, therefore, are not fully comparable.
The deletion of HPR2 and, to a greater extent, of HPR1 decreased net CO2 uptake. Moreover, the CO2 compensation point, which reflects the balance of photosynthetic CO2 uptake and photorespiratory CO2 release (von Caemmerer, 2000), also displayed a response to the deletion of HPR1. The deletion of HPR2 had no clear effect on CO2 compensation point in plants grown under the shorter photoperiod but resulted in significant changes when plants were exposed to a 14-h photoperiod. The deletion of both enzymes produced the largest effects. Notably, the double mutant flowered very early when grown under 14 h of daily light. Leaves of the double mutant were small under these conditions and prohibited the acquisition of comparable data.
While some of the observed changes were only marginally significant, our data nevertheless indicate that the deletion of HPR2 alone impairs photosynthetic CO2 exchange. This becomes even more obvious in the double mutant, where these impairments were always additive to the negative effects caused by the deletion of HPR1. The results obtained after deletion of HPR2 alone or in combination with the deletion of HPR1 suggest an active role of HPR2 in photorespiratory metabolism under temperate conditions.
DISCUSSION
Photorespiratory hydroxypyruvate is produced within the peroxisomal matrix, where it exclusively originates from Ser transamination by Ser:glyoxylate aminotransferase (AGT1) (Liepman and Olsen, 2001). This is evident from the lethality of both chemical and T-DNA insertion mutations in the AGT1 gene in combination with the normalization of the mutants observed under elevated CO2 (Somerville and Ogren, 1980; Liepman and Olsen, 2001). Both effects are typical features of photorespiratory mutants (Blackwell et al., 1988; Somerville, 2001; Boldt et al., 2005; Voll et al., 2006; Schwarte and Bauwe, 2007).
The general assumption of an exclusively peroxisome-localized photorespiratory conversion of hydroxypyruvate to glycerate by HPR1 may not be comprehensive. The hpr1 mutants are unique among the photorespiratory mutants in that they show only relatively little air-sensitivity. This was suspected from earlier work (Murray et al., 1989) and was substantiated by the results of our analyses (Figures 2A, 2B, 8, and 9). These studies, based on the identification of HPR2 and the use of genetically defined knockout mutants, show that photorespiratory carbon flux can efficiently elude one of the core reactions of the photorespiratory cycle. In contrast with the reported impermeability of the peroxisomal matrix for hydroxypyruvate (Heupel et al., 1991; Reumann et al., 1994), our data also provide indirect evidence that this metabolite can relatively easily equilibrate with the cytosol when no HPR1 is present within the peroxisome. This conclusion arises from the only minor effects on plant growth observed with the hpr1 mutant in comparison with the detrimental effects of the additional deletion of HPR2 (Figure 2B). It remains to be examined how this relates to the suggested peroxisomal channeling of photorespiratory intermediates (Reumann, 2000), but it is not unlikely that such equilibration also occurs in wild-type plants, although on a lower steady state concentration level.
The capacity of this cytosolic bypass is apparently high enough to nearly fully match the needs of photorespiratory 2-phosphoglycolate recycling in Arabidopsis, at least under medium to long photoperiods. Arabidopsis is adapted to temperate environments (Shindo et al., 2007), and the plants used in our experiments were kept under temperate conditions. In this context, it is interesting that light-regulated alternative splicing of one pre-mRNA results in a peroxisomal and a cytosolic HPR1 form in Cucurbita species, which are adapted to warmer environments than Arabidopsis (Mano et al., 1999). These authors hypothesized that the cytosolic form of HPR1 could be important for metabolizing hydroxypyruvate leaked from the peroxisomes, but a possible relation to photorespiratory metabolism has not, to our knowledge, been reported in these plants. It is also remarkable that an orthologous protein has been described as hydroxyphenylpyruvate reductase in the rosmarinic acid biosynthetic pathway of flame nettle (Solenostemon scutellarioides) (Kim et al., 2004). The reported specific activity with 3,4-dihydroxyphenylpyruvate, however, is only ∼1% of the hydroxypyruvate-dependent specific activity of Arabidopsis HPR2. It could well be that HPR2 contributes to secondary metabolism in these warm-habitat plants of the mint family, but its major function is clearly photorespiration-related.
