Abstract
Mechanisms regulating legume root nodule development are still poorly understood, and very few regulatory genes have been cloned and characterized. Here, we describe EFD (for ethylene response factor required for nodule differentiation), a gene that is upregulated during nodulation in Medicago truncatula. The EFD transcription factor belongs to the ethylene response factor (ERF) group V, which contains ERN1, 2, and 3, three ERFs involved in Nod factor signaling. The role of EFD in the regulation of nodulation was examined through the characterization of a null deletion mutant (efd-1), RNA interference, and overexpression studies. These studies revealed that EFD is a negative regulator of root nodulation and infection by Rhizobium and that EFD is required for the formation of functional nitrogen-fixing nodules. EFD appears to be involved in the plant and bacteroid differentiation processes taking place beneath the nodule meristem. We also showed that EFD activated Mt RR4, a cytokinin primary response gene that encodes a type-A response regulator. We propose that EFD induction of Mt RR4 leads to the inhibition of cytokinin signaling, with two consequences: the suppression of new nodule initiation and the activation of differentiation as cells leave the nodule meristem. Our work thus reveals a key regulator linking early and late stages of nodulation and suggests that the regulation of the cytokinin pathway is important both for nodule initiation and development.
INTRODUCTION
Legumes play a crucial role in both ecological and agricultural systems by their capacity to establish a symbiosis with nitrogen-fixing bacteria called rhizobia. This symbiosis involves the formation of a specific organ, the root nodule, which provides the proper microenvironment for nitrogen fixation by bacteroids and nutrient exchange between both partners. The process relies on their mutual recognition via molecular signals and activation of the plant symbiotic program to form rhizobium-infected nodules.
Important progress has been achieved in the past years toward understanding the initial stages of this complex developmental process. Several genes have been identified that play key roles in the perception and transduction of the bacterial Nod factors (NFs), lipo-chito-oligosaccharidic signals essential for triggering the symbiotic genetic program in specific legume hosts (for recent reviews, see Oldroyd and Downie, 2006, 2008; Jones et al., 2007). Three of these genes, NSP1, NSP2, and ERN, encode transcription factors (Oldroyd and Long, 2003; Kalo et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Middleton et al., 2007). Downstream from NF signaling, another putative transcriptional regulator, called NIN, is essential for triggering nodule organogenesis (Schauser et al., 1999; Borisov et al., 2003; Marsh et al., 2007).
The formation of nodule primordia also requires the action of endogenous signals, notably auxin and cytokinins (for reviews, see Ferguson and Mathesius, 2003; Frugier et al., 2008). Indeed, activation by a gain-of-function mutation of a cytokinin receptor (LHK1) is necessary and sufficient to induce spontaneous nodule formation in Lotus japonicus, in a process that requires the GRAS domain transcriptional regulator NSP2 (Tirichine et al., 2007). By contrast, loss of function of this cytokinin receptor reduces nodulation both in Medicago truncatula and L. japonicus (Gonzalez-Rizzo et al., 2006; Murray et al., 2007). Two response regulator genes involved in cytokinin signaling, Mt RR1 and Mt RR4, have also been found to be induced during early stages of nodulation (Gonzalez-Rizzo et al., 2006; Lohar et al., 2006) and are proposed to define a common pathway together with Mt NIN, allowing crosstalk between plant cytokinins and bacterial NFs (Gonzalez-Rizzo et al., 2006).
This work on cytokinins has provided some of the first insights into the underlying mechanisms that are responsible for the production of the nodule meristem. In contrast with the NF signaling pathway, only a few genes encoding regulators of nodule development have been identified. The formation of a nodule necessitates the coordination of plant cell and bacterial cell differentiation, leading to nodules containing differentiated bacteroids capable of nitrogen fixation. A Krüppel-like C2H2 zinc finger protein, Mt ZPT2-1, is expressed in vascular tissues of Sinorhizobium meliloti–infected roots and nodules and is necessary for the differentiation of bacteroids and of the nitrogen-fixing zone within the nodule (Frugier et al., 2000). A CAAT binding transcription factor, Mt HAP2.1, is necessary for nodule meristem maintenance as well as for bacterial release from infection threads (Combier et al., 2006). The expression of this gene is confined to the nodule meristem, notably because of posttranscriptional controls mediated by the microRNA miR169 (Combier et al., 2006) and differential splicing (Combier et al., 2008).
While these two transcriptional regulators are important for the processes underlying nodule formation, it is clear that many other components will also be necessary. Genetic dissection has revealed a number of loci important for nodule formation, infection thread growth, and bacterial release (Benaben et al., 1995; Schauser et al., 1998; Szczyglowski et al., 1998; Kawaguchi et al., 2002; Kuppusamy et al., 2004; Veereshlingam et al., 2004; Bright et al., 2005; Morandi et al. 2005; Starker et al., 2006; Arrighi et al., 2008; Teillet et al., 2008). However, to date, there are very few loci that have been cloned to reveal the underlying mechanisms inherent to these processes. One exception is ign (for ineffective greenish nodules), a non-nitrogen fixing and early senescing mutant showing abnormal symbiosome formation. This mutant is affected in a plasma membrane–located ankyrin repeat protein (Kumagai et al., 2007).
Nodulation is a tightly regulated process that ensures appropriate levels of nitrogen fixation to meet the needs of the plant without incurring excessive yield penalties. Autoregulation of nodulation (AON) allows the number of nodules to be systemically controlled by the plant, by signal exchanges between the shoots and the roots (for review, see Oka-Kira and Kawaguchi, 2006). AON involves both the nodule density and the size of the nodulation zone. Mutants defective in AON (called nts in Glycine max, har1 in L. japonicus, sunn in M. truncatula, and sym29 in pea [Pisum sativum]; Krusell et al., 2002; Nishimura et al., 2002b; Searle et al., 2003; Schnabel et al., 2005) form five to ten times more nodules than the corresponding wild-type plant, even in the presence of an excess of nitrate, which normally suppresses nodulation. These mutants are affected in a CLAVATA1–like leucine-rich repeat receptor-like kinase, active in the shoots and for which the corresponding ligand(s) remain(s) to be discovered. Nodule number is also regulated by ethylene locally in the root, as the M. truncatula sickle (skl) mutant is insensitive to ethylene and shows supernodulation (Penmetsa and Cook, 1997), a phenotype also observed in an L. japonicus line expressing a mutated ethylene receptor gene (Nukui et al., 2004). Finally, the light-insensitive L. japonicus astray mutant, affected in the HY5 bZIP transcription factor, shows a twofold increase in the number of nodules, which appear in a wider zone of the root (Nishimura et al., 2002a).
The ethylene response factor (ERF) family is one of the largest families of plant-specific transcription factors (Nakano et al., 2006), originally named from proteins able to bind the ethylene-responsive element motif (Riechmann and Meyerowitz, 1998). While many ERFs have been associated with plant responses to biotic and abiotic stresses (Gutterson and Reuber, 2004), at least five ERFs have been shown to be involved in the control of organ development, cell division, or differentiation (Wilson et al., 1996; van der Graaff et al., 2000; Banno et al., 2001; Boutilier et al., 2002; Chuck et al., 2002; Kirch et al., 2003; Marsch-Martinez et al., 2006). In addition, one ERF, called ERN (for ERF required for nodulation) has recently been demonstrated to be central for NF signal transduction (Middleton et al., 2007). Indeed, a deletion mutant in the corresponding gene, designated bit1-1, is defective in NF induction of early nodulin genes, infection thread development, and nodule meristem establishment. ERN and two other closely related ERFs, designated ERN2 and 3, were also found to bind to the NF-box, a cis-element driving NF induction of the early nodulation marker gene Mt ENOD11, using a yeast one-hybrid screen (Andriankaja et al., 2007). Finally, an ERF protein distinct from ERNs was also recently shown to be a positive factor of early nodulation stages in L. japonicus (Asamizu et al., 2008).
In this article, we report on a symbiotic ERF that we discovered is required for the differentiation of functional Fix+ nodules, and we call this protein EFD (for ethylene response factor required for nodule differentiation). EFD also seems to participate in an ethylene-independent feedback inhibition of nodulation process and regulates the expression of the primary cytokinin response regulator Mt RR4. We therefore propose that the symbiotic roles of EFD may be mediated by Mt RR4 through a modulation of the cytokinin pathway.
RESULTS
EFD Is an ERF Transcription Factor That Functions in Nodule Development and Regulation
An ERF transcription factor (originally named MtC50408) was initially identified as being upregulated in M. truncatula nodules using macroarray analyses (El Yahyaoui et al., 2004) and suppression subtractive hybridization approaches (Godiard et al., 2007). To assess the function of this ERF transcription factor, we used a high-throughput reverse genetic resource available for M. truncatula and screened a fast neutron bombardment deletion-TILLING mutant collection using oligonucleotides flanking 2.5 kb from within the promoter region to the end of the coding region. A mutant designated efd-1 was identified, containing a 1.5 kb deletion covering nucleotides −656 to +915 (Figure 1A), including the TATA box, the start codon, and a large segment of the AP2/ERF domain. This deletion completely abolishes the production of the EFD transcript, as verified by quantitative RT-PCR (Q-RT-PCR) analysis (see Supplemental Table 1 online).
Figure 1.
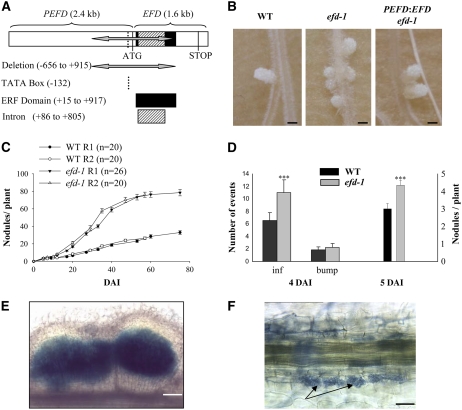
Symbiotic Phenotype of the efd-1 Mutant.
(A) The efd-1 deletion encompasses 1571 bp within the promoter (PEFD) and the coding region (EFD), including the ERF domain.
(B) Appearance of 27-d-old nodules. From left to right: the wild type (Fix+ and elongated), efd-1 (Fix− and more spherical), and efd-1 complemented by a PEFD:EFD construct (Fix+ and elongated). All three samples were collected from the same experiment. Bars = 1 mm.
(C) Time course of S. meliloti–induced nodule production in wild-type M. truncatula versus the efd-1 mutant. Two biological repetitions are represented (R1 and R2). Error bars represent se.
(D) Number of infections, bumps, and nodules counted at 4 DAI (left panel; n = 10) or 5 DAI (right panel; n = 29) in wild-type M. truncatula versus efd-1. Statistically significant differences are indicated by asterisks (Mann and Whitney test, P < 0.001). Error bars represent se.
(E) Example of a multilobe nodule (5 DAI) found in the efd-1 mutant: nonsectioned nodule induced by S. meliloti hemA-lacZ, stained in blue following a β-galactosidase assay. Bar = 160 μm.
(F) The efd-1 mutant exhibits a normal mycorrhization phenotype. Interaction between M. truncatula roots and G. intradices at 22 DAI. Arrows indicate arbuscules. Bar = 20 μm.
The efd-1 mutant produces more numerous nodules than wild type plants following S. meliloti inoculation (about threefold more at 40 d after inoculation [DAI], with plants grown in pouches; statistically significant with P < 0.001, Mann and Whitney test) (Figures 1B and 1C). An increase in the nodule number was already detectable at 5 DAI in the efd-1 mutant (Figure 1D) in the same restricted region as in wild-type roots. This increased nodule density was accompanied by numerous infection threads particularly in the epidermis (Figure 1D), showing wild-type structures. Nodule primordia were more frequently infected by several infection threads in the efd-1 mutant than in the wild type, leading to broader or multilobed nodules with several meristems (found in 6 out of 42 efd-1 nodules at 7 DAI versus none out of 49 wild-type nodules; and in 15 out of 108 efd-1 nodules versus 3 out of 73 wild-type nodules at 21 DAI) (see an example in Figure 1E).
