Abstract
Nodulation is tightly regulated in legumes to ensure appropriate levels of nitrogen fixation without excessive depletion of carbon reserves. This balance is maintained by intimately linking nodulation and its regulation with plant hormones. It has previously been shown that ethylene and jasmonic acid (JA) are able to regulate nodulation and Nod factor signal transduction. Here, we characterize the nature of abscisic acid (ABA) regulation of nodulation. We show that application of ABA inhibits nodulation, bacterial infection, and nodulin gene expression in Medicago truncatula. ABA acts in a similar manner as JA and ethylene, regulating Nod factor signaling and affecting the nature of Nod factor–induced calcium spiking. However, this action is independent of the ethylene signal transduction pathway. We show that genetic inhibition of ABA signaling through the use of a dominant-negative allele of ABSCISIC ACID INSENSITIVE1 leads to a hypernodulation phenotype. In addition, we characterize a novel locus of M. truncatula, SENSITIVITY TO ABA, that dictates the sensitivity of the plant to ABA and, as such, impacts the regulation of nodulation. We show that ABA can suppress Nod factor signal transduction in the epidermis and can regulate cytokinin induction of the nodule primordium in the root cortex. Therefore, ABA is capable of coordinately regulating the diverse developmental pathways associated with nodule formation and can intimately dictate the nature of the plants' response to the symbiotic bacteria.
INTRODUCTION
The availability of nitrogen is crucial for plant growth, and legumes have evolved the capability to interact with nitrogen-fixing rhizobial bacteria to provide a ready source of this nutrient. Legumes develop a specialized organ, the nodule, where the rhizobia are fed dicarboxylic acids in exchange for ammonia generated through the reduction of molecular dinitrogen. The fixation of nitrogen is energetically expensive, and as a result this process is carefully regulated by the plant either through regulating the number of nodules that form or by regulating the level of fixation within nodules. Therefore, a balance is maintained between the nitrogen levels required and the availability of a carbon source to ensure the optimization of plant growth.
The formation of nitrogen-fixing nodules involves two major developmental programs: root epidermal responses that prepare the cell for bacterial infection and the reactivation of cell division within the cortex leading to the nodule primordia. Epidermal responses are associated with the perception of Nod factor, a signaling molecule generated by rhizobial bacteria, and include root hair deformation, the activation of calcium spiking, early nodulin (ENOD) gene induction, and infection thread initiation (Oldroyd and Downie, 2008). Components in the signaling pathway responsible for Nod factor perception have been defined in the last several years. Lysine motif receptor-like kinases have been identified as strong candidates for the Nod factor receptor (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006). Following perception of Nod factor at the plasma membrane, the signal is transduced via an additional receptor-like kinase to the activation of calcium spiking in the nuclear region (Ehrhardt et al., 1996; Endre et al., 2002; Stracke et al., 2002). This requires components of the nuclear pore and cation channels located on the nuclear membrane (Ane et al., 2004; Imaizumi-Anraku et al., 2005; Kanamori et al., 2006; Riely et al., 2007; Saito et al., 2007). Calcium spiking is perceived by a Ca2+/calmodulin-dependent protein kinase (CCaMK) (Levy et al., 2004; Mitra et al., 2004; Tirichine et al., 2006) and the information transduced via two GRAS domain transcriptional regulators, NSP1 and NSP2, and an ERF transcription factor, ERN1 (Kalo et al., 2005; Smit et al., 2005; Middleton et al., 2007).
Epidermal responses to Nod factor are coordinated with the activation of the nodule primordia in the cortex. Indeed, the application of high concentrations of Nod factor or gain-of-function mutations in CCaMK are sufficient to induce cortical cell division and nodule formation (Yang et al., 1994; Gleason et al., 2006; Tirichine et al., 2006). Recent studies examining gain-of-function and loss-of-function mutations in a cytokinin receptor highlight the essential role of cytokinins in the activation of the nodule primordia (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007). In Arabidopsis thaliana, cytokinins are known to mediate cell cycle arrest in pericycle cells (Li et al., 2006), thus inhibiting lateral root initiation (Laplaze et al., 2007). An analogous function has been found in Medicago truncatula (Gonzalez-Rizzo et al., 2006), and this indicates that changes in cytokinins will alter the balance of nodules and lateral roots, a proposal suggested many years ago (Nutman, 1948). It is likely that Nod factor perception at the epidermis leads to localized increases in cytokinin levels that then induce nodule organogenesis in the cortex (Hirsch and Fang, 1994; Fang and Hirsch, 1998; Oldroyd, 2007). In addition to cytokinins, several other hormones, such as auxin, gibberellins, and brassinosteroids, also have positive functions in nodule formation (Mathesius et al., 1998; Ferguson et al., 2005; Prayitno et al., 2006), and this suggests that changes in these hormone levels will be a prerequisite for nodule initiation.
In contrast with these plant hormones with positive functions in nodule inception, a number of plant hormones have been found to negatively regulate nodulation. Ethylene is the best characterized of these negative regulators, although jasmonic acid (JA), salicylic acid, and abscisic acid (ABA) have all been shown to negatively regulate the levels of nodulation (Cho and Harper, 1993; Penmetsa and Cook, 1997; Suzuki et al., 2004; Nakagawa and Kawaguchi, 2006; Sun et al., 2006). Ethylene has been shown to regulate nodulation at multiple levels: it negatively regulates the sensitivity of the plant to Nod factor and in so doing inhibits root hair deformation, calcium spiking, bacterial infection, the expression of nodulin genes, and the number of nodules formed (Oldroyd et al., 2001). In addition, ethylene has also been proposed to regulate the growth of infection threads and dictate the positioning of nodules on the root (Heidstra et al., 1997; Penmetsa and Cook, 1997). These studies on the role of ethylene in nodulation have been facilitated by the identification of skl, an ethylene-insensitive mutant of M. truncatula that shows hypernodulation (Penmetsa and Cook, 1997). Like ethylene, JA is also able to regulate the plants' sensitivity to Nod factor and as such regulates many aspects of the nodulation process. JA has also been shown to regulate the nature of Nod factor–induced calcium spiking with the ability to define the period between calcium spikes (Sun et al., 2006).
ABA has diverse effects on plant development in Arabidopsis, acting as an endogenous signal in seed germination, a regulator of leaf development and root growth, as well as a root-to-shoot signal that controls the closure of stomata (Fedoroff, 2002; De Smet et al., 2006). Besides the effects on primary root growth, ABA also inhibits lateral root initiation through interplay with auxin (Brady et al., 2003; De Smet et al., 2003, 2006). In contrast with Arabidopsis, ABA in M. truncatula promotes lateral root formation (Liang and Harris, 2005), indicating that the specifics of ABA action can differ between plant species. It has recently been shown that ABA can regulate nodulation in Lotus japonicus, and the ABA signaling gene, LATD, regulates nodule development in M. truncatula, although the mechanisms by which it achieves this have not been characterized (Suzuki et al., 2004; Liang et al., 2007). In this study, we use genetics and physiology to define the nature of ABA action on the nodulation process. We show that ABA acts at multiple levels in this process, regulating both Nod factor signaling in the epidermis and cytokinin-induced cell division in the cortex. We show that this is the result of multiple independent sites of action of ABA in the regulation of nodulation. Therefore, ABA has the capability of fine-tuning the process of nodulation, dictating at multiple levels the nature of this response. We propose that ABA achieves this through its regulation of the Nod factor signaling pathway, and its interplay with cytokinin and as such can dictate the responses to the two master regulators of both the epidermal and cortical nodulation processes.
RESULTS
ABA Regulates Lateral Root Emergence, Nodule Number, and Infection Frequency
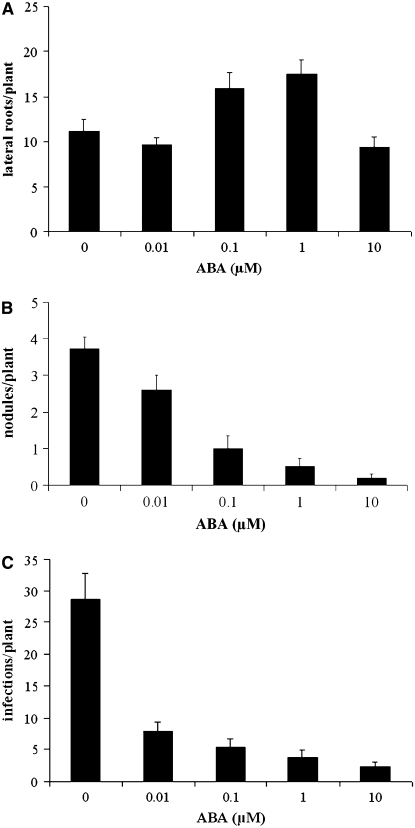
It has previously been shown that ABA inhibits nodulation in legumes (Cho and Harper, 1993; Suzuki et al., 2004; Liang and Harris, 2005). To better understand the nature of this regulation, we initiated studies to assess the concentrations of ABA under our growth conditions that led to reduced nodulation efficiency in M. truncatula. We grew M. truncatula cv Jemalong line A17 plants on 0, 0.01, 0.1, 1, and 10 μM ABA and quantified nodulation 21 d after inoculation with Sinorhizobium meliloti strain 1021 (pXLGD4). Consistent with previous studies in L. japonicus, Trifolium repens, and M. truncatula (Suzuki et al., 2004; Liang et al., 2007), nodulation was abolished at 1 and 10 μM ABA (Figure 1B). These studies also confirmed that ABA plays a positive role on lateral root growth in M. truncatula (Figure 1A), which has been previously reported (Liang et al., 2007).
Figure 1.
ABA Inhibits Nodulation and Promotes Lateral Roots in M. truncatula.