Unlike all other photorespiratory enzymes, both HPR1 and HPR2 are dispensable for survival in normal air; however, at least one of these enzymes must be present in Arabidopsis for plants to grow to maturity. HPR1 plays the dominant role under many, if not all, natural conditions. Although the contribution of HPR2 to photorespiratory metabolism may be small under temperate environmental conditions, it could become much more important under other conditions.
We hypothesize that the extent to which photorespiratory hydroxypyruvate flux is diverted from the peroxisome via the cytosol in varying natural environments might be dynamically regulated by the availability of NADH in the context of peroxisomal redox homeostasis. Larger photorespiratory flux rates require larger amounts of NADH for hydroxypyruvate reduction, which are provided to the peroxisome by the oxidation of malate delivered from chloroplasts and mitochondria (Scheibe et al., 2005; Reumann and Weber, 2006). This process of intraperoxisomal NADH generation is seen as the rate-limiting step in the conversion of Ser to glycerate (Yu and Huang, 1986). That is, all conditions that shift peroxisomal redox homeostasis to a more oxidized equilibrium and hence to lower peroxisomal NADH levels would restrain the HPR1 reaction and divert a fraction of photorespiratory carbon flux via HPR2 (and possibly via cytosolic HPR1 in Cucurbita species). Interestingly, an Arabidopsis double mutant lacking both isoforms of peroxisomal malate dehydrogenase was recently reported to be viable in normal air conditions with photosynthetic rates only slightly lower than those in the wild type (Cousins et al., 2008). It is tempting to speculate that most hydroxypyruvate is diverted to the cytosol in this mutant.
It appears that HPR2 is the major component of the extraperoxisomal hydroxypyruvate-reducing pathway in Arabidopsis. The hpr1 hpr2 double mutation is not lethal, however, and the double knockout plants are able to slowly grow in normal air (Figures 2B and 9). This suggests that minor contributions can be made by additional enzymes. In contrast with an earlier report on spinach glyoxylate reductase, which had small activity with hydroxypyruvate (Kleczkowski et al., 1986), two recently identified Arabidopsis glyoxylate reductases, one cytosolic and one plastidial, do not show HPR activity (Hoover et al., 2007; Simpson et al., 2008). More likely candidate enzymes for these minor activities would be other 1-hydroxacid dehydrogenases with relaxed substrate specificity, for example lactate dehydrogenase (Betsche, 1981), or the two putative Arabidopsis proteins At1g12550 and At2g45630. These latter proteins share ∼50% sequence similarity with HPR2 and also harbor a d-isomer–specific 2-hydroxyacid dehydrogenase domain.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotypes Columbia (Col-0) and Landsberg erecta were used as wild-type references. SALK lines SALK067724 (hpr1-1), SALK143584 (hpr1-2), and SALK105875 (hpr2-1) and gene trap line GT.89166 (hpr2-2) were obtained from the Nottingham Arabidopsis Stock Centre (Sundaresan et al., 1995; Alonso et al., 2003). Unless otherwise stated, plants were grown in normal air (380 to 400 μL/L CO2) and in air with elevated CO2 (1%) in controlled environment chambers (Conviron; 10-h photoperiod, 22/18°C, ∼130 μmol·m−2·s−1 irradiance) on a 4:1 mixture of soil (Type Mini Tray; Einheitserdewerk Uetersen) and vermiculite and were regularly watered with 0.2% Wuxal liquid fertilizer (Aglukon).
Isolation of Knockout Mutants and RT-PCR
To verify the insertion, leaf DNA was PCR-amplified (1 min at 94°C, 1 min at 58°C, 1.5 min at 72°C; 35 cycles) with primers specific for the left border (R175 for SALK lines and R576 for gene trap lines; details for all primers are shown in Supplemental Table 4 online) or the right border (R409) and HPR1- or HPR2-specific primers (R379 and R380 for HPR1; R666 and R668 for HPR2). The resulting fragments were then sequenced. Zygosity was examined by PCR amplification of leaf DNA with primer combinations R379 and R380 (for hpr1) and R666 and R668 (for hpr2). The knockout of HPR1 and HPR2 in homozygous plants was verified by RT-PCR using 2.5 μg of leaf RNA for cDNA synthesis (Nucleospin RNA plant kit [Macherey-Nagel] and RevertAid cDNA synthesis kit [MBI Fermentas]) and the primer combination R663 (sense) and R664 (antisense), yielding a 478-bp PCR product for HPR1, or the primer combination R666 (sense) and R668 (antisense), resulting in the expected 942-bp PCR fragment for HPR2 in wild-type plants but not in the homozygous knockout mutants. Before PCR analysis, cDNA amounts were calibrated according to signals from 432-bp fragments obtained by PCR amplification of the constitutively expressed 40S ribosomal protein S16 gene, with primers R176 (sense) and R177 (antisense). Gels were stained with ethidium bromide.
