Abstract
AUXIN BINDING PROTEIN1 (ABP1) has long been characterized as a potentially important mediator of auxin action in plants. Analysis of the functional requirement for ABP1 during development was hampered because of embryo lethality of the null mutant in Arabidopsis thaliana. Here, we used conditional repression of ABP1 to investigate its function during vegetative shoot development. Using an inducible cellular immunization approach and an inducible antisense construct, we showed that decreased ABP1 activity leads to a severe retardation of leaf growth involving an alteration in cell division frequency, an altered pattern of endocycle induction, a decrease in cell expansion, and a change in expression of early auxin responsive genes. In addition, local repression of ABP1 activity in the shoot apical meristem revealed an additional role for ABP1 in cell plate formation and cell shape. Moreover, cells at the site of presumptive leaf initiation were more sensitive to ABP1 repression than other regions of the meristem. This spatial context-dependent response of the meristem to ABP1 inactivation and the other data presented here are consistent with a model in which ABP1 acts as a coordinator of cell division and expansion, with local auxin levels influencing ABP1 effectiveness.
INTRODUCTION
Recent research has led to significant advances in our understanding of the molecular mechanism by which the growth regulator auxin is perceived by the plant and the signal transduced into a molecular output (Badescu and Napier, 2006; Quint and Gray, 2006). In particular, this work has led to the identification of the TRANSPORT INHIBITOR RESPONSE PROTEIN1 (TIR1) F-box factor as an auxin receptor whose binding with auxin leads to an SCF ubiquitin-ligase catalyzed degradation of auxin/indole-3-acetic acid (Aux/IAA) transcriptional repressors (Dharmasiri et al., 2005a; Kepinski and Leyser, 2005). Mutational analysis has revealed that loss of functional TIR1 and related AFB genes leads in a combinatorial fashion to progressively more severe developmental and physiological affects, confirming that auxin signal transduction via the TIR1 receptor family represents a major pathway for auxin function (Dharmasiri et al., 2005b). However, even quadruple mutants in which all TIR1/AFB genes are mutated can form functional (although abnormal) plants. Although it is difficult to dismiss the possibility that in these mutants there is some residual TIR1/AFB function, the data suggest that auxin may also act via the function of an additional receptor (Badescu and Napier, 2006); indeed, classical data on auxin receptor biology support this idea. Recent results derived from the analysis of other plant growth factors, such as abscisic acid, also indicate that plants use a surprising variety of molecular mechanisms to perceive and transduce growth factor signals (Verslues and Zhu, 2007). Some of these may resemble standard signal transduction motifs identified in other organisms, whereas others seem to be unique to plants.
With respect to auxin, one potential receptor in addition to TIR1 is AUXIN BINDING PROTEIN1 (ABP1). A substantial body of evidence demonstrates that this protein binds auxin at physiologically relevant levels, that it can discriminate between physiologically active and inactive forms of auxin, and that its activity mediates the activation of ion fluxes across the plasma membrane in response to auxin (Venis et al., 1992; Rück et al., 1993; Thiel et al., 1993; Leblanc et al., 1999b; Bauly et al., 2000). Such alteration of ion fluxes could mediate changes in cell expansion, accounting for the long-reported effect of auxin on growth of shoot tissues. Thus, ABP1 has been shown to mediate part of the auxin-induced swelling response of maize (Zea mays) coleoptile protoplasts and of epidermal protoplasts from elongating pea (Pisum sativum) internodes (Steffens et al., 2001; Yamagami et al., 2004). However, there are still question marks over the endogenous function of ABP1. First, the mechanism by which binding of auxin to ABP1 might lead to signal transduction (e.g., altered ion channel activity, altered secretary pathway, or altered gene expression) remains unclear. Second, the outcome of altered ABP1 expression in the intact plant is poorly documented. Inducible overexpression of ABP1 leads to an increased responsiveness of some leaf tissues to auxin at particular stages of development. Thus, excised segments from specific stages of developing leaves displayed increased component cell size after induction of ABP1 overexpression, although overall leaf size and shape in the intact plant was not affected (Jones et al., 1998). The only reported outcome of loss of ABP1 activity in intact plants described how mutation of the single gene copy of ABP1 in Arabidopsis thaliana led to an embryo-lethal phenotype (Chen et al., 2001). Malformed embryos (composed of relatively small and misshapen cells) aborted at the globular stage, seemingly unable to form a polar embryo structure. These data indicate that ABP1 is essential for early embryogenesis, but the embryo lethality precluded any interpretation of the role of ABP1 during postembryonic development. Since auxin clearly plays a fundamental role at many stages of the plant life cycle, our lack of knowledge on the outcome of loss of ABP1 activity during postembryonic development is a major gap in our understanding of auxin and ABP1 function.
One approach to this problem is to generate plants in which the activity of ABP1 can be conditionally repressed. In previous work, ABP1 activity was blocked in BY2 suspension cultured cells by overexpression of a recombinant antibody fragment (scFv) directed against ABP1 (David et al., 2007). This cellular immunization approach suggested that ABP1 plays a critical role in auxin's regulation of the cell cycle, whereas most previous work had focused on the potential role of ABP1 in cell growth. These recent data were obtained from the analysis of in vitro cell cultures, raising the question of their relevance to the intact plant.
In this article, we report on the outcome of conditional repression of ABP1 activity during shoot postembryonic development using both antisense and cellular immunization approaches, the latter being performed in both Arabidopsis and tobacco (Nicotiana tabacum). These experiments indicate that ABP1 is required for the maintenance of shoot and leaf growth. In addition, by restricting the spatial and temporal parameters of ABP1 inactivation (via use of a microinduction technique) we show that ABP1 is required for the coordination of cell division and cell expansion in a highly context-dependent manner. In particular, during the very early stages of leaf formation even transient loss of ABP1-mediated coordination of cell plate formation and cell expansion has drastic downstream consequences for growth of the whole plant. These data demonstrate the requirement for ABP1 during shoot postembryonic development and accentuate the importance of developmental context in understanding the function of a gene product involved in growth factor perception.
RESULTS
Ethanol-Inducible Inactivation of ABP1 Leads to Repression of Plant Growth
Our previous work described the generation of a number of transgenic lines of tobacco BY2 cells containing constructs that encoded single chain fragment variable regions (scFv12) derived from a hybridoma line expressing the well-characterized anti-ABP1 monoclonal antibody mAb12 (Leblanc et al., 1999b; David et al., 2007). Analysis of these transgenic cell lines showed that the induced scFv12 protein interacted in vivo with ABP1 and led to a block on cell division.
To establish a system to conditionally downregulate ABP1 activity in the intact plant, we generated a number of transgenic Arabidopsis lines containing scFv12 constructs in a vector designed to allow induction of the encoded protein by exposure of the plants to ethanol vapor (Roslan et al., 2001). The scFv12 constructs were modified to direct the encoded protein either to the apoplast (SS12S construct) or to be retained within the endoplasmic reticulum (SS12K construct). In the experiments described below, very similar phenotypes were observed with both constructs, consistent with our data from BY2 cells (David et al., 2007). In addition, conditional ABP1 antisense transgenic Arabidopsis plants were also generated using the same expression system. After appropriate selection, a number of transgenic lines homozygous for each transgene were identified. Individual plants were then germinated either in vitro or on soil and exposed at selected time points to ethanol vapor (as described in Methods).
As shown in Figures 1A and 1B, exposure of p35S:AlcR>pAlcA:SS12K (further referred to as AtSS12K) and p35S:AlcR>pAlcA:ABP1AS (further referred to as AtABP1AS) plants to ethanol led to a rapid accumulation of the scFv12 mRNA and ABP1 antisense RNAs, respectively, with transcripts being detectable within 2 h and rising to a maximum level after 24 h. Protein gel blot analysis revealed a detectable accumulation of the scFv12 protein in the AtSS12K plants after 8 h (data not shown) and strong accumulation after 24 h (Figure 1C). In antisense plants, 3 to 4 d of exposure to ethanol led to loss of detection of ABP1, even using n-butyl alcohol solubilized membrane fractions that are enriched in ABP1 (Figure 1D). Pull-down experiments using an anti-ABP1 antibody revealed that the scFv12 protein generated within the plant tissue was associated with endogenous ABP1 protein (C. Perrot-Rechenmann, unpublished data). These observations are consistent with our previous work in which the scFv12 construct was expressed in tobacco BY2 cells and where specific interaction of the scFv12 protein with endogenous ABP1 was also demonstrated (David et al., 2007). Taken together, our data indicate that following exposure to ethanol vapor, the AtSS12K plants rapidly accumulated the scFv12 protein and that this cellular antibody interacted with endogenous ABP1.
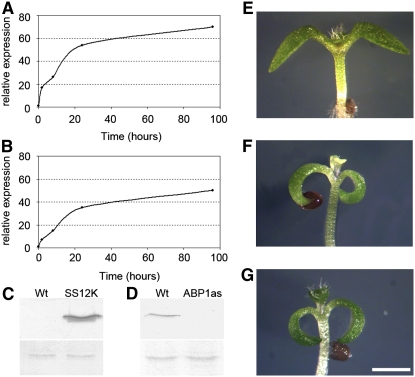
Figure 1.
Conditional Repression of ABP1 Activity in Arabidopsis.
(A) and (B) Expression kinetics of scFv12 (A) and ABP1 antisense (B) mRNA accumulation after exposure to ethanol in AtSS12K and AtABP1AS lines, respectively. Data are expressed as the ratio between induced and noninduced samples.
(C) Immunodetection of the recombinant antibody scFv12 in a microsomal fraction of wild-type and AtSS12K plants induced with ethanol for 24 h. Top blot shows signal detected with an antibody against scFv12. Bottom panel shows protein loading detected with Ponceau S staining prior to incubation with the specific antibody.