Plants were grown on differing concentrations of ABA and assayed for lateral root initiation (A), nodule formation (B), and S. meliloti infection events (C). Increasing ABA concentrations promote lateral root formation up to 1 μM ABA but suppress nodulation and S. meliloti infection. Infections were measured following lacZ staining, and both infection foci and foci with infection threads were counted as infection events. Ten to twelve plants were analyzed for each ABA concentration, with different plants being used to assay lateral roots, nodulation, and rhizobial infection. Error bars represent se.
The generation of a nodule involves a number of processes that could be regulated by ABA, including the reinitiation of cortical cell division to form a nodule primordia and the facilitation of bacterial infection through epidermal and cortical cells. We assessed whether ABA could regulate bacterial infection of the epidermis by quantifying the number of infection events in plants grown at different concentrations of ABA. Six days after inoculation with S. meliloti, we quantified the number of visible infection threads and microcolonies of bacteria in the center of a root hair curl. Figure 1C shows that infection frequency decreased with increasing concentrations of ABA, indicating that ABA regulates both nodule formation and bacterial infection.
ABA Inhibits Rhizobial and Nod Factor–Induced Gene Expression
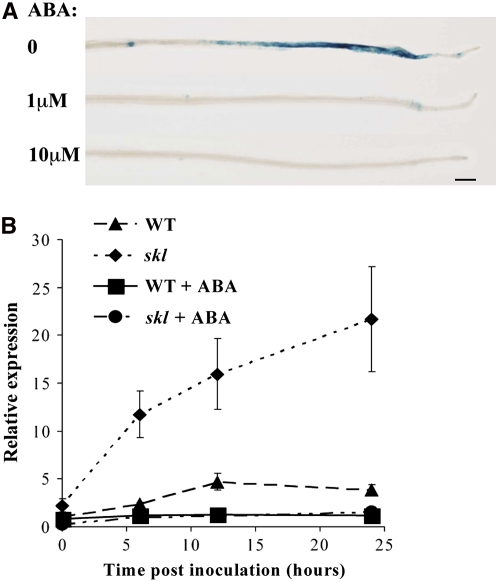
RIP1 and ENOD11 induction represent some of the earliest markers for the response to Nod factor and rhizobia (Cook et al., 1995; Gamas et al., 1996; Journet et al., 2001). Induction of these genes requires the Nod factor signal transduction pathway and as such provides markers for the regulation of this signaling pathway. To assess the effects of ABA on Nod factor–induced gene expression, we used M. truncatula plants stably transformed with β-glucuronidase (GUS) regulated by the ENOD11 promoter (Journet et al., 2001). Five-day-old plants were transferred to liquid medium containing 0, 1, and 10 μM ABA. Following 24 h of ABA treatment, 1 nM Nod factor was added to the medium, and after 12 h of Nod factor treatment, the roots were tested for GUS activity. Plants without Nod factor treatment show GUS activity only in root caps and the root vasculature, while Nod factor activates ENOD11 in epidermal cells within a specific region of the root (Figure 2A) (Journet et al., 2001). Pretreatment with 10 μM ABA inhibited Nod factor induction of ENOD11, while 1 μM ABA pretreatment led to reduced ENOD11 induction (Figure 2A). To assess the effects of ABA on RIP1 induction, we grew M. truncatula on 0 and 10 μM ABA and used quantitative real-time RT-PCR (qRT-PCR) to measure RIP1 expression at 0, 6, 12, and 24 h after inoculation with S. meliloti. We found that ABA abolishes S. meliloti induction of RIP1 expression in wild-type plants (Figure 2B). Taken together, these results indicate that ABA negatively regulates early gene expression induced by Nod factor and rhizobia and implies that ABA inhibition of nodulation may be through the regulation of the Nod factor signal transduction pathway.
Figure 2.
ABA Suppresses Nod Factor and S. meliloti–Induced Gene Expression.
(A) and (B) ABA effect on early nodulin gene expression was assayed using transgenic plants containing ENOD11:GUS (A) or qRT-PCR of RIP1 (B).
(A) Pretreatment with ABA (concentrations indicated) inhibits ENOD11 induction following 1 nM Nod factor treatment. The blue color represents GUS expression.
(B) Pretreatment with 10 μM ABA abolished S. meliloti induction of RIP1 in both wild-type and ethylene-insensitive skl plants. Error bars represent se.
The Balance of ABA and Nod Factor Concentrations Dictates the Nature of Nod Factor Signal Transduction
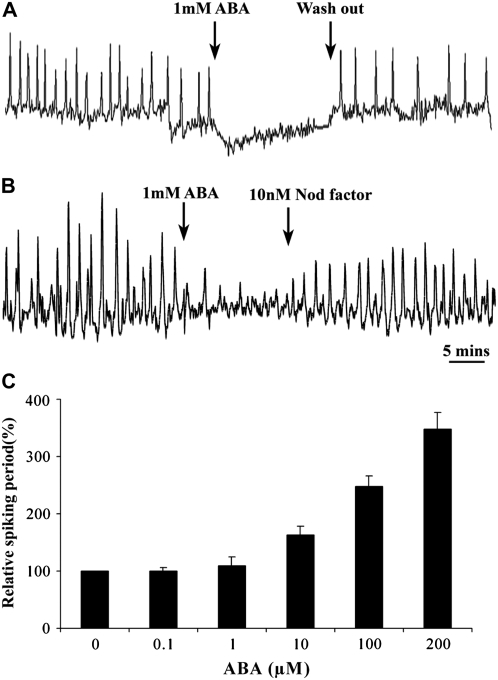
Oscillations of calcium (calcium spiking) act as a secondary messenger in the Nod factor signal transduction pathway (Ehrhardt et al., 1996; Oldroyd and Downie, 2004). To better understand ABA's affect on Nod factor signal transduction, we determined the role of ABA on the regulation of calcium spiking, using an M. truncatula line stably transformed with the calcium reporter cameleon YC2.1 (Miwa et al., 2006). We treated plants with 0.1, 1, 10, 100, 200, and 1000 μM ABA following 1 nM Nod factor induction of calcium spiking. Twenty cells were analyzed for each concentration. Treatment with 1 mM ABA completely inhibited Nod factor–induced calcium spiking, with calcium spiking returning upon the removal of ABA (Figure 3A). Analogous to what we have previously reported for JA (Sun et al., 2006), lower concentrations of ABA in the micromolar range caused a lengthening of the period between individual calcium spikes leading to a reduction in the frequency of the spiking response (Figure 3C). Similar to our studies with JA, higher concentrations of ABA were required for inhibition of calcium spiking than were required for the inhibition of nodulation and nodulin gene expression. However, in the nodulation and gene expression assays, the plants were pretreated with ABA, while the calcium spiking assay required an immediate response to ABA. To further validate these observations, we pretreated plants overnight with 0.1 and 1 μM ABA (concentrations sufficient to inhibit nodulation) and found that this pretreatment abolished the ability of Nod factor to induce calcium spiking (10 cells analyzed for 0.1 μM ABA and eight cells analyzed for 1 μM ABA). Our findings indicate that ABA, along with JA and ethylene, can regulate both the occurrence and nature of Nod factor–induced calcium spiking.
Figure 3.
ABA Modulates Nod Factor–Induced Calcium Spiking.
(A) and (B) Representative calcium traces of M. truncatula root hair cells preinduced with 1 nM Nod factor and secondarily treated with 1 mM ABA. Treatment with ABA inhibits calcium spiking, and this can be recovered following ABA washout in continuous 1 nM Nod factor (A) or by raising the Nod factor concentration to 10 nM (B). Calcium measurements were generated from cameleon transformed plants, and the y axes represent the ratio of cyan fluorescent protein: yellow fluorescent protein (CFP:YFP) in arbitrary units. Error bars represent se.
(C) While 1 mM ABA inhibits calcium spiking, lower concentrations of ABA cause a lengthening of the interval between individual calcium spikes. The period between calcium spikes was averaged from 20 min of spiking in 20 cells per treatment, and this period was standardized relative to no ABA treatment.
Both ethylene and JA affect the threshold concentrations of Nod factor that are necessary to activate calcium spiking (Oldroyd et al., 2001; Sun et al., 2006). This suggests that Nod factor is in competition with these negative regulators for the activation of its signal transduction pathway. To assess whether ABA plays a similar role, we tested whether increasing Nod factor concentrations could overcome the inhibition by ABA. We initiated calcium spiking in cells using 1 nM Nod factor and then blocked calcium spiking with 1 mM ABA. Secondarily raising the Nod factor concentration to 10 nM led to a recovery of calcium spiking in 18 out of 20 cells (Figure 3B). This result implies that the activation of Nod factor signaling is balanced between Nod factor and ABA concentrations.