Purification and Identification of HPR2
Approximately 500 g of Arabidopsis hpr1-1 leaves was homogenized in 1 liter of ice-cold extraction buffer (25 mM HEPES-KOH, 1 mM EDTA, 1 mM MgCl2, 1 mM KCl, 10 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, and 10% glycerol, pH 7.6). All subsequent steps followed a previously published procedure (Kleczkowski and Randall, 1988). The final HPR2 fraction, eluted from a 1.6- × 7-cm 2′,5′-ADP-Sepharose 4B column (GE Healthcare) with a linear NADPH concentration gradient (0 to 2 mM NADPH), was further analyzed by SDS-PAGE and nanoliquid chromatography tandem mass spectrometry analysis as described elsewhere (Boldt et al., 2005).
Heterologous Expression of HPR1 and HPR2
cDNA, produced as described above, from whole leaf RNA was PCR-amplified with the primer pairs R686 (sense) and R687 (antisense) to isolate HPR1 and R659 (sense) and R660 (antisense) to isolate HPR2. These primers (details listed in Supplemental Table 4 online) were also used to introduce restriction sites (XhoI, SpeI, and KpnI) for later excision and cloning into the expression vector. The resulting fragments, encoding the entire HPR1 and HPR2 sequences, were first cloned into pGEM-T Easy (Promega) and then, after sequence confirmation, ligated in-frame into the expression vector pBAD/HisA (Invitrogen). Recombinant HPR1 and HPR2 were isolated using nickel-nitrilotriacetic acid agarose affinity chromatography (HisTrap; GE Healthcare).
Enzyme Assays
One hundred milligrams of leaf tissue was homogenized in 1.0 mL of extraction buffer containing 25 mM HEPES-KOH (1 mM MgCl2, 1 mM KCl, 10 mM β-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride, pH 7.6). The supernatant obtained after centrifugation at 4°C at 20,000g for 20 min was used for the determination of hydroxypyruvate and glyoxylate reductase activity, according to published procedures (Tolbert et al., 1970; Husic and Tolbert, 1987). Protein concentrations were determined according to Bradford (1976), using BSA as a standard.
Immunological Studies
SDS-PAGE of whole leaf protein extracts and protein gel blotting experiments were performed according to standard protocols. The production of antibodies against HPR1 and HPR2, both purified and semipurified from spinach (Spinacia oleracea), has been described previously (Kleczkowski et al., 1986; Kleczkowski and Randall, 1988).
Metabolite Extraction, Gas Chromatography–Time of Flight–Mass Spectrometry Analysis, and Enzymatic Determination
Metabolite extraction and analysis were performed essentially as described (Lisec et al., 2006). The abundance of 13C was determined as described previously (Roessner-Tunali et al., 2004; Sweetlove et al., 2006). The leaf discs were taken from 5-week-old plants, growing in normal air, and incubated in 10 mM MES-KOH solution, pH 6.5, 10 mM [U-13C]Glc for 2 h at an irradiance of 80 μmol·m−2·s−1. The molecular accumulation of isotope was determined as described by Sienkiewicz-Porzucek et al. (2008).
Standard and Radiogasometric Measurement of CO2 Exchange
Standard gas-exchange analyses (net photosynthetic rates and CO2 compensation points) were performed using a Licor-6400 gas-exchange system (Li-Cor) and fully developed rosette leaves from plants grown in normal air conditions to developmental stage 5.1, as described above. Measurements were performed at a photosynthetic photon flux density of 250 μmol·m−2·s−1, respectively, supplied by an in-built red/blue LED light source, 380 μL/L CO2, 21% O2, and a leaf temperature of 25°C.