To examine root nodulation responses at late time points under optimal physiological conditions, we used plants grown under aeroponic conditions (see Methods). A similar difference in the total nodule number was found (on average 46 for efd-1 versus 26 for wild-type plants at 21 DAI [n = 9 roots]). Epidermal and cortical infection threads infections were 5.6- and 2.7-fold more frequent, respectively, in efd-1 than in wild-type roots. In addition, a striking observation was that cortical cell divisions were much more abundant in the efd-1 mutant, being frequently associated with early infection structures (versus associated with later stages of infection in wild-type roots). Indeed, they often accompanied root hair curls (58 out of 106 counted curls versus none in wild-type plants; see an example in Supplemental Figure 1 online) and epidermal infection threads (67 out of 104 counted epidermal infections versus 1 out of 18 in wild-type plants). Cortical cell divisions without any associated infection structures were also observed in efd-1 (on average 7.4 per efd-1 plant, n = 9) but not in wild-type plants.
The second significant feature we noted in efd-1 nodules was their abnormal development. They were less elongated than wild-type nodules (Figure 1B) and white (therefore defective in leghemoglobin production). They were shown to be defective in nitrogen fixation (Fix− phenotype) by an acetylene reduction assay (16.2 ± 3.3 [se] arbitrary units versus nondetectable for wild-type and efd-1 nodules, respectively). This explained the reduced growth of efd-1 aerial parts and the chlorotic aspect of their leaves after 4 weeks in the absence of external combined nitrogen. In the presence of ammonium nitrate, the growth of efd-1 plants was similar to that of wild-type plants. We also tested the capacity of efd-1 to undergo symbiotic interactions with the arbuscular mycorrhizal fungus Glomus intraradices and found that efd-1 behaves like wild-type M. truncatula (Myc+ phenotype; Figure 1F).
After backcrossing to wild-type M. truncatula, we found a strict correlation between an abnormal nodulation phenotype and homozygosity in efd-1 mutants, which could be distinguished from heterozygous lines by PCR analysis using genomic DNA (125 individuals examined). To confirm that this altered symbiotic behavior resulted from a monogenic recessive mutation in EFD, we complemented the mutation in efd-1 via Agrobacterium rhizogenes–mediated root transformation using EFD expressed under the control of its own promoter (PEFD; 1.0- and 2.4-kb fragments). A normal number of nodules was restored, and elongated nodules similar to the wild type (both visually and by microscopy study) were recovered in 10 (out of 44) plants transformed with PEFD:EFD constructs (Figure 1B). In addition, we tested the symbiotic behavior of M. truncatula roots transformed with an EFD RNA interference (RNAi) construct expressed under the control of the 35S cauliflower mosaic virus promoter. A significant increase in nodulation (Mann and Whitney test, P < 0.001) and in the number of infection threads (Mann and Whitney test, P < 0.05) was also observed (see Supplemental Figure 2 online) in these roots, corresponding to a weaker efd mutant allele.
We can thus conclude that EFD participates in the negative regulation of infections and nodule initiations and that it also plays a positive role in the formation of functional Fix+ nodules. By contrast, EFD is not involved in mycorrhizal symbiotic interactions.
EFD Overexpression Studies Confirm That EFD Negatively Regulates S. meliloti Infections and Nodulation
The role of EFD was further tested by overexpressing EFD alone or fused to a VP16 transcriptional activator domain, which allows a transcription factor to be active without any associated cofactors (Wilde et al., 1994). Following A. rhizogenes–mediated root transformation, the overexpression of EFD in roots was confirmed by Q-RT-PCR with both constructs (12- and 14-fold on average with P35S:EFD:VP16 and P35S:EFD, respectively).
In a nodulation time course, we observed a statistically significant (Mann and Whitney test, P < 0.001) threefold reduction in the number of nodules with the EFD:VP16 construct compared with roots expressing the VP16 domain alone (Figures 2A and 2B), accompanied by a 3.5-fold reduction in the number of infection threads in roots (Figure 2B). A detailed examination of infections showed that the ratio of epidermal versus cortical infection threads was similar in wild-type and EFD-overexpressing roots (see Supplemental Table 2 online). More moderate but qualitatively similar nodulation results were obtained when overexpressing EFD without a VP16 fusion (see Supplemental Figure 3 online), suggesting that a protein interacting with EFD within a transcriptional complex might exist.
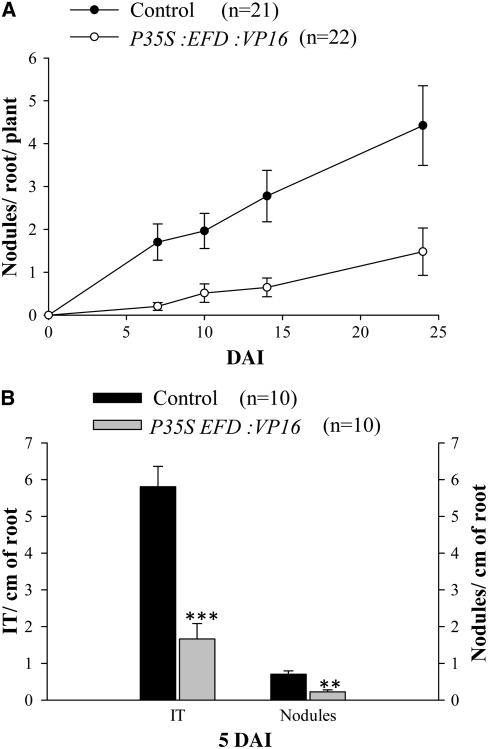
Figure 2.
The Number of Infections and Nodules Is Strongly Decreased in Roots Overexpressing EFD Fused to the VP16 Activator Domain.
(A) One-month-old transgenic roots expressing P35S:EFD:VP16 or P35S:VP16 (Control) constructs were inoculated by wild-type S. meliloti. Nodules were counted until 23 DAI. Graphs represent the average of two biological repetitions, and error bars represent se.
(B) Histograms of infection threads (IT) and nodules number per root at 5 DAI, determined on a third biological repetition. Error bars represent se. Asterisks indicate statistically significant differences (Mann and Whitney test, P < 0.001 [***] and P < 0.01[**]).
These results indicate that EFD is a negative regulator of nodulation and S. meliloti infections within the root and confirms that the enhanced nodulation phenotype of the efd-1 mutant is not simply a consequence of a lack of nitrogen fixation.
A Positive Role of EFD in Nodule Differentiation Revealed by Microscopy Observations of the efd-1 Mutant
To characterize the efd-1 symbiotic phenotype at the cellular level, we examined nodule sections. At 5 DAI, infection threads were much more numerous and branched in efd-1 nodules than in wild-type nodules and were found within a region containing many highly vacuolized cells that were not observed in wild-type nodules (cf. Figures 3A and 3B). Moreover, at this stage, symbiosome formation was observed in the proximal region of wild-type nodules but not of efd-1 nodules. Indeed symbiosome formation was first observed at 7 DAI in efd-1, in much fewer cells than in the wild-type control, with a disorganized distribution of bacteroids (cf. Figures 3C and 3D). At 10 and 20 DAI, efd-1 nodules showed a wider infection zone II (cf. Figures 3E and 3F) and a strong reduction in the number of infected cells within the zone III (nitrogen-fixing zone in wild-type nodules). Most noninvaded cells of the zone III were crossed by highly branched infection threads, a phenotype not observed in wild-type nodules (cf. Figures 3G and 3H). Amyloplasts were observed in this region, but not in massive amounts, in contrast with some Fix− mutants (Vasse et al., 1990; Frugier et al., 2000).
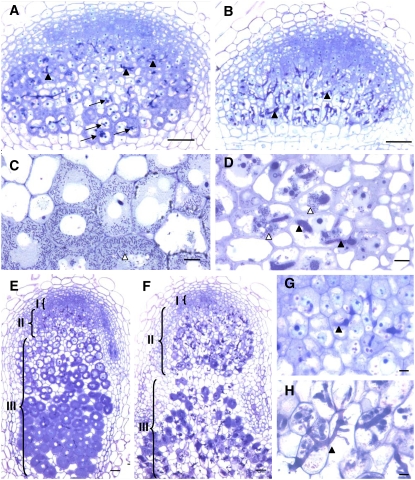
Figure 3.
Microscopy Characterization of efd-1 Nodules Reveals Defects in Symbiosome Formation and Tissue Differentiation.
(A) and (B) Four-micrometer sections of 5-d-old nodules, following inclusion in Technovit, induced by S. meliloti in wild-type M. truncatula (A) or the efd-1 mutant (B). Arrows in (A) point to bacterial release in the proximal infection region.
(C) and (D) Four-micrometer sections showing infected cells from 7-d-old nodules (zone II) induced by S. meliloti in the wild type (C) or efd-1 mutant (D) (epon inclusions).
(E) to (H) Sections of 10-d-old nodules (Technovit inclusions) induced by S. meliloti in the wild type ([E] and [G]) or efd-1 mutant ([F] and [H]). (G) and (H) are a close-up of the zone II region shown in (E) and (F). Black and white arrowheads show infection threads and released bacteria, respectively. Brackets in (E) and (F) indicate nodule zones I, II, and III. Bars = 10 μm in (C), (D), (G), and (H) and 50 μm in (A), (B), (E), and (F).
The structure of infection threads from wild-type and efd-1 nodules examined by electron microscopy (EM) studies is shown in Figures 4A and 4B. Infection threads were again observed to be more numerous and branched in efd-1 nodules, but their wall and matrix were similar to the wild type. Plant cells from nodule zones II and III exhibited an altered cytoplasm in efd-1, with a modified endoplasmic reticulum correlated with an accumulation of small vesicles (Figures 4C and 4D). The bacteroid release structures (infection droplets) were often found to be much larger in efd-1 than in wild-type nodules and present not only in zone II but also in zone III, which was not observed in the wild-type control (Figures 4E and 4F). The size of released bacteroids was normal, but the symbiosome membrane was often more difficult to see. Most bacteroids found in efd-1 nodules corresponded to the first stages of differentiation (types 1 and 2) (Vasse et al., 1990), but a few type 3 and 4 bacteroids were also detected (the type 4 corresponding to the nitrogen-fixing form). These type 4 bacteroids were often Y-shaped and not regularly oriented around a central vacuole as in wild-type zone III cells (Figures 4E and 4F). At 20 DAI, many invaded cells contained degenerating bacteroids, suggesting an early senescence process.
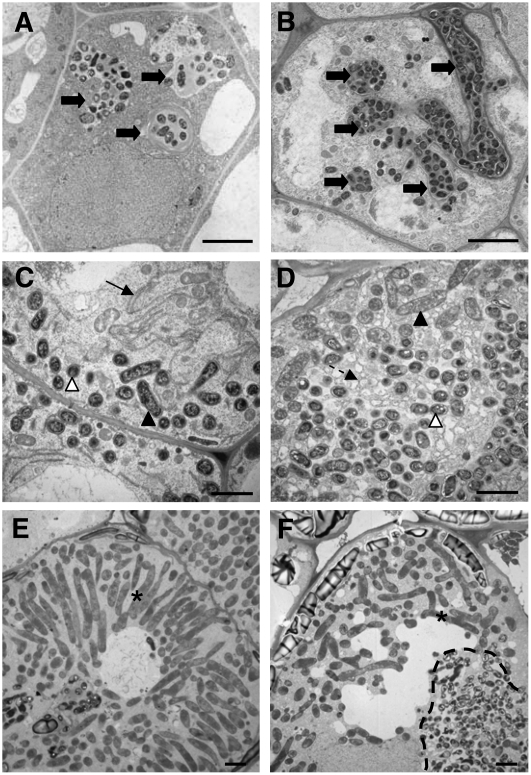
Figure 4.
EM Characterization of Zones II and III Cells from Wild-Type M. truncatula and efd-1 10-d-Old Nodules.
Wild-type nodule ([A], [C], and [E]); efd-1 nodule ([B], [D], and [F]). (A) and (B) The arrows show infection threads, which are more numerous and branched in efd-1 nodules. Bars = 5 μm.
(C) and (D) The arrow shows the typical endoplasmic reticulum observed in wild-type nodules, whereas numerous small vesicles are found in efd-1 nodules (broken arrow). The white and black arrowheads point to bacteroids, types 1 and 2, respectively.