ABA and Ethylene Function Independently in the Regulation of Lateral Root Initiation, Nodulation, and Nod Factor Signal Transduction
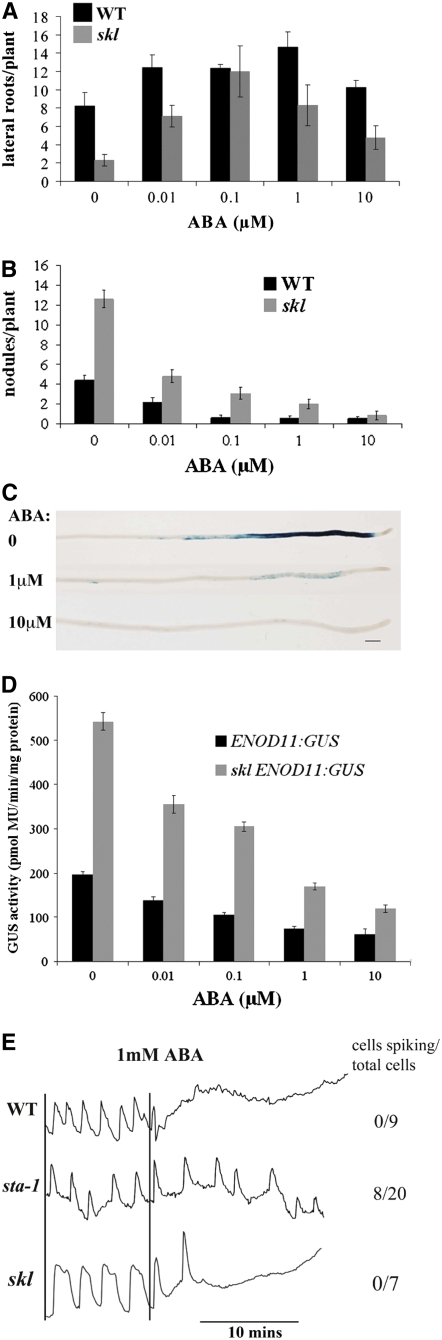
The regulation of nodulation by ABA is similar in nature to the effects seen by the negative regulators ethylene and JA (Penmetsa and Cook, 1997; Oldroyd et al., 2001; Sun et al., 2006). Ethylene and JA act in combination to regulate nodulation and Nod factor signal transduction (Sun et al., 2006). To assess the combinatorial nature of ABA regulation, we used the ethylene insensitive mutant skl (Penmetsa and Cook, 1997) to assess the affect of ethylene on ABA regulation of nodulation. We grew both wild-type and skl plants on 0, 0.01, 0.1, 1, and 10 μM ABA, scoring lateral root emergence and on separate plants scoring nodulation 21 d after inoculation with S. meliloti. ABA can regulate both lateral root emergence and nodulation in skl in a similar manner to the regulation of these processes in wild-type plants (Figures 4A and 4B). However, skl shows hypernodulation and reduced lateral root emergence caused by ethylene insensitivity. The effects of ABA are superimposed upon these differences, such that higher concentrations of ABA are necessary in skl plants to completely abolish nodulation or activate lateral root emergence to levels comparable to the wild type (Figures 4A and 4B).
Figure 4.
ABA and Ethylene Act Independently in the Suppression of Nodulation.
(A) and (B) The ethylene-insensitive mutant skl shows ABA induction of lateral roots (A) and ABA suppression of nodulation (B) in a manner similar to that observed in the wild type. However, the lateral root and nodule number is affected by the ethylene-insensitive nature of skl, and the ABA effects are superimposed upon this preexisting state.
(C) ENOD11:GUS induction by 1 nM Nod factor in skl (all roots) is suppressed following ABA addition, at the concentrations indicated.
(D) Quantification of GUS in wild-type plants carrying ENOD11:GUS and skl plants with ENOD11:GUS. skl shows higher levels of ENOD11 induction by 1 nM Nod factor, which is suppressed by ABA treatment.
(E) Representative traces of cells pretreated with 1 nM Nod factor for 30 min prior to calcium imaging. Addition of 1 mM Nod factor leads to inhibition of calcium spiking in all wild-type and skl cells, but sta-1 is partially insensitive to ABA treatment. The calcium imaging was generated following microinjection of Oregon green/Texas red, and the y axes represent the change in florescence of Oregon green versus Texas red in arbitrary units. Error bars represent se.
The formation of a nodule involves the coordination of a number of processes, and to simplify the analysis of the interrelationship between ABA and ethylene regulation, we focused on the effects on Nod factor signal transduction using calcium spiking and gene expression as markers of this signaling pathway. One and 10 μM ABA treatment reduced ENOD11 expression in both skl and wild-type plants (Figures 2A and 4C). Similar to the nodulation responses, ENOD11:GUS activation in skl is stronger than the wild type, and its inhibition by ABA was slightly less effective in skl than in wild-type plants (Figure 4D). We also tested the effects of ethylene on ABA regulation of Nod factor–induced RIP1 expression using qRT-PCR. As previously shown, RIP1 shows higher levels of induction in skl compared with the wild type in the absence of ABA (Figure 2B) (Oldroyd et al., 2001). We found that RIP1 induction by S. meliloti was suppressed by ABA to equivalent levels in both skl and wild-type plants. We found comparable results for the regulation of calcium spiking, with 1 mM ABA inhibiting calcium spiking in all skl cells analyzed (Figure 4E). These data indicate that ABA can regulate the Nod factor signaling pathway in the absence of the ethylene response, suggesting that these two negative regulators function as parallel pathways with little crosstalk.
Suppressing ABA Signaling in M. truncatula Enhances Nodulation
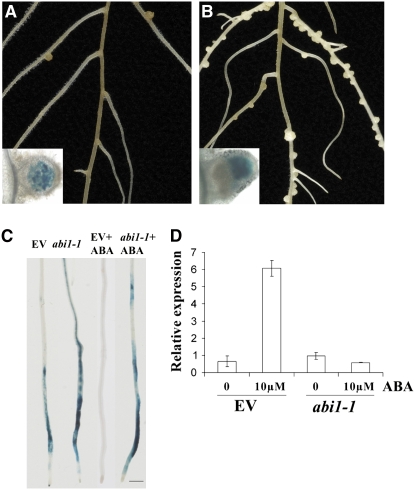
Our work indicated that the external application of ABA inhibits nodulation responses. To support these physiological studies, we used the dominant negative allele abi1-1 from Arabidopsis to genetically regulate ABA signaling in M. truncatula. ABI1 encodes a protein phosphatase of the type IIC class (Leung et al., 1994), and the abi1-1 allele dominantly suppresses ABA signaling (Hagenbeek et al., 2000; Gampala et al., 2001; Wu et al., 2003). We overexpressed the Arabidopsis abi1-1 allele in M. truncatula using Agrobacterium rhizogenes transformation (Boisson-Dernier et al., 2001). Transformed roots were initially selected using kanamycin, secondarily selected using the dsRED reporter and a subset of roots validated for transformation by PCR of abi1-1. To assess whether abi1-1 can regulate ABA signaling in M. truncatula, we used the ABA-responsive gene RD22 (Yamaguchishinozaki et al., 1992) (represented by TC95843 of M. truncatula). We grew the transformed plants on medium containing 0 and 10 μM ABA for 5 d and then analyzed the expression levels of RD22 by qRT-PCR. While plants transformed with the empty vector were responsive to ABA treatment with RD22 induction, plants transformed with abi1-1 were no longer able to induce RD22 following ABA treatment (Figure 5D). This indicates that abi1-1 is able to inhibit ABA signaling in M. truncatula.
Figure 5.
Arabidopsis abi1-1 Induces Insensitivity to ABA and Hypernodulation in M. truncatula.
(A) and (B) Transformation of abi1-1 from Arabidopsis leads to a hypernodulation phenotype (B) that was not observed when M. truncatula was transformed with the empty vector (A). The insets indicate nodules from these plants stained with X-gal to reveal bacterial infection inside the nodules.
(C) abi1-1 transformation leads to enhanced Nod factor induction of ENOD11:GUS and makes this response insensitive to 10 μM ABA treatment.
(D) qRT-PCR shows that the abi1-1–transformed roots are also insensitive to ABA treatment for induction of RD22, a marker of ABA signaling. EV, empty vector. Error bars represent se.
To assess the effect of abi1-1 on nodulation, we counted nodules on roots transformed with abi1-1 versus roots transformed with the empty vector 21 d after inoculating with S. meliloti. abi1-1 transformed roots showed 29 ± 2.9 nodules per plant compared with only 8.6 ± 2.4 nodules per plant in the empty vector controls (Figures 5A and 5B). It is commonly observed that excessive numbers of nodules form in plants with ineffective nodules (no nitrogen fixation). We used acetylene reduction and a bacterial strain that constitutively expresses β-galactosidase to ensure that the hypernodulation phenotype we had observed in the abi1-1 transformed roots was not the result of ineffective bacterial infection or nitrogen fixation. These tests show that the abi1-1 transformed roots form normal nodules infected by bacteria (Figure 5B, inset) and are able to reduce acetylene at levels comparable to the empty vector control (2.2 ± 0.3 acetylene reduction activity (ARA) units in the abi1-1 transformed plants versus 1.5 ± 0.2 ARA units in the empty vector controls).
We used abi1-1 transformation to further validate our earlier observations on ABA regulation of Nod factor signaling. We transformed the abi1-1 and empty vector constructs into the pENOD11:GUS transgenic line. We treated the transformed plants with 0 and 10 μM ABA for 24 h, followed by a 12 h treatment with 0 and 1 nM Nod factor. ABA was able to inhibit Nod factor–induced ENOD11 in the empty vector transformed roots, but the abi1-1–transformed roots were resistant to ABA treatment (Figure 5C), further validating that abi1-1 abolishes ABA signaling in M. truncatula. Furthermore, in the absence of ABA, ENOD11 induction by Nod factor was greater in the abi1-1–transformed roots compared with the empty vector controls as measured by GUS quantification (384.6 ± 14.2 pM MU/min/mg protein in the abi1-1 transformed plants compared with 208.9 ± 10.3 pM MU/min/mg in the empty vector controls). These results provide genetic proof that ABA negatively regulates nodulation and Nod factor signaling.