CO2 exchange was radiogasometrically measured in the second half of the photoperiod (after 4 h of illumination) at 25°C and at growth irradiance with fully developed rosette leaves from 10-week-old plants grown in an 8-h-light/16-h-dark cycle with a photon flux density of 200 μmol·m−2·s−1. Details have been described elsewhere (Pärnik and Keerberg, 2007).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: At1g68010 (HPR1), At1g79870 (HPR2), and At2g09990 (40S ribosomal protein S16).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Photorespiratory Cycle Spans Four Cellular Compartments, Including the Cytosol.
Supplemental Table 1. True Photosynthesis and Decarboxylation Rates of Metabolite Pools with Different Turnover Half-Times Measured at 21% Oxygen.
Supplemental Table 2. Purification Scheme for the Isolation of Arabidopsis HPR2 from hpr1 Knockout Plants.
Supplemental Table 3. Alteration of the Leaf Contents of Selected Metabolites in the Individual hpr1-1 and hpr2-1 Knockout Plants and the hpr1-1 hpr2-1 Double Mutant.
Supplemental Table 4. Primers used for PCR amplification of genomic DNA and cDNA.
Supplementary Material
Acknowledgments
We are grateful to Thomas Hartwig and to Ralf Boldt and Üner Kolukisaoglu for initial contributions to this project. This work would not have been possible without the mutant lines provided by the Nottingham Arabidopsis Stock Centre and the support received from the Deutsche Forschungsgemeinschaft (Grant BA 1177/7 to H.B.) and the Estonian Science Foundation (Project 5989 to T.P. and O.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Hermann Bauwe (hermann.bauwe@uni-rostock.de).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Baxter, C.J., Liu, J.L., Fernie, A.R., and Sweetlove, L.J. (2007). Determination of metabolic fluxes in a non-steady-state system. Phytochemistry 68 2313–2319. [DOI] [PubMed] [Google Scholar]
- Betsche, T. (1981). L-Lactate dehydrogenase from leaves of higher plants—Kinetics and regulation of the enzyme from lettuce (Lactuca sativa L). Biochem. J. 195 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell, R.D., Murray, A.J.S., Lea, P.J., Kendall, A., Hall, N.P., Turner, J.C., and Wallsgrove, R.M. (1988). The value of mutants unable to carry out photorespiration. Photosynth. Res. 16 155–176. [DOI] [PubMed] [Google Scholar]
- Boldt, R., Edner, C., Kolukisaoglu, Ü., Hagemann, M., Weckwerth, W., Wienkoop, S., Morgenthal, K., and Bauwe, H. (2005). D-Glycerate 3-kinase, the last unknown enzyme in the photorespiratory cycle in Arabidopsis, belongs to a novel kinase family. Plant Cell 17 2413–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes, D.C., Zayed, A.M., Ascenzi, R., McCaskill, A.J., Hoffman, N.E., Davis, K.R., and Gorlach, J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cousins, A.B., Pracharoenwattana, I., Zhou, W., Smith, S.M., and Badger, M.R. (2008). Peroxisomal malate dehydrogenase is not essential for photorespiration in Arabidopsis but its absence causes an increase in the stoichiometry of photorespiratory CO2 release. Plant Physiol. 148: 786–795. [DOI] [PMC free article] [PubMed]
- Crabtree, B., and Newsholme, E.A. (1987). A systematic approach to describing and analyzing metabolic control systems. Trends Biochem. Sci. 12 4. [Google Scholar]
- Heupel, R., Markgraf, T., Robinson, D.G., and Heldt, H.W. (1991). Compartmentation studies on spinach leaf peroxisomes: Evidence for channeling of photorespiratory metabolites in peroxisomes devoid of intact boundary membrane. Plant Physiol. 96 971–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover, G.J., Van Cauwenberghe, O.R., Breitkreuiz, K.E., Clark, S.M., Merrill, A.R., and Shelp, B.J. (2007). Characteristics of an Arabidopsis glyoxylate reductase: General biochemical properties and substrate specificity for the recombinant protein, and developmental expression and implications for glyoxylate and succinic semialdehyde metabolism in planta. Can. J. Bot. 85 883–895. [Google Scholar]
- Husic, D.W., and Tolbert, N.E. (1987). NADH:hydroxypyruvate reductase and NADPH:glyoxylate reductase in algae: Partial purification and characterization from Chlamydomonas reinhardtii. Arch. Biochem. Biophys. 252 396–408. [DOI] [PubMed] [Google Scholar]