(E) and (F) Type 4 bacteroids (asterisks) found in zone III of wild-type nodules and in the proximal region (zone III-like) of efd-1 nodules; note in efd-1 the absence of a radial organization of bacteroids, in contrast with wild-type nodules, and the presence of a bacterial release structure (dotted line) next to type 4 bacteroids, generated from an infection thread crossing the cell. Bars = 2 μm in (C) to (F).
We examined the expression of S. meliloti nodF, bacA, and nifH marker genes, previously used to distinguish bacteroid differentiation stages and to characterize Fix− mutants (Starker et al., 2006). Indeed, the nodF gene, involved in NF synthesis, was shown to be expressed in most Fix− mutants, in contrast with bacA (required for S. meliloti survival and differentiation following release from infection threads) and nifH (required for nitrogen fixation). Although nodF expression was similar in the wild type and in efd-1 nodules, the abundance of bacA transcripts was approximately twofold higher and that of nifH transcripts 2.8-fold lower in efd-1 nodules (see Supplemental Figure 4 online). These results are unlikely to be affected by the number of bacteroids since transcript levels were normalized using the pnp gene expression, which is similar in zones II and III (Becker et al., 2004; Naya et al., 2007). This suggests that bacteroids are able to survive in efd-1 nodules but unable to activate their genetic program responsible for nitrogen fixation.
We thus conclude that EFD is required for the proper development of nodule zones II and III, including symbiosome formation and bacteroid differentiation processes.
EFD Expression Is Found in Nodule Primordia and Nodule Zone II but Is Not Associated with Infection Threads
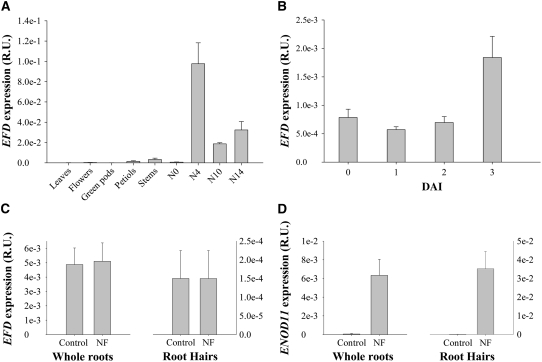
To further define the role of EFD in nodulation, we determined the pattern of EFD expression. Q-RT-PCR analysis of various organs of M. truncatula revealed that EFD is mainly expressed in root nodules, at a higher level in immature nodules (Figure 5A). When we examined EFD expression in roots at 1, 2, and 3 DAI with S. meliloti (Figure 5B), a 2.3-fold induction compared with noninoculated roots was observed at 3 DAI (P < 0.05, following Cumming et al., 2007). By contrast, purified NF at 10−8 M, a concentration at which Mt ENOD11 is clearly upregulated (Figure 5D), could not activate EFD expression in whole roots or isolated root hairs (Figures 5C and 5D). EFD was not induced by S. meliloti in M. truncatula mutants affected in NF perception (nfp-1, allele C31) and early signaling (nsp1-1, allele B85) (see Supplemental Table 3 online). In contrast with Mt ENOD11, no induction of EFD was detected in the hcl-1 mutant (allele B56), which is impaired in nodule development and S. meliloti infection, but in which some cortical cell divisions are triggered.
Figure 5.
Q-RT-PCR Analyses of EFD Expression in Different Tissues.
(A) EFD expression in M. truncatula leaves, flowers, green pods, petiols, stems, roots (N0), and nodules at different developmental stages (4, 10, and 14 DAI).
(B) EFD expression in wild-type S. meliloti–inoculated roots of M. truncatula at 0, 1, 2, and 3 DAI.
(C) EFD expression in NF- or water-treated roots and root hairs (18 h treatment).
(D) Mt ENOD11 expression in NF- or water-treated roots and root hairs (18 h treatment).
All data are from at least three biological repetitions and are normalized by EF1-α expression. Error bars represent se. R.U., relative units.
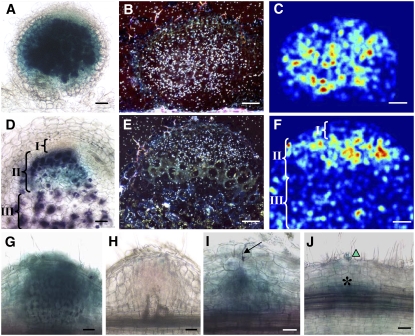
These results suggested that EFD expression is not activated during very early stages of the nodulation process. The fact that EFD expression is stronger in young than mature nodules also suggested a preferential expression in nodule zone I and/or II, which become relatively less important as the nodule grows. To precisely determine the tissue localization of EFD transcripts, we isolated the EFD promoter by BAC library screening (see Methods) and generated fusions with the β-glucuronidase (GUS) reporter gene. The expression pattern of two promoter segments of 2.4 and 1.0 kb was examined in A. rhizogenes–transformed M. truncatula roots (Figure 6). Both fusions gave similar results, which were validated by in situ hybridizations performed with a 35S-labeled antisense EFD probe. EFD expression was found to be distributed in the central region of immature nodules but not in the apical (meristem) region (Figures 6A to 6C) and then confined to the distal part of zone II in differentiated nodules (Figures 6D to 6F). No signal was detected when doing in situ hybridizations with a control sense EFD probe (see Supplemental Figure 5 online).
Figure 6.
EFD Is Expressed in the Apical Zone II of the Nodule and in Nodule Primordia.
(A) to (F) Localization of EFD mRNA in wild-type 4-d-old ([A] to [C]) and 10-d-old ([D] to [F]) nodules, as determined by PEFD:GUS fusion ([A] and [D]; blue) or in situ hybridization ([B], [C], [E], and [F]); hybridization signals appear as white dots in (B) and (E) or false color representation following digitization in (C) and (F) (Diatrack software) where the strongest signals are indicated by yellow-orange and the weakest by dark-blue colors. S. meliloti hemA:lacZ bacteria are stained in purple in (A) and (D). Brackets in (D) and (F) indicate nodule zone I, II and III.
(G) and (H) Localization of EFD mRNA, based on the PEFD:GUS fusion, in 4- and 10-d-old nodules induced by an infection-defective exoA mutant of S. meliloti.
(I) PEFD:GUS expression (blue color) in a wild-type nodule primordium beneath a developing infection thread (arrow) containing S. meliloti hemA:lacZ bacteria (purple).
(J) PEFD:GUS expression associated with early cortical cell divisions (asterisk) before the elongation of infection threads; note the presence of a curled root hair (triangle), containing S. meliloti bacteria (purple).
Bars = 50 μm.
The nodule zone II corresponds to the infection zone, but it also represents the first tissue below the meristem in which plant cell and bacterial differentiation take place. To determine whether EFD expression is directly associated with the infection process, we then examined empty nodules induced by an infection-defective exoA mutant of S. meliloti (Yang et al., 1994). A very clear PEFD:GUS expression was detected in 5-d-old exoA nodules (Figure 6G), and this expression disappeared in 10-d-old exoA nodules, a stage at which the meristem is no longer active (Figure 6H). This indicated that EFD expression in nodules requires an active meristem but is not associated with S. meliloti infection per se. This was consistent with the PEFD:GUS expression observed at a distance from infection threads in young wild-type nodule primordia (Figure 6I). This expression was first detected at the beginning of nodule primordium formation, associated with cortical cell divisions (Figure 6J). Using these promoter:GUS fusions, we confirmed that NFs did not induce EFD expression (see Supplemental Figure 6 online). We also took advantage of these fusions to look for EFD expression in noninoculated roots. We thus found that PEFD:GUS was expressed in primary root tips and lateral root primordia (see Supplemental Figure 6 online).
Finally, in view of the reported regulation of some ERFs by ethylene (Guo and Ecker, 2004) and considering the involvement of EFD in the negative regulation of nodulation (Figure 1), we tested the response of EFD to this phytohormone. We could not detect any activation of EFD expression by 50 μM 1-aminocyclopropane-1-carboxilate (ACC; ethylene precursor). Moreover, the ethylene inhibitor aminoethoxy-vinyl glycine (AVG) (10 μM) did not change EFD activation by S. meliloti, and EFD was still induced by S. meliloti in the ethylene-insensitive skl mutant (see Supplemental Figure 7 online).
These results suggest that EFD expression is triggered by nodule primordium formation but is not activated during earlier stages of the nodulation process. It then requires meristem activity in nodules.
EFD Is a Member of Group Va of the AP2/EREBP Family
Following Nakano's classification (2006), the Arabidopsis thaliana AP2/ERF superfamily, defined as possessing the AP2/ERF domain (59 amino acids), comprises three major families: the ERF family (a single AP2/ERF domain; currently 122 genes, including the DREB protein genes), the AP2 family (two AP2/ERF domains; 18 genes), and the RAV family (one AP2/ERF domain and one B3 domain; six genes). The alignment of the AP2/ERF domain of all Arabidopsis ERF proteins reveals seven invariant residues and a set of eight that are found in >95% of the ERF proteins (Nakano et al., 2006). These 15 residues are all found in the AP2/ERF domain of EFD (see Supplemental Figure 8 online). The seven residues involved in direct contact with DNA and the seven Ala residues involved in the stability of the ERF domain (Allen et al., 1998) are also perfectly conserved in EFD, which thus clearly belongs to the AP2/ERF superfamily.
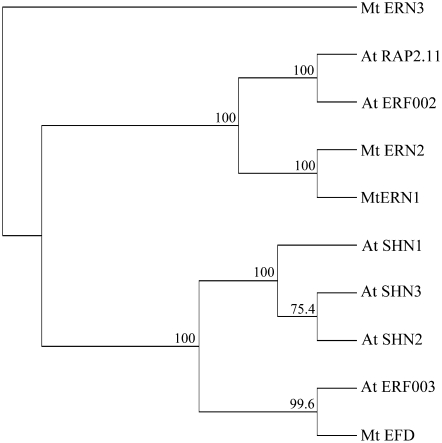
The ERF family has been itself subdivided into 12 groups by a phylogenetic analysis based on the AP2/ERF domains (Nakano et al., 2006). We found that, as the three ERFs involved in NF signaling ERN1, ERN2, and ERN3, EFD belongs to group V (Figure 7). However, EFD is related to a different subgroup (Va) together with Arabidopsis ERF#003 (At5g25190, unknown function) and At SHN1, 2, and 3, which regulate the accumulation of epidermal wax (Aharoni et al., 2004). Two differences should be noted within the AP2/ERF domain, concerning two amino acids (a Trp and a Lys residue) directly involved in DNA binding, and which are conserved in the EFD but not in the ERN1-3 subgroup (see Supplemental Figure 8 online).
Figure 7.
Phylogenetic Tree of Group V ERFs from Arabidopsis and M. truncatula.
All Arabidopsis proteins from ERF group V, as described by Nakano et al. (2006), were aligned with M. truncatula group V ERF proteins (ERNs and EFD). Alignment was done with entire proteins. Indicated bootstrap values were determined from 1000 iterations.
The analysis of motifs outside the AP2/ERF domain reveals other differences between these ERFs. The ERN1-3 subgroup exhibits CMV-3 and CMV-4 motifs (Nakano et al., 2006; Middleton et al., 2007), whereas the EFD subgroup shares CMV-1 and CMV-2 motifs with unknown function. Finally, EFD and At SHN1, 2, 3 genes possess an intron at a conserved position, which is not found in the ERN1-3 subgroup.
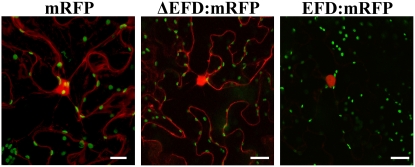
EFD Localizes to the Nucleus
To assess the subcellular location of EFD, fusions were generated at the N and C termini of EFD with the red fluorescent protein (RFP) marker and placed under the control of a 35S promoter. These constructs, as well as a control fusion protein where the AP2/ERF domain was deleted, were transformed into Nicotiana benthamiana, and the subcellular protein localization was examined by fluorescence microscopy. Figure 8 shows that the fluorescent signal was exclusively found in the nucleus when using the entire EFD (both for the N- or C-terminal fusion), whereas the deleted fusion protein or the RFP protein alone gave a cytoplasmic signal. This experiment suggests that the EFD AP2/ERF domain plays an important role in targeting EFD to the nucleus.