(D) Immunodetection of ABP1 in enriched microsomal fractions from wild-type and AtABP1AS plants induced with ethanol for 3 d. Top blot shows signal detected with an antibody against ABP1. Bottom blot shows protein loading detected with Ponceau S staining prior to incubation with the specific antibody.
(E) to (G) Apices of wild-type (E), AtSS12K (F), and AtABP1AS (G) 5-d-old seedlings grown in vitro in the presence of 5% ethanol vapor. Cotyledon epinasty and decreased leaf growth are apparent in (F) and (G) compared with (E). Bar = 2.5 mm.
Subsequent to ethanol exposure, both scFv12 expressing lines and ABP1 antisense lines showed a dramatic reduction in growth and development of young seedlings compared with induced control seedlings (Figures 1E to 1G). Four-day-old AtSS12K seedlings exposed to ethanol exhibited small and epinastic cotyledons and the absence or delayed emergence of pale primary leaves that did not develop further (Figure 1F). Similar phenotypes were observed in seedlings in which the ABP1 antisense construct was induced by exposure of the plant to ethanol (Figure 1G). No effect of ethanol was observed in wild-type plants (Figure 1E) or in plants expressing a β-glucuronidase (GUS) reporter gene construct under the same ethanol-inducible system (see Supplemental Figure 1A online). Thus, using two different approaches to inducibly decrease endogenous ABP1 activity produced essentially the same phenotype.
Germination and growth of seedlings on soil allowed a controlled exposure to ethanol (and thus repression of ABP1 activity) at various time points. Induction with ethanol at very early stages of seedling development led to the phenotype as described in Figure 1, leading to death of the plant. When older (12-d-old) AtSS12K or AtABP1AS plants were exposed to ethanol vapor, down-curling of leaves occurred accompanied by growth arrest within a few days (Figures 2C and 2D), whereas induced wild-type and AlcAGus control plants or noninduced AtSS12K plants showed no alteration in growth (Figures 2A and 2B; see Supplemental Figures 1B to 1D online). Extended exposure to ethanol caused drastic growth defects characterized by the formation of small, curled leaves (Figures 2C to 2G; see Supplemental Figure 1E online). Thus, when plants that had formed five visible leaves under noninducing conditions were exposed to ethanol, the 6th leaf reached a length of 9 to 11 mm over the subsequent 14 d, whereas in the same time the 6th leaf from a plant kept under noninducing conditions attained a length of 27 to 30 mm. In addition to this general reduction in growth, leaf morphology was also affected. Preexisting leaves induced to repress ABP1 activity exhibited warped and shorter laminas, whereas leaves that appeared subsequent to repression of ABP1 activity displayed a decrease both in lamina width and length. In addition, these leaves were epinastic (Figure 2H). Measurements of auxin content in shoots of ABP1 inactivated plantlets revealed that the induced phenotype was not correlated with a global modification of auxin content (see Supplemental Figure 2 online). Organization of leaf vasculature was almost normal, although rosette leaves with decreased growth exhibited a reduced number of veins (Figure 2I) (Alonso-Peral et al., 2006). Exposure of AtSS12K plants to ethanol vapor at later stages of development also led to a decrease in plant growth, and exposure during flowering led to sterility (see Supplemental Figure 3 online). Measurement of rosette diameter indicated a significant repression of growth in AtSS12K plants after repression of ABP1 activity (Figure 2J).
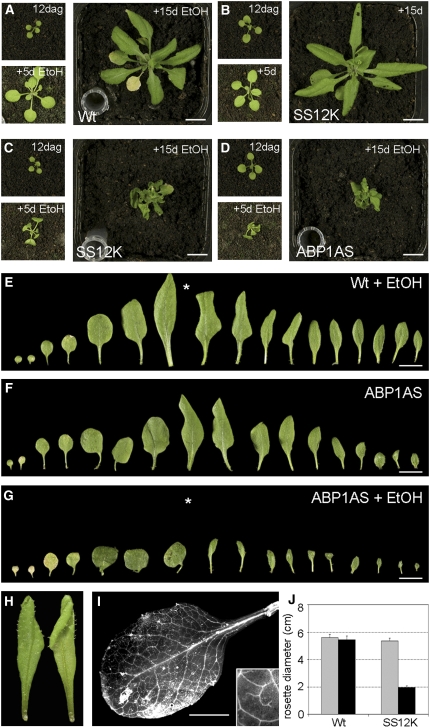
Figure 2.
Leaf Growth Decreases after Inactivation of ABP1.
(A) to (D) Compared with ethanol (EtOH)-induced wild-type (A) and noninduced AtSS12K (B) plants, ethanol-induced AtSS12K (C) and ethanol-induced AtABP1AS (D) plants show reduced leaf growth. In each panel, the top left image shows plant growth at 12 d after germination (dag) (time of initial exposure to ethanol), and the bottom left image shows the same plant 5 d later, with the larger right-hand image showing plant growth 15 d after ethanol exposure. Bars = 1 cm.
(E) to (G) Successive leaves of Arabidopsis rosettes from ethanol-induced wild type (E), noninduced AtABP1AS (F), and ethanol-induced AtABP1AS (G) plants. Exposure to ethanol vapor was performed after emergence of leaf 5 (indicated with asterisk in [E] and [G]). Bars = 1 cm.
(H) An ethanol-induced AtABP1AS leaf showing epinasty. Abaxial view (left-hand image) and adaxial view (right-hand image).
(I) Vasculature of ethanol-induced AtSS12K leaf, cleared with chloral hydrate solution and unrolled. Detail of venation is shown in inset. Bar = 2 mm.
(J) Quantitation of rosette diameter (cm) of wild-type and AtSS12K plants grown for 12 d and then exposed (black bars) or not exposed (gray bars) to ethanol for a further 15 d (error bar indicates se; n = 8).
Previous investigations of the effect of overexpressing ABP1 indicated that although there was little change in leaf growth rate or form, the cell size in certain regions of the leaf was altered (Jones et al., 1998). To investigate whether the observed leaf growth phenotype in the ABP1 inactivated plants was related to an altered internal structure of the leaf we performed a histological and scanning electron microscopy analysis.
Histological analysis of leaves from ethanol-exposed AtSS12K plants indicated that all expected cell types were present and that these were arranged in an appropriate fashion compared with the wild type (Figures 3A to 3E) (i.e., leaf adaxial/abaxial differentiation appeared normal). The severe epinasty of ethanol-induced AtSS12K leaves is clearly visible in transverse sections (Figures 3C and 3D). Some relatively minor changes in histology were apparent, for example, the occurrence of some smaller epidermal cells (Figure 3E). Occasionally, the distal tip of the induced AtSS12K leaves exhibited a lack of vascular tissue (see Supplemental Figure 4A online), indicating that the mid-rib did not extend to the tip of these leaves. In addition, scanning electron microscopy analysis revealed the occurrence of double vascular mid-ribs at the tips of a number of induced AtSS12K or AtABP1AS leaves and twisted veins (see Supplemental Figures 4B and 4C online).
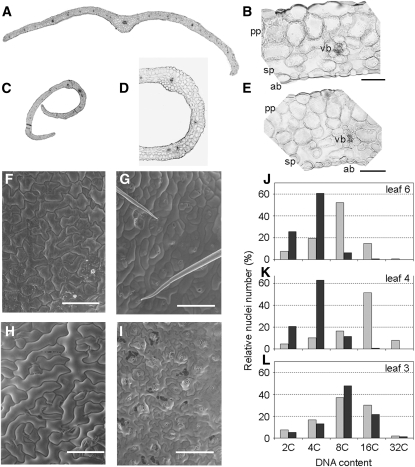
Figure 3.
Repression of ABP1 Leads to Altered Cell Size and Nuclear C Content.
(A) Cross section of leaf number 9 from a wild-type plant 15 d after exposure to ethanol.
(B) Detailed histology of section shown in (A).
(C) Cross section of leaf number 5 from an AtSS12K plant 15 d after exposure to ethanol.
(D) and (E) Detailed histology of section shown in (C). Ab, Abaxial face; pp, palisade parenchyma; sp, spongy parenchyma; vb, vascular bundle. Bars = 50 μm in (B) and (E), 200 μm in (D), and 500 μm (A) and (C).
(F) to (I) Scanning electron microscopy analysis of ethanol-induced wild-type ([F] and [H]) and AtABP1AS ([G] and [I]) leaves. Images of the adaxial leaf surface ([F] and [G]) and abaxial surface ([H] and [I]) are shown. All images were taken in the median region of leaf number 5 from the respective plants that had been exposed to ethanol prior to leaf formation and during seven subsequent days. Bars = 100 μm.
(J) to (L) DNA content analysis of wild-type (gray bars) and AtABP1AS (black bars) leaves from plants induced with ethanol vapor after the emergence of leaf 4. Measurements were performed on leaf 6 (J), which emerged subsequent to ethanol exposure; on leaf 4 (K), which had emerged just prior to ethanol treatment; and on leaf 3 (L), which had emerged prior to ethanol exposure. This experiment was performed three times with similar results.
Scanning electron microscopy analysis of the induced AtABP1AS leaves indicated that epidermal cell size and shape were significantly altered compared with induced control leaves of similar age (Figures 3F to 3I, Table 1). This analysis was somewhat constrained by the extreme curvature in the induced AtABP1AS or AtSS12K leaves, which made it difficult to view the leaf surface. Cell surface measurements were performed on the medial part of each leaf analyzed, both on the adaxial and the abaxial surfaces. Comparison of wild-type leaves with AtABP1AS leaves of similar age (leaf number 5) that were formed subsequent to ethanol exposure showed that the mean cell surface area was over threefold smaller in the adaxial epidermis of the AtABP1AS leaves and over fourfold smaller in the abaxial epidermis of the AtABP1AS leaves (Table 1). This reduction in mean cell surface area was accompanied by much slower growth of the leaves and by epinasty (decreased abaxial growth relative to the adaxial growth). With respect to the number of cells per leaf, there was approximately half the number of epidermal cells in an ethanol-induced AtABP1AS leaf compared with a wild-type leaf of equivalent size (comparison of leaf number 5 and 9 from AtABP1AS and wild-type plants, respectively; Table 1). Thus, both tissue expansion and cell division were altered in leaves of ABP1-inactivated plants.