- Kim, K.H., Janiak, V., and Petersen, M. (2004). Purification, cloning and functional expression of hydroxyphenylpyruvate reductase involved in rosmarinic acid biosynthesis in cell cultures of Coleus blumei. Plant Mol. Biol. 54 311–323. [DOI] [PubMed] [Google Scholar]
- Kleczkowski, L.A., Givan, C.V., Hodgson, J.M., and Randall, D.D. (1988). Subcellular location of NADPH-dependent hydroxypyruvate reductase activity in leaf protoplasts of Pisum sativum L. and its role in photorespiratory metabolism. Plant Physiol. 88 1182–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski, L.A., and Randall, D.D. (1988). Purification and characterization of a novel NADPH(NADH)-dependent hydroxypyruvate reductase from spinach leaves. Comparison of immunological properties of leaf hydroxypyruvate reductases. Biochem. J. 250 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski, L.A., Randall, D.D., and Blevins, D.G. (1986). Purification and characterization of a novel NADPH(NADH)-dependent glyoxylate reductase from spinach leaves. Comparison of immunological properties of leaf glyoxylate reductase and hydroxypyruvate reductase. Biochem. J. 239 653–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleczkowski, L.A., Randall, D.D., and Edwards, G.E. (1991). Oxalate as a potent and selective inhibitor of spinach (Spinacia oleracea) leaf NADPH-dependent hydroxypyruvate reductase. Biochem. J. 276 125–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman, A.H., and Olsen, L.J. (2001). Peroxisomal alanine:glyoxylate aminotransferase (AGT1) is a photorespiratory enzyme with multiple substrates in Arabidopsis thaliana. Plant J. 25 487–498. [DOI] [PubMed] [Google Scholar]
- Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., and Fernie, A.R. (2006). Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protocols 1 387–396. [DOI] [PubMed] [Google Scholar]
- Lorimer, G.H., and Andrews, T.J. (1981). The C2 chemo- and photorespiratory carbon oxidation cycle. In The Biochemistry of Plants, Vol. 8, M.D. Hatch and N.K. Boardman, eds (New York: Academic Press), pp. 329–374.
- Mano, S., Hayashi, M., Kondo, M., and Nishimura, M. (1997). Hydroxypyruvate reductase with a carboxy-terminal targeting signal to microbodies is expressed in Arabidopsis. Plant Cell Physiol. 38 449–455. [DOI] [PubMed] [Google Scholar]
- Mano, S., Hayashi, M., and Nishimura, M. (1999). Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17 309–320. [DOI] [PubMed] [Google Scholar]
- Murray, A.J.S., Blackwell, R.D., and Lea, P.J. (1989). Metabolism of hydroxypyruvate in a mutant of barley lacking NADH-dependent hydroxypyruvate reductase, an important photorespiratory enzyme activity. Plant Physiol. 91 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi, A., Sulpice, R., Gibon, Y., and Fernie, A.R. (2008). The enigmatic contribution of mitochondrial function in photosynthesis. J. Exp. Bot. 59 1675–1684. [DOI] [PubMed] [Google Scholar]
- Pärnik, T., and Keerberg, O. (2007). Advanced radiogasometric method for the determination of the rates of photorespiratory and respiratory decarboxylations of primary and stored photosynthates under steady-state photosynthesis. Physiol. Plant. 129 34–44. [Google Scholar]
- Pärnik, T.R., Voronin, P.Y., Ivanova, H.N., and Keerberg, O.F. (2002). Respiratory CO2 fluxes in photosynthesizing leaves of C3 species varying in rates of starch synthesis. Russ. J. Plant Physiol. 49 729–735. [Google Scholar]
- Reumann, S. (2000). The structural properties of plant peroxisomes and their metabolic significance. Biol. Chem. 381 639–648. [DOI] [PubMed] [Google Scholar]
- Reumann, S., Heupel, R., and Heldt, H.W. (1994). Compartmentation studies on spinach leaf peroxisomes. 2. Evidence for the transfer of reductant from the cytosol to the peroxisomal compartment via a malate shuttle. Planta 193 167–173. [Google Scholar]
- Reumann, S., and Weber, A.P. (2006). Plant peroxisomes respire in the light: Some gaps of the photorespiratory C2 cycle have become filled—others remain. Biochim. Biophys. Acta 1763 1496–1510. [DOI] [PubMed] [Google Scholar]