Figure 8.
Nuclear Localization of EFD:RFP Fusion Protein.
Leaves of N. benthamiana were A. tumefaciens transformed with the following constructs, expressed under the control of the 35S promoter: from left to right, reporter mRFP protein alone; mRFP protein fused to a deleted form of EFD lacking the putative DNA binding domain (ΔEFD; N-terminal fusion); full-size EFD fused to mRFP reporter protein (N-terminal fusion). The mRFP reporter protein is detected as a red signal, while green spots correspond to plastids. Note that wild-type EFD is found exclusively in the nucleus in contrast with the mRFP reporter protein alone or to EFD deleted for the ERF domain. Bars = 40 μm.
Possible Targets of the EFD Transcription Factor Revealed by Transcriptome Analyses
As a complementary approach to characterize the efd-1 phenotype and to look for candidate target genes directly or indirectly controlled by the EFD transcription factor, we performed microarray analyses, first by comparing efd-1 and wild-type nodules, at 4 and 10 DAI. We used Mt16KOLI1Plus microarrays representing 16,470 M. truncatula EST clusters (Küster et al., 2007). In the following sections, genes are considered as differentially expressed when they show at least a twofold ratio and an adjusted P value ≤ 0.05.
At 4 DAI, no gene was found to be less or more expressed in efd-1 nodules than in the wild type when taking into account the 0.05 threshold for adjusted P values. By contrast, at 10 DAI, 225 genes were downregulated in efd-1 compared with wild-type nodules and 34 genes upregulated (see Supplemental Table 4 online). Among the transcripts showing a decreased expression in efd-1, several late nodulin genes related to the nitrogen fixation process were found (e.g., encoding leghemoglobins and Gln and Asn synthases), as well as other nodulin genes of unknown function (Mt N19, Mt N20, and Mt N31). By contrast, several early nodulin marker genes were either weakly or not affected (e.g., Mt ENOD2, Mt ENOD16, and Mt ENOD40), while others (e.g., Mt ENOD11 and Mt LEC4) were more expressed in efd-1 10-d-old nodules.
A striking observation was the downregulation of a set of 75 NCR/CCP (for nodule-specific cysteine-rich/cysteine cluster proteins) genes (Fedorova et al., 2002; Mergaert et al., 2003) and of one GRP (glycine-rich protein) gene in efd-1 10-d-old nodules. This may be linked to the presumed role of NCRs and GRPs in bacteroid differentiation (Mergaert et al., 2006; Alunni et al., 2007). Among the 34 upregulated genes in efd-1 10-d-old nodules, genes coding for peptidases and Cys proteinases were identified, some of which are highly homologous to the SAG2-like senescence-specific genes (Noh and Amasino, 1999), such as As NODf32, activated at the onset of nodule senescence (Naito et al., 2000). This is highly reminiscent of Cys proteinases found by cDNA-amplified fragment length polymorphism in the senescent zone IV in M. truncatula (Van de Velde et al., 2006). These microarray results were validated by Q-RT-PCR analyses on 48 genes, of which 40 gave qualitatively similar results (see Supplemental Table 5 online).
To look more specifically for genes likely to be regulated by EFD, we used three criteria to be met simultaneously: (1) a reduced expression in efd-1 nodules versus wild-type nodules at 4 and 10 DAI, (2) an increased expression in P35S:EFD:VP16 transgenic roots compared with empty vector–transformed roots (microarray analyses; see Supplemental Table 6 online), and (3) an expression profile similar to EFD pattern in nodules of wild-type M. truncatula induced by various strains of S. meliloti (microarray analyses; S. Moreau and P. Gamas, unpublished data). We found only one gene, Mt RR4, which clearly met these three criteria among the 34 genes activated in P35S:EFD:VP16 roots. Q-RT-PCR analyses confirmed that Mt RR4 expression was strongly decreased in efd-1 nodules (∼16-fold at 4 DAI and sixfold at 10 DAI; see Supplemental Table 5 online) and upregulated in P35S:EFD:VP16 roots (4.2-fold, using pools of 31 control plants and 54 EFD-overexpressing plants). Importantly, in situ hybridizations showed that Mt RR4 and more generally the type-A response regulator gene family are expressed in nodule zone II, consistent with the EFD expression pattern (see Supplemental Figure 9 online). Other nodulin genes expressed in nodule zone II, such as Mt N6 (Mathis et al., 1999), Mt ENOD11 (Boisson-Dernier et al., 2005), and Mt MMPL1 (Combier et al., 2007), were not found to be similarly affected in the efd-1 and P35S:EFD:VP16 samples (see Supplemental Table 5 online).
EFD Activates the Expression of Mt RR4, a Gene Encoding a Response Regulator That Controls the Cytokinin Signaling Pathway
Mt RR4 is homologous to ARR4 from Arabidopsis, which encodes a type-A response regulator, induced by cytokinins and involved in the negative control of the cytokinin pathway (To et al., 2004). The crucial role of cytokinins in nodulation prompted us to confirm if Mt RR4 could be transcriptionnally activated by EFD. We first verified that Mt RR4 is indeed a cytokinin response gene. Using Q-RT-PCR analyses, we observed a 5- to 10-fold transcriptional activation after 1 to 3 h of root treatment with 10−7 M benzyl amino purine (BAP) (see Supplemental Figure 10 online). Moreover, this induction took place in the presence of 100 μM cycloheximide, establishing that Mt RR4 is a primary cytokinin response gene. Using the same samples, we did not find EFD to be regulated by BAP (see Supplemental Figure 10 online).
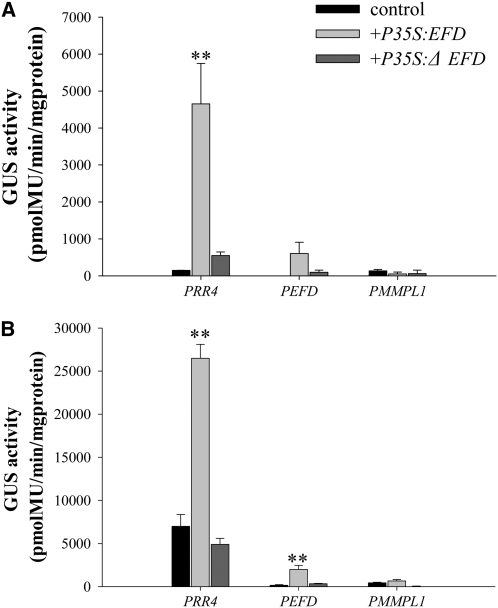
To test a potential role of EFD in the regulation of Mt RR4 expression, we cotransformed into N. benthamiana the GUS reporter gene fused to a 1132-bp fragment of Mt RR4 promoter, along with a P35S:EFD construct fused either to a RFP reporter protein or to a HA tag. As a negative control, to detect any possible nonspecific trans-activation effect, we used a Mt MMPL1 promoter:GUS fusion not controlled by EFD (previous array analyses) and showing a very low basal level of expression in these tissues (Combier et al., 2007). Quantitative analyses of GUS activity in N. benthamiana leaf extracts revealed that the Mt RR4 promoter was clearly activated (on average 8.5-fold on three biological repetitions) by either EFD:RFP (Figures 9A and 9B) or EFD:HA fusions (see Supplemental Figure 11 online). A deletion of the EFD AP2/ERF domain abolished this trans-activation. Similar experiments performed with ERN1, 2, and 3 (ERFs involved in NF signaling) did not reveal any trans-activation of Mt RR4 (F. de Carvalho Niebel, personal communication). Finally, we also tested whether EFD is able to regulate its own expression and found that EFD was indeed able to trans-activate its own promoter (on average sixfold on three biological repetitions; Figures 9A and 9B), suggesting a positive feedback loop for EFD.
Figure 9.
Trans-Activation of PRR4, PEFD, and PMMPL1 in N. benthamiana.
(A) GUS activity at 24 h following cotransformation.
(B) GUS activity at 48 h following cotransformation.
For both (A) and (B), leaves of N. benthamiana were cotransformed by A. tumefaciens with PRR4:GUS, PEFD:GUS, or PMMPL1:GUS plus either P35S:EFD:RFP or P35S:ΔEFD:RFP (ΔEFD is EFD deleted for its putative DNA binding domain). Controls correspond to leaves of N. benthamiana transformed with PRR4:GUS, PEFD:GUS, or PMMPL1:GUS alone. GUS activity was measured using 10 μg of total protein extracts at 24 (A) and 48 (B) h after transformation using three biological repetitions. Error bars represent se. PRR4 was only significantly activated by EFD at 24 and 48 H and PEFD at 48H (P < 0.01 for both, following Cumming et al., 2007). PMMPL1 was not significantly activated.
We can thus conclude that the expression of the primary cytokinin response gene Mt RR4 and of the EFD gene itself is directly or indirectly controlled by EFD.
DISCUSSION
We have identified a transcription factor involved in the rhizobium-legume symbiotic interaction, EFD, that belongs to the large AP2/ERF family. A null mutant of EFD is severely affected in its capacity to differentiate functional Fix+ nodules and shows increased number of nodules compared with a wild-type line. These experiments coupled with RNAi and overexpression approaches support a role of EFD both in the regulation of nodule number and nodule differentiation. Transcriptomic studies and trans-activation assays allowed us to demonstrate that Mt RR4, encoding a type-A response regulator of cytokinin signaling, is a target of EFD in S. meliloti infected roots and nodules. We propose that EFD may regulate diverse symbiotic responses through interaction with cytokinin signaling.
EFD, a New ERF Transcription Factor Linked to Symbiosis
EFD is different in terms of sequence from the three previously described M. truncatula ERFs involved in rhizobium-legume symbiotic interactions (ERN1, 2, 3; Andriankaja et al., 2007; Middleton et al., 2007), even though they belong to the same ERF group (group V). It is therefore expected that different target genes are controlled by EFD and ERN transcription factors. Indeed, ERN1, 2, and 3 bind to the promoter of the early nodulin gene Mt ENOD11 (Andriankaja et al., 2007), whereas there is no indication that this is the case for EFD. Conversely, the response regulator gene Mt RR4 appears to be controlled by EFD, but not by ERN proteins (F. de Carvalho Niebel, personal communication).
Another important difference between EFD and ERN genes is their expression pattern. The three ERN genes are constitutively expressed in root hairs, upregulated by NF treatment, and moderately regulated (up or down) in nodules (Andriankaja et al., 2007; Middleton et al., 2007). By contrast, EFD expression is not detected in root hairs, is not induced by NF treatment (up to 48 h), and is strongly activated in nodule primordia and infection zone II. The EFD-inducing signal during symbiosis is likely not ethylene as EFD is not induced by an ACC treatment and is still induced by S. meliloti in the ethylene-insensitive skl mutant. The nature of this signal remains to be identified.
EFD is also different from Lj ERF1 (Asamizu et al., 2008), considering different criteria: Lj ERF1 belongs to a distinct ERF group (group IX in the classification of Nakano et al., 2006); Lj ERF1 is upregulated much earlier and only in root epidermis following Mesorhizobium loti inoculation (without any expression in nodules); Lj ERF1 seems to be a positive regulator of early nodulation processes, based notably on RNAi and overexpression experiments (Asamizu et al., 2008).
EFD Is a Negative Regulator of Nodule Initiation and Is Required for Late Stages of Nodule Development
Two distinct symbiotic phenotypes, early and late, have been revealed by the characterization of the efd-1 knockout mutant, RNAi studies, and overexpression of activated EFD. The early phenotype is a significant increase in the knockout and knockdown lines in the number of S. meliloti infections and nodules compared with wild-type M. truncatula. This increase is detected well before the onset of nitrogen fixation in wild-type lines, ruling out at this stage an indirect effect linked to the defect in nitrogen fixation, observed in other Fix− mutants, such as the sst1-2 L. japonicus mutant defective in a sulfate transporter (Krusell et al., 2005). This opens the possibility that EFD participates in the negative regulation of nodulation, together with other (e.g., ethylene-dependent) mechanisms. Our observations suggest that the efd-1 mutation first leads to local effects on nodule density. Further experiments will be required to determine whether EFD may also exert a systemic effect, as in AON (Oka-Kira and Kawaguchi, 2006).