Table 1.
Surface Cell Area of AtABP1AS Leaves with and without Repression of ABP1
| Adaxial Leaf Surface
|
Abaxial Leaf Surface
|
||||
|---|---|---|---|---|---|
| Genotype | Wild Type | ABP1AS | Wild Type | Wild Type | ABP1AS |
| Leaf number | 5 | 5 | 9 | 5 | 5 |
| Mean cell surface area (μm2) | 1810 | 527 | 280 | 3251 | 707 |
| sd | 98 | 34 | 15 | 903 | 43 |
| n | 202 | 327 | 186 | 159 | 270 |
| Calculated number of cells · mm−2 | 552 | 1897 | 3575 | 307 | 1414 |
The cell surface areas were calculated from scanning electron microscopy images of leaf number 5 or 9 of either the wild type or AtABP1AS in plants exposed to ethanol. Values are given as means, with sd calculated from sample size (n).
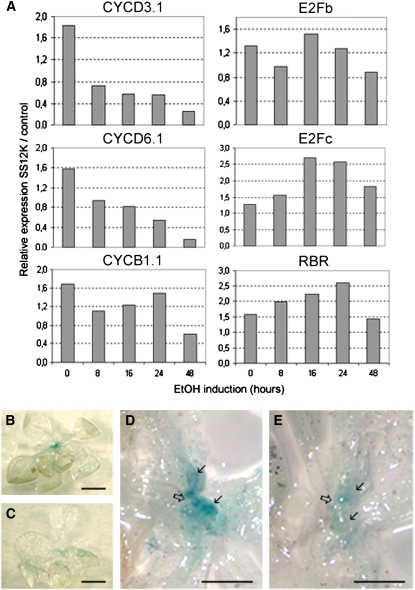
To investigate further the effect of ABP1 inactivation on cell division, we analyzed the expression of core cell cycle regulators (Figure 4A). ABP1 inactivation was followed by a rapid decrease in mRNA accumulation of D-type CYCLINS (Figure 4A), which are early regulators of the G1/S transition (Menges et al., 2006). In the same time frame, the transcriptional activator E2Fb was not significantly changed, whereas the transcriptional repressor E2Fc was transiently increased, as were transcripts of the retinoblastoma-related gene, which is thought to act at the interface between regulators of the cell cycle and the cell cycle process itself. Accumulation of the G2/M marker CYCB1.1 mRNA was not significantly affected within the first 24 h of ABP1 inactivation but tended to decrease later. As CYCB1.1 is also strictly regulated at the protein level and is often used as a quantitative marker of cell division, we introgressed the pCYCB1.1:DboxCYCB1.1-GUS reporter (Colon-Carmona et al., 1999) into the SS12K line. After 48 h of ethanol induction, GUS staining was decreased in young leaves and at the apical meristem, consistent with a decrease in cell division frequency (Figures 4B to 4E).
Figure 4.
Inactivation of ABP1 Alters Cell Division.
(A) Kinetics of cell cycle regulator gene expression following ABP1 inactivation. Quantitative RT-PCR (qRT-PCR) data for the indicated cell cycle genes were normalized with respect to ACTIN2-8 and then expressed relative to AlcAGus controls treated under the same conditions for the indicated times. The analysis was performed with two biological repeats, each in duplicate with similar results.
(B) to (E) GUS staining of SS12K seedlings 12 d after germination expressing DboxCYCB1;1-GUS. Seedlings were exposed to ethanol vapor for 48 h ([C] and [E]) or maintained under control conditions ([B] and [D]). Open arrows indicate the apex and solid arrows the emerging leaves. Bars = 2.5 cm in (A) and (B) and 700 μm in (C) and (D).
Normally during Arabidopsis leaf development, there is a progressive entry into endoreduplication, with developmentally older leaves gradually containing more cells with higher nuclear C values. This increased nuclear C value generally correlates with a larger cell size (Sugimoto-Shirasu and Roberts, 2003). We therefore investigated the outcome of reduced ABP1 activity on nuclear C value in control and AtSS12K leaves before or after exposure to the inducer (Figures 3J to 3L). In this experiment, plants were grown under noninducing conditions until leaf number 5 became visible (∼1 mm length). The plants were then exposed to ethanol vapor and the plants allowed to grow for a week until appearance of leaf number 9. The C value for nuclei extracted from leaf numbers 3, 4, and 6 were then obtained. Leaf number 3 of each genotype tested (the wild type or AtSS12K) had a similar spectrum of DNA content/nucleus after exposure of the plant to ethanol (Figure 3L), with C values ranging from 2C to 32C, with a median value of 8C. However, both leaves 4 and 6 (which developed during the exposure of the plant to ethanol) showed an altered spectrum of DNA content/nucleus after induced repression of ABP1 activity (Figures 3J and 3K). In both cases, AtSS12K leaves had a maximum of 4C content and ∼20% of 2C cells, whereas wild-type leaves contained more cells with a higher C content. For example, wild-type leaf 4 contained a large proportion of 16C cells, with >10% of the cells having a 32C content. These data show that after ethanol induction, the nuclei in the AtSS12K leaves maintained a much lower C value than developmentally equivalent leaves formed under noninducing conditions, suggesting that ABP1 is necessary to promote endoreplication. As described above, the cells formed in these induced leaves were smaller than cells in developmentally equivalent leaves; thus, there was a correlation between mean nuclear C value and cell size.
Local Repression of ABP1 in the Shoot Apical Meristem Reveals Its Context-Specific Role in the Coordination of Cell Division and Growth
Using the inducible expression system described above in Arabidopsis allowed us to analyze the overall response of plants to ABP1 inactivation during postembryonic development. To refine this analysis and to investigate any potential temporal or spatial specificity in the plant response to decreased ABP1 activity, we exploited a microinduction system developed in tobacco that allows transient and localized expression of gene constructs in different regions of the shoot apical meristem (SAM) of intact plants (Pien et al., 2001; Wyrzykowska and Fleming, 2003).
We particularly focused on the SAM since auxin has been implicated in the earliest events of leaf initiation in this tissue (Reinhardt et al., 2003), and we were interested in investigating whether ABP1 is involved in mediating these auxin-associated events. We made use of previously described SS12S and SS12K constructs to provide tetracycline-dependent repression of ABP1 activity via induction of scFv12. This approach was previously used successfully to impair ABP1 activity in tobacco BY2 cells (David et al., 2007). Tobacco R7 plants, in which the Tet repressor protein is constitutively expressed at a high level (Jones et al., 1998), were transformed with the appropriate constructs and independent homozygous lines were selected (Figure 5A). In the absence of exogenous inducer (anhydydrotetracycline [AhTet]) transcription of the scFv12 sequence was repressed, whereas supply of AhTet led to transcriptional derepression and expression of the scFv12 gene (Figure 5A). Quantitative RT-PCR analysis of RNA extracted from either AhTet-treated or control leaf discs of a NtSS12S7 line revealed that transcripts encoding the scFv12 protein accumulated within 2 h of AhTet induction, reaching a maximum at ∼16 h, before decreasing over the subsequent 12 h, so that 72 h after induction transcripts were no longer detectable (Figure 5B). Protein gel blot analysis indicated that the scFv12 protein was detectable 16 h after induction in microsomal membrane fractions for SS12K transgenes and in the soluble protein fraction for the SS12S transgene (Figure 5C). These data are consistent with our previous investigations using the microinduction approach in which a transient accumulation of the target mRNA and protein occurs, with the dynamics of gene expression being specific for the target gene (Pien et al., 2001; Wyrzykowska et al., 2002).
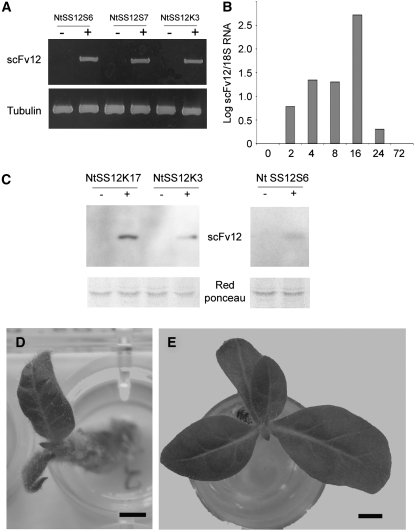
Figure 5.
Conditional Inactivation of ABP1 in Tobacco.
(A) RT-PCR analysis of scFv12 expression in leaf discs taken from independent transgenic lines of tobacco (NtSS12S6, NtSS12S7, and NtSS12K3) either treated (+) or not treated (−) with AhTet. Primers to amplify a tobacco tubulin gene were used as an amplification control.
(B) qRT-PCR analysis of scFV12 RNA extracted from apices of NtSS12S6 plants that had been induced locally with AhTet for various times. Signal is expressed relative to that obtained using primers for 18S rRNA. The analysis was performed with two biological repeats, each in triplicate, with similar results.
(C) Protein gel blot analysis of protein extracted from NtSS12K17, NtSS12K3, and NtSS12S6 leaves induced with (+) or without (−) AhTet. Microsomal fractions were used for the analysis of NtSS12K17 and NtSS12K3 extracts, whereas the soluble fraction was used for the NtSS12S6 extract. Blots were incubated with an antibody against scFv12. Ponceau S staining is shown as a loading control.
(D) Overview of a tobacco SS12S6 apex 4 weeks after local induction with AhTet at the I1 position of the SAM. Bar = 2 mm.