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., and Fernie, A.R. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali, U., Liu, J.L., Leisse, A., Balbo, I., Perez-Melis, A., Willmitzer, L., and Fernie, A.R. (2004). Kinetics of labelling of organic and amino acids in potato tubers by gas chromatography-mass spectrometry following incubation in C-13 labelled isotopes. Plant J. 39 668–679. [DOI] [PubMed] [Google Scholar]
- Rontein, D., Rhodes, D., and Hanson, A.D. (2003). Evidence from engineering that decarboxylation of free serine is the major source of ethanolamine moieties in plants. Plant Cell Physiol. 44 1185–1191. [DOI] [PubMed] [Google Scholar]
- Schauer, N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24 447–454. [DOI] [PubMed] [Google Scholar]
- Scheibe, R., Backhausen, J.E., Emmerlich, V., and Holtgrefe, S. (2005). Strategies to maintain redox homeostasis during photosynthesis under changing conditions. J. Exp. Bot. 56 1481–1489. [DOI] [PubMed] [Google Scholar]
- Schwarte, S., and Bauwe, H. (2007). Identification of the photorespiratory 2-phosphoglycolate phosphatase, PGLP1, in Arabidopsis. Plant Physiol. 144 1580–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, C., Bernasconi, G., and Hardtke, C.S. (2007). Natural genetic variation in Arabidopsis: Tools, traits and prospects for evolutionary ecology. Ann. Bot. (Lond.) 99 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow, J.N., and Day, D.A. (2001). Respiration and photorespiration. In Biochemistry and Molecular Biology of Plants, B.B. Buchanan, W. Gruissem and R.L. Jones, eds (Rockville, MD: American Society of Plant Physiologists), pp. 676–728.
- Sienkiewicz-Porzucek, A., Nunes-Nesi, A., Sulpice, R., Lisec, J., Centeno, D.C., Carillo, P., Leisse, A., Urbanczyk-Wochniak, E., and Fernie, A.R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147 115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, J.P., Di Leo, R., Dhanoa, P.K., Allan, W.L., Makhmoudova, A., Clark, S.M., Hoover, G.J., Mullen, R.T., and Shelp, B.J. (2008). Identification and characterization of a plastid-localized Arabidopsis glyoxylate reductase isoform: Comparison with a cytosolic isoform and implications for cellular redox homeostasis and aldehyde detoxification. J. Exp. Bot. 59 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C.R. (2001). An early Arabidopsis demonstration resolving a few issues concerning photorespiration. Plant Physiol. 125 20–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville, C.R., and Ogren, W.L. (1980). Photorespiration mutants of Arabidopsis thaliana deficient in serine-glyoxylate aminotransferase activity. Proc. Natl. Acad. Sci. USA 77 2684–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9 1797–1810. [DOI] [PubMed] [Google Scholar]
- Sweetlove, L.J., Lytovchenko, A., Morgan, M., Nunes-Nesi, A., Taylor, N.L., Baxter, C.J., Eickmeier, I., and Fernie, A.R. (2006). Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 103 19587–19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert, N.E. (1997). The C2 oxidative photosynthetic carbon cycle. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 1–25. [DOI] [PubMed] [Google Scholar]
- Tolbert, N.E., Yamazaki, R.K., and Oeser, A. (1970). Localization and properties of hydroxypyruvate and glyoxylate reductases in spinach leaf particles. J. Biol. Chem. 245 5129–5136. [PubMed] [Google Scholar]
- Voll, L.M., Jamai, A., Renné, P., Voll, H., McClung, C.R., and Weber, A.P.M. (2006). The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 140 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Caemmerer, S. (2000). Biochemical models of leaf photosynthesis. In Techniques in Plant Science, B. Osmond, ed (Collingwood, Australia: CSIRO Publishing), pp. 1–165.
- Wingler, A., Lea, P.J., Quick, W.P., and Leegood, R.C. (2000). Photorespiration: metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci. 355 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C., and Huang, A.H. (1986). Conversion of serine to glycerate in intact spinach leaf peroxisomes: Role of malate dehydrogenase. Arch. Biochem. Biophys. 245 125–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.