The EFD-dependent regulation may be triggered by nodule primordium formation and related to the early EFD expression detected in these primordia. We showed that this regulation clearly influences the number of epidermal infections, even though EFD is not expressed in close proximity to infection threads or in root hairs. This represented the earliest difference that we detected in the symbiotic behavior between efd-1 and wild-type plants (or EFD-overexpressing and wild-type roots) and might thus be considered as the primary effect of EFD-dependent regulation. However S. meliloti–inoculated efd-1 roots also showed an excess of cortical cell divisions compared with wild-type plants, accompanying root hair curls and epidermal infection threads or without associated visible infection structures in some cases. Overall, efd-1 cortical cells thus seem more reactive to S. meliloti infections, which would represent an attractive hypothesis regarding a possible involvement of the cytokinin pathway (see below). However, an alternative hypothesis could be that the slowing down of infection thread development in efd-1 at late stages would lead to an altered coordination between infections and cortical cell divisions.
The second altered symbiotic phenotype of efd-1 is the production of Fix− nodules, in which fewer bacteroids differentiate, as shown both by EM studies and a reduced expression of the S. meliloti nifH marker gene. An increased accumulation of S. meliloti bacA transcripts is also observed, which may be related to an enlarged nodule zone II. The rare type 4 bacteroids observed are not found in regular rays well organized around a central vacuole as in wild-type nodule zone III (Timmers et al., 1999). Previous work has established that the bacteroid organization in wild-type zone III is linked to the microtubular cytoskeleton (Timmers et al., 1999), which is thus likely to be altered in efd-1 nodules.
It can thus be proposed that EFD plays a positive role in both bacterial and plant cell differentiation. Transcriptomic comparison of efd-1 and wild-type nodules showed that differences are moderate in 4-d-old nodules but strong in 10-d-old nodules, supporting a role of EFD in late stages of nodule development. Whereas the expression of early nodulin genes is not affected (Mt ENOD2 or Mt ENOD40) or even increased (Mt ENOD11, a zone II gene) in 10-d-old efd-1 nodules, nodulin genes associated with the nitrogen fixation process are poorly expressed, consistently with the Fix− phenotype. A striking observation is the downregulation in efd-1 nodules of numerous NCR/CCP genes that belong to the large family encoding Cys-rich peptides specifically found in indeterminate nodules of hologalegoid legumes (Fedorova et al., 2002; Mergaert et al., 2003; Graham et al., 2004; Silverstein et al., 2006, 2007; Alunni et al., 2007). These peptides are similar to defensins and have been proposed to be involved in bacteroid differentiation by acting as antimicrobial peptides (Mergaert et al., 2006). The downregulation of 75 NCR genes in efd-1 correlates with the bacteroid differentiation defects detected by microscopy. In addition, six plant genes related to senescence are upregulated in 10-d-old efd-1 nodules. An early senescence process is thus likely to take place in efd-1 nodules, which may explain the disappearance of the endoplasmic reticulum correlated with the appearance of numerous small vesicles observed in our EM studies. The fact that bacA transcripts are abundant in efd-1 nodules indicates, however, that S. meliloti bacteria survive after release from infection threads (Glazebrook et al., 1993), which is confirmed by our EM studies. We thus favor the hypothesis that this early senescence process is a consequence of defects in plant and/or bacteria differentiation. However, we cannot exclude, with our data, the converse hypothesis that differentiation defects result from early nodule senescence.
To our knowledge, the efd-1 mutant does not strictly resemble any of the already described Fix− M. truncatula mutants. Nevertheless, it can be noted that abnormal proliferation of infection threads has also been reported for nip (Veereshlingam et al., 2004), sym1 (TE7) (Benaben et al., 1995), and RNAi DMI2 lines (Limpens et al., 2005). Defects in bacterial release and/or bacteroid differentiation have also been observed for sym1, RNAi DMI2, and antisense Mt Zpt2-1 lines (Frugier et al., 2000). However, other features of these mutants differ from efd-1, such as a strong defense response and an early stop in nodule development observed in nip, normal nodule elongation in sym1, and strong accumulation of amyloplasts in nodules of antisense Mt zpt2-1 lines. The efd-1 mutant may resemble class 3 dnf mutants, as defined by Starker et al. (2006), but plant gene expression and nodule structure (Pislariu and Dickstein, 2007) seem to be differently affected in efd-1 than in dnf4 and dnf7.
So far, Mt ZPT2-1 and EFD represent the only two transcription factors known to participate in the coordinated differentiation process of the plant and bacterial partners taking place during late stages of nodule development.
EFD Activates a Type-A Response Regulator Gene
To explore how EFD could act during nodulation, we searched for target genes by comparing transcriptomes of wild-type and efd-1 nodules as well as EFD-overexpressing roots. Expression of the best candidate target gene, Mt RR4, was trans-activated in N. benthamiana by a P35S:EFD construct. However, such experiments do not indicate whether EFD binds directly Mt RR4 promoter.
Mt RR4 is highly similar to type-A response regulator genes, demonstrated to be primary response genes to cytokinins in Arabidopsis (for review, see Ferreira and Kieber, 2005). We confirmed that Mt RR4 is itself rapidly induced by exogenous cytokinin in a process that does not require de novo protein synthesis. Since type-A response regulator genes are thought to negatively control the cytokinin pathway (for reviews, see Ferreira and Kieber, 2005; Doerner, 2007), itself required for nodulation (Lohar et al., 2004; Gonzalez-Rizzo et al., 2006; Frugier et al., 2008), the activation of Mt RR4 by EFD provides a possible mechanism regarding the early negative role of EFD on regulation of nodule number. Hence, EFD activation at around 3 DAI may restrict cytokinin signaling and function to prevent further rhizobial infections and nodule initiation. Conversely, as mentioned above, one explanation for the increased abundance of cortical cell divisions in the efd-1 mutant could be an overactivation of the cytokinin pathway. Further experiments will be needed to test these hypotheses.
The role of cytokinins at later stages of nodule development is still poorly documented. Reporter gene fusions with ARR5, a marker gene of cytokinin pathway activation, and Mt CRE1, the main cytokinin receptor gene linked to nodulation, have revealed that the cytokinin pathway is active at later stages of the symbiotic interaction (Lohar et al., 2004, 2006). The expression of Mt CRE1 is found in dividing cells of nodule primordium and is then restricted to the apical meristematic region of mature nodules. A reasonable assumption is therefore that cytokinins are required for nodule meristem activity. Cell differentiation in nodule zone II/III may involve a fine tuning of cytokinin action, with a gradient of decreasing cytokinin activity from the meristem to underlying tissues. We propose that EFD, which is maximally expressed at the border between zones I and II, participates in the control of this gradient by regulating the cytokinin pathway via the type-A response regulator Mt RR4. Altering this gradient in efd-1 would lead to an enlarged zone II because the proximal zone II and zone III cannot form properly. Following this hypothesis, the nodule meristem would be reminiscent of shoot meristems more than of root meristems, cytokinins being thought in the latter to be required for cell differentiation rather than proliferation (Dello Ioio et al., 2007). EFD could thus be viewed as a factor that contributes to define spatially and temporally the niche for proliferating meristematic cells by turning down the cytokinin pathway. As documented for several key factors regulating development (Ferrell, 2002), the observed positive feedback regulation of EFD on its own expression is likely another important regulatory mechanism that allows EFD expression to be maintained in a defined region, thus leading to an irreversible developmental switch during nodule differentiation. The fact that EFD is also expressed in root meristems is intriguing, and further studies will be required to examine possible impacts of EFD on root development.
In conclusion, this study illustrates how high-throughput genomics tools set up for model legumes can contribute to the identification of novel and important regulators of nodule development. It is anticipated that a number of other regulators will be identified in the near future and characterized efficiently thanks to reverse genetics platforms. This will lead to the dissection of the developmental cascade controlling the formation of an organ that plays a central role for agricultural and environmental beneficial properties of legumes.
METHODS
Plant Growth and Bacterial Strains
Medicago truncatula cv Jemalong A17 was used as the wild-type reference and for backcrosses of the efd-1 mutant. Surface-sterilized seeds were placed on inverted agar plates in the dark for 3 d at 8°C and 1 d at 20°C. Germinated seeds were transferred into pouches (cytologic studies), Farhaeus agar plates (treatments), or in aeroponic caissons (kinetic studies by Q-RT-PCR and backcross segregation analysis) containing an appropriate plant growth medium, as described in the Medicago handbook (http://www.noble.org/MedicagoHandbook/). Plants in pouches were inoculated with 600 μL of Sinorhizobium meliloti suspension at an OD600 = 0.02 and placed at 25°C (light-dark photoperiod: 16 h/8 h). Plant growth and inoculation in caisson were as described by Combier et al. (2007), with the following chamber conditions: temperature, 22°C; 75% hygrometry; light intensity, 200 μE·m−2·s−1; light-dark photoperiod, 16 h/8 h. For ACC and AVG treatments, plants were grown on Farhaeus medium in plates at 20°C with the same photoperiod.
Wild-type S. meliloti RCR2011 pXLGD4 (GMI6526) and S. meliloti RCR2011 exoA pXLGD4 (GMI3072) were grown at 28°C in tryptone yeast medium supplemented with 6 mM calcium chloride and 10 μg mL−1 tetracycline. For root transformation, we used ARqua1 Agrobacterium rhizogenes as described by Boisson-Dernier et al. (2001). For Nicotiana benthamiana transient expression, we used Agrobacterium tumefaciens strains GV3101 and GV3103 (as in Andriankaja et al., 2007) grown at 28°C in Luria-Bertani medium supplemented with rifampicin (10 μg mL−1).
ACC, AVG, NF, and Cytokinin Treatments
For ACC (Sigma-Aldrich) treatment, a 50 μM solution of ACC was applied onto roots of 5-d-old seedlings. Thirty roots per time point (0, 5, 24, and 48 h after treatment) were cut and frozen before RNA extraction. The impact of AVG (Sigma-Aldrich) treatment onto EFD induction was determined on S. meliloti–induced nodules from 50 independent roots. NF treatments (18 h for Q-RT-PCR, and until 48 h for PEFD:GUS observations) and root hairs isolation were done as described by Sauviac et al. (2005). Cytokinin (BAP; Sigma-Aldrich) treatments were done as described by Gonzalez-Rizzo et al. (2006), with or without a 1 h cycloheximide 100 μM treatment (Sigma-Aldrich). Three biological repetitions were done for each of these treatments.
Identification of the efd-1 Deletion Mutant from a Fast Neutron Mutant Population
The efd-1 deletion mutant was identified using De-TILLING (deletion TILLING), a novel reverse-genetics platform that has been established in M. truncatula exploiting fast neutron mutagenesis and a highly sensitive PCR-based detection (C. Rogers and G. Oldroyd, unpublished data). A population of 60,000 M2 lines, prepared originally as 2400 DNA samples, was pooled to a set of 10 templates for PCR screening. The identification of false positives was avoided by simultaneously screening a differential pooling of these DNAs, giving a total of 20 PCR reactions. Genomic regions of 2 to 2.5 kb were identified possessing a unique restriction site adjacent to exon 1 of the EFD gene. Amplification of one of these genomic regions, a 2.55-kb target region centered on an EcoRI restriction site, revealed the presence of an ∼1.5-kb deletion mutant within the population. This was confirmed by screening a second region of 2.9 kb centered on the same restriction site. Sequencing of these amplified fragments revealed a deletion covering nucleotides −656 to +915 (1571 bp), completely removing exon 1. Subsequent PCR screening of three-dimensional DNA pools allowed the mutant to be located and recovered from within archived M2 seed stocks.
The EFD genomic region was analyzed by PCR using the same primers as in Q-RT-PCR experiments (see Supplemental Table 7 online) and the two following primers: 5′-GGGGTACCCACCCCGAACCC-3′ and 5′-TGACCTTCAAACCCAACACA-3′.