(E) Overview of a tobacco SS12S6 plant 4 weeks after local induction with AhTet at the I2 position of the SAM. Bar = 2.5 mm.
In a first series of microinduction experiments, apices were dissected to reveal the SAM. Lanolin beads impregnated with AhTet were then positioned on either the I1 position of the SAM (where leaf initiation is imminent) or on the I2 position opposite (where leaf formation does not normally occur until after leaf formation at the I1 position) (see Figure 6A for delineation of I1 and I2 sites). AhTet induction at the I1 position led to a marked retardation of apex growth (as indicated by limited leaf initiation and growth over the 4 weeks subsequent to the induction process) (Figure 5D). By contrast, microinduction of NtSS12S6 SAMs at the I2 position did not generally lead to a retardation of apex growth, with leaves being initiated and growing out in a fashion comparable to that observed in control mock-induced plants (Figure 5E). Analysis of a total of 96 apices (with 59 being induced at the I1 and 37 being induced at the I2 position) revealed a statistically significant different frequency (0.01 confidence level) of apex growth response when induction at the I1 or I2 position was compared (Table 2). Severe reduction of growth was not observed when the I1 of control Tet:GUS apices (harboring the GUS reporter gene under tetracycline-dependent transcriptional control) (Pien et al., 2001) was induced (Table 2), indicating that the observed growth retardation was linked with the induction of the scFv12 gene rather than with the induction process itself.
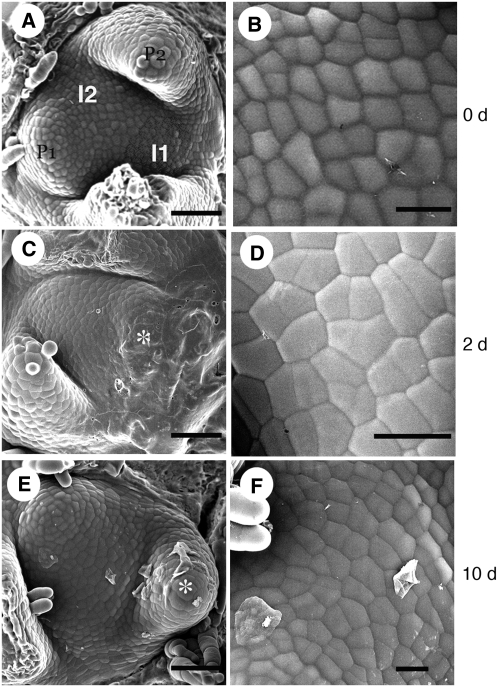
Figure 6.
Scanning Electron Microscopy Analysis of the Apex after Local Repression of ABP1.
(A) SAM prior to microinduction indicating the I1 and I2 positions on the surface. I1 is highlighted in gray.
(B) Epidermal cell pattern at the I1 position prior to microinduction.
(C) A NtSS12S6 SAM 2 d after microinduction at the I1 position. A leaf primordium (partially obscured by remnants of the AhTet containing lanolin used in the microinduction) has formed (asterisk) at the position equivalent to I1 in (A).
(D) Epidermal cell pattern of I1-derived tissue 1d after microinduction of a NtSS12S6 SAM.
(E) NtSS12S6 SAM 10 d after microinduction at the I1 position. A leaf primordium (asterisk) has formed at the position equivalent to I1 in (A), as has a subsequent primordium (equivalent to the I2 position in [A]).
(F) Epidermal cell pattern on the NtSS12S6 SAM surface just internal to the primordium derived from an I1 region induced to repress ABP1.
Bars = 40 μm in (A), (C), and (D) and 25 μm in (B), (D), and (F).
Table 2.
Differential Response of the I1 and I2 Positions in the SAM to Repression of ABP1
| Genotype | I1 or I2 Induction | Severely Retarded Growth (Two or Fewer Leaves in 4 Weeks) | Moderately Retarded Growth (Three to Four Leaves in 4 Weeks) | Normal Growth (Five to Six Leaves in 4 Weeks) | Sample Size (n) |
|---|---|---|---|---|---|
| NtSS12S6 | I1 | 34 | 19 | 6 | 59 |
| I2 | 0 | 10 | 27 | 37 | |
| Tet∷GUS | I1 | 0 | 5 | 26 | 31 |
| I2 | 0 | 0 | 18 | 18 |
SAMs of NtSS12S6 and Tet:GUS plants were induced on the I1 or I2 position, and the subsequent regeneration of the apex (quantified by leaf formation) was measured.
To further investigate the nature of this differential response to induction of scFv12, we performed scanning electron microscopy and histological analysis of induced and noninduced apices. Scanning electron microscopy analysis indicated that subsequent to scFv12 induction at the I1 position, leaf initiation still occurred; however, the rate of primordium outgrowth was drastically retarded relative to control treated apices (Figure 6). Thus, after induction at the I1 position in NtSS12S SAMs, a primordium bulge became apparent ∼2 d later (Figure 6C), but the primordium remained small even at 10 d after induction (Figure 6E). New primordia were formed subsequently at the appropriate position on the SAM, but the growth rate of these primordia was extremely slow. Measurement of cell surface areas in induced and noninduced regions on the SAM indicated that there was a slight but significant (0.05 confidence level) increase in the mean cell surface area in the induced I1 area relative to the equivalent position on a noninduced SAM (Figures 6B, 6D, and 6F, Table 3). In addition, while the I1 position is normally characterized by a regular cobblestone cell pattern (Figure 6B), within 24 h of induction of the scFv12 construct cells in this area showed a more irregular division pattern (Figure 6D).
Table 3.
Surface Cell Area at the I1 Position after Induction of NtSS12S Plants
| Genotype | Cell Surface Area (μm2) | sd | n |
|---|---|---|---|
| Wild type | 145.7 | 47.2 | 89 |
| 145.9 | 54.3 | 104 | |
| NtSS12S6 | 168.3 | 54.5 | 58 |
| 184.1 | 61.6 | 77 |
Cell surface areas were calculated from scanning electron microscopy images of SAMs of either wild-type or NtSS12S6 plants induced 24 h previously with AhTet at the I1 position. Results are given for two independent experiments for each genotype, with each experiment consisting of at least four apices. Values are given as means, with sd calculated from sample size (n).
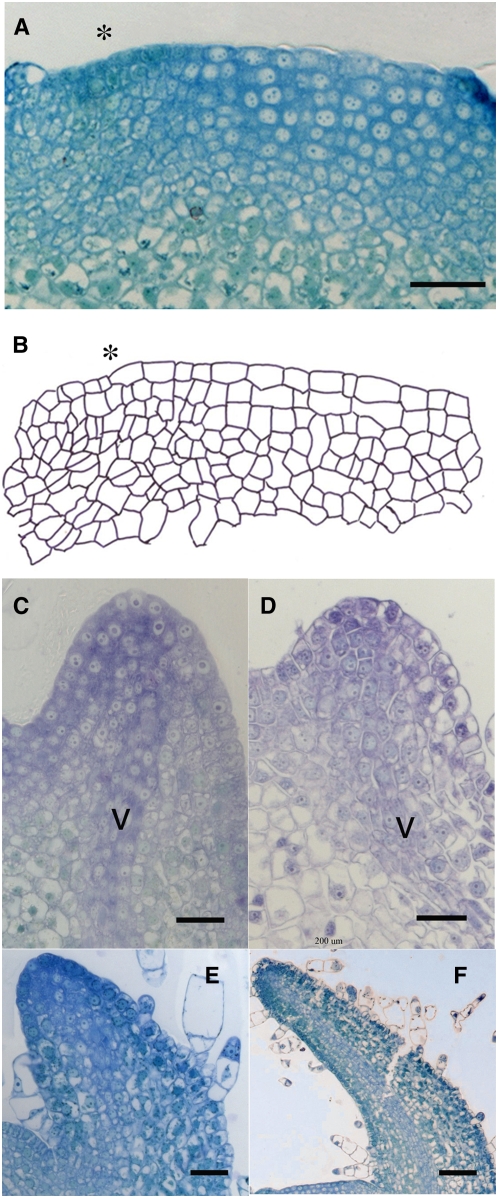
Histological analysis of the I1-induced NtSS12S SAMs revealed a loss of the normal cellular organization at the induced position prior to bulge formation (Figures 7A and 7B). The SAM is normally characterized by a distinct layered organization in which cell division pattern is constrained in the outer regions of the SAM so that new cell walls form perpendicular to the surface of the SAM. This results in the formation of cell layers toward the surface of the SAM (the tunica) below which the orientation of cell division plane is not so constrained (leading to the definition of the corpus). After induction of a NtSS12S SAM at the I1 position, this tunica organization was lost due to the occurrence of cytokinesis in abnormal planes (Figures 7A and 7B). In addition, there was a marked and significant decrease (t test: 0.01 confidence level) in cell area in these longitudinal sections at the I1 induced region relative to equivalent I1 regions of control SAMs (Table 4). At the same time, the cell cross-sectional area in the I2 region of the I1-induced SAMs was larger than the equivalent I2 region of control SAMs (t test: 0.01 confidence level; Table 4). Thus, as a result of the I1 induction of scFvABP1, there was both a disorganization of the cellular patterning at the I1 position and there was an increase in mean cell size at the I2 position (with cellular pattern remaining approximately normal).
Figure 7.
Histology of the SAM and Primordia after Local Repression of ABP1.
(A) Longitudinal section through the SAM of a NtSS12S6 plant that has been induced 24 h previously at the I1 position (asterisk). Bar = 25 μm.
(B) Outline cellular structure of the section shown in (A).
(C) Longitudinal section through a primordium derived from the I1 position of an NtSS12S6 SAM that has been induced 2d previously at this position. V, vasculature. Bar = 40 μm.
(D) Longitudinal section through a P1 primordium from a wild-type plant (∼24 h after formation from the I1 position of a wild-type SAM). Bar = 20 μm.