A Novel Genetic Component That Modulates ABA Sensitivity in M. truncatula
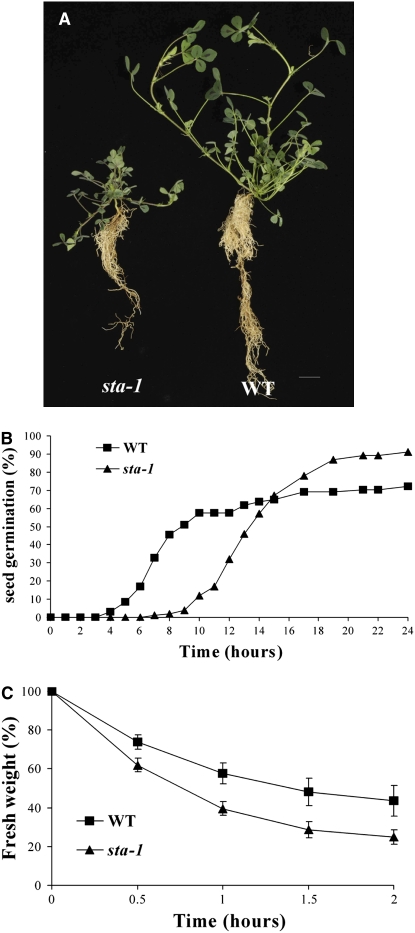
A genetic screen that we undertook in M. truncatula for nodulation defective mutants revealed a mutant, generated by fast neutron mutagenesis, with reduced nodulation efficiency. This mutant shows many fewer nodules than wild-type plants: 11.7 ± 0.1 nodules per plant in the mutant compared with 120 ± 6.8 nodules per plant in the wild type in soil-grown plants 3 months after inoculation with S. meliloti. The mutant behaves as a single recessive locus (segregation ratio of 3.4:1), and allelism tests with known nodulation mutants indicate that this is a novel locus (Table 1; due to the poor success rate of crosses onto this mutant, allelism tests were performed always with this mutant as the male parent). In addition to the nodulation phenotype, the mutant shows a number of pleiotropic defects (Figure 6A), the most obvious being dwarf stature and a reduced root system that occurred in the absence of rhizobial infection. These phenotypes cosegregate with the nodulation defect in 79 mutant plants in a segregating population of 266 individuals, indicating that they are caused by the same mutation.
Table 1.
Allelism Tests of sta-1 with Known Nodulation Mutants of M. truncatula
| nfp-1 | dmi1-3 | dmi2-1 | dmi3-3 | nsp1-1 | nsp2-2 | hcl-1 | lin-2 | nin-1 | |
|---|---|---|---|---|---|---|---|---|---|
| sta-1 | + (15/4) | + (29/5) | + (13/4) | + (23/5) | + (17/5) | + (22/5) | + (15/5) | + (7/5) | + (4/6) |
The plus sign indicates that nodules were present on progeny of the cross; number of plants scored/number of crosses is shown in parenthesis.
Figure 6.
sta-1 Shows Defects in Plant Growth, Seed Germination, and Stomatal Behavior.
(A) sta-1 shows pleiotropic effects for root and shoot growth revealed by a reduced stature of the plant.
(B) sta-1 shows delayed seed germination compared with the wild type. The number of germinated seeds was measured using radicle emergence as a marker relative to the time since removing the seeds from 4°C. Two hundred seeds are analyzed per treatment.
(C) The fresh weight of detached leaves was used to measure relative water loss that provides an indication of stomatal aperture. sta-1 shows more rapid water loss, indicating an alteration in its stomatal behavior. Error bars represent se.
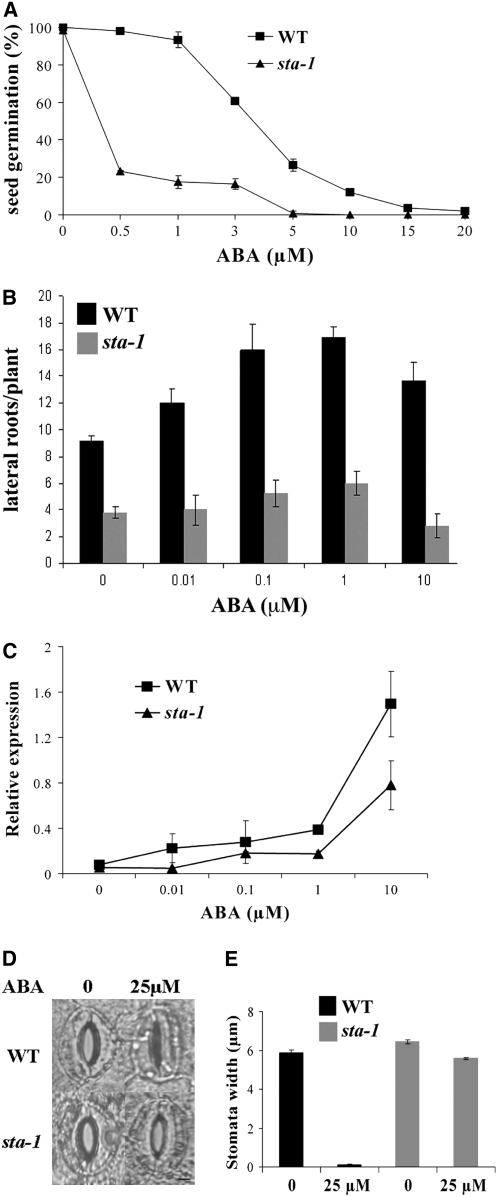
Further characterization of this mutant revealed a number of phenotypes indicative of a defect in ABA signaling. The mutant shows delayed seed germination (Figure 6B) and enhanced water loss (Figure 6C), both of which have been previously shown to be present in ABA defective mutants (Schroeder et al., 2001; Gubler et al., 2005; De Smet et al., 2006). We analyzed the responsiveness of this mutant to a range of plant hormones (10−6 and 10−9 M GA3, 10−7 and 10−8 M benzyl-amino purine (BAP), 10−6 M ABA, 10−6 M 24-epibrassinolide, 10−6 M 1-aminocyclopropane-1-carboxylic acid, 10−7 M indole-3-acetic acid, 10−4 M salicylic acid, and 10−6 M JA) and found defects only in the response to ABA. For reasons described below, we chose to call this locus SENSITIVITY TO ABA (STA); therefore, the mutant will be referred to as sta-1. We assessed the sensitivity of sta-1 to ABA treatment for a number of plant developmental responses. sta-1 is hypersensitive to ABA for seed germination, showing complete inhibition of seed germination at low ABA concentrations (Figure 7A). By contrast, sta-1 was less sensitive to ABA for the induction of lateral roots (Figure 7B) and was insensitive to ABA for stomatal closure (Figures 7D and 7E). In the stomata assay, we found that wild-type stomata all closed in response to 25 μM ABA, while no stomata closed at this ABA concentration in sta-1 leaves. To further confirm the ABA sensitivity phenotype of sta-1, we assessed the induction of RD22 by ABA. qRT-PCR revealed that sta-1 shows reduced sensitivity to ABA for the activation of RD22 (Figure 7C). The phenotypic characterization of sta-1 indicates that this mutant is altered in its sensitivity to ABA with hypersensitivity to ABA for seed germination and reduced sensitivity to ABA for lateral root initiation, RD22 induction, and guard cell physiology.
Figure 7.
sta-1 Is Altered in Its Sensitivity to ABA.
(A) sta-1 shows hypersensitivity to ABA for seed germination. In this assay the number of seeds germinated was analyzed at 24 h after removal from 4°C, a time point where the maximum number of seeds have germinated in each line. The data points are presented as the percentage of seeds germinated relative to germination in the absence of ABA. Two hundred seeds are analyzed per treatment.
(B) to (E) In contrast with seed germination, sta-1 is insensitive to ABA for lateral root initiation (B) and induction of RD22 (C) as well as stomatal closure ([D] and [E]). In (D) and (E), the plants were pretreated to ensure maximal stomatal opening and then secondarily treated with buffer alone (0) or with buffer containing 25 μM ABA. In wild-type plants, all stomata closed in the ABA treatment, while in sta-1 mutant leaves, all stomata remained open.
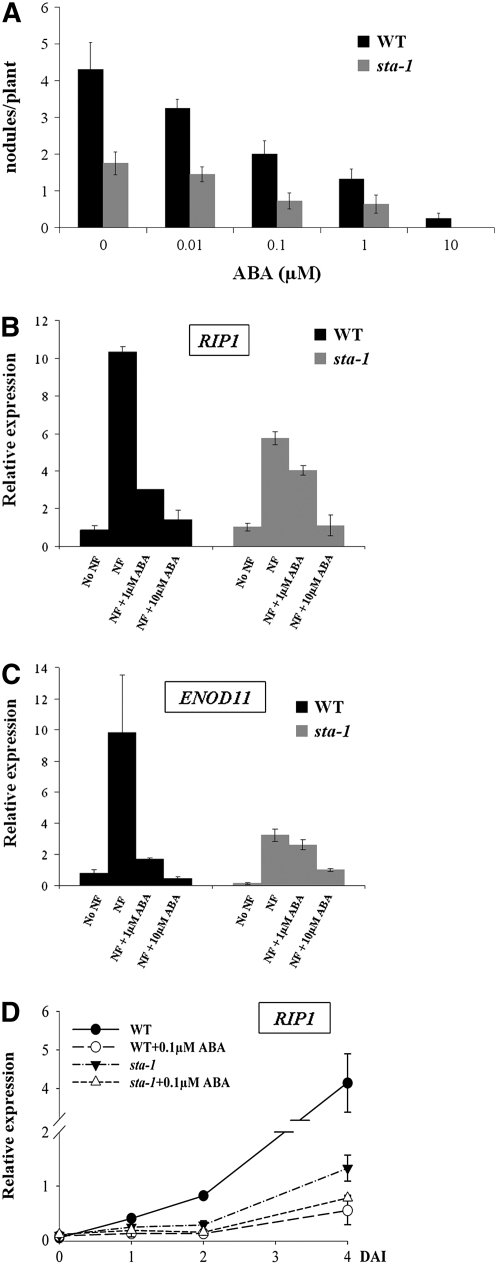
The STA locus appears to regulate the plants' sensitivity to ABA with opposite effects depending on the developmental process studied. Considering that ABA is a negative regulator of nodulation, we propose that the reduced nodulation in sta-1 is the result of a hypersensitivity to ABA for the regulation of nodulation. To further characterize sta-1, we tested a number of nodulation responses in this mutant to ABA. Nodulation levels are lower in sta-1 compared with the wild type, and treatment with ABA abolished nodulation in sta-1 (Figure 8A). To test whether sta-1 affects Nod factor signaling, we assayed RIP1 and ENOD11 induction using qRT-PCR and the activation of calcium spiking. sta-1 showed reduced levels of induction of RIP1 and ENOD11 by Nod factor (Figures 8B and 8C) and RIP1 by rhizobia (Figure 8D). Because of this low level of gene induction, it is difficult to judge the effect of ABA in sta-1. However, it appears that the suppression of RIP1 and ENOD11 induction by ABA was reduced in sta-1 compared with the wild type (Figures 8B to 8D). Furthermore, ABA was less effective at suppressing Nod factor–induced calcium spiking in sta-1 plants compared with the wild type (Figure 4E). This work suggests that sta-1 shows reduced sensitivity to ABA for the regulation of Nod factor signaling in epidermal cells.