Phenotypic Characterization of efd-1
S. meliloti spot inoculation was done as described by Mathis et al. (1999). Histological studies of nodules were performed after fixation in 2.5% of glutaraldehyde buffered in 0.1 M sodium phosphate buffer, pH 7.2, dehydration in an alcohol series, and embedding in Technovit 7100 resin (Hereaus Kulzer). Sections (4 μm thick) were observed after counterstaining in a 0.02% aqueous toluidine blue solution. For EM studies, following the glutaraldehyde fixation step, nodules were postfixed in 1% osmium phosphate buffer solution and embedded in epon as described by Vasse et al. (1993). Ultrathin sections stained with uranyl acetate and lead citrate were examined using a JEM 2100 electron microscope.
For mycorrhizal observations, germinating seeds of efd-1 and A17 were placed on Farhaeus plates for 10 d and then transferred on M medium (Bécard and Fortin, 1988) plates (supplemented with 0.5% phytagel) with pouch paper and inoculated 9 d later by Glomus intraradices as described by Boisson-Dernier et al. (2005). Mycorrhizal coloration was done at 22 DAI with black ink (Sheaffer) as described by Chabaud et al. (2002).
Nitrogenase activity was assayed by the acetylene reduction technique (Hardy et al., 1968). Nitrogen fixation ability was measured on 10 individual wild-type or efd-1 plants at 24 DAI. Data were calculated as a ratio between ethylene and acetylene peak heights given in percentage.
Plasmid Constructs and A. rhizogenes Transformation
To generate the P35S:EFD:VP16 construct, we first introduced the VP16 domain in the pPex vector (Combier et al., 2007). VP16 was amplified on PFP101HAVP16 (from PZP200; Hajdukiewicz et al., 1994) with the primers 5′-AGGATCCCCAACGATGAAAAGCTTGGC-3′ and 5′-GATCCGCTCTAGAGATATCC-3′ and introduced in pGEM-T (Promega) between BamHI and XbaI sites. We amplified the EFD coding sequence using Pfx polymerase (Invitrogen) and primers 5′-ACTCGAGATGGCAAGACCACAACAACGTTATAG-3′ and 5′-AGGATCCAGATGAACCAACAGAACAAAG-3′ and inserted it in pPex-VP16 between XhoI and BamHI sites. To generate the P35S:EFD construct, we inserted the XhoI-BamHI fragment from pPex-EFD:VP16 plasmid into pPex between XhoI and BamHI sites. Finally, we added into these two plasmids, at their KpnI site, the DsRED gene from the pRed Root vector (Limpens et al., 2004).
For RNAi EFD construct, we used pPex-RNAi described by Combier et al. (2006). Using Pfx polymerase and primers 5′-TCAGTCCGCTCGAGCCAAACAACAACAACCACCA-3′ and 5′-TTGGGAAGCTTAGATGAACCAACAGAACAAAG-3′ (sense cloning) and 5′-AAGGAAAAAAGCGGCCGCCCAAACAACAACAACCACCA-3′ and 5′-CCTTTAAGACTAGTAGATGAACCAACAGAACAAAG-3′ (antisense cloning), we amplified a 290-bp region of the EFD gene outside of the ERF domain that showed no similarity with other known M. truncatula genes. We cloned these fragments between XhoI and HindIII (sense) and NotI and SpeI (antisense) sites.
To generate the PRR4:GUS construct, we amplified an 1132-bp fragment from the CR962134.2 genomic BAC clone using Pfx polymerase and primers 5′-GGGGTACCCCCGAGAAAATAACT-3′ and 5′-CATGCCATGGCACTCTCTTTGAAGAAAAAAAAGA-3′ and inserted it between KpnI and NcoI sites of the pPex-GUS vector (Combier et al., 2007).
To generate the PEFD:GUS construct, since the EFD gene sequence was not available, we screened, using standard procedures, a 4.5× fraction of the M. truncatula mth2 BAC library (http://www.medicago.org/genome/) by hybridization on high-density filters. A 175-bp genomic EFD DNA fragment obtained by PCR amplification with primers 5′-CCAAACAACAACAACCACCA-3′ and 5′-TGACCTTCAAACCCAACACA-3′ was used as a probe and allowed to identify four BAC clones. We sequenced the mth2-4K7 BAC using primers listed in Supplemental Table 7 online. We then made two constructs with PEFD: the first one was based on a 1072-bp sequence amplified using Pfx polymerase and primers 5′-GGGGTACCCACCCCGAACCC-3′ and 5′-CATGCCATGGGATGATGAAACAAAAAAAACGTG-3′ and inserted between KpnI and NcoI sites of pPex-GUS; the second one was based on a 2.4-kb fragment, obtained by adding a 1375-bp sequence (from −1067 to −2441 of ATG) upstream of the first 1072-bp sequence, amplified by Pfx polymerase and primers 5′-GGGGTACCAATCATATTCGATGTGTATGAGAC-3′ and 5′-GGGGTACCTATATAACCGTTACC-3′ and inserted in KpnI of the pPex-GUS containing the 1072-bp PEFD fragment.
The efd-1 complementation construct was generated using PEFD:GUS constructs. The GUS gene was deleted by NcoI and NotI digestion and replaced by EFD:Myc from the overexpressing EFD construct in pPex.
All these constructs were checked by sequencing, introduced into A. rhizogenes strain ARqua1 by electroporation, and used for M. truncatula root transformation as described by Boisson-Dernier et al. (2001) and Combier et al. (2007). All A. rhizogenes root transformations were done in at least three biological repetitions, except for the efd-1 complementation.
Plasmid Constructs and Transient Expression in N. benthamiana
The plasmid constructs were made in two steps. First, we cloned EFD and EFD deleted from the ERF domain (ΔEFD) in the gateway vector pKAs207 (kindly provided by L. Deslandes [LIPM, Toulouse] and corresponding to pDONR207 in which an AscI kanamycin cassette has been introduced) and then we did an LR recombination with destination vectors PAM-PAT35S-GWY-mRFP/mRFP-GWY/GWY-3HA/3HA-GWY (provided by L. Deslandes) following the manufacturer's instructions (Invitrogen). For this step, we amplified the EFD coding sequence (588 bp) with or without the STOP codon, with Pfx polymerase and primers 5′-AGGCGCGCCTACCATGGCAAGACCACAACAACGTTAT-3′ and 5′-AGGCGCGCCCAGATGAACCAACAGAACAAAGCTC-3′ (within STOP codon) or 5′-AGGCGCGCCCCTAAGATGAACCAACAGAACAAAG-3′ (without STOP codon). For ΔEFD, we amplified a 390-bp fragment using Pfx polymerase and primers 5′-AGGCGCGCCTACCATGCCAAATGGACCACAATCTTCTTCA-3′ and 5′-AGGCGCGCCCAGATGAACCAACAGAACAAAGCTC-3′ (within STOP codon) or 5′-AGGCGCGCCCCTAAGATGAACCAACAGAACAAAG-3′ (without STOP codon).
For bacteria preparation and N. benthamiana infiltration, we used the protocol described by Andriankaja et al. (2007). The subcellular localization of mRFP-fused proteins was analyzed by fluorescence confocal microscopy (Leica AOBS SP2) from 30 to 72 h after infiltration. Each construct was observed in three independent infiltrated leaves and at least in three biological repetitions. Fluorescence was collected at 565 to 620 nm for mRFP and 630 to 730 nm for chloroplasts. For transactivation studies, leaf discs were collected at 24 and 48 h after inoculation on independent infiltrated leaves using at least three biological repetitions. For each point, two discs were used directly for histochemical GUS assay and two were frozen in liquid nitrogen for enzymatic GUS assays on protein extracts.
Histochemical and Fluorometric GUS Assays
Histochemical GUS staining (using 5-bromo-4-chloro-3-indolyl-β-glucuronic acid; MP Biomedicals) and double staining for both GUS and β-galactosidase activities after inoculation with S. meliloti strain carrying a constitutive hemA-lacZ fusion (Ardourel et al., 1994) were performed as described by Boisson-Dernier et al. (2005). For simple β-galactosidase assays, we used X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; MP Biomedicals) instead of Magenta-Gal. Roots and nodule sections (50 μm thick) were prepared in 4% agarose with a vibrating microtome (Leica VT 1000S), and stained samples were observed with a Zeiss Axiophot light microscope. Observations were done at least on 10 independent roots transformed by A. rhizogenes and at least two biological repetitions, at different times after mock or S. meliloti inoculation.
For GUS quantitative assay, leaf discs of N. benthamiana were ground in liquid nitrogen and total proteins were extracted in GUS buffer (50 mM potassium phosphate buffer, pH 7.5, 10 mM 2-mercaptoethanol, 10 mM Na2EDTA, 0.1% sodium lauryl-sarcosine, and 0.1% Triton X-100). Protein concentrations were normalized with Bradford reagent (Bio-Rad). Enzymatic reactions were performed using 10 μg of total protein extract with 4-methylumbelliferyl-β-d-glucuronide (Biosynth) as substrate. Fluorescence was measured using a microtiter fluorimeter (FL600; Bio-Tek) and measurements read every 30 min (during 4 h). Enzyme activity was calibrated with a dilution series of 4-methylumbelliferone (Sigma-Aldrich).
In Situ Hybridizations
In situ hybridizations were performed for EFD using a riboprobe made from the full-size cDNA as described by de Billy et al. (2001) on wild-type 4- and 10-d-old nodules harvested from plants grown in aeroponic caissons. For Mt RR4, in situ hybridizations were performed following the procedure described by Valoczi et al. (2006) and Boualem et al. (2008). The following primers were used for generating the Mt RR4–specific probe (251 nucleotides long): 5′-AATGTGGGAAGCCAAGACAC-3′ and 5′-CGGTGCCGGTCATTTAAG-3′. For the probe (182 nucleotides long) corresponding to the whole type-A response regulator gene family, the primers were 5′-ATGCTTTTGTTCCGGGTTTA-3′ and 5′-CGGTGCCGGTCATTTAAG-3′.
Q-RT-PCR Analysis
RNA samples were isolated using the SV total RNA extraction kit (Promega) according to the manufacturer's recommendations. The absence of DNA contamination was verified by PCR with EF1-α primers, and RNA quality was checked using a Bioanalyzer (Agilent Technologies). Reverse transcription was performed on 1 μg of RNA using the superscript reverse transcriptase II (Invitrogen) and anchored oligo(dT) for plant cDNAs synthesis and random hexamers for bacterial cDNAs. We used 80 pg per sample of human desmin RNA as external standard to check the efficiency of the reverse transcription and EF1-α (MtGI8: TC106470) or pnp (SMc00324) as internal Q-RT-PCR standard for the analysis of plant or bacterial gene expression, respectively. Quantitative PCR was performed on a Lightcycler (Roche Diagnostics), with the Light Cycler Fast Start Reaction Mix MasterPLUS SYBR Green according to the manufacturer's recommendations. Cycling conditions were as follow: 95°C for 8 min, 45 cycles at 95°C for 5 s, 60°C for 7 s, and 72°C for 15 s. The specificity of primer pairs (see Supplemental Table 8 online) was confirmed by sequencing PCR amplicon and analysis of dissociation curves (65 to 99°C). Each reaction was performed on a 1:16 (v/v) cDNA dilution with technical replicates. The data shown represent means of values obtained from two or three independent biological replicates.
Q-RT-PCR analysis was also conducted on 384-well plates for validation of microarray results using primers shown in Supplemental Table 8 online and an ABI 7900HT thermocycler (Applied Biosystems) following manufacturer conditions. Cycling conditions were as follow: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min.
Microarray Studies
RNA was extracted from nodules by the Trizol method (Invitrogen) and purified using Microcon-30 column (Millipore). When analyzing P35S:EFD:VP16 roots, RNA was prepared using the SV total RNA extraction kit (Promega) and amplified with the BD SMART mRNA amplification kit (BD Biosciences). Sixteen micrograms for the nodules and 5 μg for the root were used to synthesize Cy3- and Cy5-labeled cDNA (Hohnjec et al., 2005). Mt16kOLI1Plus microarrays (Hohnjec et al., 2005; Tellström et al., 2007) were used, the design of which can be viewed at http://www.ebi.ac.uk/arrayexpress (accession number A-MEXP-138). Three independent biological replicates were performed with a dye swap for each. Hybridization of targets, image acquisition, and analysis were performed precisely as described by Hohnjec et al. (2005), using an ASP hybridization station (Amersham Biosciences) and ImaGene 5.5 software (Biodiscovery).