(E) Longitudinal section through a primordium derived from the I1 position of an NtSS12S6 SAM that has been induced 10 d previously at this position. Bar = 30 μm.
(F) Longitudinal section through a P4 primordium from a wild-type plant (∼10 d after formation of the primordium from the I1 position of a wild-type SAM). Bar = 80 μm.
Table 4.
Longitudinal Cross-Sectional Cell Area after Induction of scFv12 in the SAM
| Genotype | Position and Induction | Mean Cross Section Area | sd | n |
|---|---|---|---|---|
| NtSS12K3 | I1 (induced) | 124.69 | 37.83 | 43 |
| I2 (noninduced) | 265.88 | 70.89 | 37 | |
| NtSS12S7 | I1 (induced) | 122.53 | 37.03 | 24 |
| I2 (noninduced) | 231.36 | 32.59 | 18 | |
| I1 (induced) | 118.06 | 44.27 | 28 | |
| I2 (noninduced) | 224.40 | 52.04 | 25 | |
| Wild type | I1 (noninduced) | 145.35 | 43.17 | 214 |
| I2 (noninduced) | 155.22 | 48.40 | 110 |
Cell areas at the I1 and the I2 position were calculated from images of longitudinal sections of SAMs of either wild-type, NtSS12S7, or NtSS12K3 plants induced 24 h previously with AhTet at the I1 position. Values are given as means, with sd calculated from sample size (n).
As described above, despite the induction of a disorganized cellular pattern at the I1 position in the NtSS12S SAMs, a delayed leaf initiation still occurred. Although the histology of the induced organs appeared relatively normal with, for example, clear differentiation of vascular tissue, cellular organization at the distal tip of the primordia appeared abnormal (Figure 7C). In particular, although in normal primordia the elongated cells characteristic of the emerging vasculature extend to the tip of the primordium (Figure 7D), in the tissue derived from the induced portion of the NtSS12S SAMs overt vascular differentiation seemed to terminate below the tip of the primordium (Figure 7C). After initiation, the subsequent growth of the leaves derived from the induced NtSS12S SAM tissue was greatly decreased. Thus, while control leaf primordia underwent a process of extensive elongation (Figure 7F), leaves derived from NtSS12S induced SAM tissue remained relatively short (Figure 7E).
To further analyze these cellular defects, we performed qRT-PCR analysis using dissected tobacco apices, both with and without suppression of ABP1 activity. We focused on a number of cell cycle marker genes known to act at various phases of the cell cycle (see Supplemental Figure 5 online). As already observed in Arabidopsis leaves (Figure 4), transcript levels for D-type cyclins rapidly decreased after inactivation of ABP1 but in these experimental conditions was followed by a partial recovery in transcript level. Histones H1 and H4 and PCNA (all S-phase markers) showed a relative rise in transcript level 24 h after repression of ABP1 activity, as did a marker for the M phase of the cell cycle (CYCLIN B). Interestingly, transcripts for a RETINOBLASTOMA RELATED PROTEIN (whose activity links D-type cyclins and the G1/S phase transition) showed a temporal transcript pattern similar to the S phase marker genes and distinct from the D-type cyclin genes.
To investigate the spatial pattern underlying these changes in cell cycle gene expression, we performed a series of in situ hybridizations of tobacco apices in which ABP1 activity had been suppressed. In control apices, the ABP1 transcript is uniformly expressed (see Supplemental Figures 6A and 6B online) and the meristem shows a characteristic pattern for the NTH15 (KNOX-like) marker gene in which transcripts are absent from the I1 position (see Supplemental Figure 6C online). The general pattern of cell cycle gene expression in the SAM was little changed after repression of ABP1 activity (data not shown). However, after suppression of ABP1 activity at the I1 site, two regions of decreased NTH15 transcript level become apparent (see Supplemental Figure 6D online), suggesting that the timing of leaf initiation was altered so that two regions within the meristem underwent commitment to leaf formation.
The primordia originating from the induced I1 of NtSS12S apices were rounder than wild-type primordia, and this was associated with an altered pattern of vasculature (see Supplemental Figure 7 online). Thus, instead of the clearly defined single mid-rib observed in control tobacco leaves, two or more parallel veins formed toward the center of the induced leaves (see Supplemental Figure 7A online). This alteration in vascular pattern and leaf shape was reminiscent of that reported in other plants to result from supply of the auxin transport inhibitor naphthylphthalamic acid (NPA) (Mattsson et al., 1999). After applying NPA to the I1 position of wild-type SAMs, we observed the formation of small, rounded leaves with an abnormal vascular pattern very similar to that observed after local induction (and subsequent outgrowth of the primordium) at the I1 position of SAMs of NtSS12S apices (see Supplemental Figure 7B online). Our efforts to visualize PIN protein distribution (which can be used as an indicator of auxin flux pattern) were inconclusive, probably reflecting poor cross-reactivity of the antibodies used with the endogenous tobacco proteins (data not shown).
To investigate whether induction of NtSS12S leaves at later stages of development led to changes in leaf form, microinductions were performed along the flanks of otherwise normally formed leaf primordia. These inductions led to a range of growth response varying from a waviness along the leaf lamina to downward curling of the leaf lamina, reminiscent of the phenotype observed in Arabidopsis (see Supplemental Figure 7C online). In all these cases there was no overt abnormality in the pattern of leaf vasculature.
Repression of ABP1 Leads to Rapid Changes in Auxin Signaling-Related Transcript Levels
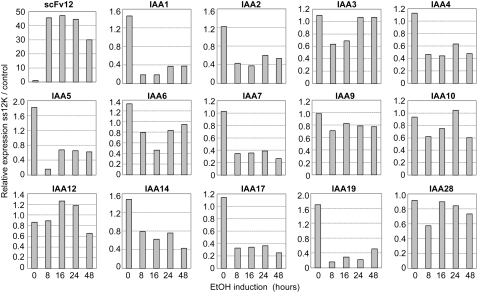
The mechanism by which ABP1 triggers the responses reported both in this and other articles remains obscure (Napier et al., 2002). One possibility is that it is primarily an electrophysiological response to auxin that does not per se require altered gene expression. At the same time, one might expect there to be an eventual crosstalk with gene expression responses known to be triggered by auxin (most probably via a TIR1-like receptor pathway). To investigate this possibility, we performed an analysis of the transcriptional response of Arabidopsis leaf tissue at various times after ABP1 inactivation in the SS12K line. In particular, we performed qRT-PCR analysis of an array of Aux/IAA genes related to auxin signaling via the TIR1 pathway. As shown in Figure 8, downregulation of ABP1 activity generally led to a decrease in transcript level for a large spectrum of Aux/IAA genes. Accumulation of transcripts was diversely affected, ranging from a seven- to eightfold decrease within the first hour of ABP1 inactivation for IAA1, IAA5, or IAA19 to no significant alteration for IAA9 or IAA28. In a second series of experiments, plants were treated with a high concentration of exogenous auxin after suppression of ABP1 activity and the response of the SS12K plantlets analyzed with respect to the same spectrum of Aux/IAA genes. In such conditions, a subset of Aux/IAA genes showed increased auxin responsiveness (see Supplemental Figure 8A online), whereas others exhibited a similar response to control plants (see Supplemental Figure 8B online). Taking into account the effect of ABP1 inactivation on the steady state level of Aux/IAA transcripts (Figure 8), the absolute transcript level for most Aux/IAAs was, however, reduced in comparison to controls even after auxin treatment.
Figure 8.
Response of Aux/IAA Transcript Levels to Inactivation of ABP1 in Arabidopsis Leaves.
Kinetics of Aux/IAA transcript levels following ABP1 inactivation. qRT-PCR data for the indicated Aux/IAA genes were normalized with respect to ACTIN2-8 and then graphed as the expression of Aux/IAAs in SS12K samples relative to AlcAGus controls exposed to ethanol for the same time. The analysis was performed with two biological repeats, each in triplicate, with similar results.
These data are important since they indicate that ABP1 is required to maintain appropriate expression of Aux/IAA genes in leaf tissue. In addition to the auxin/TIR1-mediated derepression of these genes, ABP1 also plays a role (Mockaitis and Estelle, 2008).
DISCUSSION
ABP1 has long been identified as a potentially important mediator of auxin action. Understanding its role in the plant requires characterization of the loss-of-function phenotype during development. However, constitutive loss of ABP1 activity is embryo-lethal, thus precluding any firm conclusions on the requirement of ABP1 during postembryonic development (Chen et al., 2001). In this article, we provide data characterizing the role of ABP1 during vegetative plant growth. These results were obtained using both a novel cellular immunization technique to conditionally repress ABP1 activity and an antisense approach to conditionally decrease ABP1 protein level. Both approaches led to broadly similar phenotypes, suggesting that in both cases the phenotypes observed were indeed due to a loss of ABP1 function. In addition, by combining techniques to conditionally suppress ABP1 activity with a microinduction method to allow spatial/temporal control of gene expression, we show that the requirement for ABP1 activity in the intact plant is highly context dependent and that the outcome of loss of ABP1 activity in shoots involves aspects of both cell division and expansion. As well as providing key data on the endogenous function of ABP1, our results demonstrate the importance of temporal and spatial context in understanding the role of mediators of plant development.