Figure 8.
sta-1 Is Insensitive to ABA for Nod Factor–Induced Gene Induction.
(A) Nodulation of sta-1 is greatly reduced compared with wild-type plants and suppressed by ABA treatment.
(B) qRT-PCR demonstrating that sta-1 shows reduced sensitivity to ABA for the regulation of Nod factor (1 nM for 12 h) induced RIP1 expression. sta-1 shows reduced levels of RIP1 induction compared with wild-type plants, but the suppression of RIP1 by ABA is also greatly reduced in sta-1.
(C) sta-1 shows reduced levels of Nod factor (1 nM) induced ENOD11 expression, as measured by qRT PCR, and reduced suppression of this gene induction by ABA.
(D) sta-1 shows reduced levels of S. meliloti induction of RIP1 and reduced suppression of this gene induction by ABA.
For (B) to (D), 10 plants were used for each treatment and four biological repeats were performed. DAI, days after inoculation; NF, Nod factor. Error bars represent se.
ABA Regulates Cytokinin Induction of Nodulation Processes
Since all our results indicate that ABA is a negative regulator of nodulation, it was very surprising to see that sta-1, with a reduced nodulation phenotype, was insensitive to ABA for the regulation of Nod factor responses in epidermal cells. We decided that it was important to validate that the nodulation response in sta-1 was a function of the altered ABA sensitivity in this mutant. To test this, we inhibited ABA signaling in sta-1 by transformation with abi1-1. The introduction of abi1-1 rescued the nodulation defect of sta-1, resulting in the formation of 6.4 ± 1.7 nodules per plant compared with only 1.0 ± 0.5 nodule per plant in sta-1 transformed with the empty vector. This result indicates that the nodulation defect of sta-1 is the result of hypersensitivity to ABA.
Considering that sta-1 shows reduced sensitivity to ABA for the regulation of Nod factor signaling, the nodulation process that shows ABA hypersensitivity in sta-1 must be after Nod factor–induced calcium spiking. To validate this, we transformed sta-1 with a CCaMK gain-of-function mutation that activates spontaneous nodulation using A. rhizogenes (Gleason et al., 2006). We found that sta-1 could suppress CCaMK-induced spontaneous nodulation (0/27 plants showed spontaneous nodules on sta-1, compared with 22/41 plants showing spontaneous nodules in dmi3 plants). This reveals that the site of STA action that allows ABA regulation of nodulation must be downstream of CCaMK and calcium spiking.
Cytokinins are key activators of nodule initiation in the root cortex, and the regulation of cytokinin action by ABA may be one mechanism to explain sta-1. A gain-of-function mutation in the cytokinin receptor LHK1 of L. japonicus results in spontaneous nodule formation. We used the L. japonicus line snf2 that carries the LHK1 gain-of-function mutation to test whether ABA was able to abolish cytokinin-induced nodulation. When snf2 was grown in the absence of ABA, we saw that six out of 13 plants generated spontaneous nodules, consistent with previous reports, while treatment with 1 μM ABA completely abolished spontaneous nodulation in all 14 plants analyzed. This result indicates that ABA is able to regulate the cytokinin induction of nodule initiation in the root cortex.
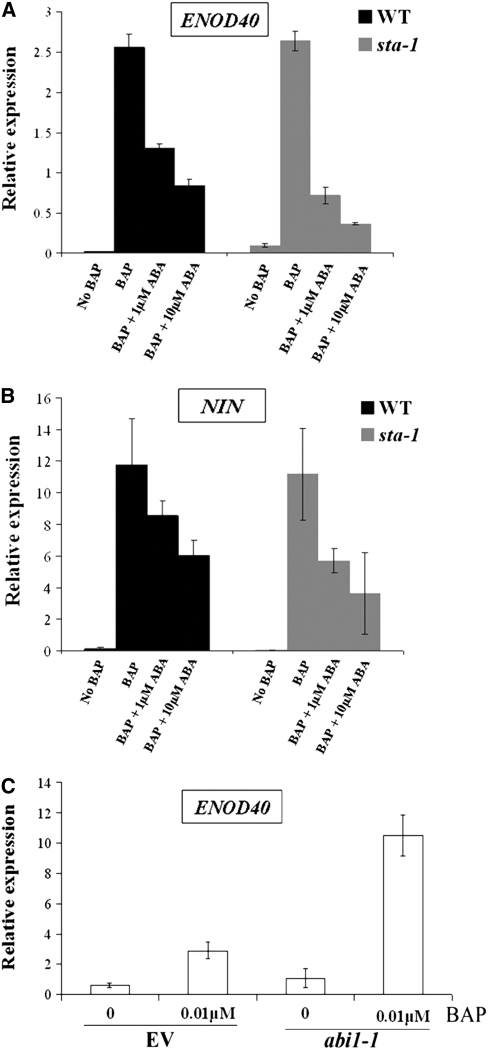
Cytokinins have previously been shown to activate ENOD40, a gene that is expressed in the root cortex during the initiation of the nodule primordium (Fang and Hirsch, 1998; Mathesius et al., 2000; Murray et al., 2007). We used ENOD40 induction by cytokinin to assess the ability of ABA to regulate cytokinin-induced nodulation responses in M. truncatula. We found that 10 μM ABA suppressed the cytokinin induction of ENOD40 by 3.04-fold relative to plants without ABA treatment (Figure 9A). In addition, ABA treatment also reduced cytokinin induction of NIN (Figure 9B), a gene critical for nodule organogenesis (Stracke et al., 2002). Furthermore, wild-type M. truncatula plants transformed with abi1-1 showed a much greater induction of ENOD40 following cytokinin treatment compared with plants transformed with the empty vector (Figure 9C). These experiments show that ABA can regulate cytokinin-induced nodulation gene expression. Taken together with the ABA suppression of snf2-induced spontaneous nodulation, this reveals that ABA can suppress cytokinin-induced nodulation processes.
Figure 9.
ABA Can Suppress Cytokinin-Induced Nodulation Responses.
(A) Treatment with 0.01 μM cytokinin (BAP) for 10 d induces ENOD40 in M. truncatula, and this is blocked upon treatment with ABA. sta-1 shows increased levels of ABA suppression of ENOD40 induction by cytokinin.
(B) Treatment with 0.01 μM BAP induces NIN in M. truncatula, and this is blocked upon treatment with ABA. sta-1 shows increased levels of ABA suppression of NIN induction by cytokinin.
(C) Wild-type M. truncatula plants were transformed with abi1-1 or empty vector (EV) and tested for cytokinin (BAP) induction of ENOD40. abi1-1–transformed plants showed much higher levels of ENOD40 induction by cytokinin compared with EV transformed plants.
For (A) and (B), 10 plants were used for each treatment and four biological repeats were performed. All error bars represent se.
We tested whether sta-1 was altered in its sensitivity to ABA for cytokinin induction of nodulation genes. ABA suppressed the cytokinin induction of ENOD40 (Figure 9A) and NIN (Figure 9B) to a greater extent in sta-1 than in wild-type plants, suggesting that sta-1 may be hypersensitive to ABA for the regulation of cytokinin-induced nodulation processes.
DISCUSSION
Nodulation in legumes is both dependent upon and regulated by plant hormones. Here, we show that ABA regulation is intricately woven into the nodulation process, with ABA having independent modes of action in the suppression of the nodulation process. As such, ABA, and most likely ethylene and JA, can rapidly and completely abolish the plants responses to the symbiotic bacteria.
We have previously shown that ethylene and JA are negative regulators of the Nod factor signaling pathway acting at or upstream of calcium spiking (Oldroyd et al., 2001; Sun et al., 2006). It has previously been suggested that ABA may regulate nodulation responses in root hair cells (Suzuki et al., 2004), and consistent with this, we found that external application of ABA is also able to abolish both Nod factor–induced calcium spiking and gene expression. Furthermore, the effects of ABA on calcium spiking are similar to what we have previously observed with JA (Sun et al., 2006): high concentrations of ABA abolish calcium spiking, while lower concentrations impact the period between calcium spikes. Interestingly, increasing the Nod factor concentration overwhelmed ABA inhibition allowing a recovery in calcium spiking, suggesting that the activation of Nod factor signaling is balanced between the ABA and Nod factor concentrations. In addition to these physiological studies, we found that blocking ABA signaling using the Arabidopsis abi1-1 allele led to enhanced Nod factor–induced gene expression. These results indicate that ABA, like ethylene and JA, can inhibit Nod factor signaling at or above calcium spiking. The similarities in the mode of inhibition imply that ethylene, JA, and ABA may have a similar mode of action in the regulation of Nod factor signaling. However, unlike ethylene and JA that act combinatorially, ABA appears to function independently of ethylene, since the ethylene-insensitive mutant skl shows normal regulation of nodulation by ABA.