Data files were processed using EMMA2 array analysis software (Küster et al., 2007). Microarray data were normalized by Lowess normalization with a floor value of 20 and t-statistics used to identify differentially regulated genes. Adjusted P values were determined to carry out multiple-comparison corrections using the Benjamini and Hochberg method, which controls the false discovery rate (Benjamini and Hochberg, 1995; Reiner et al., 2003).
Phylogenetic Analysis of ERF Proteins
Full-length amino acid sequences of selected ERF proteins were aligned using ClustalW (http://clustalw.genome.ad.jp/) and analyzed with the PHYLIP software package (http://www.csc.fi/molbio/progs/phylip/doc/main.html). The tree was calculated on the basis of 1000 bootstraps and parsimony calculations using randomized inputs and 100× jumbling. The resulting 100 data sets were combined for a consensus tree. The resulting tree file was displayed using TreeView.
Accession Numbers
Sequence data for the proteins used for phylogenetic analyses can be found in the GenBank/EMBL data libraries under the following accession numbers: Mt ERN1, EU038802; Mt ERN2, EU038803; Mt ERN3, EU038804; At ERF002, At5g19790; At Rap2.11, NP_197480; At SHN1, At1g15360; At SHN2, At5g11190; At SHN3, At5g25390; At ERF003, At5g25190. Bacterial genes: bacA, SM-b20999; nifH, SMa0825; nodF, SMa0852; pnp, SMc00324. Mt RR4 EST, TC103991 (MTGI8 database); Mt ENOD11 gene, AJ297721. Results of gene expression profiling experiments are accessible at ArrayExpress under accession number E-TABM-393. The EFD name has priority (VandenBosch and Frugoli, 2001) and has been registered at the GenBank database (EU251063).
Supplemental Data
The following materials can be found in the online version of this article.
Supplemental Figure 1. Cortical Cell Divisions Are Frequently Associated with Root Hair Curls in the efd-1 Mutant but Not in Wild-Type Plants.
Supplemental Figure 2. A Weak Supernodulant Phenotype Is Exhibited by EFD RNAi Roots.
Supplemental Figure 3. A Decrease in Nodulation Is Exhibited by EFD-Overexpressing Roots.
Supplemental Figure 4. Q-RT-PCR Analyses of Bacterial Symbiotic Gene Expression in efd-1 versus Wild-Type Nodules.
Supplemental Figure 5. In Situ Hybridization on Nodule Sections with a Control 35S-Labeled EFD Sense Probe.
Supplemental Figure 6. EFD Is Expressed in Root Tips and Root Primordia and Is Not Induced by Purified Nod Factors.
Supplemental Figure 7. EFD Expression Is Not Induced by Ethylene.
Supplemental Figure 8. Alignment of Group V ERF Proteins from M. truncatula and Arabidopsis.
Supplemental Figure 9. Mt RR4 and EFD Are Expressed in Nodule Zone II.
Supplemental Figure 10. Mt RR4 Is a Primary Cytokinin Response Gene, and EFD Transcription Is Not Regulated by Cytokinins.
Supplemental Figure 11. Trans-Activation of PRR4, PEFD, and PMMPL1 in N. benthamiana by P35S:EFD:HA.
Supplemental Table 1. Q-RT-PCR Analysis of EFD Expression in Roots of Wild-Type and the efd-1 Mutant of M. truncatula Inoculated by Wild-Type S. meliloti.
Supplemental Table 2. Infection Thread Formation Is Decreased in EFD-Overexpressing Roots.
Supplemental Table 3. Q-RT-PCR Analysis of EFD and Mt ENOD11 Expression in Roots of Early Symbiotic M. truncatula nfp-1 (C31 Allele), nsp1-1 (B85), and hcl-1 (B56) Mutants Inoculated by Wild-Type S. meliloti.
Supplemental Table 4. Genes Differentially Expressed between Wild-Type and efd-1 Nodules as Determined by Microarray Analyses.
Supplemental Table 5. Q-RT-PCR Validation of Genes Identified by Mt16KOLI1Plus Microarrays as Down- or Upregulated in efd-1 Nodules Compared with Wild-Type Nodules.
Supplemental Table 6. Genes Differentially Expressed between P35S:EFD Transgenic M. truncatula Roots and Empty Vector Transformed Roots, with (4 DAI) and without S. meliloti Inoculation, as Determined by Microarray Analyses.
Supplemental Table 7. Primers Used for PEFD Sequencing.
Supplemental Table 8. Primers Used in Q-RT-PCR.
Supplementary Material
Acknowledgments
We thank Helge Küster (Bielefeld University, Germany) for providing 16K+ microarrays and accompanying procedures and Jérôme Gouzy for complementary information on gene annotation. We also thank Delphine Capela (Laboratoire des Interactions Plantes Micro-Organismes [LIPM]), Frédéric Debéllé (LIPM), Laurent Deslandes (LIPM), Alain Jauneau (IFR40, Toulouse), and Thomas Ott (LIPM) for their help with acetylene reduction assay, cloning vectors, BAC library screening, confocal microscopy, and phylogenetic analyses, respectively. We are grateful to Martin Crespi (Institut des Sciences du Végétal) and Jean Dénarié (LIPM, Toulouse) for useful comments on the manuscript and to Fernanda de Carvalho Niebel (LIPM) for fruitful discussions and sharing unpublished data. We wish to acknowledge the contribution of José Garcia (LIPM) for plant production and Christelle Latorre (LIPM) for some Q-RT-PCR analyses. M. truncatula wild-type A17 seeds were provided by Jean-Marie Prospéri (Institut National de la Recherche Agronomique). This work was supported by the FP6 Grain Legumes Integrated Project. Tatiana Vernié and Julie Plet were supported by a doctoral grant from EU-CNRS (fonds social européen) and from the French Research Ministry, respectively.
The author responsible for distribution of the materials integral to the findings presented in this article in accordance with policy described in the Instructions for Authors (www.plantcell.org) is: Pascal Gamas (pascal.gamas@toulouse.inra.fr).
Online version contains Web-only data.
References
- Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 2463–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, M.D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17 5484–5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alunni, B., Kevei, Z., Redondo-Nieto, M., Kondorosi, A., Mergaert, P., and Kondorosi, E. (2007). Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol. Plant Microbe Interact. 20 1138–1148. [DOI] [PubMed] [Google Scholar]
- Andriankaja, A., Boisson-Dernier, A., Frances, L., Sauviac, L., Jauneau, A., Barker, D.G., and de Carvalho-Niebel, F. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19 2866–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel, M., Demont, N., Debelle, F., Maillet, F., de Billy, F., Prome, J.C., Denarie, J., and Truchet, G. (1994). Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi, J.F., Godfroy, O., de Billy, F., Saurat, O., Jauneau, A., and Gough, C. (2008). The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. USA 105 9817–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu, E., Shimoda, Y., Kouchi, H., Tabata, S., and Sato, S. (2008). A positive regulatory role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus. Plant Physiol. 147 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno, H., Ikeda, Y., Niu, Q.W., and Chua, N.H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard, G., and Fortin, J.A. (1988). Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108 211–218. [DOI] [PubMed] [Google Scholar]
- Becker, A., et al. (2004). Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant Microbe Interact. 17 292–303. [DOI] [PubMed] [Google Scholar]
- Benaben, V., Duc, G., Lefebvre, V., and Huguet, T. (1995). TE7, an inefficient symbiotic mutant of Medicago truncatula Gaertn. cv Jemalong. Plant Physiol. 107 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approch to multiple testing. J. Roy. Statist. Soc. Ser. B. Methodological 57 289–300. [Google Scholar]
- Boisson-Dernier, A., Andriankaja, A., Chabaud, M., Niebel, A., Journet, E.P., Barker, D.G., and de Carvalho-Niebel, F. (2005). MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol. Plant Microbe Interact. 18 1269–1276. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., Garcia, F., Becard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Borisov, A.Y., Madsen, L.H., Tsyganov, V.E., Umehara, Y., Voroshilova, V.A., Batagov, A.O., Sandal, N., Mortensen, A., Schauser, L., Ellis, N., Tikhonovich, I.A., and Stougaard, J. (2003). The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 131 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boualem, A., Laporte, P., Jovanovic, M., Laffont, C., Plet, J., Combier, J.P., Niebel, A., Crespi, M., and Frugier, F. (2008). MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J. 54 876–887. [DOI] [PubMed] [Google Scholar]
- Boutilier, K., Offringa, R., Sharma, V.K., Kieft, H., Ouellet, T., Zhang, L., Hattori, J., Liu, C.M., van Lammeren, A.A., Miki, B.L., Custers, J.B., and van Lookeren Campagne, M.M. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright, L.J., Liang, Y., Mitchell, D.M., and Harris, J.M. (2005). The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant Microbe Interact. 18 521–532. [DOI] [PubMed] [Google Scholar]
- Chabaud, M., Venard, D., Defaux-Petras, A., Bécard, G., and Barker, D.G. (2002). Targeted inoculation of Medicago truncatula in vitro root cultures reveals MtENOD11 expression during early stages of infection by arbuscular mycorrhizal fungi. New Phytol. 156 265–273. [DOI] [PubMed] [Google Scholar]
- Chuck, G., Muszynski, M., Kellogg, E., Hake, S., and Schmidt, R.J. (2002). The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298 1238–1241. [DOI] [PubMed] [Google Scholar]
- Combier, J.P., de Billy, F., Gamas, P., Niebel, A., and Rivas, S. (2008). Trans-regulation of the expression of the transcription factor MtHAP2–1 by a uORF controls root nodule development. Genes Dev. 22 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier, J.P., Frugier, F., de Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., Vernie, T., Ott, T., Gamas, P., Crespi, M., and Niebel, A. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier, J.P., Vernié, T., de Billy, F., El Yahyaoui, F., Mathis, R., and Gamas, P. (2007). The MtMMPL1 early nodulin is a novel member of the matrix metalloendoproteinase family with a role in Medicago truncatula infection by Sinorhizobium meliloti. Plant Physiol. 144 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming, G., Fidler, F., and Vaux, D.L. (2007). Errors bars in experimental biology. J. Cell Biol. 177 7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy, F., Grosjean, C., May, S., Bennett, M., and Cullimore, J.V. (2001). Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol. Plant Microbe Interact. 14 267–277. [DOI] [PubMed] [Google Scholar]
- Dello Ioio, R., Linhares, F.S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., and Sabatini, S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17 678–682. [DOI] [PubMed] [Google Scholar]
- Doerner, P. (2007). Plant meristems: Cytokinins–the alpha and omega of the meristem. Curr. Biol. 17 R321–R323. [DOI] [PubMed] [Google Scholar]
- El Yahyaoui, F., Küster, H., Ben Amor, B., Hohnjec, N., Puhler, A., Becker, A., Gouzy, J., Vernie, T., Gough, C., Niebel, A., Godiard, L., and Gamas, P. (2004). Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 136 3159–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova, M., van de Mortel, J., Matsumoto, P.A., Cho, J., Town, C.D., VandenBosch, K.A., Gantt, J.S., and Vance, C.P. (2002). Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol. 130 519–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson, B.J., and Mathesius, U. (2003). Signaling interactions during nodule development. J. Plant Growth Regul. 22 47–72. [Google Scholar]
- Ferreira, F.J., and Kieber, J.J. (2005). Cytokinin signaling. Curr. Opin. Plant Biol. 8 518–525. [DOI] [PubMed] [Google Scholar]
- Ferrell, J.E., Jr. (2002). Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14 140–148. [DOI] [PubMed] [Google Scholar]
- Frugier, F., Kosuta, S., Murray, J.D., Crespi, M., and Szczyglowski, K. (2008). Cytokinin: Secret agent of symbiosis. Trends Plant Sci. 13 115–120. [DOI] [PubMed] [Google Scholar]
- Frugier, F., Poirier, S., Satiat-Jeunemaitre, B., Kondorosi, A., and Crespi, M. (2000). A Kruppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev. 14 475–482. [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J., Ichige, A., and Walker, G.C. (1993). A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 7 1485–1497. [DOI] [PubMed] [Google Scholar]
- Godiard, L., Niebel, A., Micheli, F., Gouzy, J., Ott, T., and Gamas, P. (2007). Identification of new potential regulators of the Medicago truncatula-Sinorhizobium meliloti symbiosis using a large-scale suppression subtractive hybridization approach. Mol. Plant Microbe Interact. 20 321–332. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, M.A., Silverstein, K.A., Cannon, S.B., and VandenBosch, K.A. (2004). Computational identification and characterization of novel genes from legumes. Plant Physiol. 135 1179–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., and Ecker, J.R. (2004). The ethylene signaling pathway: New insights. Curr. Opin. Plant Biol. 7 40–49. [DOI] [PubMed] [Google Scholar]