ABP1 Is Required during Postembryonic Development and Acts at the Interface of Cell Division and Expansion in Leaf Morphogenesis
Conditional repression of ABP1 activity led to a rapid repression of shoot growth. This repression was lethal in very young seedlings, whereas older plants were able to maintain a limited level of growth and produced the same number of leaves as control plants. Leaf size and shape were both altered, revealing that ABP1 is essential for appropriate leaf growth and development. The dwarf phenotype of the induced AtABP1AS and AtSS12K plants resulted from a decreased number of cells, a reduced level of endoreplication, and a decrease in cell size. The decreased number of cells indicates that cell division was impaired, and a role for ABP1 in influencing cell division was also suggested by our data showing that following loss of ABP1 activity, there was a relatively rapid fall in transcripts encoding cyclin D proteins. D-type Cyclins are associated with the control of entry into the cell cycle; thus, decreased expression of these transcripts following decreased ABP1 activity is consistent with ABP1 being required for entry into the cell cycle (Maughan et al., 2006; David et al., 2007). These data are consistent with results obtained in tobacco BY2 cell suspension cultures, which showed that ABP1 was essential for the G1/S transition (David et al., 2007).
Endoreduplication can be understood as a truncated cell cycle in which the mitotic phase is skipped, and it is believed that the regulatory machinery required for the G1/S transition and DNA replication is shared by both the mitotic cell cycle and endocycle (Vlieghe et al., 2007). Thus, we hypothesize that the inhibition of the endocycle reported here after ABP1 inactivation might also reflect an inhibition of reentry into the cell cycle. Endoreplication has been proposed to be part of a mechanism allowing cell expansion, thus contributing to leaf growth (Vlieghe et al., 2007), and in mature leaves of Arabidopsis, a positive correlation has been shown between endopolyploidy and cell size (Melaragno et al., 1993). However, although we can describe a correlation between repression of ABP1 activity in leaves, reduced endoreduplication, and lack of cell expansion, inferring a causal relationship is problematical (Sugimoto-Shirasu and Roberts, 2003). For example, we observed that inactivation of ABP1 in leaves that had already undergone polyploidization also decreased further leaf growth, indicating that ABP1 is involved in a mechanism controlling cell expansion independent of endoreplication. An involvement of ABP1 in the control of cell expansion in the leaf is in accordance with the previously reported increase in auxin-dependent cell expansion of ABP1 overexpressing tobacco leaves (Jones et al., 1998). Leaf morphogenesis requires the control of both cell proliferation and final cell size, and the data in this article demonstrate that altered ABP1 activity can lead to changes in both of these cellular processes (Figure 9). Among the input signals that can influence the balance between cell proliferation and cell expansion, auxin is known to be a major regulator (Davies, 1987), with exogenous auxin tending to promote cell division/inhibit cell expansion at high concentrations and lower concentrations tending to stimulate cell expansion but not cell division. One possibility is that ABP1 mediates a differential response of cell division and expansion to differential local levels of auxin. This concept is further explored in our analysis of the role of ABP1 in the SAM, described in the next section.
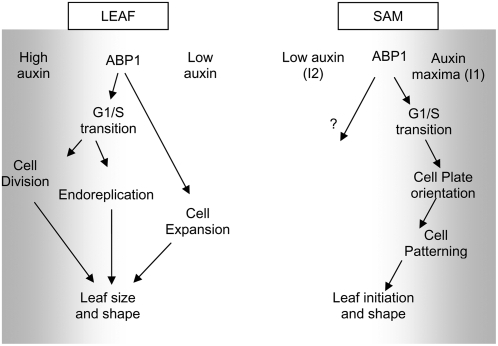
Figure 9.
Model for a Context-Dependent Role of ABP1 in the Shoot.
Depending on the local auxin concentration (gray shading) and developmental context of the tissue, ABP1 may mediate different cellular outputs of division frequency, orientation, and expansion.
ABP1 Is Required during Early Leaf Initiation and Acts on Cellular Patterning and Cell Growth
In an early step of leaf initiation, founder cells are recruited from the peripheral zone located at the flanks of the SAM. The outer region of the SAM is characterized by an ordered pattern of cell division, forming the tunica, which surrounds an inner mass of cells, the corpus. Transient repression of ABP1 activity at the I1 site of the SAM (the position of presumptive leaf formation) led to disruption of this cellular patterning. Cell size was not massively changed; rather, there was a change in cell shape and the positioning of the new cell plates laid down as cells in this region underwent cytokinesis. Interestingly, these data are reminiscent of the altered cellular patterning reported in the ABP1 knockout mutant (Chen et al., 2001). Repression of ABP1 activity after leaf initiation did not lead to a significant alteration of tissue organization, suggesting that the influence of ABP1 on cell patterning might be highly context dependent. Both the embryo and the SAM are distinguished by the presence of relatively undifferentiated cells, suggesting that a role of ABP1 in coordinating cell plate formation might be limited to this cell type.
Although repression of ABP1 led to a cellular patterning defect at the I1 position of the SAM, the same manipulation at the I2 position (where leaf initiation is due to occur subsequent to that at the I1) did not have any outcome on cellular patterning or growth of the tissue, indicating that not all cells in the peripheral zone are equally responsive to altered ABP1 activity. Although we cannot discount the possibility of differential posttranscriptional activation of ABP1, recent investigations into the role of auxin in leaf initiation have led to the paradigm that local, transient maxima of auxin level occur at the site of leaf initiation (I1), whereas relative depletion of auxin occurs at the distant I2 position (de Reuille et al., 2006; Jönsson et al., 2006; Smith et al., 2006). Our observations on the differential sensitivity of the I1 and I2 sites on the SAM to the same repression of ABP1 activity are consistent with this paradigm (i.e., a loss of ABP1 activity only leads to a phenotype in a region where a relatively high level of auxin is expected to occur).
Although unambiguous support for this paradigm will require direct quantitation of auxin levels within the SAM (a technically challenging requirement), our data do provide evidence that ABP1 mediated response maxima correlate with the auxin maxima, itself tied with leaf initiation. The observation that repression of ABP1 activity at the I1 site led to an altered cellular patterning is also consistent with the idea that an influence of ABP1 on cell division might only be apparent in regions of relatively high auxin level. In the ABP1 knockout mutant, the phenotype was also characterized by a loss of appropriate cellular patterning during early stages of embryo development when relatively high levels of auxin are predicted to occur (Chen et al., 2001). One possibility is that in regions of relatively high auxin concentration (e.g., early stage embryos and meristems) ABP1 is involved in both cell growth and cytokinesis, whereas in regions of relatively low auxin concentration, its function is more limited to cell expansion (Figure 9).
Interestingly, the altered cellular patterning at the I1 position induced by repression of ABP1 was associated with a prolonged suppression of expression of the KNOTTED-1-like transcription factor NTH15. The exact relationship between auxin and the key developmental regulators of meristem function, such as the KNOTTED class of homeodomain transcription factors, has been the subject of a number of investigations. The data reported here add to this body of literature, but the mechanism of interaction remains obscure. Our results indicating a link between ABP1 and expression of early auxin-responsive genes of the Aux/IAA family provide one potential way by which ABP1 and transcriptional control of developmental regulators might interact.
Although ABP1 clearly has the potential to influence plant function via non-nuclear targets, the data presented here indicate the interaction of ABP1 activity and auxin-associated gene expression. The mechanism by which ABP1 affects expression of Aux/IAA genes needs to be elucidated. The absence of effect of ABP1 inactivation on the level of free auxin within the same time frame (see Supplemental Figure 2 online) indicates that the decrease in Aux/IAA transcripts does not result from a change in auxin content but rather from an altered regulation of theses genes at the transcriptional or posttranscriptional level. The data presented here show that decreased ABP1 activity leads to an array of altered transcript levels for genes associated with the TIR1 auxin signaling pathway. The contribution of ABP1 to the fine-tuning of early auxin-responsive gene regulation suggests a crosstalk with the SCFTIR1 pathway, and elucidating how ABP1 interacts with this pathway will be an important challenge of the coming years.
Altered Cellular Patterning via Repression of ABP1 Activity Leads to Downstream Events That Alter Plant Growth and Form
The data presented in this article demonstrate how different the outcome of altered parameters of the cell cycle, cycle division pattern, and cell expansion can be, depending on the timing and position of the manipulation. Repression of ABP1 activity at the I1 position of the SAM led to an altered pattern of cell division, but the initial step of leaf morphogenesis, corresponding to the formation of a primordium bulge, still occurred. Thus, early leaf initiation occurs by a cell division–independent mechanism (consistent with our previous observations with phragmoplastin and the cell wall protein expansin) (Wyrzykowska and Fleming, 2003). However, further growth of the primordium was delayed until partial recovery of ABP1 activity and restoration of cell division and cell expansion. The subsequent initial stages of vascular differentiation in the young leaf primordium appeared to be disrupted. It seems that the earliest steps in vascular differentiation require a capacity of the tissue to form particular cellular patterns (presumably to align the cellular elements that will form the vasculature). If this is disrupted, as in the case following local repression of ABP1 at the I1 position, vascular formation does not occur at the expected position. Nevertheless, the surrounding tissue possesses the ability to respond to this situation to form a new pattern of vascular bundles, leading to the situation observed occasionally in Arabidopsis and at a more regular and extreme level in tobacco of more than one mid-vein being formed. This alteration of vascular pattern has an outcome for the growth and form of the whole leaf, leading to the formation of rounder leaves of limited growth potential. Thus, a transient alteration in cell division pattern at a precise and early stage of leaf formation associated with a protein implicated in auxin perception has a serious long-term outcome on the development of the whole organ.
Published data clearly implicate auxin flux as having a major role in vascular patterning (Scarpella et al., 2006), and the data shown here indicate a close resemblance between primordia derived from NPA-treated tissue (in which auxin transport is inhibited) and scFvABP1-induced tissue. However, although ABP1 could have a direct role in mediating auxin's influence on vascular patterning, it is more likely that it has an indirect influence. As outlined above, if vascular differentiation requires a specific cellular patterning, then loss of ABP1 activity would (indirectly) prevent or disrupt the interpretation of an auxin pattern into appropriate differentiation. The observed changes in leaf shape would thus be an indirect, downstream event of organ response to an initial relatively local change in cellular patterning. Whether the reported changes in vasculature resulting from altered expression of some cell cycle genes (e.g., Dewitte et al., 2007) also reflect an indirect outcome awaits further investigation.