Considering the absolute importance of Nod factor signaling for nodule initiation, the suppression of Nod factor signaling by ABA was sufficient to explain the inhibition of nodulation. This was supported by an absolute correlation between the inhibition of Nod factor signaling and the inhibition of both bacterial infection and nodule initiation in our physiological tests and the abi1-1 studies. However, during the course of this research, we found a novel mutant that causes both a reduction in nodule formation and an alteration in ABA sensitivity. Interestingly, the effects of sta-1 on ABA sensitivity were dictated by the developmental response studied, with hypersensitivity for ABA during seed germination, but insensitivity to ABA for lateral root initiation and stomatal closure. Considering that ABA is a negative regulator of nodulation, we can only conclude that the reduction in nodules in sta-1 must be the result of hypersensitivity to ABA for nodulation responses. We were therefore very surprised when sta-1 showed a reduced sensitivity to ABA for the suppression of the Nod factor signaling pathway. From this work we concluded that the important site for sta-1 action in nodulation must be at a stage after the initiation of Nod factor signaling.
The formation of a nodule requires the activation and coordination of two separate developmental processes: the initiation of bacterial infection in the epidermis and the reactivation of cell division in the root cortex to form the nodule primordia (Oldroyd and Downie, 2008). While these processes are intimately interlinked, they can be separated genetically. Gain-of-function mutations in CCaMK and the cytokinin receptor LHK1 cause spontaneous nodule formation without the activation of root hair deformation or infection thread formation (Gleason et al., 2006; Tirichine et al., 2006, 2007). Conversely, loss-of-function mutations in LHK1 cause a block in the activation of the nodule primordia without inhibiting bacterial infection (Murray et al., 2007). This indicates that while the cortical and epidermal programs are normally coordinated, they can be discriminated experimentally. We have previously suggested that epidermal responses leading to the initiation of bacterial infection are mostly controlled by Nod factor signaling, while cortical responses leading to nodule primordia formation are mostly the function of cytokinin and auxin signaling (Oldroyd and Downie, 2008). While the epidermal and cortical processes can be separated, it seems likely that Nod factor recognition in epidermal cells activates cytokinin signaling in the cortex, most likely by increasing cytokinin levels (Oldroyd, 2007).
ABA is known to interfere with auxin and cytokinin in cell division during plant growth in Arabidopsis (Wang et al., 1998; De Smet et al., 2006; Li et al., 2006; Laplaze et al., 2007). We therefore chose to test how ABA affects cytokinin-induced cortical cell division during nodulation. We found that ABA treatment abolished spontaneous nodulation activated by the LHK1 gain-of-function snf2 and could suppress ENOD40 and NIN induction by external cytokinin application. The fact that the reduced nodulation mutant sta-1 is insensitive to ABA for the regulation of Nod factor responses in epidermal cells, but hypersensitive to ABA for cytokinin induction of ENOD40 and NIN in cortical cells, highlights the importance of ABA regulation of cytokinin responses in the cortex during the regulation of nodulation. This indicates that ABA can suppress both Nod factor signaling in the epidermis and cytokinin activation of cell division in the cortex. As such, ABA can coordinately suppress the two signals currently known to be sufficient for nodule initiation.
Legumes show opposite responses to cytokinins and ABA for lateral root initiation to what has been reported in Arabidopsis (De Smet et al., 2003, 2006; Liang and Harris, 2005; Li et al., 2006; Laplaze et al., 2007). In legumes, cytokinins inhibit lateral root formation, while promoting nodule initiation. An alternative way of looking at this is that cytokinins inhibit cell division in the pericycle, the predominant cell type involved in lateral root initiation, and promote it in the cortex, the predominant cell type involved in the formation of the nodule. By contrast, ABA promotes lateral root formation and inhibits nodule inception, thus having opposite effects to cytokinin on cell division in the pericycle and cortex. It has long been observed that nodule initiation is associated with a suppression of lateral root formation (Nutman, 1948). This is most likely the result of changes in the ratio of cytokinins and ABA in legume roots during nodule initiation and their opposite effects on the inception of cell division associated with nodules and lateral roots. However, alternative explanations for the link between nodulation and lateral roots are also possible.
The LATD gene of M. truncatula is required for the establishment and maintenance of root meristems and is also required for normal development of symbiotic nodules (Bright et al., 2005). LATD functions downstream of ABA, and latd mutants have been shown to have reduced sensitivity to ABA for seed germination and stomatal closure. Unlike the sta1 mutation, the latd mutation has no effect on nodule initiation. Exogenous ABA can rescue the root meristem defects of latd mutants; the nodule development defect, however, cannot be rescued by ABA because the inhibitory effect of ABA on nodule initiation masks any later positive effect of ABA on nodule morphogenesis (Liang et al., 2007). These observations indicate that ABA plays a positive role in later stages of nodule development, in contrast with the negative role it plays in earlier stages of nodule development, as described in this work. Thus, ABA plays diverse roles at multiple stages of nodule formation, and the ABA sensitivity genes LATD and STA appear to specifically regulate ABA responses at different stages of the nodulation process.
Nodulation is regulated through multiple mechanisms. There is a shoot-derived signal that suppresses nodule number and requires the legume CLV1 homolog: SUNN in M. truncatula, HAR1 in L. japonicus, and NARK in soybean (Glycine max; Krusell et al., 2002; Nishimura et al., 2002; Penmetsa et al., 2003; Searle et al., 2003; Schnabel et al., 2005). Strikingly, the ABA-to-cytokinin ratio in the phloem is higher in wild-type soybean compared with nark mutants (Caba et al., 2000), and the sunn mutant has been shown to have altered auxin responses (van Noorden et al., 2006). These alterations in hormones are likely to be a factor that dictates the shoot regulation of nodule number and highlights the possibility that the shoot-derived signal produced by SUNN/HAR1/NARK is an alteration in hormone levels directed to the roots. The use of the stress hormones ABA, JA, and ethylene to regulate nodulation directly links the level of nodulation to the environmental status of the plant. We have shown that ABA can regulate multiple signaling pathways associated with nodule formation. This intimate interlinking of nodule development with ABA provides a mechanism to rapidly and decisively suppress nodulation according to the environmental and developmental status of the plant.
METHODS
Plant Growth Conditions and Bacterial Strains
Sterilized Medicago truncatula seeds were germinated overnight and plated on buffered nodulation medium (Ehrhardt et al., 1992) with 1.2% agar, with a filter paper over the surface of the media and allowed to grow for a minimum of 3 d before inoculation. Seeds of cv Jemalong line A17 were used as the wild type. Plants were grown in a growth chamber at 20°C with 16 h of light per day. During transformation studies, plants were left on selective media with 25 μg/mL kanamycin for 14 d. After that, roots lacking dsRed fluorescence were removed and the plants transferred to growth pouches. Sinorhizobium meliloti strain 1021 was grown in liquid Luria broth overnight in the presence of 500 μg/mL streptomycin selection. Bacteria were pelleted at 23,340g for 10 min and resuspended in 10 mM MgSO4. This bacterial suspension was diluted 1:50 in 10 mM MgSO4 and inoculated onto plants by flooding the roots with the inoculum. Where appropriate, the bacterial inoculum was diluted to a specific concentration after quantification at OD600. Nod factor preparations were isolated as described (Ehrhardt et al., 1996). S. meliloti strain Rm1021 carrying the vector pXLGD4, which contains lacZ driven by the hemA promoter, was used to visualize bacterial infection events (Penmetsa and Cook, 1997). ENOD11:GUS was generated as previously described (Journet et al., 2001), and this was crossed into the skl mutant (Penmetsa and Cook, 1997). sta-1 was identified from a screen of fast neutron mutagenized M. truncatula Jemalong, in which plants were cocultured for 6 weeks with S. meliloti and Glomus hoi. The sta-1 mutant was identified as having a nodulation defect, but no alteration in mycorrhization.
Assessment of Infection Frequency
To quantify the number of infection events, plants were grown on buffered nodulation medium (BNM) with differing concentrations of ABA (Sigma-Aldrich). At 6 d after inoculation, roots from eight plants per treatment were fixed for 1 h in 1.25% glutaraldehyde and 200 mM sodium cacodylate, washed twice in 200 mM sodium cacodylate, and stained overnight in 200 mM sodium cacodylate, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 0.08% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. Roots were destained in a 1:5 dilution of household bleach for 5 min and imaged using a Canon Optiphot microscope. The total number of infection events was counted.
GUS Assay
Plants carrying the ENOD11 promoter driving the expression of GUS were grown on medium for 7 d. Six plants were transferred to liquid medium containing either 1 μM ABA, 10 μM ABA, or medium alone and left for 24 h in the dark. They were then treated with 1 nM Nod factor for 12 h in liquid medium. The plants were fixed in 0.3% formaldehyde and 0.1 M potassium phosphate, pH 7.0, for 1 h on ice. GUS staining was performed overnight at 37°C with 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 5 mM EDTA, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.1 M potassium phosphate, pH 7.0. The experiments were repeated three times with similar results.
GUS activity was measured by the fluorimetric assay as described (Jefferson et al., 1987). The transgenic roots were ground in liquid nitrogen and homogenized in GUS extraction buffer for total protein extraction. Enzymatic reactions were performed using 1 μg of total protein extract with 4-methylumbelliferyl-β-d-glucuronide as substrate (Sigma-Aldrich). GUS activities were measured using a microtiter fluorimeter.