- Gutterson, N., and Reuber, T.L. (2004). Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 7 465–471. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Hardy, R.W., Holsten, R.D., Jackson, E.K., and Burns, R.C. (1968). The acetylene-ethylene assay for N(2) fixation: Laboratory and field evaluation. Plant Physiol. 43 1185–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann, A.B., Lombardo, F., Miwa, H., Perry, J.A., Bunnewell, S., Parniske, M., Wang, T.L., and Downie, J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142 1739–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohnjec, N., Vieweg, M.F., Puhler, A., Becker, A., and Küster, H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 137 1283–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.M., Kobayashi, H., Davies, B.W., Taga, M.E., and Walker, G.C. (2007). How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalo, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kawaguchi, M., Imaizumi-Anraku, H., Koiwa, H., Niwa, S., Ikuta, A., Syono, K., and Akao, S. (2002). Root, root hair, and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 15 17–26. [DOI] [PubMed] [Google Scholar]
- Kirch, T., Simon, R., Grunewald, M., and Werr, W. (2003). The DORNROSCHEN/ENHANCER OF SHOOT REGENERATION1 gene of Arabidopsis acts in the control of meristem cell fate and lateral organ development. Plant Cell 15 694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell, L., et al. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell, L., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420 422–426. [DOI] [PubMed] [Google Scholar]
- Kumagai, H., Hakoyama, T., Umehara, Y., Sato, S., Kaneko, T., Tabata, S., and Kouchi, H. (2007). A novel ankyrin-repeat membrane protein, IGN1, is required for persistence of nitrogen-fixing symbiosis in root nodules of Lotus japonicus. Plant Physiol. 143 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy, K.T., Endre, G., Prabhu, R., Penmetsa, R.V., Veereshlingam, H., Cook, D.R., Dickstein, R., and Vandenbosch, K.A. (2004). LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 136 3682–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küster, H., et al. (2007). Development of bioinformatic tools to support EST-sequencing, in silico- and microarray-based transcriptome profiling in mycorrhizal symbioses. Phytochemistry 68 19–32. [DOI] [PubMed] [Google Scholar]
- Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T., and Geurts, R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens, E., Ramos, J., Franken, C., Raz, V., Compaan, B., Franssen, H., Bisseling, T., and Geurts, R. (2004). RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J. Exp. Bot. 55 983–992. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P., Schaff, J.E., Laskey, J.G., Kieber, J.J., Bilyeu, K.D., and Bird, D.M. (2004). Cytokinins play opposite roles in lateral root formation, and nematode and Rhizobial symbioses. Plant J. 38 203–214. [DOI] [PubMed] [Google Scholar]
- Lohar, D.P., Sharopova, N., Endre, G., Penuela, S., Samac, D., Town, C., Silverstein, K.A., and VandenBosch, K.A. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, J.F., Rakocevic, A., Mitra, R.M., Brocard, L., Sun, J., Eschstruth, A., Long, S.R., Schultze, M., Ratet, P., and Oldroyd, G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsch-Martinez, N., Greco, R., Becker, J.D., Dixit, S., Bergervoet, J.H., Karaba, A., de Folter, S., and Pereira, A. (2006). BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol. Biol. 62 825–843. [DOI] [PubMed] [Google Scholar]
- Mathis, R., Grosjean, C., de Billy, F., Huguet, T., and Gamas, P. (1999). The early nodulin gene MtN6 is a novel marker for events preceding infection of Medicago truncatula roots by Sinorhizobium meliloti. Mol. Plant Microbe Interact. 12 544–555. [DOI] [PubMed] [Google Scholar]
- Mergaert, P., Nikovics, K., Kelemen, Z., Maunoury, N., Vaubert, D., Kondorosi, A., and Kondorosi, E. (2003). A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132 161–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., Mausset, A.E., Barloy-Hubler, F., Galibert, F., Kondorosi, A., and Kondorosi, E. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. USA 103 5230–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton, P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi, D., Prado, E., Sagan, M., and Duc, G. (2005). Characterisation of new symbiotic Medicago truncatula (Gaertn.) mutants, and phenotypic or genotypic complementary information on previously described mutants. Mycorrhiza 15 283–289. [DOI] [PubMed] [Google Scholar]
- Murray, J.D., Karas, B.J., Sato, S., Tabata, S., Amyot, L., and Szczyglowski, K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104. [DOI] [PubMed] [Google Scholar]
- Naito, Y., Fujie, M., Usami, S., Murooka, Y., and Yamada, T. (2000). The involvement of a cysteine proteinase in the nodule development in Chinese milk vetch infected with Mesorhizobium huakuii subsp. rengei. Plant Physiol. 124 1087–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 411–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naya, L., Ladrera, R., Ramos, J., Gonzales, E.M., Arrese-Igor, C., Minchin, F.R., and Becana, M. (2007). The response of carbon metabolism and antioxidant defenses of alfalfa nodules to drought stress and to the subsequent recovery of plants. Plant Physiol. 144 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, R., Hayashi, M., Wu, G.J., Kouchi, H., Imaizumi-Anraku, H., Murakami, Y., Kawasaki, S., Akao, S., Ohmori, M., Nagasawa, M., Harada, K., and Kawaguchi, M. (2002. b). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420 426–429. [DOI] [PubMed] [Google Scholar]
- Nishimura, R., Ohmori, M., Fujita, H., and Kawaguchi, M. (2002. a). A Lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc. Natl. Acad. Sci. USA 99 15206–15210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (1999). Identification of a promoter region responsible for the senescence-specific expression of SAG12. Plant Mol. Biol. 41 181–194. [DOI] [PubMed] [Google Scholar]
- Nukui, N., Ezura, H., and Minamisawa, K. (2004). Transgenic Lotus japonicus with an ethylene receptor gene Cm-ERS1/H70A enhances formation of infection threads and nodule primodia. Plant Cell Physiol. 45 427–435. [DOI] [PubMed] [Google Scholar]
- Oka-Kira, E., and Kawaguchi, M. (2006). Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 9 496–502. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Downie, J.A. (2006). Nuclear calcium changes at the core of symbiosis signalling. Curr. Opin. Plant Biol. 9 351–357. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Downie, J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Long, S.R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod factor signaling. Plant Physiol. 131 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275 527–530. [DOI] [PubMed] [Google Scholar]
- Pislariu, C.I., and Dickstein, R. (2007). An IRE-like AGC kinase gene, MtIRE, has unique expression in the invasion zone of developing root nodules in Medicago truncatula. Plant Physiol. 144 682–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner, A., Yekutieli, D., and Benjamini, Y. (2003). Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 368–375. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 633–646. [DOI] [PubMed] [Google Scholar]
- Sauviac, L., Niebel, A., Boisson-Dernier, A., Barker, D.G., and de Carvalho-Niebel, F. (2005). Transcript enrichment of Nod factor-elicited early nodulin genes in purified root hair fractions of the model legume Medicago truncatula. J. Exp. Bot. 56 2507–2513. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Handberg, K., Sandal, N., Stiller, J., Thykjaer, T., Pajuelo, E., Nielsen, A., and Stougaard, J. (1998). Symbiotic mutants deficient in nodule establishment identified after T-DNA transformation of Lotus japonicus. Mol. Gen. Genet. 259 414–423. [DOI] [PubMed] [Google Scholar]
- Schnabel, E., Journet, E.P., de Carvalho-Niebel, F., Duc, G., and Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58 809–822. [DOI] [PubMed] [Google Scholar]
- Schauser, L., Roussis, A., Stiller, J., and Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402 191–195. [DOI] [PubMed] [Google Scholar]
- Searle, I.R., Men, A.E., Laniya, T.S., Buzas, D.M., Iturbe-Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 109–112. [DOI] [PubMed] [Google Scholar]
- Silverstein, K.A., Graham, M.A., and VandenBosch, K.A. (2006). Novel paralogous gene families with potential function in legume nodules and seeds. Curr. Opin. Plant Biol. 9 142–146. [DOI] [PubMed] [Google Scholar]
- Silverstein, K.A., Moskal, W.A., Jr., Wu, H.C., Underwood, B.A., Graham, M.A., Town, C.D., and VandenBosch, K.A. (2007). Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 51 262–280. [DOI] [PubMed] [Google Scholar]
- Smit, P., Raedts, J., Portyanko, V., Debelle, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Starker, C.G., Parra-Colmenares, A.L., Smith, L., Mitra, R.M., and Long, S.R. (2006). Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol. 140 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyglowski, K., Shaw, R., Wopereis, J., Copeland, S., Hamburger, D., Kasiborski, B., Dazzo, F.B., and de Bruijn, F. (1998). Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 7 684–697. [Google Scholar]
- Teillet, A., Garcia, J., de Billy, F., Gherardi, M., Huguet, T., Barker, D.G., de Carvalho-Niebel, F., and Journet, E.P. (2008). api, a novel Medicago truncatula symbiotic mutant impaired in nodule primordium invasion. Mol. Plant Microbe Interact. 21 535–546. [DOI] [PubMed] [Google Scholar]
- Tellström, V., Usadel, B., Thimm, O., Stitt, M., Küster, H., and Niehaus, K. (2007). The lipopolysaccharide of Sinorhizobium meliloti suppresses defense-associated gene expression in cell cultures of the host plant Medicago truncatula. Plant Physiol. 143 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers, A.C., Auriac, M.C., and Truchet, G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., Sandal, N., Madsen, L.H., Radutoiu, S., Albrektsen, A.S., Sato, S., Asamizu, E., Tabata, S., and Stougaard, J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107. [DOI] [PubMed] [Google Scholar]
- To, J.P., Haberer, G., Ferreira, F.J., Deruere, J., Mason, M.G., Schaller, G.E., Alonso, J.M., Ecker, J.R., and Kieber, J.J. (2004). Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell 16 658–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi, A., Varallyay, E., Kauppinen, S., Burgyan, J., and Havelda, Z. (2006). Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 47 140–151. [DOI] [PubMed] [Google Scholar]
- VandenBosch, K.A., and Frugoli, J. (2001). Guidelines for genetic nomenclature and community governance for the model legume Medicago truncatula. Mol. Plant Microbe Interact. 14 1364–1367. [DOI] [PubMed] [Google Scholar]
- van der Graaff, E., Dulk-Ras, A.D., Hooykaas, P.J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127 4971–4980. [DOI] [PubMed] [Google Scholar]
- Van de Velde, W., Guerra, J.C., De Keyser, A., De Rycke, R., Rombauts, S., Maunoury, N., Mergaert, P., Kondorosi, E., Holsters, M., and Goormachtig, S. (2006). Aging in legume symbiosis. A molecular view on nodule senescence in Medicago truncatula. Plant Physiol. 141 711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse, J., de Billy, F., Camut, S., and Truchet, G. (1990). Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasse, J., de Billy, F., and Truchet, G. (1993). Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 4 555–566. [Google Scholar]
- Veereshlingam, H., Haynes, J.G., Penmetsa, R.V., Cook, D.R., Sherrier, D.J., and Dickstein, R. (2004). nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol. 136 3692–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, R.J., Cooke, S.E., Brammar, W.J., and Schuch, W. (1994). Control of gene expression in plant cells using a 434:VP16 chimeric protein. Plant Mol. Biol. 24 381–388. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., de Blank, C., Meskiene, I., Hirt, H., Bakker, J., van Kammen, A., Franssen, H., and Bisseling, T. (1994). Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.