The ability of plant tissue to respond to (and cope with) altered cellular patterning is also demonstrated by the observation that the changes in cellular patterning at the I1 position of the SAM induced by local repression of ABP1 activity was to some extent compensated for by altered cell growth at the I2 position (i.e., mean cell size at the I2 position increased after the inactivation of ABP1 at the I1 position). The mechanism of this local growth compensation is unclear, but the paradigm of autoregulated auxin fluxes provides an attractive mechanism by which a local change in cell growth and patterning involving altered auxin signaling might automatically lead to altered signal flux in a region at a distance from the initial perturbation to elicit a compensatory growth response. The application of recent modeling approaches to understanding dynamic changes in auxin flux and their influence on leaf primordia formation (de Reuille et al., 2006; Jönsson et al., 2006; Smith et al., 2006) and vascular development (Rolland-Lagan and Prusinkiewicz, 2005; Scarpella et al., 2006) might help to test this hypothesis.
In conclusion, the data reported here show that ABP1 activity is required during postembryonic development. In particular, they suggest that the role of ABP1 is highly context dependent and that, depending on auxin level and differentiation state, ABP1 can act on the cell cycle, endocycle, cell plate formation, and cell expansion and contributes to the control of auxin-related gene expression. The mechanism by which ABP1 acts in the coordination of these critical cellular parameters is a matter for urgent consideration.
METHODS
Plant Material and Growth Conditions
The Columbia-0 ecotype of Arabidopsis thaliana was used for transformation and experimental controls. Line R7 of Nicotiana tabacum, cv Wisconsin, overexpressing the Tet repressor, was used for transformation and for controls. Seeds were surface sterilized, stratified for 3 d at 4°C, and germinated on plates containing half-strength Murashige and Skoog (MS) basal salt mixture, buffered at pH 5.8 with 2.5 mM 2-(N-morpholino) ethanesulfonic acid and 0.9% vitro agar (Kalys). Square Petri dishes were used and placed in a vertical position at 22°C under constant light at 99 μmol·m−2·s−1 intensity. Culture in soil was performed in greenhouses for plant selection and seed production and in environmental growth chambers at 22°C constant, 16 h daylight, and 65% hygrometry for inductions and phenotype studies.
Induction of scFv and ABP1 Antisense Expression
For Arabidopsis, ethanol inductions on in vitro cultures were performed by adding 500 μL of 5% ethanol in a pinholed microtube placed at the bottom of the plate. Plates were sealed with Parafilm to keep ethanol vapor within the plate. For culture on soil, a microtube containing 2 mL of 95% ethanol was placed in the soil and each pot was placed within an open 9-cm diameter, 30-cm-high methyl polymethacrylate pipe to canalize and maintain ethanol vapor.
In tobacco, for Ahtet inductions at the tissue level prior to RNA extraction and protein gel blot analysis, leaf strips were incubated overnight in half-strength MS medium, pH 5.6, containing 10 μM anhydrotetracycline. Microinductions with AhTet were performed as described (Pien et al., 2001). Briefly, lanolin/paraffin paste with 10 mg/mL Ahtet or controls with DMSO/lanolin/paraffin paste without Ahtet were applied to the surface of apices or primordia dissected to reveal the SAM and young leaf primordia. After manipulation, apices were grown on half-strength MS medium, pH 5.6, in a growth chamber (16-h-light/8-h-dark cycle at 24°C, 100 μmol·m−2·s−1) before transfer to multiwell dishes for further growth, as described by Pien et al. (2001).
DNA Constructs and Transformation
ScFv constructs were cloned into the binary vector pBin-Hyg-Tx and used for transformation of an R7 tobacco line (Jones et al., 1998; David et al., 2007). Single-chain Fv 12 constructs containing the murine κ light-chain signal peptide coding sequence at the 5′-end (SS prefix) and with or without a sequence encoding a KDEL extension at the 3′-end for secretion (S suffix) or retention in the ER (K suffix) (David et al., 2007) were cloned into a pACN vector, downstream of the pAlcA promoter (Roslan et al., 2001) (Syngenta). A full-length ABP1 cDNA construct was cloned in an antisense orientation into the same vector. For controls, the UIDA coding sequence was also introduced into pACN. Each pAlcA:geneX:nos-ter cassette was excised by HindIII digestion and cloned into a pBinSRNA binary vector (Syngenta). Clones exhibiting an inverted orientation of the cassette in comparison with p35S:AlcR:nos-ter were selected for plant transformation.
Arabidopsis transformation was performed by floral dip (Clough and Bent, 1998). T1 and further progeny were selected on half-strength MS medium containing 50 mg·L−1 kanamycin.
Tobacco transformation was performed by agroinfiltration of leaf disks as described (Horst et al., 1985) and cultured on half-strength MS in the presence of 500 mg·L−1 cefotaxime, 50 mg·L−1 kanamycin, 4.4 μM 6-benzylaminopurine, and 0.05 μM NPA for shoot bud regeneration. Vegetative buds were transferred onto fresh medium without cytokinin for rooting.
Microscopy
For light microscopy, leaves were embedded into Technovit 7100, following the manufacturer's instructions (Heraeus Kulzer) after formaldehyde acetic acid fixation and progressive dehydration. Histological observations were performed on semithin sections stained with toluidine blue with a Reichert polyvar microscope (Reichert-Jung) or Olympus BX51. Digital images were captured using a charge coupled device and imported into Adobe Photoshop.
For scanning electron microscopy of Arabidopsis, leaves were treated with 70% ethanol and were analyzed with a Hitachi S-3000N in ESED mode. Samples were slowly frozen at −12°C under partial vacuum on the Peltier stage before observation. Cell surface measurement was made using Image J 1.034s software (NIH). For tobacco, cryo-scanning electron microscopy was as previously described (Fleming et al., 1999). Briefly, samples were rapid frozen in liquid nitrogen, cryo-splutter coated with platinum (7 nM), and then viewed with a Hitachi S-900 in-lens field emission microscope. Images were obtained at 140K and an accelerating voltage of 10 kV using the secondary electron signal and then recorded digitally with a Gatan Digi-Scan interface before export to Adobe Photoshop.
qRT-PCR
RNA was obtained using a Qiagen RNeasy kit and digested with RNase free DNase on the column following the manufacturer's instructions (Qiagen). First-strand cDNAs were synthesized from 5 μg of total RNA using Superscript II reverse transcriptase and oligo(dT) primers (Invitrogen) or a specific forward ABP1 primer to check the antisense construct, ABP1-F 5′-GCTCCAGGTTCAGAGACACC-3′, according to the manufacturer's instructions. qRT-PCR analyses were performed using SYBR Green QPCR master mix (Roche) with specific primers (see Supplemental Tables 1 and 2 online) (Menges et al., 2006). PCR cycling conditions for amplification were 95°C for 10 min and then 50 cycles of 95°C for 5 s, 62°C for 5 s, and 72°C for 15 s followed by 0.1°C·s−1 ramping up to 95°C for fusion curve characterization. All data were normalized with respect to ACTIN2 to ACTIN8 for Arabidopsis samples or 18SrRNA for tobacco. For kinetics of ABP1 inactivation, data were expressed as the ratio toward the control sample treated in the same condition. Two to three biological repeats were analyzed in triplicates.
Statistical analyses were performed following the formula of Pfaffl (2001) on triplicates for each of three biological samples.
Protein Analysis
Proteins were extracted from shoots of 8-d-old Arabidopsis seedlings induced or not induced with ethanol or from tobacco leaf strips by grinding at 4°C with a mortar and sand in the extraction buffer (50 mM TrisHCl, pH 7.5, 1 mM EDTA, 100 μM MgCl2.·6H20, 5 mM sodium ascorbate, 500 mM sucrose, and 200 μM AEBSF protease inhibitor). Microsomal fractions were prepared and treated as described (David et al., 2007). After SDS-PAGE electrophoresis and transfer onto Hybond-C extra nitrocellulose membrane (GE Heathcare Europe), loading and protein transfer were controlled by transient red Ponceau staining. Protein gel blot analysis was performed as described previously (Leblanc et al., 1999a), using antipeptide rabbit polyclonal E-tag antibodies for detection of scFv12 and the mAb34 mouse monoclonal antibody for ABP1.
Flow Cytometry
For DNA content analysis, nuclei were extracted from fresh tissue by thin chopping in Galbraith's medium as described (Coba de la Pena and Brown, 2001). Nuclei were stained with 50 μg/mL propidium iodide and injected into an EPICS Elite ESP cytometer (Beckman-Coulter) as described by Coba de la Pena and Brown (2001). Measurements were achieved using a minimum of 10,000 nuclei/sample.
Accession Number
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession number AT4G02980 (Arabidopsis ABP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Ethanol Induction on AlcAGus Control Plants and Leaf Growth Decreases after Inactivation of ABP1.
Supplemental Figure 2. Analysis of IAA Accumulation in Col-0 and SS12K Shoots after Various Times of Ethanol Induction as Indicated.
Supplemental Figure 3. Effect of ABP1 Inactivation during Vegetative Growth and Flowering.
Supplemental Figure 4. Leaf Abnormalities Resulting from ABP1 Inactivation in Arabidopsis.
Supplemental Figure 5. Cell Cycle Transcript Response to Repression of ABP1 in Tobacco Shoot Apex.
Supplemental Figure 6. Local Repression of ABP1 Activity Leads to Altered NTH15 Transcript Pattern.
Supplemental Figure 7. Form and Vasculature of Leaves Derived from an ABP1-Repressed I1 Position on the SAM.
Supplemental Figure 8. Altered Response of Aux/IAA Genes to Auxin after Inactivation of ABP1.
Supplemental Table 1. List of Primers Used for qRT-PCR Analysis in Arabidopsis.