Acetylene Reduction Assay
Nitrogenase activity of inoculated roots was determined by assaying acetylene reduction, using roots harvested from BNM media plates 21 d after inoculation (Somasegaran and Hoben, 1994). Nodules from control vector transgenic roots and the abi1-1 transgenic roots were assayed for nitrogenase activity. For each, samples consisting of more than three nodules were measured for acetylene reduction in 2-mL sealed vials in the presence of 10% acetylene. Three biological replicates were measured. After 2 h of incubation in acetylene, a sample (5 to 10 μL) was injected into a Photovac 10S Plus gas chromatograph that used a photo-ionizing detector. Samples were compared with a standard curve generated against a 5 ppm ethylene standard, and nitrogenase activity was expressed as ARA units [ethylene products/(time(h)*nodule numbers)].
Expression Analysis by qRT-PCR
Total RNA was extracted from roots using either TRIzol reagent (Invitrogen) or RNeasy plant mini kits (Qiagen) following the manufacturer's protocol. Ribonucleic acid was quantified using a spectrophotometer (Nanodrop-1000; NanoDrop Technologies) and checked for quality by gel electrophoresis. The RNA was treated with DNA-free (Ambion) according to the manufacturer's instructions. cDNA was prepared from 500 ng of total RNA using the SuperScript II first-strand synthesis system for RT-PCR (Life Technologies, Invitrogen) using oligo(dT) primers according to the manufacturer's instructions. qRT-PCR was performed using an ABI PRISM 7500 HT sequence detection system (Applied Biosystems) with SDS 2.2 collection software (Applied Biosystems) and using SYBR Green to monitor double-stranded DNA synthesis. Reactions were performed in a 96-well plate using the SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) and 50 nM of each gene-specific primer. An initial denaturation step of 94°C for 2 min was followed by 40 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Primers used were as follows: RIP1, 5′-TAGGGAAAAACGCATTGGAG-3′, 5′-ACAACAGGGCCTTTGCATAC-3′; CLATHRIN (TC107843), 5′-CGTGAAGGTTGCTGGGTTAT-3′, 5′-AGTACCTGGCATTCGTTTGG-3′; MtEF1α, 5′-ATTCCAAAGGCGGCTGCATA-3′, 5′-CTTTGCTTGGTGCTGTTTAGATGG-3′; RD22, 5′-CAAATGTGGGTGTAGGGAAAGGAG-3′, 5′-TCAGTGGCAGCATAGTTGTAAGC-3′; NIN, 5′-TGGACAAGGAATAGTAGGAACAG-3′, 5′-CTCAATGGAATGGCAACAGC-3′; ENOD11, 5′-TAGGGCTTGCTGATAAATCTC-3′, 5′-TAATTGGAGGCTTGTAAGTAGG-3′; and ENOD40, 5′- CAATCACTCTATCTATGTAGCACTG-3′, 5′- CTCAAAGGAAGACAACACCATC-3′. After the completion of PCR amplification, a dissociation curve was run to check for genomic DNA contamination. For this, PCR products were denatured at 95°C, annealed at 60°C, and gradually heated to 95°C over the course of 20 min. No DNA contamination was detected in any reactions. Primer–dimer formation was estimated by running a control without template DNA. Results were expressed as a threshold cycle (CT) value. Two replicate reactions were run for each sample, and their CT values averaged. These averaged values were used in all subsequent calculations. Gene expression was normalized to that of the control by subtracting the CT value of the control from the CT value of the gene under investigation to give ΔCT. Fold induction was calculated by normalizing the data for each time series to that of the 0 h sample; the ΔCT value for that sample was then subtracted from the ΔCT of the other samples to give a ΔΔCT value. The data were plotted as 2−ΔΔCT. The data from three independent experiments were averaged and plotted using standard error. Clathrin was used as a control for Figure 2B, while EF1α was used as a control in all other qRT-PCR experiments.
Calcium Spiking Experiments
Analysis of calcium spiking was performed as described by Ehrhardt et al. (1996). All plants used were grown overnight on BNM. Micropipettes were pulled from filamented capillaries on a pipette puller (model 773; Campden Instruments). These were loaded with Oregon green dextran (10,000 MW; Invitrogen) and injections performed using iontophoresis with currents generated from a cell amplifier (model Intra 767; World Precision Instruments) and a stimulus generator made to our specifications (World Precision Instruments). Cells were analyzed on an inverted epiflorescence microscope (model TE2000; Nikon) using a monochromator (model Optoscan; Cairn Research) to generate specific wavelengths of light. Images were captured with a CCD camera (model ORCA-ER) and fluorescent data analyzed using Metaflor (Molecular Devices).
Lines expressing the calcium reporter cameleon (Miwa et al., 2006) were analyzed on the same inverted epiflorescent microscope (model TE2000). During image capture, the image was split using the Optosplit (Cairn Research), and each image was passed through a filter for either CFP or YFP emissions prior to exposure on the CCD chip. The ratio of CFP:YFP was assayed using Metaflor (Molecular Devices).
Agrobacterium rhizogenes Root Transformation and Inoculation
The full-length sequence of Arabidopsis thaliana abi1-1 was cloned into the vector PK7WG2R (derived from PK7FWG2R and pB7WG2,0) using Gateway technology (Invitrogen). The vector contains a 35S promoter region with kanamycin selection and the dsRed reporter gene. The primers used were AtABI1-FOR (5′-TTACCGTTAATGGAGGAAGT-3′) and AtABI1-REV (5′-CTCTGCCTCAGTTCAAGG-3′). Both empty vector and abi1-1 were used to transform the A. rhizogenes ARqua1 strain. Transformed M. truncatula roots were prepared as described (Boisson-Dernier et al., 2001). Three weeks later, transgenic roots were inoculated with S. meliloti. Nodule numbers were measured 21 d after inoculation.
Stomatal Closure Assay and Water Loss Analysis
Stomatal responses to ABA treatment and water loss analysis were performed according to Leung et al. (1997), Pei et al. (1997), and Pandey and Assmann (2004). Leaves of 5-week-old wild-type and sta-1 plants were incubated in KCl-MES buffer (50 mM KCl and 10 mM MES, pH 6.12) under continuous white light (250 μmol m−2 s−1) for 90 min to open the stomata. Epidermal strips with opened stomata from one-half of a pretreated leaf were transferred to the KCl-CaCl2 MES buffer (50 mM KCl, 1 mM CaCl2, and 10 mM MES, pH 6.12) with 25 μM ABA (Sigma-Aldrich) to test ABA sensitivity and strips from the other leaf half transferred to the assay buffer without ABA. After incubation for 2 h under light (250 μmol m−2 s−1), the stomatal aperture width was observed for 50 randomly selected stomata. Each assay was repeated three times. The data represent means ± se (n = 150). For the water loss assay, 10 leaves were detached from wild-type and sta-1 plants grown in the identical conditions and weighed in plastic Petri dishes immediately and then placed into a laboratory bench and weighed at indicated time intervals. The water loss rate was measured as a percentage of initial fresh weight. Four repeats were taken.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: Arabidopsis abi1-1 (X77116), M. truncatula ENOD40 (X80264), M. truncatula RIP1 (U16727), M. truncatula ENOD11 (AJ297721), and M. truncatula RD22 (TIGR TC accession number TC95843).
Acknowledgments
We thank Jens Stougaard and Plant Bioscience Limited for providing the L. japonicus snf2 seed and Etienne-Pascal Journet and David Barker for providing the M. truncatula ENOD11:GUS and skl ENOD11:GUS lines. This work was supported by the Biotechnology and Biological Science Research Council as a David Philips Fellowship, a grant in aid, and by BB/C513669 to G.E.D.O. Support for J.M.H. came from National Science Foundation Grant IBN-0212992 and USDA CSREES Hatch funding to the Vermont Agricultural Experiment Station, Project VT-HO-1202 to J.M.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Giles E.D. Oldroyd (giles.oldroyd@bbsrc.ac.uk).