Supplemental Table 2. List of Primers Used for RT-PCR Analysis in Tobacco.
Supplementary Material
Acknowledgments
We thank K. Ljung for auxin content measurements. We acknowledge A. Jones for the R7 line; M. Lopez-Vernaza, A. Eschstruth, and F. Ouedrago for technical assistance; O. Catrice for help in flow cytometry; and S. Domenichini for scanning electron microscopy. N.B. was funded by the Ministère de la Recherche et de la Technologie; J.W. and D.C. received ACCY postdoctoral fellowships, and both labs were supported by the ACCY European Research Training Network HPRN-CT-2002-00334. During part of this work, C.P.-R. was supported by the Centre National de la Recherche Scientifique and a French ACI programme “Biologie du développement et physiologie integrative,” and A.J.F. was supported by a START Fellowship from the Swiss National Science Foundation in lab space generously provided by N. Amrhein.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Catherine Perrot-Rechenmann (catherine.rechenmann@isv.cnrs-gif.fr) and Andrew J. Fleming (a.fleming@sheffield.ac.uk).
Online version contains Web-only data.
References
- Alonso-Peral, M.M., Candela, H., del Pozo, J.C., Martinez-Laborda, A., Ponce, M.R., and Micol, J.L. (2006). The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis. Development 133 3755–3766. [DOI] [PubMed] [Google Scholar]
- Badescu, G.O., and Napier, R.M. (2006). Receptors for auxin: Will it all end in TIRs? Trends Plant Sci. 11 217–223. [DOI] [PubMed] [Google Scholar]
- Bauly, J.M., Sealy, I.M., Macdonald, H., Brearley, J., Droge, S., Hillmer, S., Robinson, D.G., Venis, M.A., Blatt, M.R., Lazarus, C.M., and Napier, R.M. (2000). Overexpression of auxin-binding protein enhances the sensitivity of guard cells to auxin. Plant Physiol. 124 1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J.G., Ullah, H., Young, J.C., Sussman, M.R., and Jones, A.M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Coba de la Pena, T., and Brown, S. (2001). Flow cytometry. In Plant Cell Biology: A Practical Approach, 2nd ed, C. Hawes and B. Satiat-Jeunemaître, eds (Oxford, UK: Oxford University Press), pp. 85–106.
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20 503–508. [DOI] [PubMed] [Google Scholar]
- David, K.M., Couch, D., Braun, N., Brown, S., Grosclaude, J., and Perrot-Rechenmann, C. (2007). The auxin-binding protein 1 is essential for the control of cell cycle. Plant J. 50 197–206. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1987). Plant Hormones and Their Role in Plant Growth and Development. (Dordrecht, The Netherlands: Kluwer Academie).
- de Reuille, P.B., Bohn-Courseau, I., Ljung, K., Morin, H., Carraro, N., Godin, C., and Traas, J. (2006). Computer simulations reveal properties of the cell-cell signaling network at the shoot apex in Arabidopsis. Proc Natl. Acad. Sci. USA 103 1627–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte, W., Scofield, S., Alcasabas, A.A., Maughan, S.C., Menges, M., Braun, N., Collins, C., Nieuwland, J., Prinsen, J., Sundaresan, V., and Murray, J.A.H. (2007). Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc. Natl. Acad. Sci. USA 104 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., and Estelle, M. (2005. a). The F-box protein TIR1 is an auxin receptor. Nature 435 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri, N., Dharmasiri, S., Weijers, D., Lechner, E., Yamada, M., Hobbie, L., Ehrismann, J.S., Jurgens, G., and Estelle, M. (2005. b). Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9 109–119. [DOI] [PubMed] [Google Scholar]
- Fleming, A.J., Caderas, D., Wehrli, E., McQueen-Mason, S., and Kuhlemeier, C. (1999). Analysis of expansin-induced morphogenesis on the apical meristem of tomato. Planta 208 166–174. [Google Scholar]
- Horst, R.T., Fry, J.E., Hoffmann, N., Eichholtz, D., Rogers, S.G., and Fraley, R.T. (1985). A simple and general method for transferring genes into plants. Science 227 1229–1231. [DOI] [PubMed] [Google Scholar]
- Jones, A.M., Im, K.H., Savka, M.A., Wu, M.J., Dewitt, N.G., Shillito, R., and Binns, A.N. (1998). Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282 1114–1117. [DOI] [PubMed] [Google Scholar]
- Jönsson, H., Heisler, M.G., Shapiro, B.E., Meyerowitz, E.M., and Mjolsness, E. (2006). An auxin-driven polarized transport model for phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski, S., and Leyser, O. (2005). The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435 446–451. [DOI] [PubMed] [Google Scholar]
- Leblanc, N., David, K., Grosclaude, J., Pradier, J.M., Barbier-Brygoo, H., Labiau, S., and Perrot-Rechenmann, C. (1999. b). A novel immunological approach establishes that the auxin-binding protein, Nt-abp1, is an element involved in auxin signaling at the plasma membrane. J. Biol. Chem. 274 28314–28320. [DOI] [PubMed] [Google Scholar]
- Leblanc, N., Perrot-Rechenmann, C., and Barbier-Brygoo, H. (1999. a). The auxin-binding protein Nt-ERabp1 alone activates an auxin-like transduction pathway. FEBS Lett. 449 57–60. [DOI] [PubMed] [Google Scholar]
- Mattsson, J., Sung, Z.R., and Berleth, T. (1999). Responses of plant vascular systems to auxin transport inhibition. Development 126 2979–2991. [DOI] [PubMed] [Google Scholar]
- Maughan, S.C., Murray, J.A., and Bogre, L. (2006). A greenprint for growth: signalling the pattern of proliferation. Curr. Opin. Plant Biol. 9 490–495. [DOI] [PubMed] [Google Scholar]
- Melaragno, J.E., Mehrotra, B., and Coleman, A.W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges, M., Samland, A.K., Planchais, S., and Murray, J.A. (2006). The D-type cyclin CYCD3;1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell 18 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis, K., and Estelle, M. (2008). Auxin receptors and plant development: A new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24 55–80. [DOI] [PubMed] [Google Scholar]
- Napier, R.M., David, K.M., and Perrot-Rechenmann, C. (2002). A short history of auxin-binding proteins. Plant Mol. Biol. 49 339–348. [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pien, S., Wyrzykowska, J., McQueen-Mason, S., Smart, C., and Fleming, A. (2001). Local expression of expansin induces the entire process of leaf development and modifies leaf shape. Proc. Natl. Acad. Sci. USA 98 11812–11817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint, M., and Gray, W.M. (2006). Auxin signaling. Curr. Opin. Plant Biol. 9 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Pesce, E.R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., Traas, J., Friml, J., and Kuhlemeier, C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426 255–260. [DOI] [PubMed] [Google Scholar]
- Rolland-Lagan, A.G., and Prusinkiewicz, P. (2005). Reviewing models of auxin canalization in the context of leaf vein pattern formation in Arabidopsis. Plant J. 44 854–865. [DOI] [PubMed] [Google Scholar]
- Roslan, H.A., Salter, M.G., Wood, C.D., White, M.R., Croft, K.P., Robson, F., Coupland, G., Doonan, J., Laufs, P., Tomsett, A.B., and Caddick, M.X. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28 225–235. [DOI] [PubMed] [Google Scholar]
- Rück, A., Pälme, K., Venis, M.A., Napier, R.M., and Felle, R.H. (1993). Patch-clamp analysis establishes a role for an auxin binding protein in the auxin stimulation of plasma membrane current in Zea mays protoplasts. Plant J. 4 41–46. [Google Scholar]
- Scarpella, E., Marcos, D., Friml, J., and Berleth, T. (2006). Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.S., Guyomarc'h, S., Mandel, T., Reinhardt, D., Kuhlemeier, C., and Prusinkiewicz, P. (2006). A plausible model of phyllotaxis. Proc. Natl. Acad. Sci. USA 103 1301–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens, B., Feckler, C., Palme, K., Christian, M., Bottger, M., and Luthen, H. (2001). The auxin signal for protoplast swelling is perceived by extracellular ABP1. Plant J. 27 591–599. [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu, K., and Roberts, K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6 544–553. [DOI] [PubMed] [Google Scholar]
- Thiel, G., Blatt, M.R., Fricker, M.D., White, I.R., and Millner, P. (1993). Modulation of K+ channels in Vicia stomatal guard cells by peptide homologs to the auxin-binding protein C terminus. Proc. Natl. Acad. Sci. USA 90 11493–11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venis, M.A., Napier, R.M., Barbier-Brygoo, H., Maurel, C., Perrot-Rechenmann, C., and Guern, J. (1992). Antibodies to a peptide from the maize auxin-binding protein have auxin agonist activity. Proc. Natl. Acad. Sci. USA 89 7208–7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues, P.E., and Zhu, J.K. (2007). New developments in abscisic acid perception and metabolism. Curr. Opin. Plant Biol. 10 447–452. [DOI] [PubMed] [Google Scholar]
- Vlieghe, K., Inzé, D., and De veylder, L. (2007). Physiological relevance and molecular control of endocycle in plants. In Cell Cycle Control and Plant Development, D. Inzé, ed (Singapore: Blackwell Publishing), pp. 227–248.
- Wyrzykowska, J., and Fleming, A. (2003). Cell division pattern influences gene expression in the shoot apical meristem. Proc. Natl. Acad. Sci. USA 100 5561–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrzykowska, J., Pien, S., Shen, W.H., and Fleming, A.J. (2002). Manipulation of leaf shape by modulation of cell division. Development 129 957–964. [DOI] [PubMed] [Google Scholar]
- Yamagami, M., Haga, K., Napier, R.M., and Iino, M. (2004). Two distinct signaling pathways participate in auxin-induced swelling of pea epidermal protoplasts. Plant Physiol. 134 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.