References
- Ane, J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303 1364–1367. [DOI] [PubMed] [Google Scholar]
- Arrighi, J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier, A., Chabaud, M., Garcia, F., Becard, G., Rosenberg, C., and Barker, D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14 695–700. [DOI] [PubMed] [Google Scholar]
- Brady, S.M., Sarkar, S.F., Bonetta, D., and McCourt, P. (2003). The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 34 67–75. [DOI] [PubMed] [Google Scholar]
- Bright, L.J., Liang, Y., Mitchell, D.M., and Harris, J.M. (2005). The LATD gene of Medicago truncatula is required for both nodule and root development. Mol. Plant Microbe Interact. 18 521–532. [DOI] [PubMed] [Google Scholar]
- Caba, J.M., Centeno, M.L., Fernandez, B., Gresshoff, P.M., and Ligero, F. (2000). Inoculation and nitrate alter phytohormone levels in soybean roots: Differences between a supernodulating mutant and the wild type. Planta 211 98–104. [DOI] [PubMed] [Google Scholar]
- Cho, M.J., and Harper, J.E. (1993). Effect of abscisic acid application on root isoflavonoid concentration and nodulation of wild-type and nodulation-mutant soybean plants. Plant Soil 152 145–149. [Google Scholar]
- Cook, D., Dreyer, D., Bonnet, D., Howell, M., Nony, E., and VandenBosch, K. (1995). Transient induction of a peroxidase gene in Medicago truncatula precedes infection by Rhizobium meliloti. Plant Cell 7 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C.H., and Zhang, H. (2003). An abscisic acid-sensitive checkpoint in lateral root development of Arabidopsis. Plant J. 33 543–555. [DOI] [PubMed] [Google Scholar]
- De Smet, I., Zhang, H., Inze, D., and Beeckman, T. (2006). A novel role for abscisic acid emerges from underground. Trends Plant Sci. 11 434–439. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Atkinson, E.M., and Long, S.R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256 998–1000. [DOI] [PubMed] [Google Scholar]
- Ehrhardt, D.W., Wais, R., and Long, S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85 673–681. [DOI] [PubMed] [Google Scholar]
- Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417 962–966. [DOI] [PubMed] [Google Scholar]
- Fang, Y., and Hirsch, A.M. (1998). Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 116 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff, N.V. (2002). Cross-talk in abscisic acid signaling. Sci. STKE 2002 RE10. [DOI] [PubMed] [Google Scholar]
- Ferguson, B.J., Ross, J.J., and Reid, J.B. (2005). Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 138 2396–2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamas, P., Niebel Fde, C., Lescure, N., and Cullimore, J. (1996). Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant Microbe Interact. 9 233–242. [DOI] [PubMed] [Google Scholar]
- Gampala, S.S., Hagenbeek, D., and Rock, C.D. (2001). Functional interactions of lanthanum and phospholipase D with the abscisic acid signaling effectors VP1 and ABI1-1 in rice protoplasts. J. Biol. Chem. 276 9855–9860. [DOI] [PubMed] [Google Scholar]
- Gleason, C., Chaudhuri, S., Yang, T., Munoz, A., Poovaiah, B.W., and Oldroyd, G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441 1149–1152. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Millar, A.A., and Jacobsen, J.V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Biol. 8 183–187. [DOI] [PubMed] [Google Scholar]
- Hagenbeek, D., Quatrano, R.S., and Rock, C.D. (2000). Trivalent ions activate abscisic acid-inducible promoters through an ABI1-dependent pathway in rice protoplasts. Plant Physiol. 123 1553–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidstra, R., Yang, W.C., Yalcin, Y., Peck, S., Emons, A.M., van Kammen, A., and Bisseling, T. (1997). Ethylene provides positional information on cortical cell division but is not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development 124 1781–1787. [DOI] [PubMed] [Google Scholar]
- Hirsch, A.M., and Fang, Y. (1994). Plant hormones and nodulation: What's the connection? Plant Mol. Biol. 26 5–9. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Anraku, H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433 527–531. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet, E.P., El-Gachtouli, N., Vernoud, V., de Billy, F., Pichon, M., Dedieu, A., Arnould, C., Morandi, D., Barker, D.G., and Gianinazzi-Pearson, V. (2001). Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14 737–748. [DOI] [PubMed] [Google Scholar]
- Kalo, P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308 1786–1789. [DOI] [PubMed] [Google Scholar]
- Kanamori, N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krusell, L., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420 422–426. [DOI] [PubMed] [Google Scholar]
- Laplaze, L., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell 19 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303 1361–1364. [DOI] [PubMed] [Google Scholar]
- Li, X., Mo, X., Shou, H., and Wu, P. (2006). Cytokinin-mediated cell cycling arrest of pericycle founder cells in lateral root initiation of Arabidopsis. Plant Cell Physiol. 47 1112–1123. [DOI] [PubMed] [Google Scholar]
- Liang, Y., and Harris, J.M. (2005). Response of root branching to abscisic acid is correlated with nodule formation both in legumes and nonlegumes. Am. J. Bot. 92 1675–1683. [DOI] [PubMed] [Google Scholar]
- Liang, Y., Mitchell, D.M., and Harris, J.M. (2007). Abscisic acid rescues the root meristem defects of the Medicago truncatula latd mutant. Dev. Biol. 304 297–307. [DOI] [PubMed] [Google Scholar]
- Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302 630–633. [DOI] [PubMed] [Google Scholar]
- Madsen, E.B., Madsen, L.H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., Sato, S., Kaneko, T., Tabata, S., Sandal, N., and Stougaard, J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425 637–640. [DOI] [PubMed] [Google Scholar]
- Mathesius, U., Charon, C., Rolfe, B.G., Kondorosi, A., and Crespi, M. (2000). Temporal and spatial order of events during the induction of cortical cell divisions in white clover by Rhizobium leguminosarum bv. trifolii inoculation or localized cytokinin addition. Mol. Plant Microbe Interact. 13 617–628. [DOI] [PubMed] [Google Scholar]
- Mathesius, U., Schlaman, H.R.M., Spaink, H.P., Sautter, C., Rolfe, B.G., and Djordjevic, M.A. (1998). Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 14 23–34. [DOI] [PubMed] [Google Scholar]
- Middleton, P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R.M., Shaw, S.L., and Long, S.R. (2004). Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. USA 101 10217–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa, H., Sun, J., Oldroyd, G.E., and Downie, J.A. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48 883–894. [DOI] [PubMed] [Google Scholar]
- Murray, J.D., Karas, B.J., Sato, S., Tabata, S., Amyot, L., and Szczyglowski, K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315 101–104. [DOI] [PubMed] [Google Scholar]
- Nakagawa, T., and Kawaguchi, M. (2006). Shoot-applied MeJA suppresses root nodulation in Lotus japonicus. Plant Cell Physiol. 47 176–180. [DOI] [PubMed] [Google Scholar]
- Nishimura, R., Hayashi, M., Wu, G.J., Kouchi, H., Imaizumi-Anraku, H., Murakami, Y., Kawasaki, S., Akao, S., Ohmori, M., Nagasawa, M., Harada, K., and Kawaguchi, M. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420 426–429. [DOI] [PubMed] [Google Scholar]
- Nutman, P.S. (1948). Physiological studies on nodule formation. I. The relation between nodulation and lateral root formation in red clover. Ann. Bot. (Lond.) XII 81–96. [Google Scholar]
- Oldroyd, G.E. (2007). Plant science. Nodules and hormones. Science 315 52–53. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Downie, J.A. (2004). Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5 566–576. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., and Downie, J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59 519–546. [DOI] [PubMed] [Google Scholar]
- Oldroyd, G.E., Engstrom, E.M., and Long, S.R. (2001). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13 1835–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S., and Assmann, S.M. (2004). The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein alpha subunit GPA1 and regulates abscisic acid signaling. Plant Cell 16 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Kuchitsu, K., Ward, J.M., Schwarz, M., and Schroeder, J.I. (1997). Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa, R.V., and Cook, D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275 527–530. [DOI] [PubMed] [Google Scholar]
- Penmetsa, R.V., Frugoli, J.A., Smith, L.S., Long, S.R., and Cook, D.R. (2003). Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 131 998–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prayitno, J., Rolfe, B.G., and Mathesius, U. (2006). The ethylene-insensitive sickle mutant of Medicago truncatula shows altered auxin transport regulation during nodulation. Plant Physiol. 142 168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu, S., Madsen, L.H., Madsen, E.B., Felle, H.H., Umehara, Y., Gronlund, M., Sato, S., Nakamura, Y., Tabata, S., Sandal, N., and Stougaard, J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425 585–592. [DOI] [PubMed] [Google Scholar]
- Riely, B.K., Lougnon, G., Ane, J.M., and Cook, D.R. (2007). The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49 208–216. [DOI] [PubMed] [Google Scholar]
- Saito, K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel, E., Journet, E.P., de Carvalho-Niebel, F., Duc, G., and Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58 809–822. [DOI] [PubMed] [Google Scholar]
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 410 327–330. [DOI] [PubMed] [Google Scholar]
- Searle, I.R., Men, A.E., Laniya, T.S., Buzas, D.M., Iturbe-Ormaetxe, I., Carroll, B.J., and Gresshoff, P.M. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 109–112. [DOI] [PubMed] [Google Scholar]
- Smit, P., Raedts, J., Portyanko, V., Debelle, F., Gough, C., Bisseling, T., and Geurts, R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308 1789–1791. [DOI] [PubMed] [Google Scholar]
- Somasegaran, P., and Hoben, H.J. (1994). Handbook for Rhizobia. The Acetylene Reduction Assay for Measuring Nitrogen Activity, P. Somasegaran and H.J. Hoben, eds (New York: Springer-Verlag), pp. 392–398.
- Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., Tabata, S., Sandal, N., Stougaard, J., Szczyglowski, K., and Parniske, M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417 959–962. [DOI] [PubMed] [Google Scholar]
- Sun, J., Cardoza, V., Mitchell, D.M., Bright, L., Oldroyd, G., and Harris, J.M. (2006). Crosstalk between jasmonic acid, ethylene and Nod factor signaling allows integration of diverse inputs for regulation of nodulation. Plant J. 46 961–970. [DOI] [PubMed] [Google Scholar]
- Suzuki, A., Akune, M., Kogiso, M., Imagama, Y., Osuki, K., Uchiumi, T., Higashi, S., Han, S.Y., Yoshida, S., Asami, T., and Abe, M. (2004). Control of nodule number by the phytohormone abscisic acid in the roots of two leguminous species. Plant Cell Physiol. 45 914–922. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441 1153–1156. [DOI] [PubMed] [Google Scholar]
- Tirichine, L., Sandal, N., Madsen, L.H., Radutoiu, S., Albrektsen, A.S., Sato, S., Asamizu, E., Tabata, S., and Stougaard, J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315 104–107. [DOI] [PubMed] [Google Scholar]
- van Noorden, G.E., Ross, J.J., Reid, J.B., Rolfe, B.G., and Mathesius, U. (2006). Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 140 1494–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15 501–510. [DOI] [PubMed] [Google Scholar]
- Wu, Y., Sanchez, J.P., Lopez-Molina, L., Himmelbach, A., Grill, E., and Chua, N.H. (2003). The abi1-1 mutation blocks ABA signaling downstream of cADPR action. Plant J. 34 307–315. [DOI] [PubMed] [Google Scholar]
- Yamaguchishinozaki, K., Koizumi, M., Urao, S., and Shinozaki, K. (1992). Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: Sequence analysis of one cDNA clone that encodes a putative transmembrane channel protein. Plant Cell Physiol. 33 217–224. [Google Scholar]
- Yang, W.C., de Blank, C., Meskiene, I., Hirt, H., Bakker, J., van Kammen, A., Franssen, H., and Bisseling, T. (1994). Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]